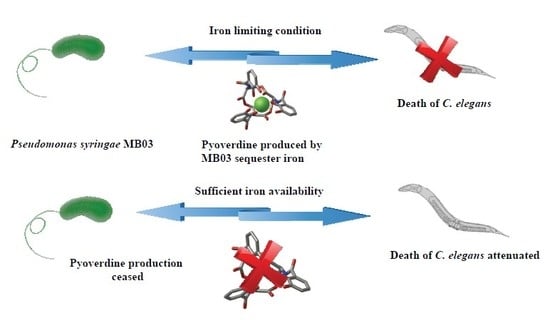
Pyoverdine-Mediated Killing of Caenorhabditis elegans by Pseudomonas syringae MB03 and the Role of Iron in Its Pathogenicity
Abstract
1. Introduction
2. Results
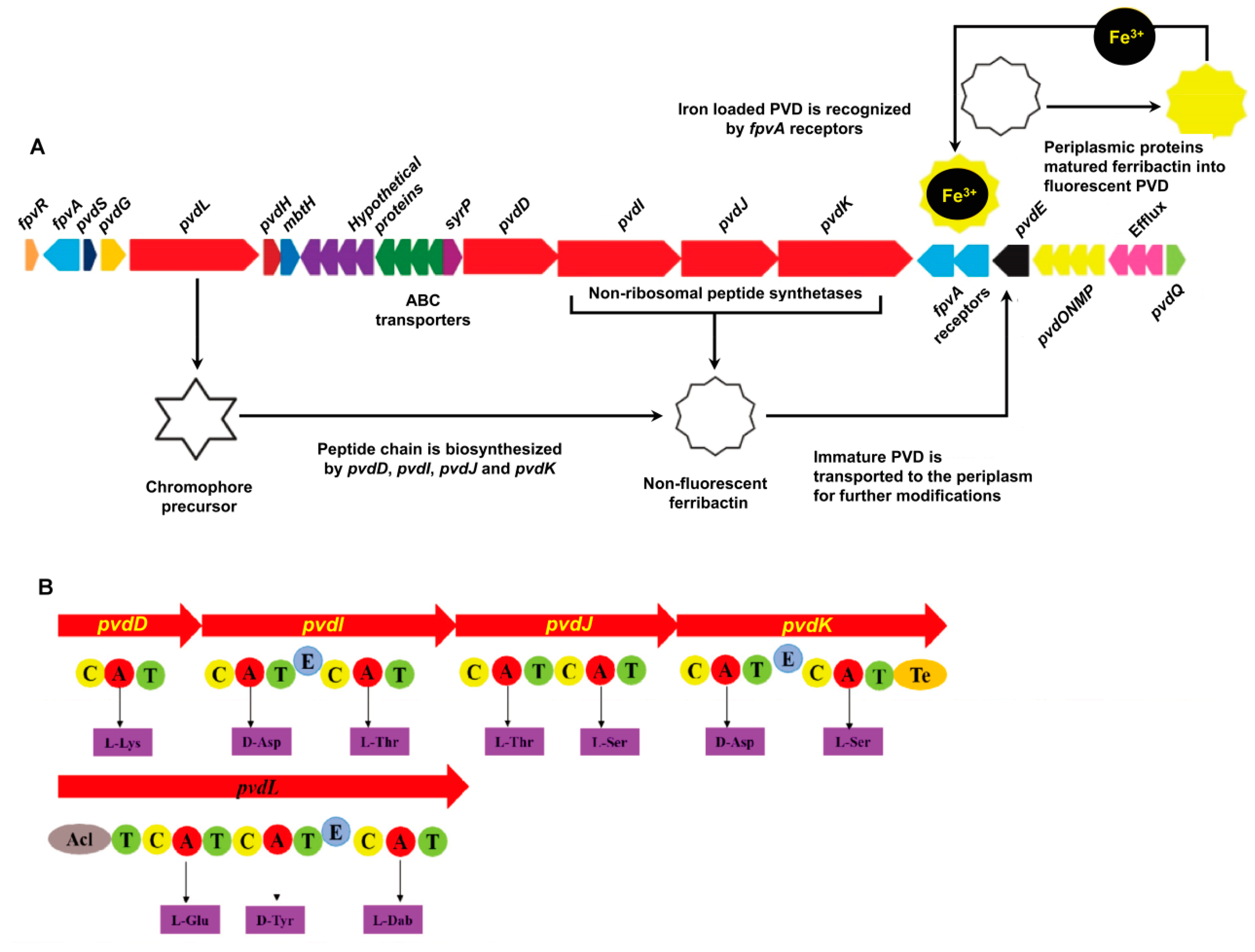
2.1. P. syringae MB03 has the Genetic Potential to Produce PVD
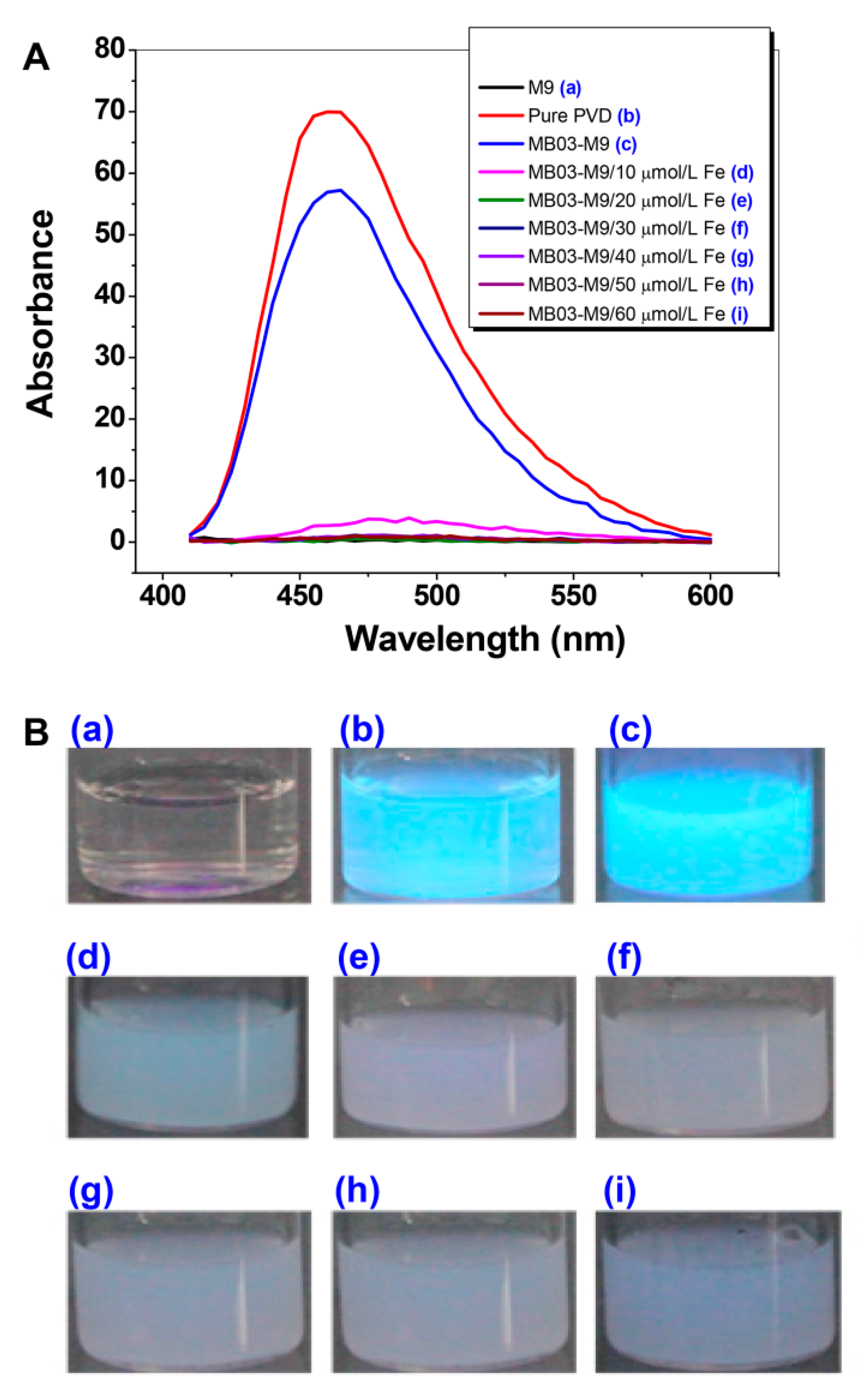
2.2. PVD is Only Produced by P. syringae MB03 under Iron-limiting Conditions
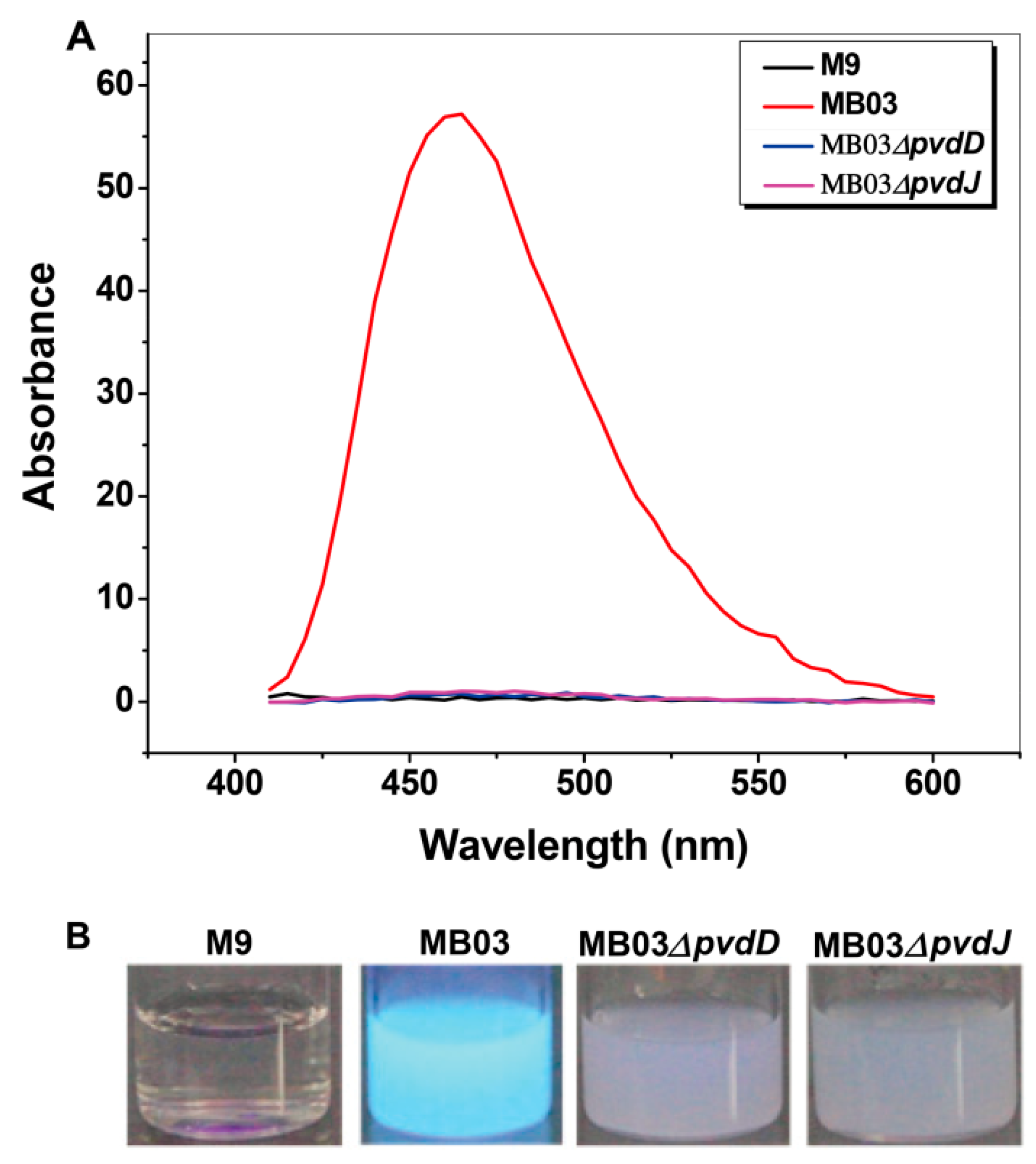
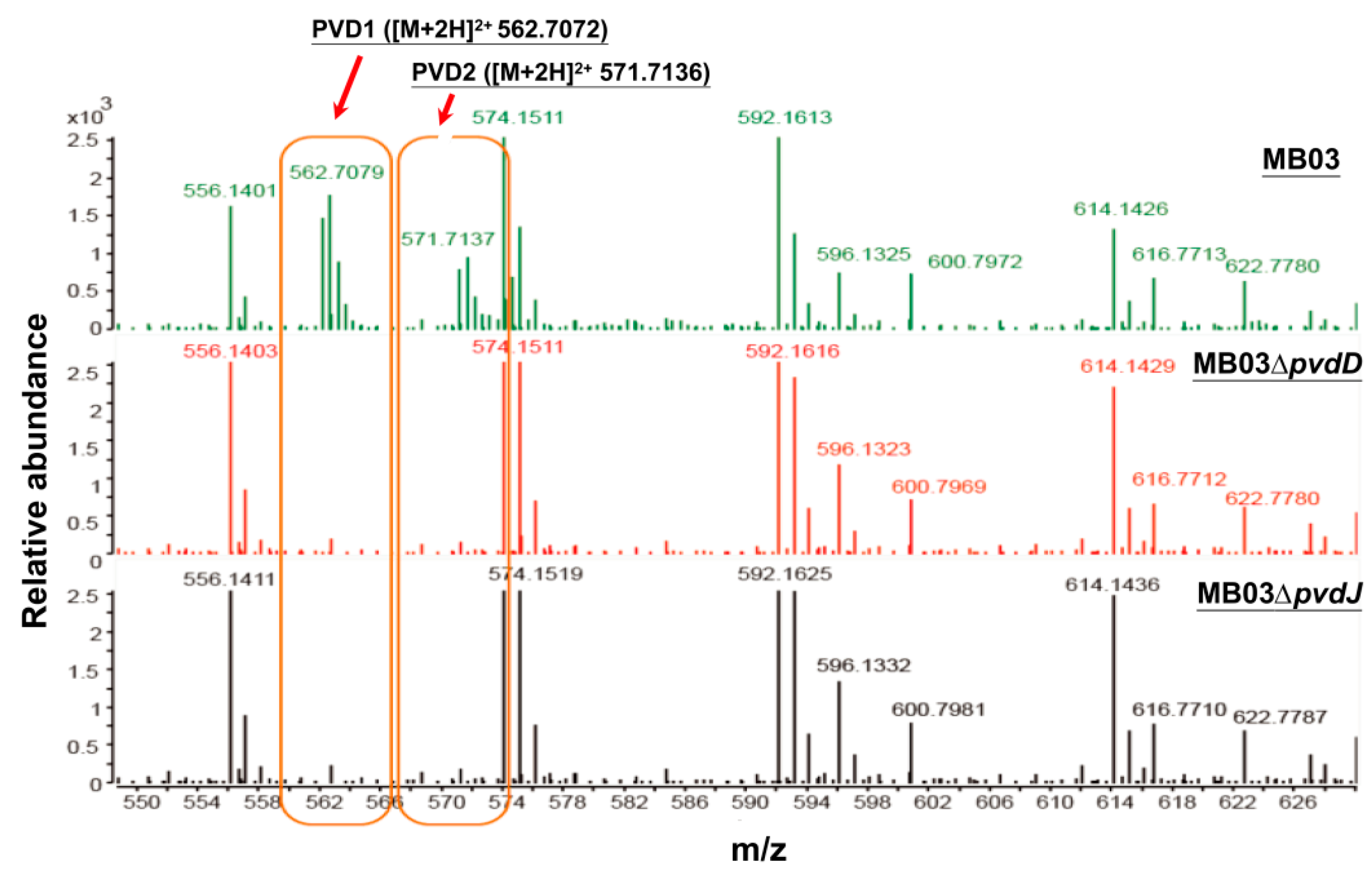
2.3. Mutants MB03∆pvdD and MB03∆pvdJ Fail to Produce PVD in MB03
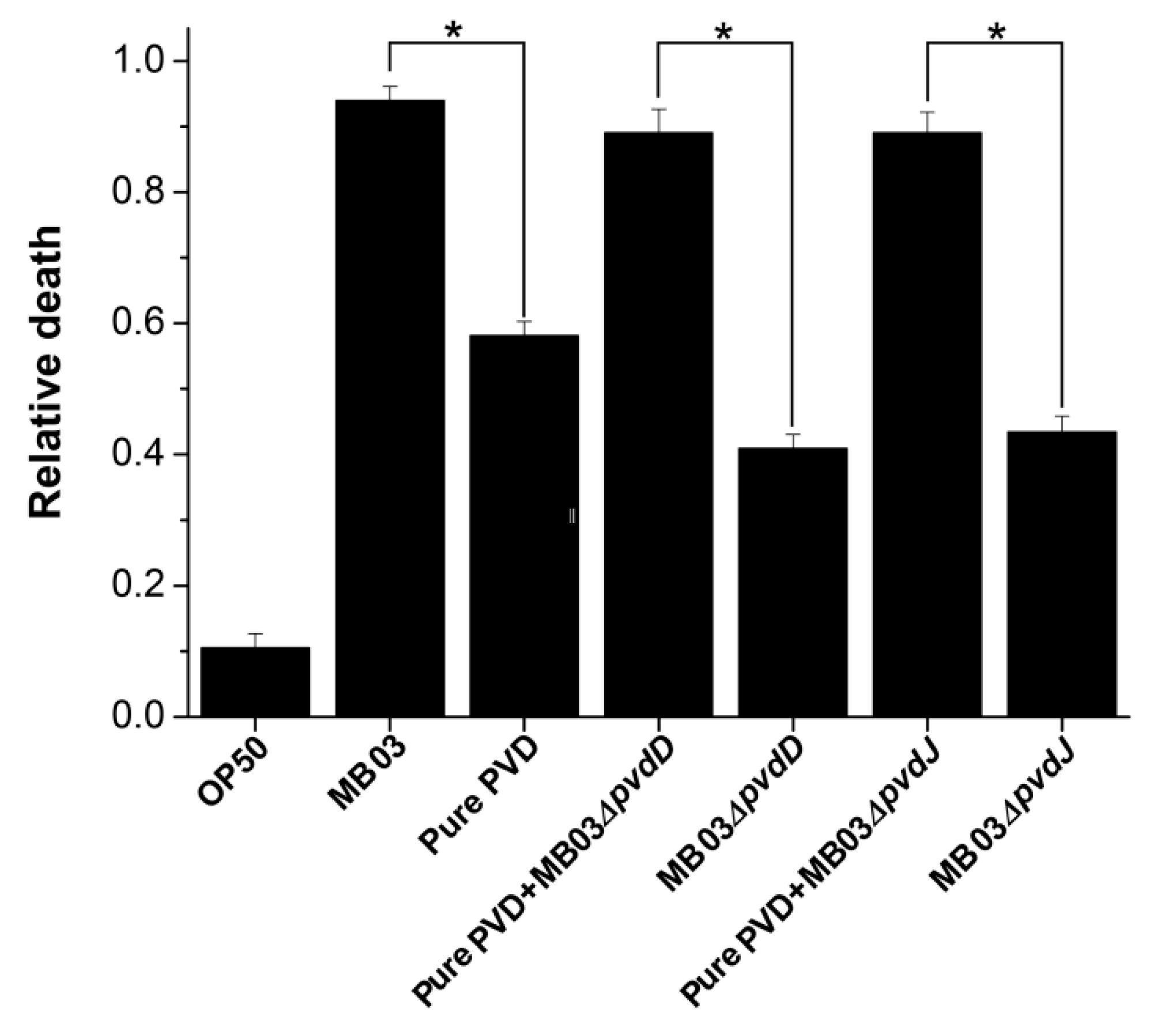
2.4. PVD from P. syringae MB03 Promotes the Death of C. elegans
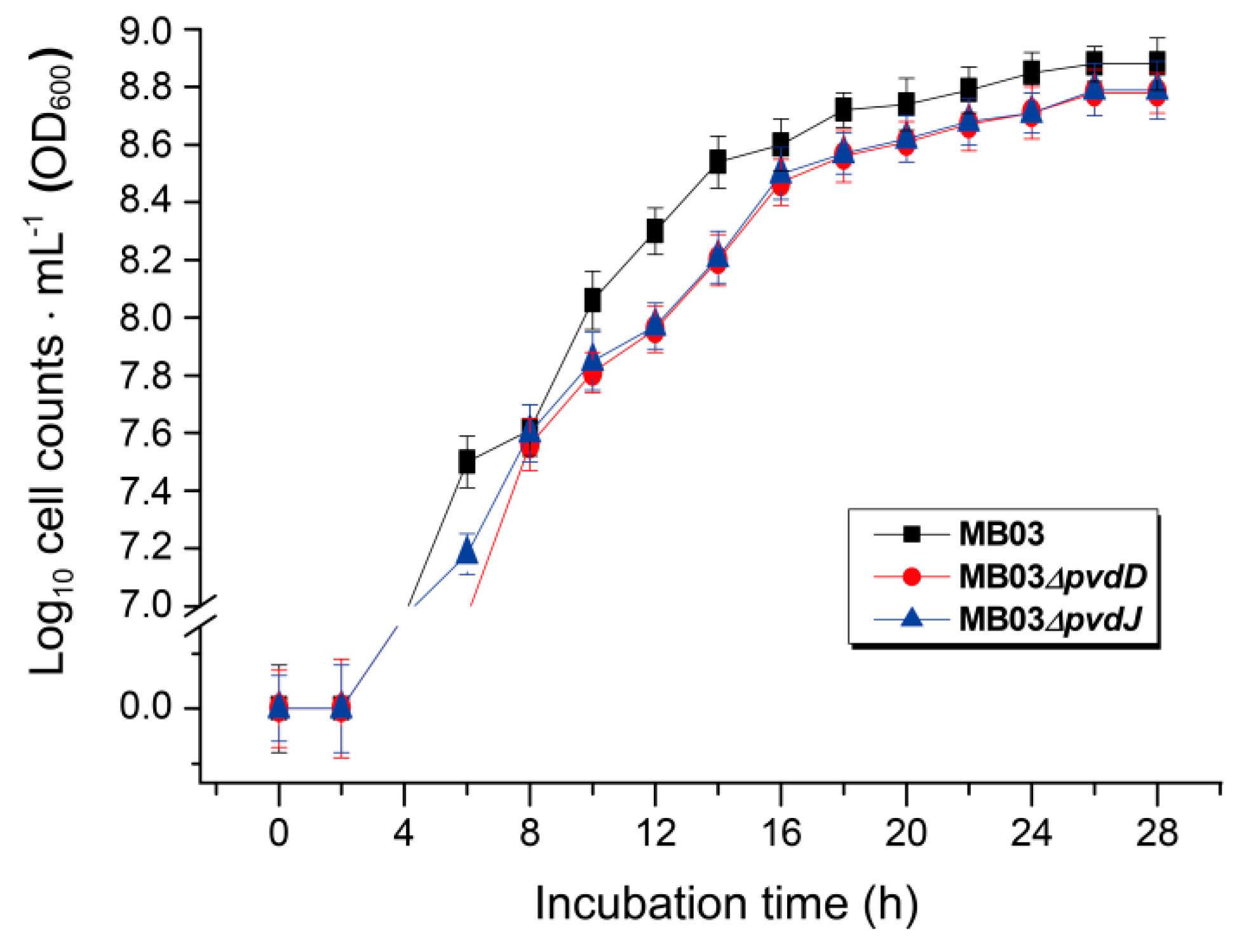
2.5. Role of Iron in the MB03–C. elegans Interaction
3. Discussion
4. Materials and Methods
4.1. Bacterial and C. elegans Strains, Plasmids, Media, Chemicals, and Culture Conditions
4.2. Direct de novo Genome Mining of MB03 to Identify PVD-encoding Biosynthetic Genes
4.3. PVD Determination Assay
4.4. Disruption of PVD Biosynthetic Genes using the RecTE Recombination System
4.5. LC-MS Analysis of Cell-Free Filtrates from MB03 and its Derivatives
4.6. Efficacy of LK Assay
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Miethke, M.; Marahiel, M.A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef]
- Ganz, T.; Nemeth, E. Iron homeostasis in host defence and inflammation. Nat. Rev. Immunol. 2015, 15, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.C.; Robinson, A.K.; Rodriguez-Quinones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Chu, B.C.; Garcia-Herrero, A.; Johanson, T.H.; Krewulak, K.D.; Lau, C.K.; Peacock, R.S.; Slavinskaya, Z.; Vogel, H.J. Siderophore uptake in bacteria and the battle for iron with the host; a bird’s eye view. Biometals 2010, 23, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.R.; Bogdan, A.R.; Miyazawa, M.; Hashimoto, K.; Tsuji, Y. Siderophores in iron metabolism: From mechanism to therapy potential. Trends Mol. Med. 2016, 22, 1077–1090. [Google Scholar] [CrossRef]
