Abstract
The ability of stem cells to divide and differentiate is necessary for tissue repair and homeostasis. Appropriate spatial and temporal mechanisms are needed. Local intercellular signaling increases expression of specific genes that mediate and maintain differentiation. Diffusible signaling molecules provide concentration-dependent induction of specific patterns of cell types or regions. Differentiation of adjacent cells, on the other hand, requires cell–cell contact and subsequent signaling. These two types of signals work together to allow stem cells to provide what organisms require. The ability to grow organoids has increased our understanding of the cellular and molecular features of small “niches” that modulate stem cell function in various organs, including the small intestine.
1. Introduction
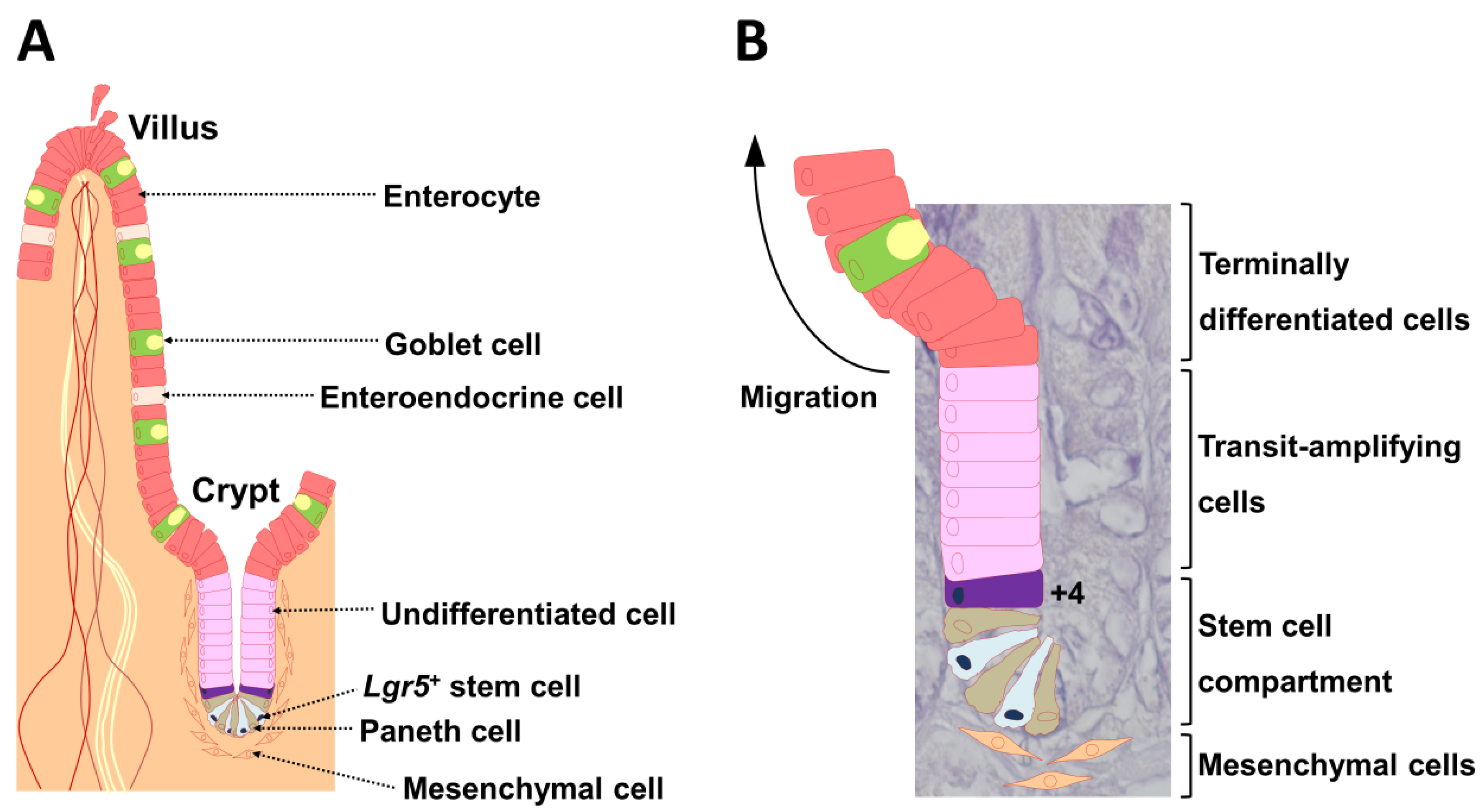
The intestinal epithelium undergoes rapid and continuous self-renewal, cell commitment, and cell differentiation along the crypt-villus axis throughout postnatal life. Cheng and Leblond [1] were the first to describe crypt base columnar cells at the crypt bottom that interact with Paneth cells. Paneth cells are intestinal epithelial cells that control stem cells via production of bactericidal products [2] and expression of critical niche signals such as epidermal growth factor, transforming growth factor-α, Wnt3, and delta-like ligand 4 (Dll4), which is the ligand for Notch [3]. Stem cells present at the crypt bottom produce a temporary group of undifferentiated cells that divide rapidly while moving toward the intestinal lumen (Figure 1A) [4]. Subepithelial mesenchymal cells are close to crypts (Figure 1). During migration, these cells differentiate into cells of various lineages; they cease division and differentiate when they reach the top of the crypts (Figure 1A) [4]. Goblet cells, enteroendocrine cells, and absorptive cells, which are also intestinal epithelial cells, move toward the gut lumen, and the Paneth cells move to the crypt bottom (Figure 1A) [5,6]. Intestinal stem cells (ISCs) comprise two functionally distinct populations. Marked leucine-rich repeat-containing G-protein coupled receptor 5 (Lgr5)+ cells persist for the lifetime in mice, whereas their progeny includes all differentiated cell lineages of the epithelium (Figure 1B) [7]. Thus, Lgr5+ cells represent cycling and long-lived multipotent stem cells. In contrast, B-cell-specific Moloney murine leukemia virus integration site 1 (Bmi1) marks quiescent ISCs, which are located at position +4 from the base of the crypt (Figure 1B). Bmi1+ cells are insensitive to Wnt signaling, contribute weakly to homeostatic regeneration, and are resistant to injury from high-dose radiation [8].
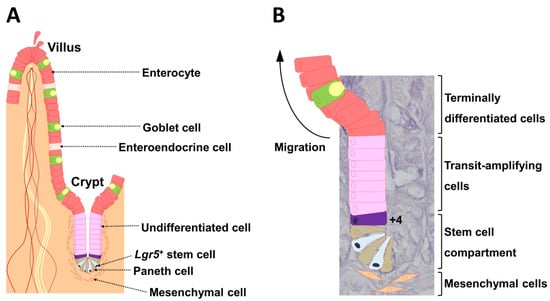
Figure 1.
The crypt-villus unit of the small intestinal epithelium. (A) The epithelium is mainly composed of enterocytes, goblet cells, enteroendocrine cells, and Paneth cells. Lgr5+ stem cells reside at the very bottom of the crypt (position +1) and are usually flanked on both sides by Paneth cells. Subepithelial mesenchymal cells are localized near Lgr5+ stem cells. (B) The hierarchical organization is illustrated in a 4′,6-diamidino-2-phenylindole (DAPI)-stained crypt. Daughter cells differentiate into functionally discrete populations of intestinal epithelial cells as they migrate up from the crypt, and are extruded into the gut lumen as they reach the villus tip in the small intestine. The cell located at position +4 from the base of the crypt also undergoes self-renewal and gives rise to all differentiated cell lineages of the small intestinal epithelium. Lgr5: leucine-rich, repeat-containing G-protein coupled receptor 5 [9].
Both development of the small intestine and homeostasis of the adult gut require canonical Wnt signaling involving β-catenin and the T-cell factor/lymphoid-enhancer-binding factor family of transcription factors [10,11,12,13,14]. Proliferation in the crypts is compromised in mice lacking canonical Wnt pathway molecules [15]. The central role of Wnt signaling is highlighted by Wnt-dependent expression of numerous ISC markers, including Lgr5 [16]. Beyond its role in maintaining ISCs, Wnt signaling mediates secretory cell fate decision. Specifically, Wnt signaling plays a role in Paneth cell differentiation [17,18], and overexpression of the Wnt inhibitor dickkopf 1 leads to loss of all secretory cells [19].
