Physiological and Proteomic Dissection of the Responses of Two Contrasting Wheat Genotypes to Nitrogen Deficiency
Abstract
1. Introduction
2. Results
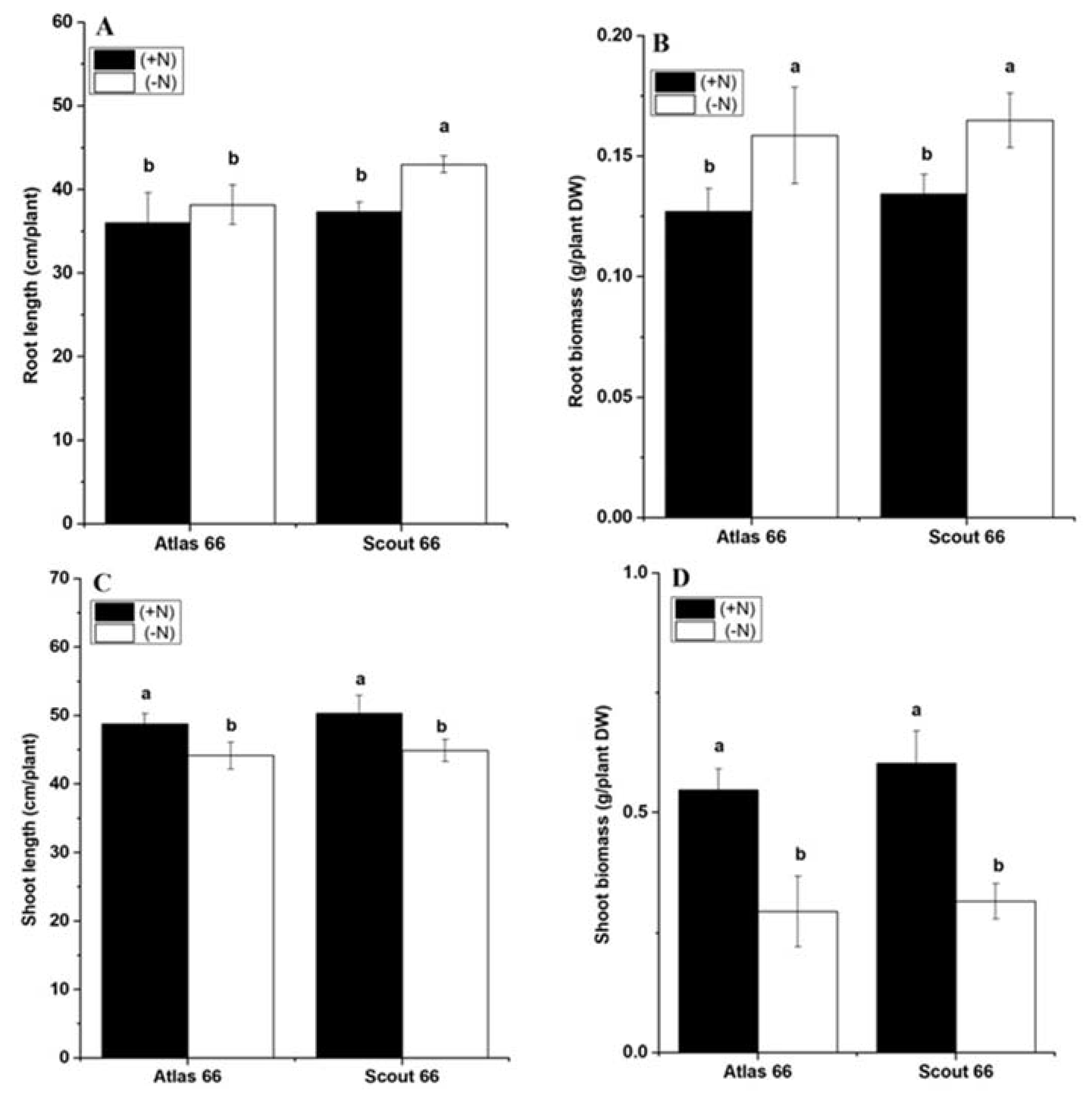
2.1. Scout 66 Showed Longer Root Length under Nitrogen Deficiency
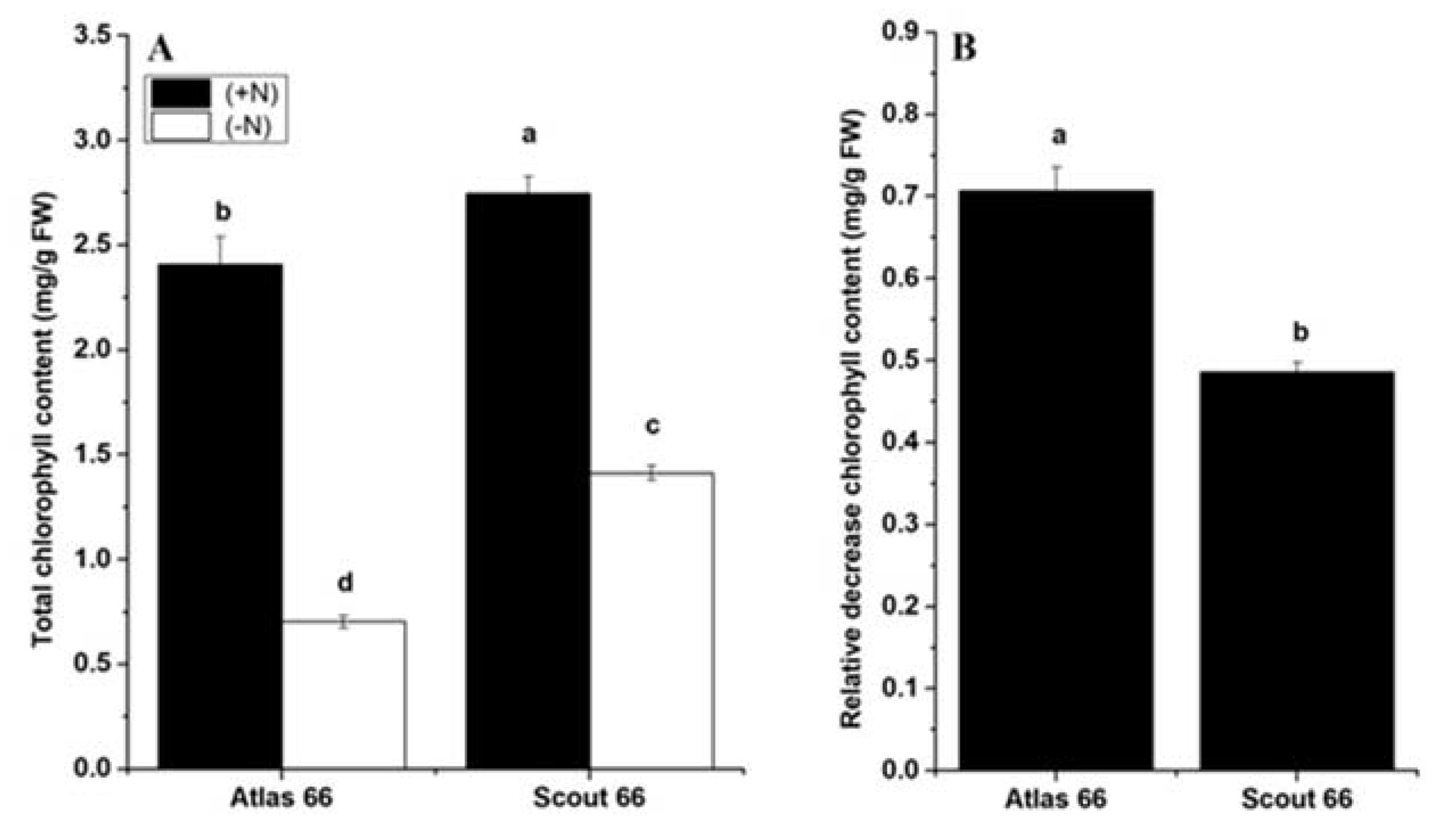
2.2. Scout 66 Contained Higher Chlorophyll Content Under Nitrogen Deficiency
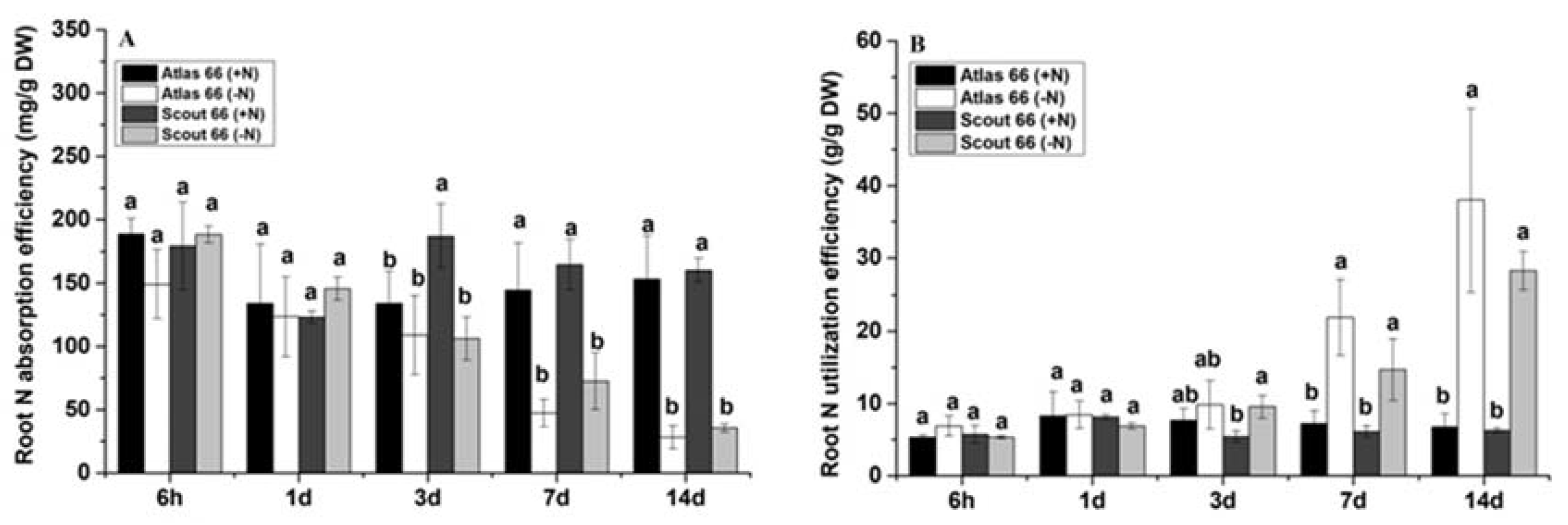
2.3. Scout 66 had Relatively Higher Nitrogen Absorption Efficiency in the Roots While Atlas 66 Showed Slightly Higher Nitrogen Utilization Efficiency in the Roots in Response to Nitrogen Deficiency
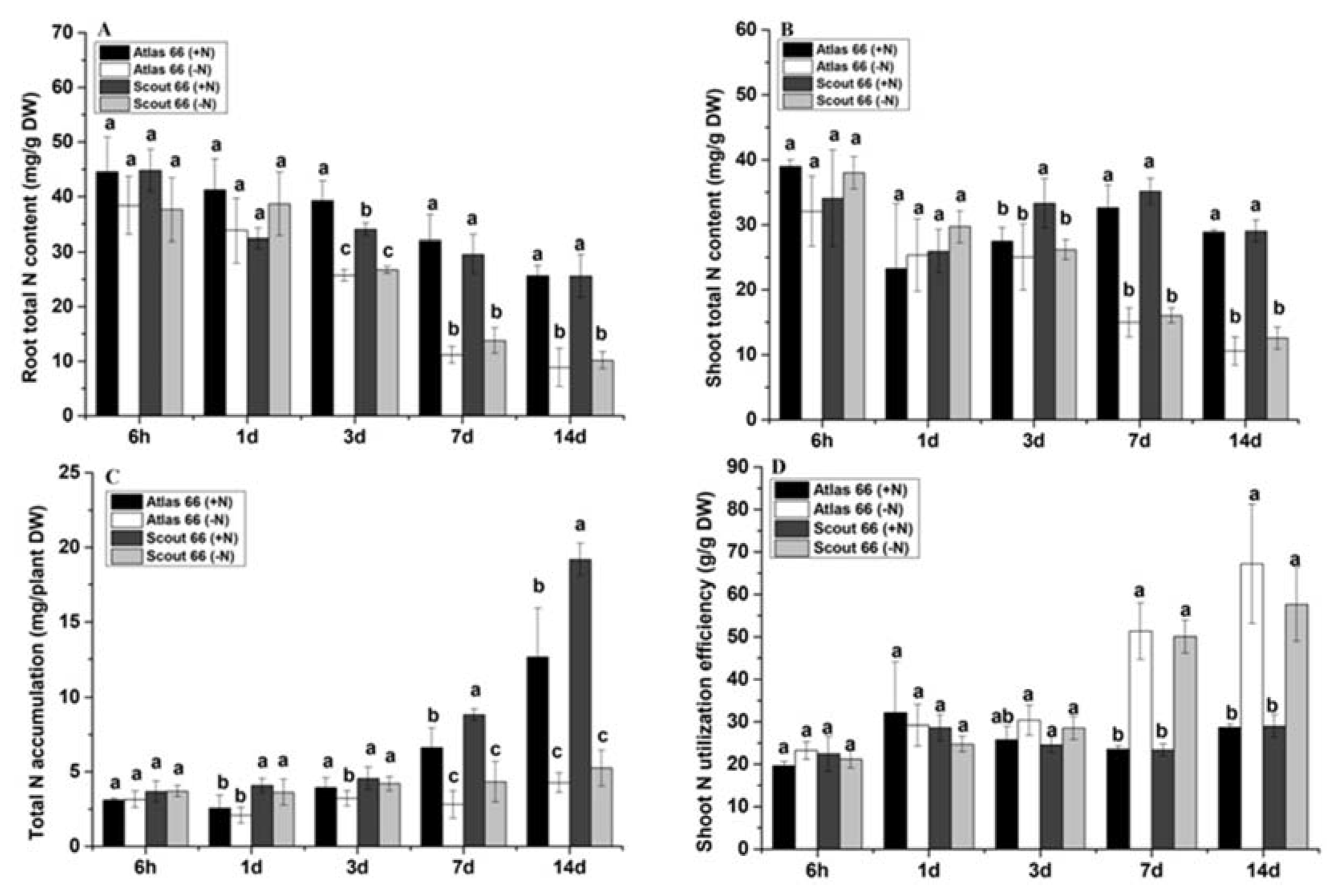
2.4. Scout 66 and Atlas 66 Showed Different Nitrogen Contents Both in the Roots and the Shoots
2.5. Scout 66 Accumulated More Nitrogen Than Atlas 66 Did
2.6. The Changes in Nitrate Concentrations of Two Wheat Cultivars in Response to Nitrogen Deficiency
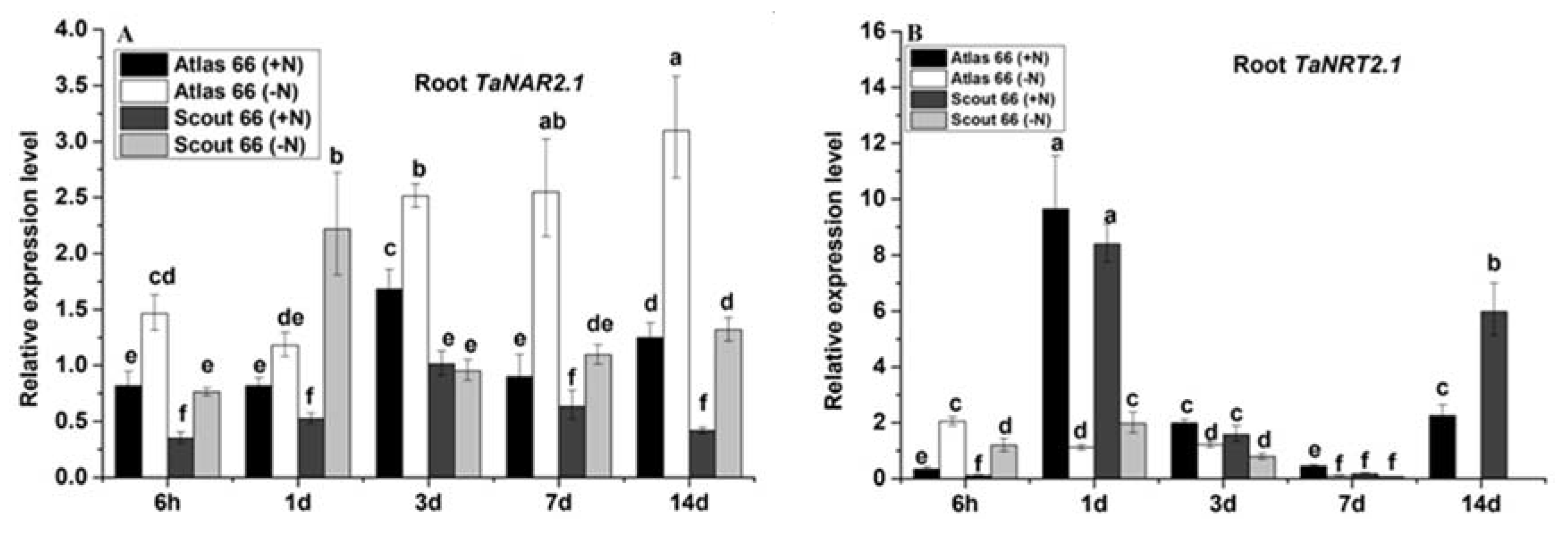
2.7. Differential Expression of Marker Genes was Induced by Nitrogen Deficiency
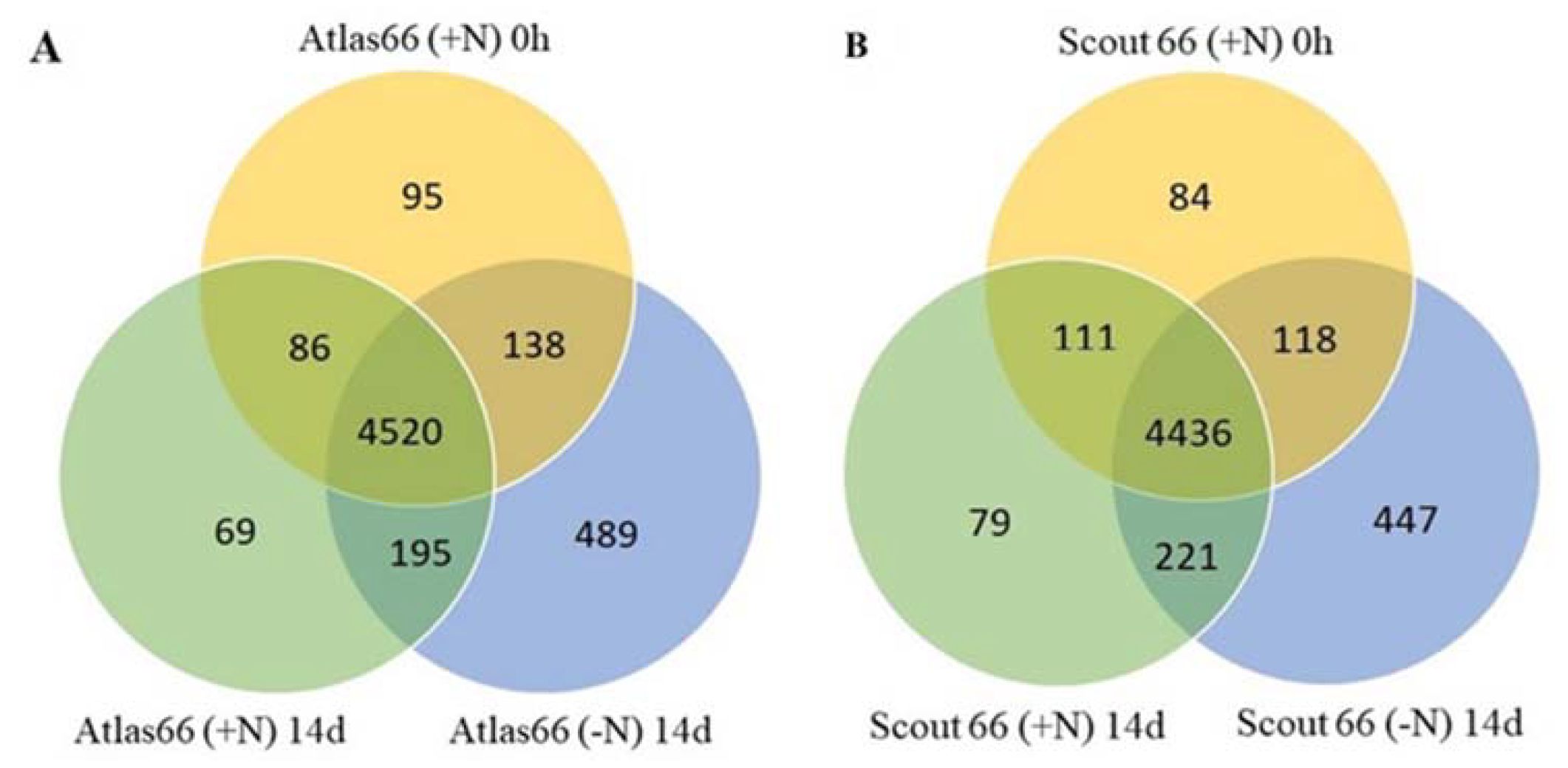
2.8. Protein Identification and Differentially Accumulated Proteins in the Two Cultivars Under Nitrogen Starvation
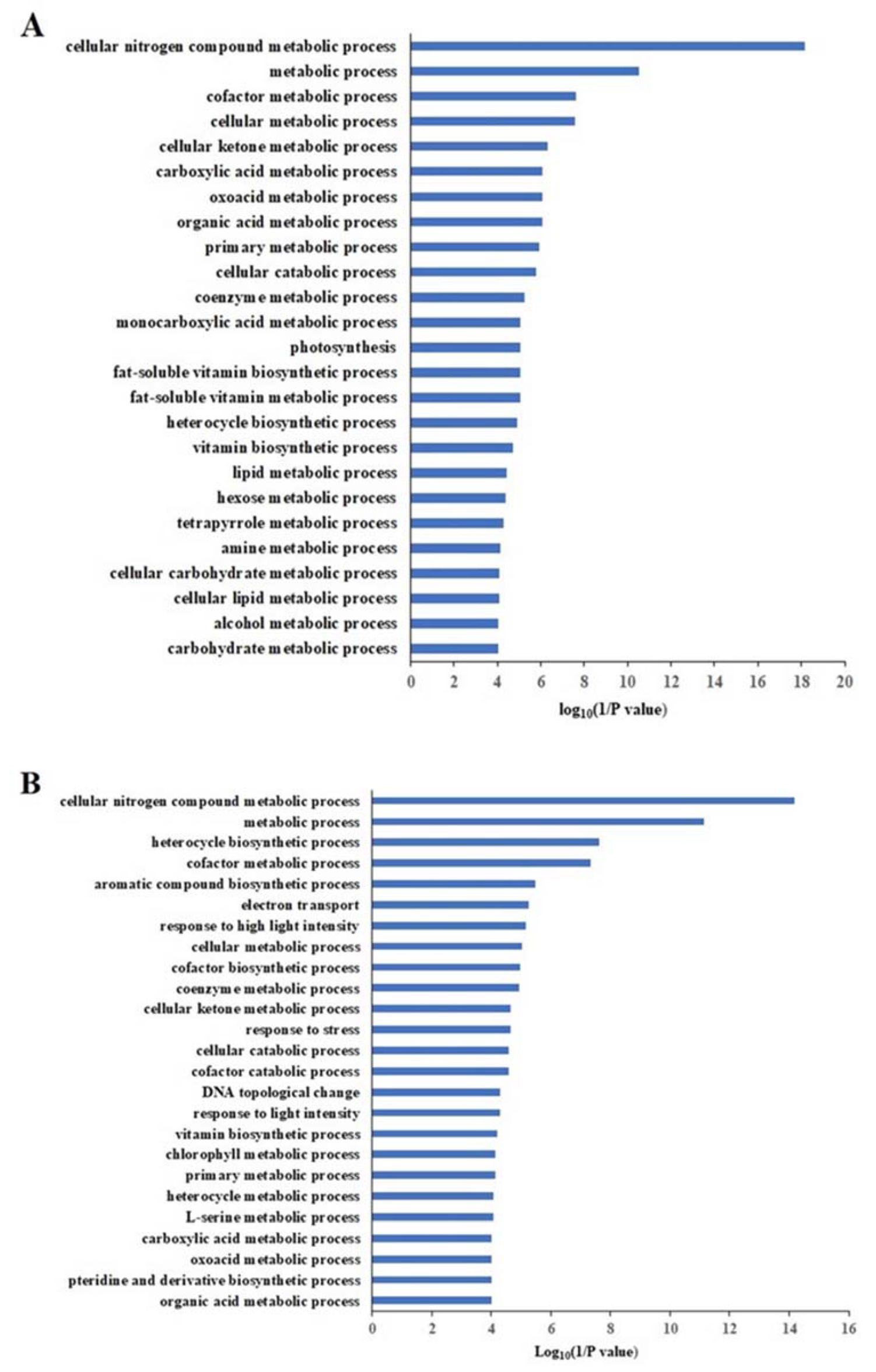
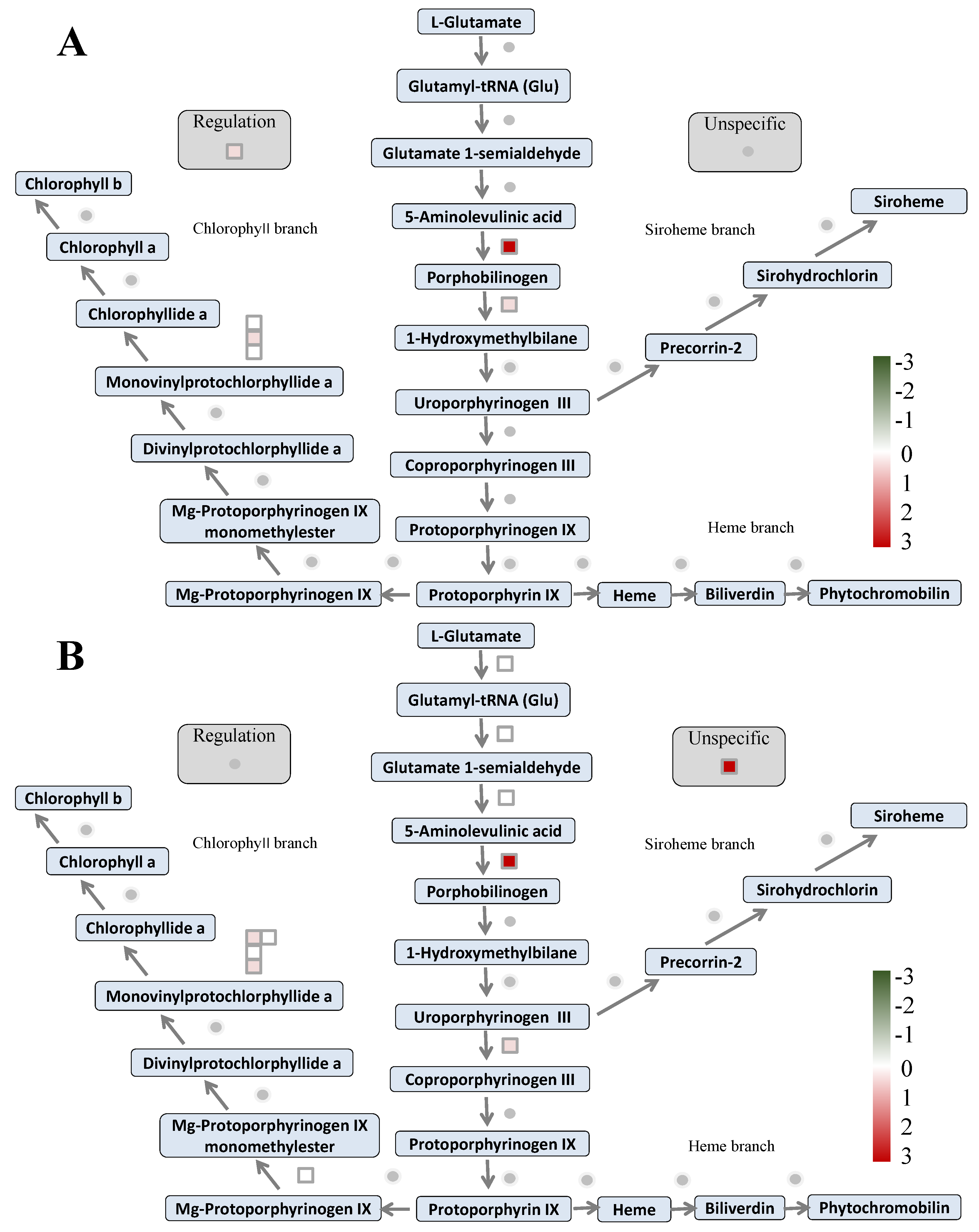
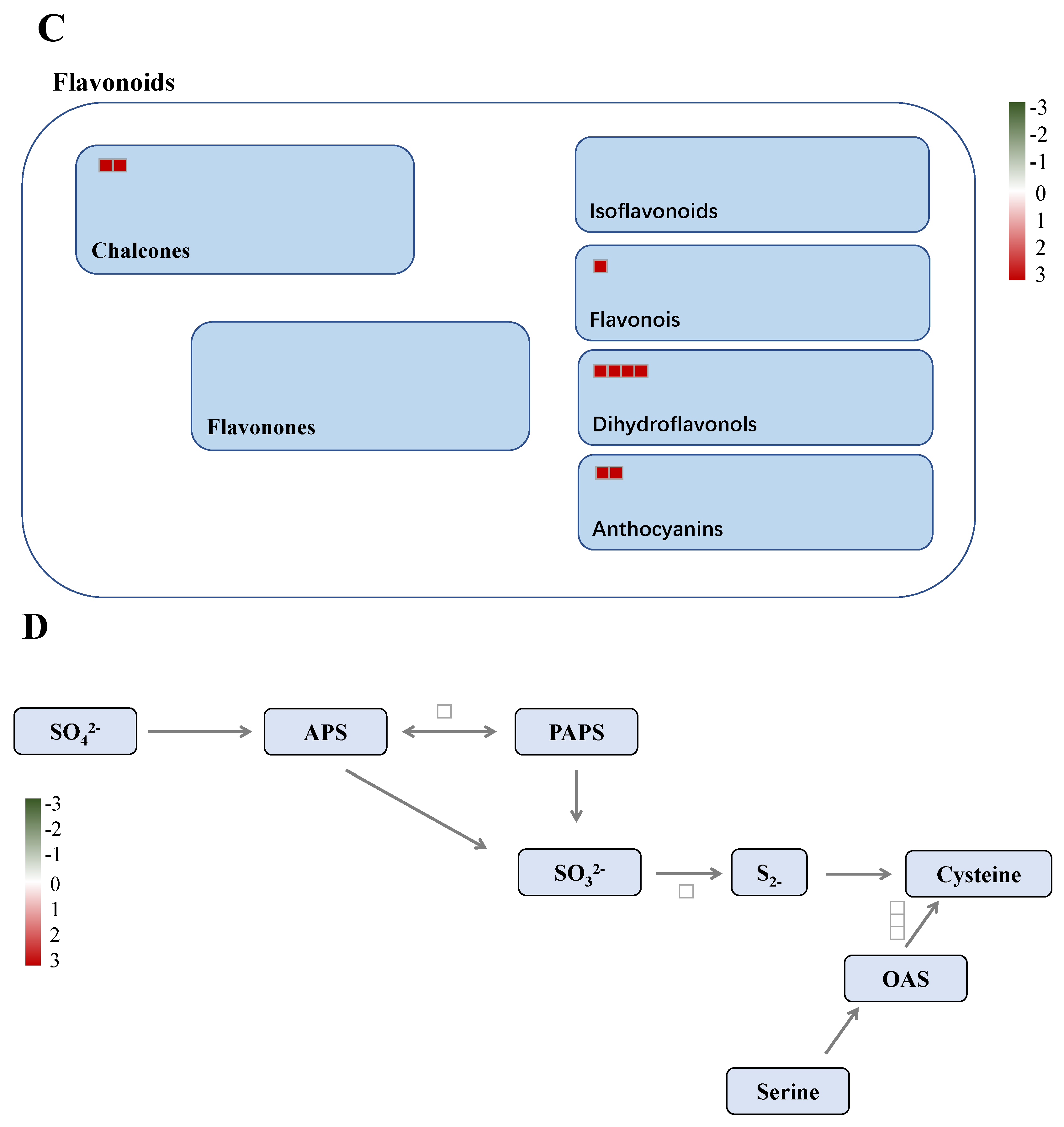
2.9. Differentially Accumulated Proteins Reveal New Aspects of the Nitrogen Deficiency Responses
2.10. Differentially Enriched Pathways Between Atlas 66 and Scout 66 Under N Deficiency
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Low Nitrogen Treatment
4.2. Physiological Assays
4.3. Determination of Total Nitrogen and Nitrate Concentrations
- Total N accumulation = root dry weight × N content of root + shoot dry weight × N content of shoot
- Root N absorption efficiency = total N accumulation/root dry weight
- N utilization efficiency = plant dry weight/total N accumulation
- Shoot N utilization efficiency = shoot dry weight/total N accumulation
- Root N utilization efficiency = root dry weight/total N accumulation
4.4. Measurement of Chlorophyll Content
4.5. Protein Extraction and Determination of Protein Concentration
4.6. Protein Digestion and Peptide Purification
4.7. LC-MS/MS Analysis
4.8. Database Search and Protein Quantification
4.9. Bioinformatics Analyses and Visualization
4.10. Expression Analysis of Different Nitrate−Transporters by Quantitative Real-Time PCR (qRT-PCR)