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637–657. [Google Scholar] [CrossRef] [PubMed]
- Koglin, A.; Walsh, C.T. Structural insights into nonribosomal peptide enzymatic assembly lines. Nat. Prod. Rep. 2009, 26, 987–1000. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sivonen, K.; Fewer, D.P. Genomic insights into the distribution, genetic diversity and evolution of polyketide synthases and nonribosomal peptide synthetases. Curr. Opin. Genet. Dev. 2015, 35, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Cezard, C.; Farvacques, N.; Sonnet, P. Chemistry and biology of pyoverdines, Pseudomonas primary siderophores. Curr. Med. Chem. 2015, 22, 165–186. [Google Scholar] [CrossRef]
- Folschweiller, N.; Gallay, J.; Vincent, M.; Abdallah, M.A.; Pattus, F.; Schalk, I.J. The interaction between pyoverdin and its outer membrane receptor in Pseudomonas aeruginosa leads to different conformers: A time-resolved fluorescence study. Biochemistry 2002, 41, 14591–14601. [Google Scholar] [CrossRef]
- Braud, A.; Hannauer, M.; Mislin, G.L.; Schalk, I.J. The Pseudomonas aeruginosa pyochelin-iron uptake pathway and its metal specificity. J. Bacteriol. 2009, 191, 3517–3525. [Google Scholar] [CrossRef] [PubMed]
- Visca, P.; Imperi, F.; Lamont, I.L. Pyoverdine siderophores: From biogenesis to biosignificance. Trends Microbiol. 2007, 15, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Baysse, C.; De Vos, D.; Naudet, Y.; Vandermonde, A.; Ochsner, U.; Meyer, J.M.; Budzikiewicz, H.; Schafer, M.; Fuchs, R.; Cornelis, P. Vanadium interferes with siderophore-mediated iron uptake in Pseudomonas aeruginosa. Microbiology 2000, 146, 2425–2434. [Google Scholar] [CrossRef] [PubMed]
- Braud, A.; Hoegy, F.; Jezequel, K.; Lebeau, T.; Schalk, I.J. New insights into the metal specificity of the Pseudomonas aeruginosa pyoverdine-iron uptake pathway. Environ. Microbiol. 2009, 11, 1079–1091. [Google Scholar] [CrossRef]
- Cornelis, P.; Matthijs, S. Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: Not only pyoverdines. Environ. Microbiol. 2002, 4, 787–798. [Google Scholar] [CrossRef]