Homeostasis of ISCs requires active Notch receptor signaling, as shown with lineage tracing in murine genetic models [20,21] and pharmacological models. Epithelial cell effects have been examined in intestinal organoid cultures [22], which exclude possible influences from Notch signaling in immune, neuronal, and mesenchymal cells. Intestinal organoids grow rapidly and show stable differentiation and are thus useful for nonmutational and genetic experiments [23,24,25]. Overall, organoids are expected to provide critical insight into human and murine ISC signaling regulation.
Erythropoietin-producing hepatocellular carcinoma cell (Eph) receptors and Eph receptor interacting proteins (ephrins) are major players in morphogenesis, where they establish and maintain the organization of cell types or regional domains within tissues [26,27,28]. Eph receptors and ephrins are divided into two classes, in which EphA receptors bind to glycosyl phosphatidyl inositol (GPI) moiety-anchored ephrinAs, and EphB receptors bind to transmembrane ephrinBs [28,29,30]. Multiple tissues and developmental processes involve cross-talk between Wnt and Eph/ephrin signaling pathways [31,32]. Eph/ephrin-mediated cross-talk between epithelial cells controls Wnt signaling and hence spatial regulation of cells located in the crypt-villus axis [4,33]. Cell migration is mediated by Eph receptors and ephrin ligands in many instances [34,35,36]. Eph receptors and ephrins, which are both membrane-bound, only interact through direct cell–cell contact, and signaling through these molecules is bi-directional. Signaling in the receptor-expressing cell is known as forward signaling, and signaling in the ligand-expressing cell is called reverse signaling [27,34,35,37].
The Hippo pathway and its effectors, yes-associated protein (YAP) and yorkie, play a role in intestinal regeneration following tissue injury in mice and Drosophila melanogaster, respectively. Activation of the Hippo pathway blocks YAP activity in ISCs via phosphorylation, which induces YAP to remain in the cytoplasm where it interacts with β-catenin and blocks canonical Wnt signaling [38]. Conflicting studies have described the importance of YAP in ISC proliferation and intestinal repair in multiple animal models and experimental paradigms [38]. A recent study [39] established intestinal organoids from single cells. The ability of the intestine to regenerate, which is mediated by transient YAP1 activation, was retained in these organoids.
This review will discuss how ISC division, differentiation, and homeostasis are controlled, how stem cell signaling affects the cell’s position in the crypt-villus axis, and the importance of these observations in development.
2. The ISC Niche
2.1. R-Spondin-LGR Signaling
Wnt signaling is an evolutionarily conserved pathway that plays a pivotal role in stem cell control. The pathway is tightly controlled not only by ligands and receptors but also by various activators and inhibitors [40]. The ligands, Wnt3, -6, and -9b, are secreted mainly from epithelial cells including Paneth cells, whereas Wnt2b, -4, and -5b are secreted from mesenchymal cells in the intestine [41]. Wnt3 or Paneth cell ablation in vivo does not affect intestinal homeostasis [42,43], indicating that excess biological interactions among ISCs, Paneth cells, and mesenchymal cells maintain the ISC niche via Wnt ligands.
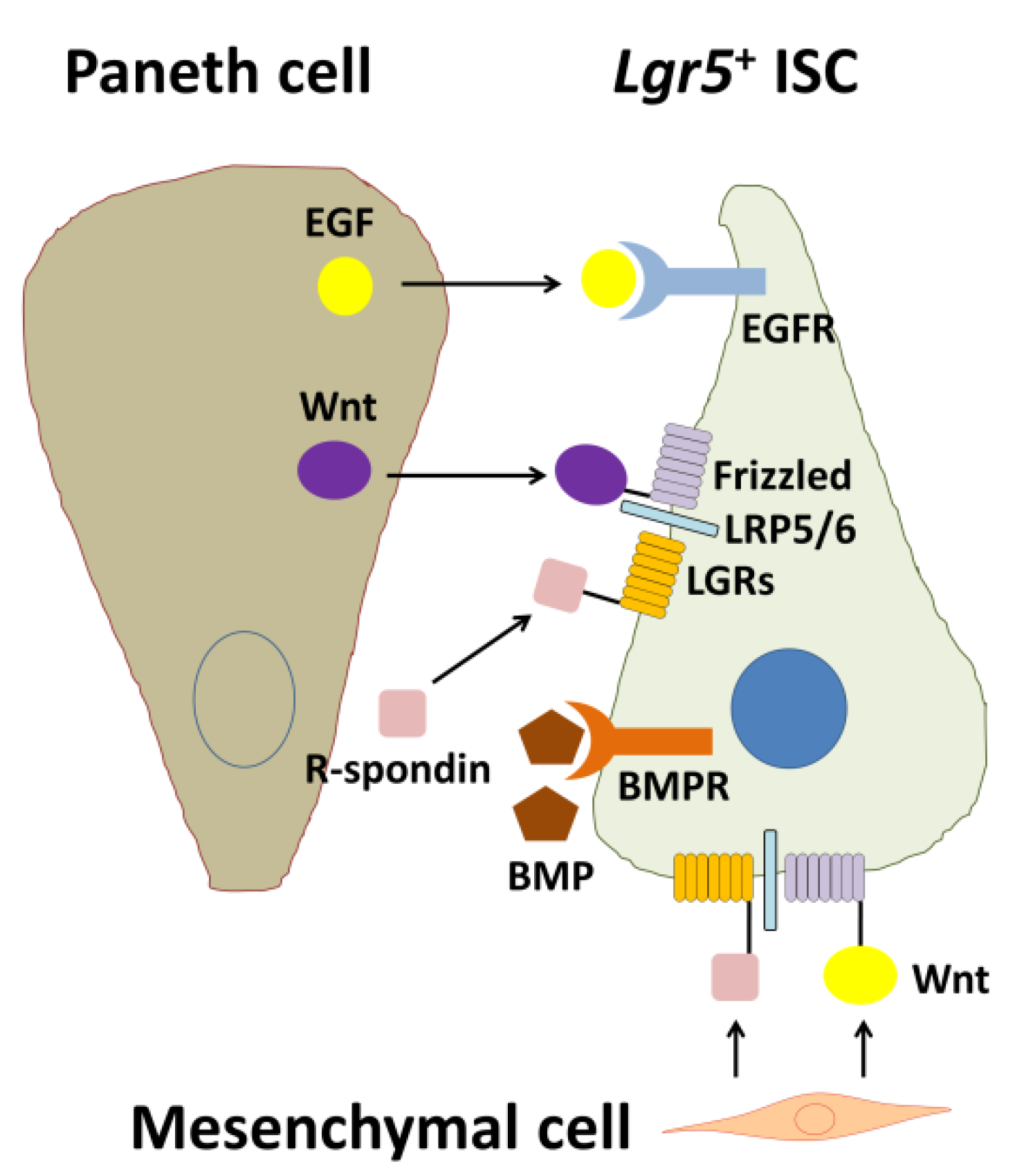
Wnt signaling engages two transmembrane receptor classes, Frizzed (Fz)-type 7-transmembrane proteins and lipoprotein receptor-related proteins 5 and 6 (LRP5/6) (Figure 2). Kazanskaya et al. [44] first identified Xenopus laevis R-spondin2 (Rspo2) as a secreted activator of Wnt signaling and showed that Rspo2 is regulated by Wnts and directly activates Wnt signaling. Xenopus Rspo2 is coexpressed with Wnts in a variety of tissues and can be ectopically activated by Wnt signaling [44]. Similarly, mouse Rspo1 expression is downregulated in mouse Wnt1 and Wnt3a mutants [45]. As Rsop2 is a secreted protein, Rspo2 may function extracellularly at the level of receptor–ligand interactions during Wnt signaling.
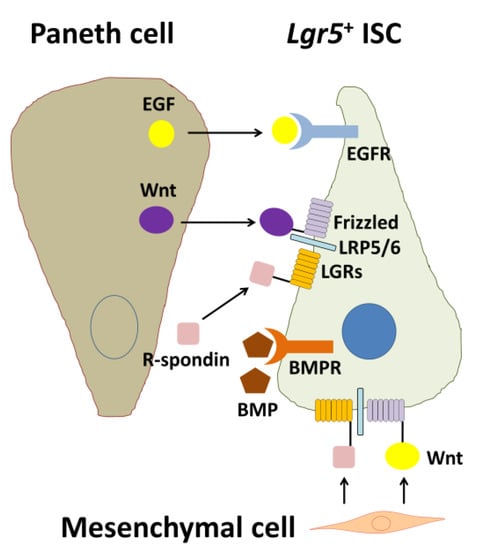
Figure 2.