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kharel, T.P.; Clay, D.E.; Clay, S.A.; Beck, D.; Reese, C.; Carlson, G.; Park, H. Nitrogen and Water Stress Affect Winter Wheat Yield and Dough Quality. Agron. J. 2011, 103, 1389–1396. [Google Scholar] [CrossRef]
- Pilar, M.; de las Heras, J. Nutrient Content in Wheat Grain and Straw Using Sludge and Compost from a Wastewater Treatment Plant as a Fertiliser. J. Sci. Food Agric. 2018, 98, 4707–4714. [Google Scholar]
- West, P.C.; Gerber, J.S.; Engstrom, P.M.; Mueller, N.D.; Brauman, K.A.; Carlson, K.M.; Cassidy, E.S.; Johnston, M.; MacDonald, G.K.; Ray, D.K. Leverage Points for Improving Global Food Security and the Environment. Science 2014, 345, 325–328. [Google Scholar] [CrossRef]
- Guo, J.H.; Liu, X.J.; Zhang, Y.; Shen, J.L.; Han, W.X.; Zhang, W.F.; Christie, P.; Goulding, K.W.T.; Vitousek, P.M.; Zhang, F.S. Significant Acidification in Major Chinese Croplands. Science 2010, 327, 1008–1010. [Google Scholar] [CrossRef]
- Rehman, H.K.; Ahmad, A.; Iqbal, M.; Gucel, S.; Ozturk, M. Nitrogen-Efficient Rice Cultivars Can Reduce Nitrate Pollution. Environ. Sci. Pollut. Res. 2011, 18, 1184–1193. [Google Scholar]
- Liang, L.Z.; Zhao, X.Q.; Yi, X.Y.; Chen, Z.C.; Dong, X.Y.; Chen, R.F.; Shen, R.F. Excessive Application of Nitrogen and Phosphorus Fertilizers Induces Soil Acidification and Phosphorus Enrichment During Vegetable Production in Y Angtze R Iver D Elta, C Hina. Soil Use Manag. 2013, 29, 161–168. [Google Scholar] [CrossRef]
- Kochian, L.V.; Hoekenga, O.A.; Pineros, M.A. How Do Crop Plants Tolerate Acid Soils? Mechanisms of Aluminum Tolerance and Phosphorous Efficiency. Annu. Rev. Plant Biol. 2004, 55, 459–493. [Google Scholar] [CrossRef] [PubMed]
- Samac, D.A.; Tesfaye, M. Plant Improvement for Tolerance to Aluminum in Acid Soils—A Review. Plant Cell Tissue Organ Cult. 2003, 75, 189–207. [Google Scholar] [CrossRef]
- Boff, T.; Espindula, L.F.; Bücker-Neto, L.; Minella, E.; Milach, S.C.K.; Da-Silva, P.R. Inheritance of Aluminum Tolerance in the Wheat Cultivar Toropi and New Findings About the Introduction of This Trait into the Brazilian Wheat Germplasm. Environ. Exp. Bot. 2019, 157, 91–99. [Google Scholar] [CrossRef]
- Yan, Y.; Zhou, W.; Liang, X.; Zhou, K.; Lin, X. Increased Bound Putrescine Accumulation Contributes to the Maintenance of Antioxidant Enzymes and Higher Aluminum Tolerance in Wheat. Environ. Pollut. 2019, 252, 941–949. [Google Scholar]