- Meyer, J.M.; Gruffaz, C.; Raharinosy, V.; Bezverbnaya, I.; Schafer, M.; Budzikiewicz, H. Siderotyping of fluorescent Pseudomonas: Molecular mass determination by mass spectrometry as a powerful pyoverdine siderotyping method. Biometals 2008, 21, 259–271. [Google Scholar] [CrossRef]
- Budzikiewicz, H. Siderophores of the human pathogenic fluorescent pseudomonads. Curr. Top. Med. Chem. 2001, 1, 1–6. [Google Scholar] [CrossRef]
- Budzikiewicz, H.; Schafer, M.; Fernandez, D.U.; Matthijs, S.; Cornelis, P. Characterization of the chromophores of pyoverdins and related siderophores by electrospray tandem mass spectrometry. Biometals 2007, 20, 135–144. [Google Scholar] [CrossRef]
- Ravel, J.; Cornelis, P. Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol. 2003, 11, 195–200. [Google Scholar] [CrossRef]
- Meyer, J.M. Pyoverdines: Pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch. Microbiol. 2000, 174, 135–142. [Google Scholar] [CrossRef]
- Budzikiewicz, H. Siderophores of the Pseudomonadaceae sensu stricto (fluorescent and non-fluorescent Pseudomonas spp.). Fortschr. Chem. Org. Naturst. 2004, 87, 81–237. [Google Scholar] [PubMed]
- Kang, D.; Kirienko, D.R.; Webster, P.; Fisher, A.L.; Kirienko, N.V. Pyoverdine, a siderophore from Pseudomonas aeruginosa, translocates into C. elegans, removes iron, and activates a distinct host response. Virulence 2018, 9, 804–817. [Google Scholar] [CrossRef] [PubMed]
- Handfield, M.; Lehoux, D.E.; Sanschagrin, F.; Mahan, M.J.; Woods, D.E.; Levesque, R.C. In vivo-induced genes in Pseudomonas aeruginosa. Infect. Immun. 2000, 68, 2359–2362. [Google Scholar] [CrossRef] [PubMed]
- Takase, H.; Nitanai, H.; Hoshino, K.; Otani, T. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect. Immun. 2000, 68, 1834–1839. [Google Scholar]
- Taguchi, F.; Suzuki, T.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. The siderophore pyoverdine of Pseudomonas syringae pv. tabaci 6605 is an intrinsic virulence factor in host tobacco infection. J. Bacteriol. 2010, 192, 117–126. [Google Scholar]