Biological interactions among an Lgr5+ intestinal stem cell (ISC), a Paneth cell, and a mesenchymal cell. EGF and Wnt are secreted from the Paneth cell and are essential for stemness of the Lgr5+ ISC. In contrast, BMP negatively regulates the stemness. For full Wnt activation, R-spondin-LGR signaling is required. In addition, Wnt and R-spondins, which are factors produced by mesenchymal cells, are also essential for maintenance of Lgr5+ ISCs. EGF: epidermal growth factor, EGFR: EGF receptor, BMP: bone morphogenetic protein, BMPR: BMP receptor, LRP5/6: lipoprotein receptor-related protein 5/6, LGRs: leucine-rich repeat-containing G-protein coupled receptors, Lgr5: leucine-rich repeat-containing G-protein coupled receptor 5 [9].
The R-spondin protein family is composed of four human paralogs (R-spondin1–4) (RSpo1–4), each of which contains a leading signal peptide, two cysteine-rich, furin-like domains, and one thrombospondin type 1 domain [44,45]. Using a transgenic mouse model, Kim et al. [46] identified a human gene, R-spondin1, with potent and specific proliferative effects on intestinal crypt cells. Examination of human R-spondin1 (hRSpo1) in mouse knock-in chimeras revealed a substantial increase in the diameter, length, and weight of the small intestine [46]. Furthermore, the small intestine of hRSpo1-knock-in chimeras revealed a marked, diffuse thickening of the mucosa and crypt epithelial hyperplasia, and a greatly expanded zone of proliferating cells [46]. At that time, the authors speculated that hRSpo1-mediated signaling is not completely dependent on the canonical Wnt/Frizzed pathway, although hRSpo1 may require a distinct frizzled receptor complex [46]. They also emphasized that identification of receptors for R-spondins is urgently necessary to understand the biology of this class of activating ligands [46].
The LGR5 protein is a member of the LGR family. LGR proteins form membrane-spanning structures with large extracellular domains that contain leucine-rich repeats and mediate ligand binding. LGR proteins also contain short cytoplasmic domains that bind heterotrimeric G proteins. The LGR family proteins can be divided into three main groups: types A, B, and C [7]. Type B proteins including, LGR4, LGR5, and LGR6, were considered orphan receptors for more than a decade, but in 2012, the secreted Wnt agonists R-spondins (RSpo1–4) were identified as endogenous ligands of these receptors, revealing the crucial role of LGR proteins in stem cell homeostasis in the gastrointestinal tract [16,47,48,49,50,51]. Gene knockout approaches have shown that LGRs 4–6 play multiple roles during embryonic development and adult tissue homeostasis [52]. The discovery of LGR stem cell markers and their R-spondin ligands has had a major beneficial impact on our understanding of stem cell biology in a range of rapidly renewing tissues [53].
2.2. Epidermal Growth Factor (EGF) Signaling
Epidermal growth factor (EGF) is a potent stimulator of proliferation of ISCs and intestinal epithelial cells throughout the crypt-villus axis upon engagement of the EGF family of receptor tyrosine kinases (ErbBs) [54]. Thus, EGF is used as a crucial component of intestinal organoid culture [22]. As ErbB family ligands and receptors are highly expressed within the ISC niche (Figure 2), these endogenous regulators control the pathway in the ISC compartment. Accordingly, Wong et al. [55] revealed that leucine-rich repeats and immunoglobulin-like domain protein 1 (Lrig1), a negative feedback regulator of the ErbB receptor family [56,57,58], is highly expressed by ISCs and controls the size of the ISC niche by regulating the amplitude of growth factor signaling. Their findings position ErbB activation as a strong inductive signal of ISC proliferation.
Bae et al. [59] investigated the relationship between Hippo and other signaling pathways including EGF signaling and the role of Mps one binder (MOB) kinase activator 1A/1B (MOB1A/B) in intestinal homeostasis. MOB1A/B is a core component of the Hippo signaling pathway, which plays a crucial role in cell proliferation, apoptosis, differentiation, and development. MOB1A/B binds to and activates large tumor suppressor 1 and 2 (LATS1/2) kinase, followed by phosphorylation of YAP. Bae et al. discovered that loss of MOB1A/B in intestinal epithelial cells reduces the expression of ISC niche factors including EGF [59]. However, other EGF receptor (EGFR) ligands, including amphiregulin (Areg) and epiregulin (Ereg), are markedly upregulated in intestinal epithelial cells of MOB1A/B-deficient mice [59]. Areg and Ereg transcription can be induced by YAP or transcriptional co-activator with PDZ-binding motif (TAZ) activity [60,61], and thus, their results support the notion that EGF signaling interacts with Hippo signaling. How EGF signaling functions in ISC maintenance and how Hippo signaling interacts with EGF signaling in intestinal epithelial cells remain to be determined.
2.3. Bone Morphogenetic Protein (BMP) Signaling
BMP signaling is active in the villus compartment (Figure 2). When BMP signaling is inhibited by Noggin (a BMP antagonist), crypt-like structures appear along the flank of the villi [62], indicating that BMP inhibition creates a crypt-permissive environment. Negative cross-talk is present between Wnt and BMP/transforming growth factor (TGF)-β signaling in the intestine. BMP2 and -4 are expressed in mature epithelial cells and mesenchymal cells [62,63]. In contrast, Noggin is expressed in the crypt region to create a BMP-low niche for ISCs. Suppression of BMP signaling induces expansion of stem and progenitor cells with increasing Wnt activity and eventually leads to development of intestinal polyposis [64]. Accordingly, Noggins such as R-spondin also constitute a key growth factor in the intestinal organoid culture medium cocktail and can be replaced by coculturing with mesenchymal cells [65]. Components of TGF-β signaling are localized in differentiated epithelial cells [66]. BMP/TGF-β-induced mothers against decapentaplegic homolog (Smad) signaling plays roles in differentiation of diverse epithelial cells, and inhibition of Smad leads to stem cell hyperplasia in humans and mice [67]. In particular, activation of BMP receptors by BMP leads to complexes between Smad1/5/8 and Smad4 to repress stemness genes in ISC nuclei [68].
A core component of MOB1A/B in the Hippo signaling pathway coordinates with BMP/TGF-β signaling, the activation of which results in inhibition of Wnt activity in the crypt region and loss of intestinal epithelial homeostasis [59]. This finding is important for understanding Wnt suppression mechanisms by Hippo and BMP/TGF-β signaling in intestinal epithelial cells and for determining which signals associate and interact with each other to influence intestinal epithelial homeostasis.