- Mohammad, K.; Dong, X.; Zheng, L.; Shen, R.; Lan, P. Can Aluminum Tolerant Wheat Cultivar Perform Better under Phosphate Deficient Conditions? Int. J. Mol. Sci. 2018, 19, 2964. [Google Scholar]
- Hu, B.; Chu, C. Nitrogen-Phosphorus Interplay: Old Story with Molecular Tale. New Phytol. 2020, 225, 1455–1460. [Google Scholar] [CrossRef]
- Anna, M.; Szponarski, W.; Dangeville, P.; Safi, A.; Dissanayake, I.M.; Saenchai, C.; Emanuel, A.; Rubio, V.; Lacombe, B.; Ruffel, S. Identification of Molecular Integrators Shows That Nitrogen Actively Controls the Phosphate Starvation Response in Plants. Plant Cell 2019, 31, 1171–1184. [Google Scholar]
- Hu, B.; Jiang, Z.; Wang, W.; Qiu, Y.; Zhang, Z.; Liu, Y.; Li, A.; Gao, X.; Liu, L.; Qian, Y. Nitrate-Nrt1. 1b-Spx4 Cascade Integrates Nitrogen and Phosphorus Signalling Networks in Plants. Nat. Plants 2019, 5, 401. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Sun, J.; Tian, Z.; Hu, H.; Michel, E.J.S.; Gao, J.; Jiang, D.; Cao, W.; Dai, T. Root Extension and Nitrate Transporter up-Regulation Induced by Nitrogen Deficiency Improves Nitrogen Status and Plant Growth at the Seedling Stage of Winter Wheat (Triticum Aestivum L.). Environ. Exp. Bot. 2017, 141, 28–40. [Google Scholar] [CrossRef]
- Tong, Y.; Zhou, J.; Li, Z.; Miller, A.J. A Two-Component High-Affinity Nitrate Uptake System in Barley. Plant J. 2005, 41, 442–450. [Google Scholar] [CrossRef]
- Andrews, M.; Raven, J.A.; Lea, P.J. Do Plants Need Nitrate? The Mechanisms by Which Nitrogen Form Affects Plants. Ann. Appl. Biol. 2013, 163, 174–199. [Google Scholar] [CrossRef]
- Kronzucker, H.J.; Siddiqi, M.Y.; Glass, A.D.M. Kinetics of No3-Influx in Spruce. Plant Physiol. 1995, 109, 319–326. [Google Scholar] [CrossRef]
- Mélanie, N.; Lacombe, B. Transporters Involved in Root Nitrate Uptake and Sensing by Arabidopsis. Front. Plant Sci. 2016, 7, 1391. [Google Scholar]
- Takatoshi, K.; Krapp, A. Plant Nitrogen Acquisition under Low Availability: Regulation of Uptake and Root Architecture. Plant Cell Physiol. 2016, 57, 707–714. [Google Scholar]
- Mathilde, O.; Chopin, F.; Leleu, O.; Smith, S.J.; Krapp, A.; Daniel-Vedele, F.; Miller, A.J. Characterization of a Two-Component High-Affinity Nitrate Uptake System in Arabidopsis. Physiology and Protein-Protein Interaction. Plant Physiol. 2006, 142, 1304–1317. [Google Scholar]