- Hirano, S.S.; Upper, C.D. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 2000, 64, 624–653. [Google Scholar] [CrossRef]
- Li, Q.; Yan, Q.; Chen, J.; He, Y.; Wang, J.; Zhang, H.; Yu, Z.; Li, L. Molecular characterization of an ice nucleation protein variant (InaQ) from Pseudomonas syringae and the analysis of its transmembrane transport activity in Escherichia coli. Int. J. Biol. Sci. 2012, 8, 1097–1108. [Google Scholar] [CrossRef]
- Feinbaum, R.L.; Urbach, J.M.; Liberati, N.T.; Djonovic, S.; Adonizio, A.; Carvunis, A.-R.; Ausubel, F.M. Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model. PLoS Pathogen. 2012, 8, e1002813. [Google Scholar] [CrossRef]
- Dubern, J.F.; Cigana, C.; De Simone, M.; Lazenby, J.; Juhas, M.; Schwager, S.; Bianconi, I.; Döring, G.; Eberl, L.; Williams, P. Integrated whole-genome screening for Pseudomonas aeruginosa virulence genes using multiple disease models reveals that pathogenicity is host specific. Environ. Microbiol. 2015, 17, 4379–4393. [Google Scholar] [CrossRef]
- Kirienko, N.V.; Kirienko, D.R.; Larkins-Ford, J.; Wählby, C.; Ruvkun, G.; Ausubel, F.M. Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe. 2013, 13, 406–416. [Google Scholar] [CrossRef]
- Zaborin, A.; Romanowski, K.; Gerdes, S.; Holbrook, C.; Lepine, F.; Long, J.; Poroyko, V.; Diggle, S.P.; Wilke, A.; Righetti, K. Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc. Natl. Acad. Sci. USA 2009, 106, 6327–6332. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Sun, Y.; Xie, L.; Yu, H.; Bashir, A.; Li, L. The pathogenicity of Pseudomonas syringae MB03 against Caenorhabditis elegans and the transcriptional response of nematicidal genes upon different nutritional conditions. Front. Microbiol. 2016, 7, 805. [Google Scholar] [CrossRef] [PubMed]
- Manan, A.; Bazai, Z.; Fan, J.; Yu, H.; Li, L. The Nif3-family protein YqfO03 from Pseudomonas syringae MB03 has multiple nematicidal activities against Caenorhabditis elegans and Meloidogyne incognita. Int. J. Mol. Sci. 2018, 19, 3915. [Google Scholar] [CrossRef] [PubMed]