2.4. Notch Signaling in ISC Homeostasis
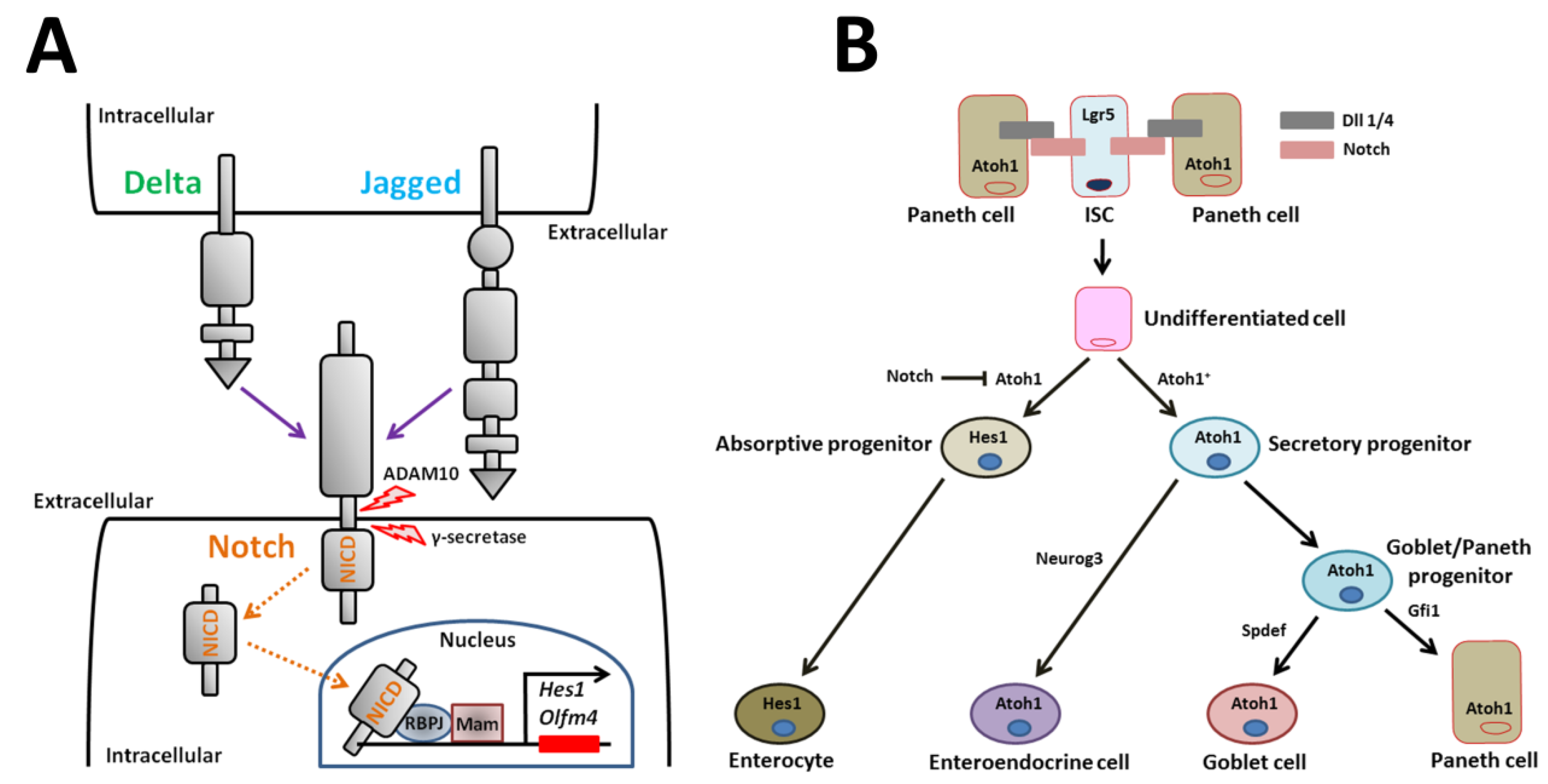
Notch receptors are single-pass transmembrane heterodimers [69,70,71,72]. Mammals express four Notch receptors (Notch1-4) encoded by different genes (Figure 3A) [71]. Five ligands for the Notch receptor, Dll1, -3, and -4 and Jagged-1 and -2, which are transmembrane Delta/Serrate/Lag2 family members, bind and activate the receptor (Figure 3A) [71,72]. Activated Notch is cleaved in several places, which leads to release of the Notch intracellular domain (NICD) fragment into the cytoplasm (Figure 3A) [72]. In the intestine, the initial cleavage event excising the extracellular receptor domain occurs via the cell surface sheddase, a disintegrin and metalloproteinase 10 (also known as ADAM10) (Figure 3A) [73]. Signals via NICD are transmitted to the nucleus following intramembrane cleavage by the γ-secretase complex (Figure 3A) [74]. NICD binds to the DNA-binding protein, recombination signal binding protein for immunoglobulin kappa J region (also known as RBPJ, CSL-CBF1, suppressor of hairless, Lag2), which changes a transcriptional repressor complex to an activator complex in cooperation with Mastermind (Figure 3A) [74]. This active complex induces transcription of various genes such as Hairy and enhancer of split (Hes) family members [69,70]. Hes1 in turn represses expression of the mouse atonal homolog 1 (Atoh1) transcription factor that is crucial for entry into the secretory lineage [75,76,77].
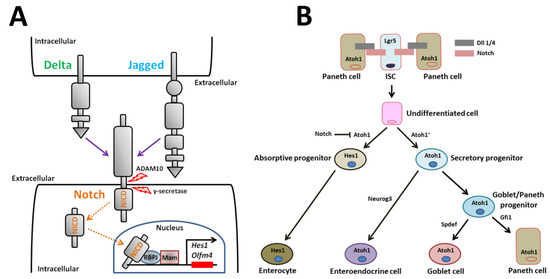
Figure 3.
Notch signaling and model for Notch regulation of small intestinal epithelial cell differentiation. (A) Transmission of Notch signaling involves engagement of two cells. The signal-sending cell expresses the Notch ligands, Delta and Jagged. The signal-receiving cell expresses Notch receptors. After ligand/receptor engagement, the Notch receptor is proteolytically cleaved. The cleaved NICD translocates to the nucleus where it recruits a transcriptional coactivator complex that activates downstream transcription of Notch target genes such as Hes1. (B) Notch signaling represses the bHLH transcriptional regulator, Atoh1, to form absorptive enterocytes. Atoh1+ secretory progenitor cells are fated towards three mature secretory cell types. Neurog3 induces enteroendocrine cell differentiation. Spdef regulates Goblet cell differentiation. Gfi1 regulates Paneth cell differentiation. ADAM10: a disintegrin and metalloproteinase 10, NICD: Notch intracellular domain, RBPJ: recombination signal binding protein for immunoglobulin kappa J region, Mam: mastermind, Hes1: Hairy and enhancer of split 1, Olfm4: olfactomedin 4, Atoh1: atonal homolog 1, Lgr5: leucine-rich repeat-containing G-protein coupled receptor 5, ISC: intestinal stem cell, Dll: delta-like ligand, Neurog3: neurogenin 3, Gfi1: growth factor independent 1, Spdef: sterile alpha motif pointed domain containing ETS transcription factor.
Notch signaling modulates small intestinal homeostasis via control of Lgr5+ ISCs and via induction of absorptive cells. Notch signaling induces cells to differentiate into one of two cell types. In the absence of Notch signaling, undifferentiated cells become secretory cells (i.e., enteroendocrine, goblet, and Paneth cells). In the presence of Notch signaling, cells differentiate into enterocytes (Figure 3B) [74]. Secretory cell fate is induced when Notch signaling is blocked by Atoh1 (Figure 3B) [78]. Atoh1 is a critical transcriptional activator that completely induces secretory cell differentiation, as shown by ablation and activation studies in murine genetic models (Figure 3B) [75,76,79]. The key Notch effector that regulates Atoh1 expression is thought to be the Notch target gene, Hes1. For instance, combined loss of both Dll1 and Hes1 results in increased secretory cell types [21,80,81]. Robinson et al. [82] recently reported that the poxvirus and zinc finger transcription factor, Kaiso, regulates the secretory cell lineage in coordination with Notch signaling in intestinal-specific Kaiso-overexpressing mice. In the mice, Kaiso inhibits the expression of Notch1 and Dll1. Consequently, Kaiso-mediated repression of the genes promotes the increase in secretory cells [82]. This finding reveals a novel signaling pathway via Kaiso in regulating Notch-mediated small intestinal homeostasis.
Disruptions in mature secretory cell homeostasis are present in murine models with intestinal-specific gene deletion. The transcription factor Neurogenin3 plays a role in differentiation of enteroendocrine cells, as shown by deletion and overexpression experiments in mice (Figure 3B) [83,84,85]. The transcription repressor, growth factor independent 1 (Gfi1), which is a zinc-finger protein family member, functions downstream of Atoh1 in intestinal secretory lineage differentiation (Figure 3B). Gfi1-null mice lack Paneth cells [86]. Another transcription factor, sterile alpha motif pointed domain containing ETS transcription factor (Spdef), when overexpressed in the small intestine, causes the expansion of goblet cells and a corresponding reduction in Paneth cells [87]. Thus, differentiation of goblet/Paneth progenitor cells into mature goblet cells is also regulated by transcription factors (Figure 3B). The colon is devoid of Paneth cells. Paneth-like goblet cells may serve the function of Paneth cells in the colon [88]. Interestingly, the transcript levels of several factors involved in goblet cell differentiation, including Atoh1, Gfi1, Spdef, and E74-like ETS transcription factor 3, are lower in colonic epithelial cells derived from Krüppel-like factor 4 mutant mice compared to controls [89]. These genes may be key players in directing the fate of goblet/Paneth progenitor cells toward mature goblet or Paneth cells.