- François, T.; le Gouis, J.; Gouache, D.; Gibon, Y.; Allard, V. Post-Flowering Nitrate Uptake in Wheat Is Controlled by N Status at Flowering, with a Putative Major Role of Root Nitrate Transporter Nrt2. 1. PLoS ONE 2015, 10, e0120291. [Google Scholar]
- The International Wheat Genome Sequencing Consortium (IWGSC). A Chromosome-Based Draft Sequence of the Hexaploid Bread Wheat (Triticum Aestivum) Genome. Science 2014, 345, 1251788. [Google Scholar] [CrossRef] [PubMed]
- Rudi, A.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J. Shifting the Limits in Wheat Research and Breeding Using a Fully Annotated Reference Genome. Science 2018, 361, eaar7191. [Google Scholar]
- Lan, P.; Li, W.; Schmidt, W. Complementary Proteome and Transcriptome Profiling in Phosphate-Deficient Arabidopsis Roots Reveals Multiple Levels of Gene Regulation. Mol. Cell. Proteom. 2012, 11, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Xuan, H.; Yang, Y.; Wang, L.; Wei, L.; Wang, Y.; Kang, G. Transcription Analysis of Genes Encoding the Wheat Root Transporter Nrt1 and Nrt2 Families During Nitrogen Starvation. J. Plant Growth Regul. 2014, 33, 837–848. [Google Scholar] [CrossRef]
- Giehl, R.F.H.; von Wirén, N.R. Nutrient Foraging. Plant Physiol. 2014, 166, 509–517. [Google Scholar] [CrossRef]
- Lie, T.A. Environmental Physiology of the Legume-Rhizobium Symbiosis. Nitrogen Fixat. Ecol. 1981, 1, 104–134. [Google Scholar]
- Crawford, N.M.; Glass, A.D.M. Molecular and Physiological Aspects of Nitrate Uptake in Plants. Trends Plant Sci. 1998, 3, 389–395. [Google Scholar] [CrossRef]
- Stitt, M. Nitrate Regulation of Metabolism and Growth. Curr. Opin. Plant Biol. 1999, 2, 178–186. [Google Scholar] [CrossRef]
- Hawkesford, M.J. Genetic Variation in Traits for Nitrogen Use Efficiency in Wheat. J. Exp. Bot. 2017, 68, 2627–2632. [Google Scholar] [CrossRef] [PubMed]
- Hawkesford, M.J.; Griffiths, S. Exploiting Genetic Variation in Nitrogen Use Efficiency for Cereal Crop Improvement. Curr. Opin. Plant Biol. 2019, 49, 35–42. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, S.A.; Ciampitti, I.A.; Slafer, G.A.; Lollato, R.P. Nitrogen Utilization Efficiency in Wheat: A Global Perspective. Eur. J. Agron. 2020, 114, 126008. [Google Scholar] [CrossRef]
- Garnett, T.; Plett, D.; Conn, V.; Conn, S.; Rabie, H.; Rafalski, J.A.; Dhugga, K.; Tester, M.A.; Kaiser, B.N. Variation for N Uptake System in Maize: Genotypic Response to N Supply. Front. Plant Sci. 2015, 6, 936. [Google Scholar] [CrossRef] [PubMed]