- Lott, J.S.; Lee, T.V. Revealing the inter-module interactions of multi-modular nonribosomal peptide synthetases. Structure 2017, 25, 693–695. [Google Scholar] [CrossRef][Green Version]
- Yin, K.; Zhang, W.; Chen, L. Pyoverdine secreted by Pseudomonas aeruginosa as a biological recognition element for the fluorescent detection of furazolidone. Biosens. Bioelectron. 2014, 51, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Beare, P.A.; For, R.J.; Martin, L.W.; Lamont, I.L. Siderophore-mediated cell signalling in Pseudomonas aeruginosa: Divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol. 2003, 47, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Lamont, I.L.; Beare, P.A.; Ochsner, U.; Vasil, A.I.; Vasil, M.L. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2002, 99, 7072–7077. [Google Scholar] [CrossRef]
- Owen, J.G.; Ackerley, D.F. Characterization of pyoverdine and achromobactin in Pseudomonas syringae pv. phaseolicola 1448a. BMC Microbiol. 2011, 11, 218. [Google Scholar]
- Redly, G.A.; Poole, K. Pyoverdine-mediated regulation of FpvA synthesis in Pseudomonas aeruginosa: Involvement of a probable extracytoplasmic-function sigma factor, FpvI. J. Bacteriol. 2003, 185, 1261–1265. [Google Scholar] [CrossRef]
- Wilson, M.J.; McMorran, B.J.; Lamont, I.L. Analysis of promoters recognized by PvdS, an extracytoplasmic-function sigma factor protein from Pseudomonas aeruginosa. J. Bacteriol. 2001, 183, 2151–2155. [Google Scholar] [CrossRef]
- Lamont, I.L.; Martin, L.W.; Sims, T.; Scott, A.; Wallace, M. Characterization of a gene encoding an acetylase required for pyoverdine synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 3149–3152. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bultreys, A.; Gheysen, I.; Maraite, H.; de Hoffmann, E. Characterization of fluorescent and nonfluorescent peptide siderophores produced by Pseudomonas syringae strains and their potential use in strain identification. Appl. Environ. Microbiol. 