In the small intestine, maintenance of stem cells and appropriate differentiation into various cell lineages require Notch and Wnt signaling. Previous studies have examined these pathways individually, but how Notch and Wnt signaling work together to maintain ISCs and modulate cell fate remains unclear. The ability to decrease Wnt signaling and thus maintain ISCs is a main function of Notch signaling [90]. Inhibition of Notch induces Lgr5+ ISCs to differentiate into secretory cells, thus depleting the stem cell pool [90], an observation that is coincident with increased Wnt pathway activity and differentiation into secretory cells [16,17,18,19]. Wnt and Notch signaling likely interact in the biology of many types of stem cells.
2.5. Eph/Ephrin Signaling in ISC Homeostasis
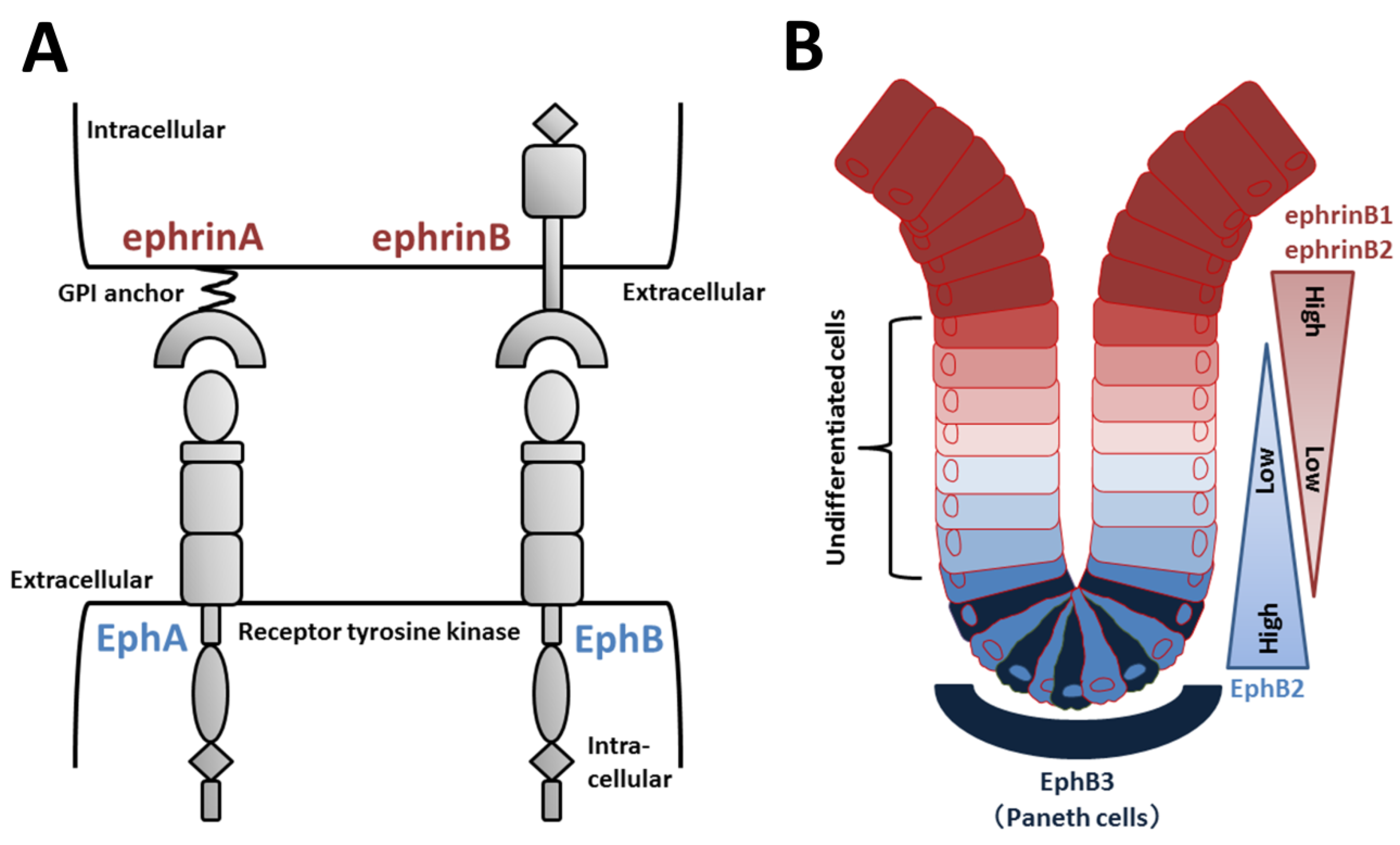
Eph receptors compose a large subfamily of receptor tyrosine kinases (Figure 4A). Nine EphA (EphA1–8 and EphA10) and five EphB (EphB1–4 and EphB6) receptors have been identified in humans [91]. Eph ligands are also divided into two types of ephrins according to their structure and binding affinity for EphA or EphB. EphrinAs (A1–A6) are GPI-linked to the plasma membrane, and ephrinBs (B1–B3) are transmembrane proteins with a short cytoplasmic tail (Figure 4A) [91]. EphA receptors generally bind to ephrinA ligands, and EphBs receptors generally bind to ephrinB ligands, although some cross-binding to members of the other type occurs [92].
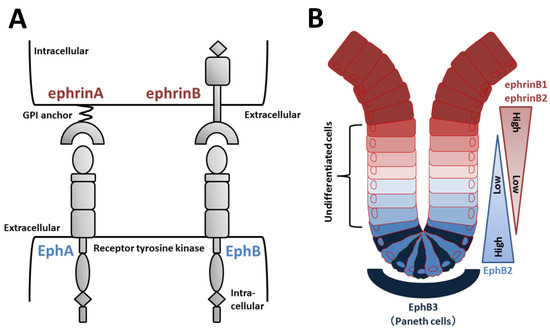
Figure 4.
Eph/ephrin structure and regulation of EphB/ephrinB signaling in intestinal crypts. (A) Domain organization of Eph receptors and ephrin ligands. EphA receptors typically bind to ephrinA (GPI-anchored) ligands, and EphB receptors bind to ephrinB ligands. (B) EphB2-positive stem and undifferentiated cells are located near the bottom of intestinal crypts, whereas ephrinB1/B2-positive differentiated epithelial cells are concentrated at the crypt-villus boundary. In the small intestine, EphB3-expressing Paneth cells are found at the crypt bottom.
Increasing evidence supports the roles of EphB/ephrinB signaling in intestinal crypts. Wnt positively regulates cell proliferation and expression of EphB2 and EphB3 [4]. Receptor-positive cells are found in intervillus pockets in the small intestine of neonatal mice and in crypts in the adult small intestine [4]. Conversely, Wnt signaling represses expression of ephrinB1 and ephrinB2 in differentiated cells, and consequently, the distribution of the ligands is largely complementary to EphB receptors [4]. In adult mice, proliferating undifferentiated cells are positive for EphB2, and Paneth cells preferentially express EphB3 (Figure 4B). Double knockout of EphB2 and EphB3 results in complementary expression of EphB receptors and ephrinB ligands to sustain the normal organization of Lgr5+ ISCs, Paneth cells, and dividing uncommitted cells in the base and differentiated cells in the apex [4,93]. EphB2 and EphB3 also control division of uncommitted cells and re-entry of quiescent cells into the cell cycle [94]. Collectively, in the presence of an increasing gradient of EphB2 receptors, ephrinB ligand-positive precursors are organized from top to bottom in crypts according to a reverse gradient of EphrinB1 and B2 expression (Figure 4B).
2.6. The Coordinated Activities of Nicotinic Acetylcholine Receptors (nAChRs), Wnt, and Hippo Signaling in ISC Homeostasis
Cholinergic transmission via acetylcholine (ACh) produces excitatory potentials in postsynaptic cells and is a major excitatory transmission pathway in the enteric nervous system [7]. ACh-mediated neurotransmission in the enteric nervous system is well-understood, but much less is known about non-neuronal ACh, which may modulate intestinal function. Experiments with crypt-villus organoids suggest that ACh is made in intestinal epithelial cells and plays a role in cell division and differentiation of Lgr5+ ISCs in the small intestine by binding to muscarinic ACh receptors (mAChRs) [24].