- Grechi, I.P.H.V.; Hilbert, G.; Milin, S.; Robert, T.; Gaudillère, J.-P. Effect of Light and Nitrogen Supply on Internal C: N Balance and Control of Root-to-Shoot Biomass Allocation in Grapevine. Environ. Exp. Bot. 2007, 59, 139–149. [Google Scholar] [CrossRef]
- Laugier, E.; Bouguyon, E.; Mauriès, A.; Tillard, P.; Gojon, A.; Lejay, L. Regulation of High-Affinity Nitrate Uptake in Roots of Arabidopsis Depends Predominantly on Posttranscriptional Control of the Nrt2. 1/Nar2. 1 Transport System. Plant Physiol. 2012, 158, 1067–1078. [Google Scholar] [CrossRef]
- Bertauski, A.F.; Swiader, J.M.; Wehner, D.J. Dry Weight Production and Nitrogen Efficiency Traits in Kentucky Bluegrass Cultivars in Nutrient Solution and Soil. Crop Sci. 1997, 37, 1548–1553. [Google Scholar] [CrossRef]
- Gojon, A.; Krouk, G.; Perrine-Walker, F.; Laugier, E. Nitrate Transceptor (S) in Plants. J. Exp. Bot. 2011, 62, 2299–2308. [Google Scholar] [CrossRef]
- Ruffel, S.; Gojon, A. Systemic Nutrient Signalling: On the Road for Nitrate. Nat. Plants 2017, 3, 1–2. [Google Scholar] [CrossRef]
- Ruffel, S.; Gojon, A.; Lejay, L. Signal Interactions in the Regulation of Root Nitrate Uptake. J. Exp. Bot. 2014, 65, 5509–5517. [Google Scholar] [CrossRef]
- Abenavoli, M.R.; Longo, C.; Lupini, A.; Miller, A.J.; Araniti, F.; Mercati, F.; Princi, M.P.; Sunseri, F. Phenotyping Two Tomato Genotypes with Different Nitrogen Use Efficiency. Plant Physiol. Biochem. 2016, 107, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Ingenbleek, Y. The Nutritional Relationship Linking Sulfur to Nitrogen in Living Organisms. J. Nutr. 2006, 136, 1641s–1651s. [Google Scholar] [CrossRef] [PubMed]
- Castro, A.; Stulen, I.; Posthumus, F.S.; de Kok, L.J. Changes in Growth and Nutrient Uptake in Brassica Oleracea Exposed to Atmospheric Ammonia. Ann. Bot. 2006, 97, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Maathuis, F.J.M. Physiological Functions of Mineral Macronutrients. Curr. Opin. Plant Biol. 2009, 12, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Eisenhut, M.; Hocken, N.; Weber, A.P. Plastidial Metabolite Transporters Integrate Photorespiration with Carbon, Nitrogen, and Sulfur Metabolism. Cell Calcium 2015, 58, 98–104. [Google Scholar] [CrossRef]