2001, 67, 1718–1727. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.M.; Neely, A.; Stintzi, A.; Georges, C.; Holder, I.A. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 1996, 64, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Nadal Jimenez, P.; Koch, G.; Papaioannou, E.; Wahjudi, M.; Krzeslak, J.; Coenye, T.; Cool, R.H.; Quax, W.J. Role of PvdQ in Pseudomonas aeruginosa virulence under iron-limiting conditions. Microbiology 2010, 156, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Truman-Rosentsvit, M.; Berenbaum, D.; Spektor, L.; Cohen, L.A.; Belizowsky-Moshe, S.; Lifshitz, L.; Ma, J.; Li, W.; Kesselman, E.; Abutbul-Ionita, I.; et al. Ferritin is secreted via 2 distinct nonclassical vesicular pathways. Blood 2018, 131, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Gourley, B.L.; Parker, S.B.; Jones, B.J.; Zumbrennen, K.B.; Leibold, E.A. Cytosolic aconitase and ferritin are regulated by iron in Caenorhabditis elegans. J. Biol. Chem. 2003, 278, 3227–3234. [Google Scholar] [CrossRef]
- Cherayil, B.J. The role of iron in the immune response to bacterial infection. Immunol. Res. 2011, 50, 1–9. [Google Scholar] [CrossRef]
- Lewis, J.P. Metal uptake in host-pathogen interactions: Role of iron in Porphyromonas gingivalis interactions with host organisms. Periodontol. 2000 2010, 52, 94–116. [Google Scholar] [CrossRef]
- Van Gijsegem, F.; Genin, S.; Boucher, C. Conservation of secretion pathways for pathogenicity determinants of plant and animal bacteria. Trends Microbiol. 1993, 1, 175–180. [Google Scholar] [CrossRef]
- Collmer, A.; Bauer, D.W. Erwinia chrysanthemi and Pseudomonas syringae: Plant pathogens trafficking in extracellular virulence proteins. Curr. Top. Microbiol. Immunol. 1994, 192, 43–78. [Google Scholar]
- Cornelis, P.; Matthijs, S.; Van Oeffelen, L. Iron uptake regulation in Pseudomonas aeruginosa. Biometals 2009, 22, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2011. [Google Scholar]