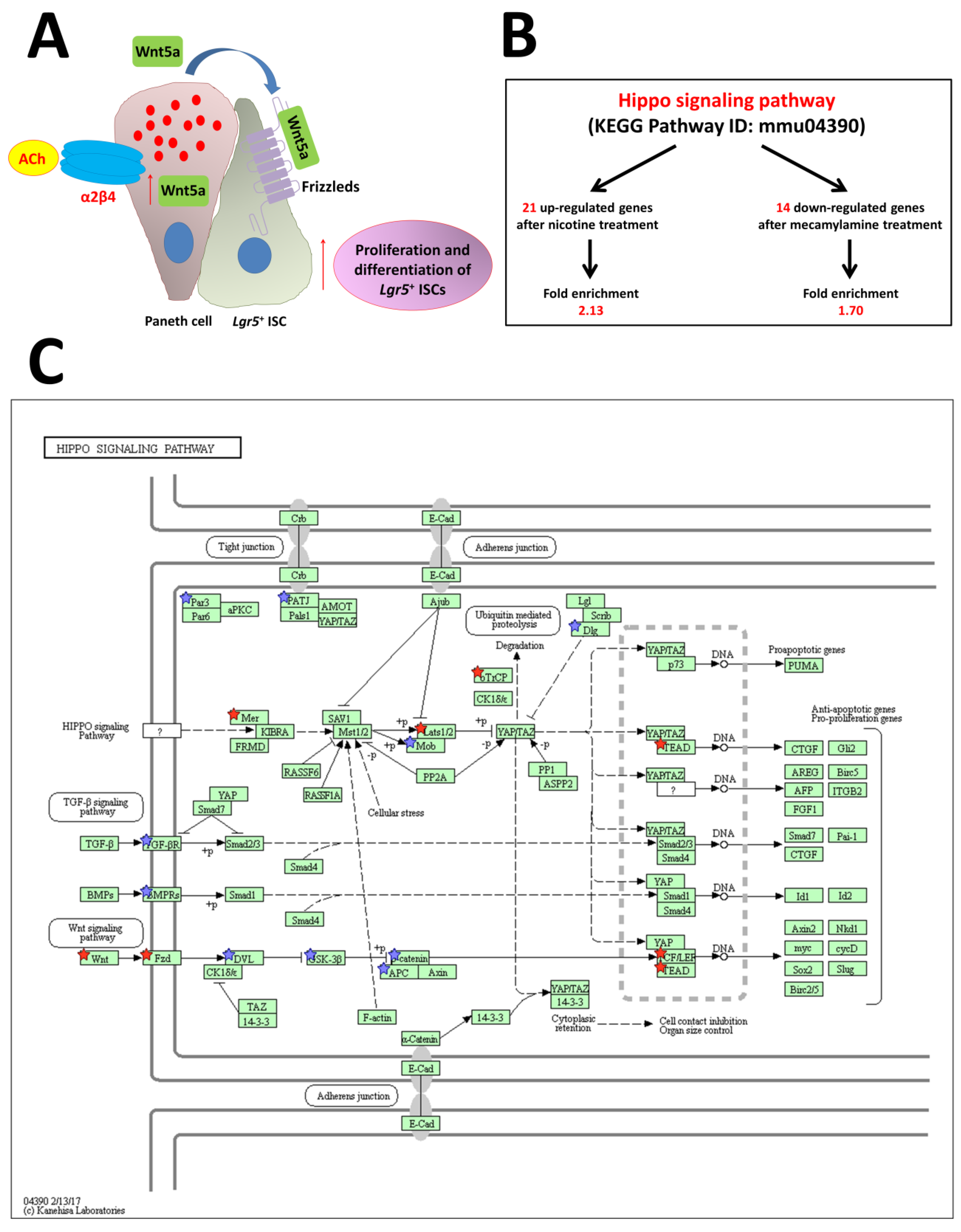
Two principal forms of ACh receptors are expressed: mAChRs and nAChRs. Experiments with nicotine and mecamylamine, which is an anti-nicotinic agent, on organoid growth and gene expression in the intestine have shown that endogenous ACh, acting via nAChRs, increases cell division and differentiation and likely modulates interactions between nAChR signaling and other types of signaling [95]. Differential expression of Wnt5a, which activates β-catenin-independent pathways, is critical for downstream nAChR signaling, as shown by our RNA-Seq experiments following stimulation with nicotine and mecamylamine [95]. nAChRs are composed of several types of subunits. Different combinations of subunits, which are variably expressed in neuronal and non-neuronal systems, produce a wide array of physiological and pharmacological effects [96,97]. nAChR subunit-specific antibodies demonstrated the presence of the α2/β4 subtype in the crypt region, suggesting important functions, such as regulation of ISC division and differentiation [95]. We also reported the co-expression of α2 and β4 subunits in Paneth cells [95]. Thus, regulation of ISCs in crypts in the normal adult mouse is associated with increased Wnt5a expression via nAChRs. Endogenous ACh binds to α2/β4 nAChRs in Paneth cells, followed by increased Wnt5a expression (Figure 5A). Wnt binds to Frizzled receptors, which induce noncanonical Wnt signaling, finally leading to increased division and differentiation of Lgr5+ ISCs (Figure 5A).
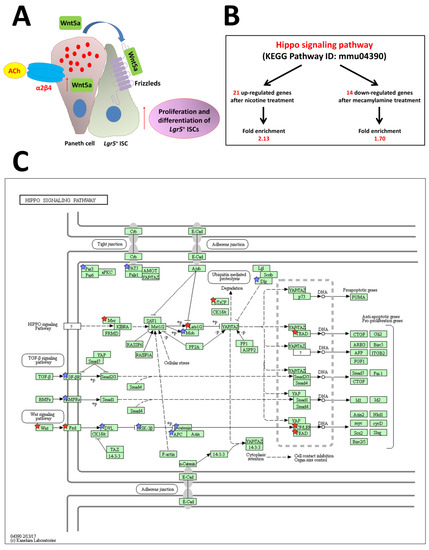
Figure 5.
Model for the proposed role of nAChR signaling pathways in ISC function. (A) Non-neuronal ACh activates the nicotinic receptor α2β4, which is localized in Paneth cells, to modulate the expression level of Wnt5a [9]. Wnt then modulates Wnt pathway activity through frizzled receptors. Eventually, proliferation and differentiation of stem cells are enhanced. (B) The stem cell regulation network, the Hippo pathway, is regulated by nAChR signaling. (C) Hippo signaling pathway (mmu04390) maps derived from Database for Annotation, Visualization, and Integrated Discovery (DAVID) analysis. The genes with red stars were upregulated by nicotine and downregulated by mecamylamine. Additionally, the genes with blue stars were upregulated by nicotine but not downregulated by mecamylamine. ACh: acetylcholine, Lgr5: leucine-rich repeat-containing G-protein-coupled receptor 5 [95].
Another network that regulates stem cells, the Hippo pathway, is upregulated after treatment with nicotine and downregulated by mecamylamine (Figure 5B,C) [95]. This result reveals that non-neuronal ACh signaling via the α2/β4 nAChR subtype is upstream of the Hippo pathway. The Hippo signaling cascade is composed of highly conserved kinases such as mammalian Ste-like kinase (hippo ortholog) and large tumor suppressor kinase (warts ortholog), as well as the downstream transcription coactivators, YAP and transcriptional co-activator with a PDZ-binding domain; mammalian homolog of Drosophila yorkie. This pathway is critical for maintenance of tissues and organ size, which occurs via regulation of tissue-specific stem cells. Many upstream events initiate the cascade, including mechanical detection of cell density and signaling through G-protein-coupled receptors that regulate Hippo pathway activity [38]. Gestational nicotine treatment has many effects on cell adhesion, which in turn affects expression of the neurexin, immunoglobulin, cadherin, and adhesion G-protein-coupled receptor superfamilies in limbic regions of the brain [98]. Non-neuronal cholinergic systems probably functionally and independently regulate or control cell functions in the mouse intestine.