- Bradstreet, R.B. Kjeldahl Method for Organic Nitrogen. Anal. Chem. 1954, 26, 185–187. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, Y. Nitrate Assay for Plant Tissues. Plant Cell 2016, 7, 1–5. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 1949, 24, 1. [Google Scholar] [CrossRef]
- Lan, P.; Li, W.; Wen, T.; Shiau, J.; Wu, Y.; Lin, W.; Schmidt, W. Itraq Protein Profile Analysis of Arabidopsis Roots Reveals New Aspects Critical for Iron Homeostasis. Plant Physiol. 2011, 155, 821–834. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Hein, M.Y.; Cox, J.; Mann, M. A Proteomic Ruler for Protein Copy Number and Concentration Estimation without Spike-in Standards. Mol. Cell. Proteom. 2014, 13, 3497–3506. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. Agrigo V2.0: A Go Analysis Toolkit for the Agricultural Community, 2017 Update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
- Daniele, M.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment Map: A Network-Based Method for Gene-Set Enrichment Visualization and Interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar]
- Jüri, R.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C. Pathway Enrichment Analysis and Visualization of Omics Data Using G: Profiler, Gsea, Cytoscape and Enrichmentmap. Nat. Protoc. 2019, 14, 482. [Google Scholar]
- Thimm, O.; Blasing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Kruger, P.; Selbig, J.; Muller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A User-Driven Tool to Display Genomics Data Sets onto Diagrams of Metabolic Pathways and Other Biological Processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative Pcr and the 2− Δδct Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




| Cultivar | Accession | Function | Abundance Ratio: (−N) 14 d/(+N) 14 d | Abundance Ratio Adj. p-value: (−N) 14 d/(+N) 14d | Abundance Ratio Variability (%): (−N) 14 d/(+N) 14 d | Metabolic Pathway |
|---|---|---|---|---|---|---|
| Atlas 66 | TraesCS7B02G314800.1 | ALA dehydratase | 100 | 9.00943820224719E-17 | 0 | Tetrapyrrole synthesis |
| TraesCS6D02G132500.2 | Porphobilinogen deaminase | 0.434 | 0.027271111 | 0.11 | ||
| TraesCS2D02G563600.1 | Protochlorophyllide reductase | 0.01 | 9.00943820224719E-17 | 0 | ||
| TraesCS2A02G590600.1 | Protochlorophyllide reductase | 0.153 | 2.55508858168777E-07 | 0.4 | ||
| TraesCS1A02G171000.1 | Protochlorophyllide reductase | 0.403 | 0.0352935604237759 | 2.4 | ||