- Bao, Z.; Cartinhour, S.; Swingle, B. Substrate and target sequence length influence RecTE(Psy) recombineering efficiency in Pseudomonas syringae. PLoS ONE 2012, 7, e50617. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Medema, M.H.; Kazempour, D.; Fischbach, M.A.; Breitling, R.; Takano, E.; Weber, T. antiSMASH 2.0-a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 2013, 41, W204–W212. [Google Scholar] [CrossRef] [PubMed]
- Rottig, M.; Medema, M.H.; Blin, K.; Weber, T.; Rausch, C.; Kohlbacher, O. NRPSpredictor2-a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011, 39, W362–W367. [Google Scholar] [CrossRef] [PubMed]
- Filloux, A.; Ramos, J.L.; Preface. Pseudomonas Methods and Protocols. Methods Mol. Biol. 2014, 1149, v. [Google Scholar] [PubMed]
- Swingle, B.; Bao, Z.; Markel, E.; Chambers, A.; Cartinhour, S. Recombineering using RecTE from Pseudomonas syringae. Appl. Environ. Microbiol. 2010, 76, 4960–4968. [Google Scholar] [CrossRef]
- Dennis, J.J.; Sokol, P.A. Electrotransformation of Pseudomonas. Methods Mol. Biol. 1995, 47, 125–133. [Google Scholar]
- Mosberg, J.A.; Lajoie, M.J.; Church, G.M. Lambda red recombineering in Escherichia coli occurs through a fully single-stranded intermediate. Genetics 2010, 186, 791–799. [Google Scholar] [CrossRef]
- Ringel, M.T.; Drager, G.; Bruser, T. PvdN enzyme catalyzes a periplasmic pyoverdine modification. J. Biol. Chem. 2016, 291, 23929–23938. [Google Scholar] [CrossRef]

| Bacterial Strains and Plasmids | Phenotypes | Sources or References |
|---|---|---|
| P. syringae | ||
| MB03 | A wild-type strain with nematocidal activity | [32] |
| MB03/RecTE | Transformed MB03 strain containing the plasmid pUCP24/recTE for the expression of the RecTE protein | This study |
| MB03∆pvdD | pvdD-disrupted mutant of MB03 | This study |
| MB03∆pvdJ | pvdJ-disrupted mutant of MB03 | This study |
| E. coli OP50 | The food source used for C. elegans N2 | Laboratory stock |
| Plasmids | ||
| pUCP24/recTE | A recombinant plasmid harboring the recTE genes | [53] |
| pKD4 | A plasmid harboring a kanamycin-resistance gene cassette | [53] |