3. Intestinal Organoid Development In Vitro
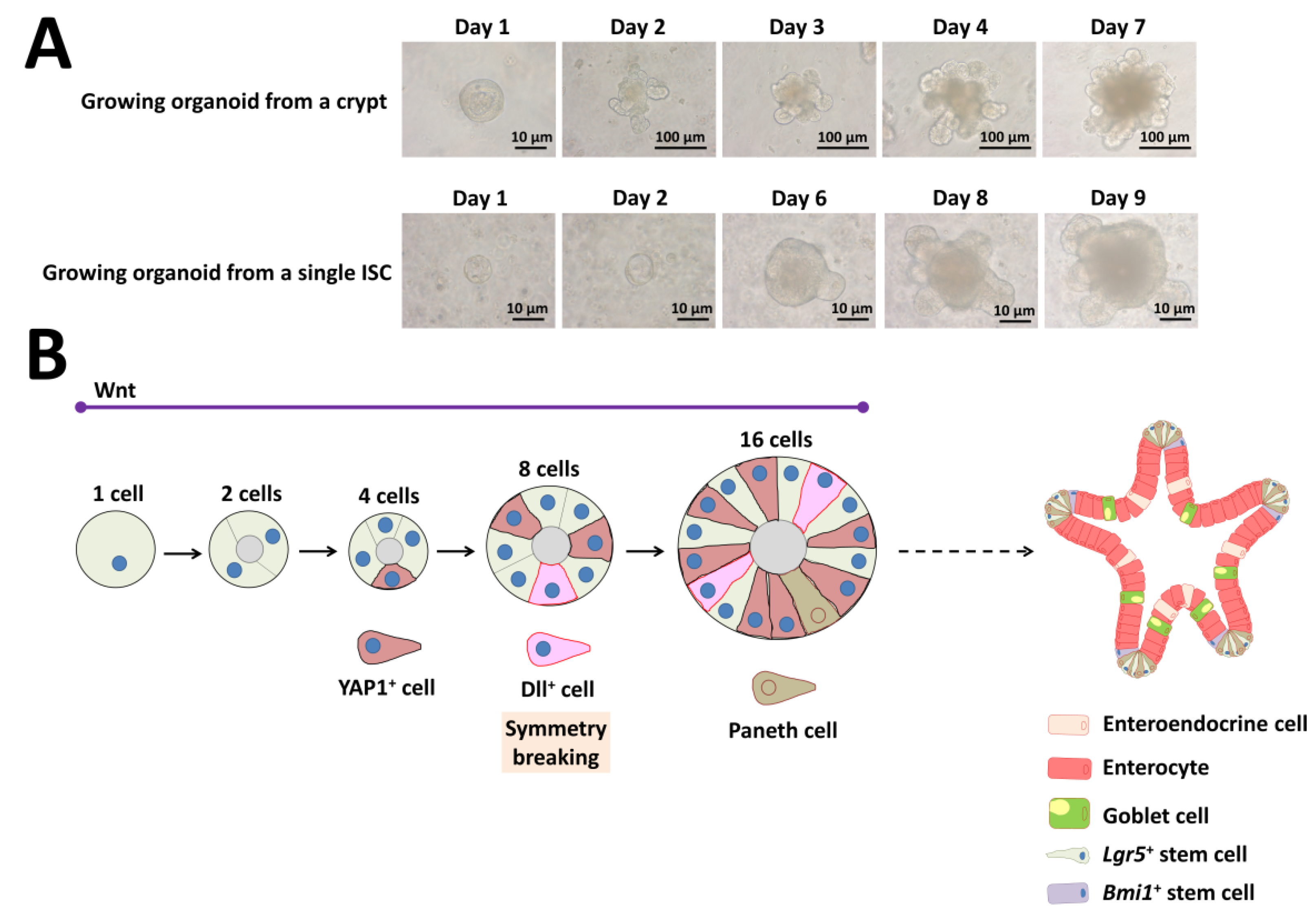
Organoids are structures that include several cell types that have differentiated from stem cells or organ progenitors. These cells self-organize via spatially restricted differentiation into various lineages and migration in a way that recapitulates the in vivo situation [99]. The mouse intestinal organoid is a typical example that first grows as a single-layered epithelium that then self-organizes into domains that are structurally similar to the in vivo intestinal crypt-villus. These organoids contain the various cell types present in the intestine that surround a cystic lumen (Figure 6) [7].
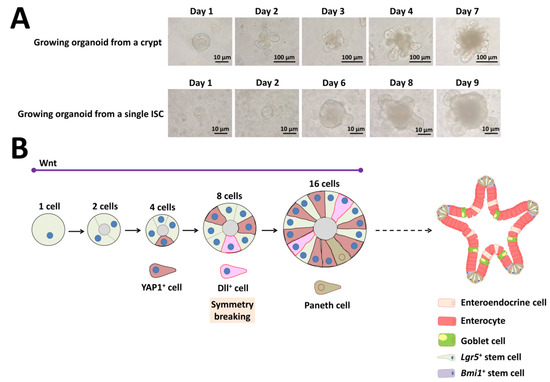
Figure 6.
Self-organization and symmetry breaking in intestinal organoid development. (A) Representative images of a growing organoid derived from a crypt or from a single ISC. (B) Model of organoid development and symmetry breaking. YAP1: yes-associated protein 1, Dll: delta-like ligand, Lgr5: leucine-rich repeat-containing G-protein-coupled receptor 5, Bmi1: B-cell-specific Moloney murine leukemia virus integration site 1.
Isolated crypts containing Lgr5+ ISCs require Matrigel, EGF, Noggin, and R-spondin 1 in serum-free medium [7]. The above three factors represent the minimal and essential stem cell maintenance factors as a cocktail for growth (Figure 6A). In addition to Matrigel support, several groups have developed non-Matrigel strategies for the culture of ISCs and intestinal organoids. Gjorevski and Lutolf [100] describe a protocol for the generation of well-defined matrices. These matrices comprise a polyethylene glycol hydrogel backbone functionalized with minimal adhesion cues, including RGD (Arg-Gly-Asp), which is sufficient for ISC expansion, and laminin-111, which is required for organoid formation. Recently, Capeling et al. [101] demonstrated that alginate, a minimally supportive hydrogel with no inherent cell instructive properties, supports growth of human intestinal organoids (HIOs) in vitro and leads to HIO epithelial differentiation that is virtually indistinguishable from Matrigel-grown HIOs. Culturing single ISCs is inefficient, with less than 2% plating efficiency, whereas up to half of ISC–Paneth cell doublets form organoids in vitro [3,7]. As Paneth cells are believed to create a niche environment and determine the future crypt site, emergence of a single Paneth cell is important for organoid development derived from a single ISC (Figure 6A).
Although researchers studying ISC biology extensively use the intestinal organoid culture system, how single ISCs give rise to a cell population with the capability of self-organization and which transcriptional program cells use are unclear. Serra and coworkers [39] clearly demonstrated that intestinal organoid formation is a regenerative process that relies on transient YAP1 activation, following a regeneration process (Figure 6B). YAP1 is a mechanosensing nuclear effector of the Hippo pathway and regulates organ growth, regeneration, and tumorigenesis [102,103]. Organoid development requires temporary cell–cell variability in YAP localization, which then leads to Notch and Dll1 lateral inhibition between the 8- and 16-cell stage (Figure 6B) [39]. These events lead to the first instance of symmetry breaking in intestinal organoid growth and initiate differentiation of the first Paneth cell (Figure 6B) [39]. Thus, single cells can induce self-organization that leads to multicellular asymmetric structures like embryos.
4. Conclusions
Stem cells can divide extensively and have the ability to produce committed cells during early mammalian development. Early in development, stem cells need to be able to divide and differentiate into various lineages in response to external signals. Stem cells have specialized internal mechanisms that allow these activities, and hence, these cells are initially present when niches can sequester, maintain, and regulate undifferentiated embryonic cells [104]. Adult stem cells in tissues also have tremendous potential. They are maintained in a steady state in which each division typically produces one replacement stem cell and one tissue cell with no apparent limit [7].
The basic architecture of the intestinal tract includes a simple epithelial layer. The small intestinal villus and its associated epithelium include enterocytes as the main cell type and secretory cell types, whereas the crypt is the site of cell division and hence the origin of all epithelial components. A complex gradient of factors maintains ISC stemness and proliferation along the crypt-villus axis. In the crypts, Paneth cells express factors that promote stem cell growth, including Wnt3a, Dll1/4, and EGF [3]. In addition, several factors, such as Wnt2b [42], Rspo1 [16,46,105], Gremlin1, Gremlin2 (BMP antagonists), and Chordin [106,107], produced by mesenchymal cells, play an essential role in the maintenance of ISCs.