| TraesCS4D02G081500.1.cds1 | Regulation | 0.448 | 0.0329941173590985 | 0.56 | ||
| Scout 66 | TraesCS7B02G314800.1 | ALA dehydratase | 100 | 8.5793991416309E-17 | 0 | |
| TraesCS1B02G075200.1 | Glu-tRNA synthetase | 0.01 | 8.5793991416309E-17 | 0 | ||
| TraesCS6A02G102500.1 | Glu-tRNA synthetase | 0.01 | 8.5793991416309E-17 | 0 | ||
| TraesCS7D02G261800.1 | GSA(Glutamate-1-semialdehyde aminotransferase) | 0.01 | 8.5793991416309E-17 | 0 | ||
| TraesCS7D02G062900.1.cds1 | Magnesium protoporphyrin IX methyltransferase | 0.01 | 8.5793991416309E-17 | 0 | ||
| TraesCS2D02G563600.1 | Protochlorophyllide reductase | 0.01 | 8.5793991416309E-17 | 0 | ||
| TraesCS2A02G590600.1 | Protochlorophyllide reductase | 0.15 | 3.40978161045871E-09 | 0.32 | ||
| TraesCS1A02G171000.1 | Protochlorophyllide reductase | 0.372 | 0.00420395928177109 | 3.03 | ||
| TraesCS1D02G168700.2 | Protochlorophyllide reductase | 0.401 | 0.00463374616837516 | 7.79 | ||
| TraesCS5D02G364100.1 | Unspecified | 100 | 8.5793991416309E-17 | 0 | ||
| TraesCS3D02G228800.1 | Uroporphyrinogen decarboxylase | 0.498 | 0.0491864992180654 | 4.99 | ||
| Atlas 66 | TraesCS6A02G017500.2 | Nitrate reductase | 0.01 | 9.00944E-17 | 0 | N metabolism |
| TraesCS2D02G388800.1 | Glutamate dehydrogenase | 3.763 | 0.023094541 | 9.7 | ||
| TraesCS5B02G437100.2 | Glutamate dehydrogenase | 100 | 9.00944E-17 | 0 | ||
| TraesCS6B02G327500.1 | Glutamine synthetase | 4.162 | 0.01132758 | 12.21 | ||
| TraesCS6A02G333900.1 | Nitrite reductase | 0.095 | 4.82064E-10 | 0.17 | ||
| TraesCS6B02G364600.1 | Nitrite reductas | 0.219 | 9.96743E-05 | 0.5 | ||
| TraesCS6D02G313100.1 | Nitrite reductase | 0.274 | 0.000867051 | 0.28 | ||
| Scout 66 | TraesCS6A02G017500.2 | Nitrite reductase | 0.01 | 8.5794E-17 | 0 | |
| TraesCS5B02G437100.2 | Glutamate dehydrogenase | 100 | 8.5794E-17 | 0 | ||
| TraesCS2D02G388800.1 | Glutamate dehydrogenase | 3.418 | 0.016723292 | 1.57 | ||
| TraesCS6B02G327500.1 | Glutamate dehydrogenase | 3.47 | 0.004060041 | 17.17 | ||
| TraesCS5B02G414000.3 | Nitrate transporter | 100 | 8.5794E-17 | 0 | ||
| TraesCS6B02G364600.1 | Nitrite reductase | 0.298 | 0.000655234 | 2.21 | ||
| TraesCS6A02G333900.1 | Nitrite reductase | 0.183 | 1.33016E-06 | 0.07 | ||
| TraesCS6D02G313100.1 | Nitrite reductase | 0.324 | 0.000403989 | 1.26 | ||
| Atlas 66 | TraesCS4A02G437900.1 | Naringenin-chalcone synthase | 11.196 | 3.95457E-05 | 48.59 | Secondary metabolism flavonoids |