| Primers | Sequence |
|---|---|
| pvdD up-F | 5′-CACGCAGTTGGTCGCCTA-3′ |
| pvdD up-R | 5′-GTTCCTATTCCGAAGTTCCCGGCAACAGGCTGAAATC-3′ |
| pvdD neo-F | 5′-GATTTCAGCCTGTTGCCGGGAACTTCGGAATAGGAAC-3′ |
| pvdD neo-R | 5′-TTCGCCATTTTTTCCCTGTCAGAAGAACTCGTCAAGAAG-3′ |
| pvdD dn-F | 5′-CTTCTTGACGAGTTCTTCTGACAGGGAAAAAATGGCGAA-3′ |
| pvdD dn-R | 5′-CGATGTGGTGCTGGGTCA-3′ |
| pvdD seq-F | 5′-TTCGCGGTTTCCGTATCGAGC-3′ |
| pvdD seq-R | 5′-GATCGGCAACGGTGCCAGTGT-3′ |
| pvdJ up-F | 5′-TGTCGGCGTACTTGGTGC-3′ |
| pvdJ up-R | 5′-GTTCCTATTCCGAAGTTCCTCTCCCTCACGGCAATCG-3′ |
| pvdJ neo-F | 5′-CGATTGCCGTGAGGGAGAGGAACTTCGGAATAGGAAC-3′ |
| pvdJ neo-R | 5′-CTCGAAGATCAGGCGCAGGAAGAACTCGTCAAGAAG-3′ |
| pvdJ dn-F | 5′-CTTCTTGACGAGTTCTTCTGACTGCGCCTGATCTTCGAG-3′ |
| pvdJ dn-R | 5′-GACCAGCCATCGGACACC-3′ |
| pvdJ seq-F | 5′-AAAATTCGCGGCTTCCGTATT-3′ |
| pvdJ seq-R | 5′-CGTAGTCGGCGTACTGAATCG-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bashir, A.; Tian, T.; Yu, X.; Meng, C.; Ali, M.; Li, L. Pyoverdine-Mediated Killing of Caenorhabditis elegans by Pseudomonas syringae MB03 and the Role of Iron in Its Pathogenicity. Int. J. Mol. Sci. 2020, 21, 2198. https://doi.org/10.3390/ijms21062198
Bashir A, Tian T, Yu X, Meng C, Ali M, Li L. Pyoverdine-Mediated Killing of Caenorhabditis elegans by Pseudomonas syringae MB03 and the Role of Iron in Its Pathogenicity. International Journal of Molecular Sciences. 2020; 21(6):2198. https://doi.org/10.3390/ijms21062198
Chicago/Turabian StyleBashir, Anum, Tian Tian, Xun Yu, Cui Meng, Muhammad Ali, and Lin Li. 2020. "Pyoverdine-Mediated Killing of Caenorhabditis elegans by Pseudomonas syringae MB03 and the Role of Iron in Its Pathogenicity" International Journal of Molecular Sciences 21, no. 6: 2198. https://doi.org/10.3390/ijms21062198
APA StyleBashir, A., Tian, T., Yu, X., Meng, C., Ali, M., & Li, L. (2020). Pyoverdine-Mediated Killing of Caenorhabditis elegans by Pseudomonas syringae MB03 and the Role of Iron in Its Pathogenicity. International Journal of Molecular Sciences, 21(6), 2198. https://doi.org/10.3390/ijms21062198