| TraesCS1A02G021100.1.cds1 | Anthocyanin 5-aromatic acyltransferase | 100 | 9.00944E-17 | 0 | ||
| TraesCS1D02G339200.1.cds1 | Dihydroflavonols | 100 | 9.00944E-17 | 0 | ||
| TraesCS4B02G335300.2 | Flavonols | 100 | 9.00944E-17 | 0 | ||
| TraesCS5D02G476400.1.cds1 | Dihydroflavonols | 100 | 9.00944E-17 | 0 | ||
| TraesCS3B02G013100.1.cds1 | Dihydroflavonols | 100 | 9.00944E-17 | 0 | ||
| TraesCS4A02G436200.1.cds1 | Naringenin-chalcone synthase | 100 | 9.00944E-17 | 0 | ||
| TraesCS3D02G001400.1.cds1 | Dihydroflavonols | 100 | 9.00944E-17 | 0 | ||
| Scout 66 | TraesCS1D02G324200.1 | Sulfite reductase (SIR) | 0.01 | 8.5794E-17 | 0 | Sulfate assimilation |
| TraesCS5B02G018700.2 | OAS-TL A(O-acetylserine sulfhydrylase, O-acetylserine (Thiol)-lyase) (CSase A)) | 0.324 | 0.000478246 | 0.56 | ||
| TraesCS7D02G096700.1 | OAS-TL A(O-acetylserine sulfhydrylase, O-acetylserine (Thiol)-lyase) (CSase A)) | 0.317 | 0.000903302 | 2.09 | ||
| TraesCS2A02G184200.1 | Adenylylsulphate kinase | 0.01 | 8.5794E-17 | 0 | ||
| TraesCS5D02G027600.1 | OAS-TL A(O-acetylserine sulfhydrylase, O-acetylserine (Thiol)-lyase) (CSase A)) | 0.408 | 0.008948742 | 2.79 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karim, M.R.; Wang, R.; Zheng, L.; Dong, X.; Shen, R.; Lan, P. Physiological and Proteomic Dissection of the Responses of Two Contrasting Wheat Genotypes to Nitrogen Deficiency. Int. J. Mol. Sci. 2020, 21, 2119. https://doi.org/10.3390/ijms21062119
Karim MR, Wang R, Zheng L, Dong X, Shen R, Lan P. Physiological and Proteomic Dissection of the Responses of Two Contrasting Wheat Genotypes to Nitrogen Deficiency. International Journal of Molecular Sciences. 2020; 21(6):2119. https://doi.org/10.3390/ijms21062119
Chicago/Turabian StyleKarim, Mohammad Rezaul, Ruonan Wang, Lu Zheng, Xiaoying Dong, Renfang Shen, and Ping Lan. 2020. "Physiological and Proteomic Dissection of the Responses of Two Contrasting Wheat Genotypes to Nitrogen Deficiency" International Journal of Molecular Sciences 21, no. 6: 2119. https://doi.org/10.3390/ijms21062119
APA StyleKarim, M. R., Wang, R., Zheng, L., Dong, X., Shen, R., & Lan, P. (2020). Physiological and Proteomic Dissection of the Responses of Two Contrasting Wheat Genotypes to Nitrogen Deficiency. International Journal of Molecular Sciences, 21(6), 2119. https://doi.org/10.3390/ijms21062119