The extracellular matrix (ECM) is a dynamic and complex environment characterized by biophysical, mechanical, and biochemical properties specific for each tissue and is able to regulate cell behavior. The ECM is an essential player in the stem cell niche, because it can directly or indirectly modulate the maintenance, proliferation, self-renewal, and differentiation of stem cells. Several ECM molecules have regulatory functions for different types of stem cells. ECM proteins and integrin receptors interact to keep ISCs within the niche [108]. Exploration of the ECM as an influencer of ISC niche formation and maintenance is ongoing [109,110,111,112]. Engineered biomaterials that mimic the in vivo characteristics of the stem cell niche will provide suitable in vitro tools for dissecting the different roles of the ECM.
The mesenchymally derived basement membrane dynamically controls morphogenesis, cell differentiation, and polarity, while also providing the structural basis for villi, crypts, and the microvasculature of the lamina propria so that crucially, tissue morphology is preserved in the absence of epithelium. Interestingly, the biomechanical work of Balbi and Ciarletta [113] supports the influence of material properties of the developing intestinal epithelium on how the villi are formed, which is closely linked to many of the signaling gradients discussed here. They demonstrated that the emergence of intestinal villi in embryos is triggered by differential growth between the epithelial mucosa and mesenchymal tissues [113]. Their proposed morpho-elastic model highlights that both the geometric and mechanical properties of the epithelial mucosa strongly influence the formation of pre-villous structures in embryos, providing useful suggestions for interpreting the dynamics of villus morphogenesis in living organisms [113]. This model may provide guidance for artificial formation of villus structures using complex signaling pathways.
Intrinsic stem cell capabilities mediate establishment of complex tissues during development and/or repair in murine [22] and D. melanogaster [114] intestines. The existence of an intrinsic process for tissue generation and repair is clear. What is less clear is whether the stem cell or the niche comes first. The Wnt pathway is present in the simplest multicellular organisms and is thus evolutionarily old [115,116]. In the earliest metazoans, Wnt appears to be an ancestral signal that breaks symmetry to divide a previously symmetric embryo into anterior and posterior domains, allowing evolutionary emergence of asymmetric organisms [117,118]. Individual cells produce genetic and nongenetic heterogeneity, which leads to specialized cell behavior [119,120,121]. Thus, single cells can break the symmetry of a population by affecting differentiation in relation to other identical cells [122]. Indeed, cell-to-cell variability in intestinal organoid development occurs owing to the combined effects of three factors: Wnt, Yap1, and Notch/Dll1 activation (Figure 6B).
Recently, Ayyaz and coworkers [123] identified a revival stem cell, termed revSC, with the use of single-cell RNA sequencing to profile the regenerating mouse intestine. revSCs are typically rare and are characterized by high expression of clusterin [123]. Radiation-induced intestinal damage, specific depletion of Lgr5+ ISCs, or treatment with dextran sodium sulfate causes revSCs, which are critical for intestinal regeneration, to temporarily express YAP1 and restore the Lgr5+ ISC pool [123]. YAP1-dependent, injury-induced revSC division may be critical for tissue repair following damage.
Funding
This research was funded by a Grant-Aid for Scientific Research (C), grant number JP17K07495. The APC was funded by JP17K07495.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (C) to Toshio Takahashi (Grant number JP17K07495) and a Grant-in-Aid for Young Scientists (B) to Akira Shiraishi (Grant number JP18K14687).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cheng, H.; Leblond, C.P. Origin, differentiation and renewal of the main epithelial cell types in the mouse small intestine V. Unitarian theory of the origin of the four epithelial cell types. Am. J. Anat. 1974, 141, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Porter, E.M.; Bevins, C.L.; Ghosh, D.; Ganz, T. The multifaceted Paneth cell. Cell 2002, 59, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Batlle, E.; Henderson, J.T.; Beghtel, H.; van den Born, M.M.W.; Sancho, E.; Huls, G.; Meeldijk, J.; Robertson, J.; van de Wetering, M.; Pawson, T.; et al. β-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/EphrinB. Cell 2002, 111, 251–263. [Google Scholar] [CrossRef]
- Potten, C.S.; Loeffler, M. Stem cells: Attribute, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development 1990, 110, 1001–1020. [Google Scholar]
- Stappenbeck, T.S.; Wong, M.H.; Saam, J.R.; Mysorekar, I.U.; Gordon, J.I. Notes from some crypt watchers: Regulation of renewal in the mouse intestinal epithelium. Curr. Opin. Cell Biol. 1998, 10, 702–709. [Google Scholar] [CrossRef]
- Takahashi, T. Organoids for drug discovery and personalized medicine. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 447–562. [Google Scholar] [CrossRef]
- Yan, K.S.; Chia, L.A.; Li, X.; Ootani, A.; Su, J.; Lee, J.Y.; Su, N.; Luo, Y.; Heilshorn, S.C.; Amieva, M.R.; et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA 2012, 109, 466–471. [Google Scholar] [CrossRef]
- Takahashi, T. Roles of nAChR and Wnt signaling in intestinal stem cell function and inflammation. Int. Immunopharmacol. 2020. Epub ahead of print. [Google Scholar] [CrossRef]
- Korinek, V.; Barker, N.; Moerer, P.; van Donselaar, E.; Huls, G.; Peters, P.J.; Clevers, H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998, 19, 379–383. [Google Scholar] [CrossRef]
- Crosnier, C.; Stamataki, D.; Lewis, J. Organizing cell renewal in the intestine: Stem cells, signals and combinatorial control. Nat. Rev. Genet. 2006, 7, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Fevr, T.; Robine, S.; Louvard, D.; Huelsken, J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol. Cell. Biol. 2007, 27, 7551–7559. [Google Scholar] [CrossRef] [PubMed]
- van der Flier, L.G.; Clevers, H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009, 71, 241–260. [Google Scholar] [CrossRef] [PubMed]
- van Es, J.H.; Haegebarth, A.; Kujala, P.; Itzkovitz, S.; Koo, B.K.; Boj, S.F.; Korving, J.; van den Born, M.; van Oudenaarden, A.; Robine, S.; et al. A critical role for the Wnt effector Tcf4 in adult intestinal homeostatic self-renewal. Mol. Cell. Biol. 2012, 32, 1918–1927. [Google Scholar] [CrossRef] [PubMed]
- van de Weterning, M.; Sancho, E.; Verweij, C.; de Lau, W.; Oving, I.; Hurlstone, A.; van der Horn, K.; Batlle, E.; Coudreuse, D.; Haramis, A.P.; et al. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 2002, 111, 241–250. [Google Scholar] [CrossRef]
- de Lau, W.; Barker, N.; Low, T.Y.; Koo, B.K.; Li, V.S.; Teunissen, H.; Kujala, P.; Haegebarth, A.; Peters, P.J.; van de Wetering, M.; et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signaling. Nature 2011, 476, 293–297. [Google Scholar] [CrossRef]
- Andreu, P.; Colnot, S.; Godard, C.; Gad, S.; Chafey, P.; Niwa-Kawakita, M.; Laurent-Puig, P.; Kahn, A.; Robine, S.; Perret, C.; et al. Crypt-restricted proliferation and commitment to the Paneth cell lineage following Apc loss in the mouse intestine. Development 2005, 132, 1443–1451. [Google Scholar] [CrossRef]
- van Es, J.H.; Jay, P.; Gregorieff, A.; van Gijn, M.E.; Jonkheer, S.; Hatzis, P.; Thiele, A.; van den Born, M.; Begthel, H.; Brabletz, T.; et al. Wnt signaling induces maturation of Paneth cells in intestinal crypts. Nat. Cell Biol. 2005, 7, 381–386. [Google Scholar] [CrossRef]
- Pinto, D.; Gregorieff, A.; Begthel, H.; Clevers, H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003, 17, 1709–1713. [Google Scholar] [CrossRef]
- Fre, S.; Hannezo, E.; Sale, S.; Huyghe, M.; Lafkas, D.; Kissel, H.; Louvi, A.; Greve, J.; Louvard, D.; Artavanis-Tsakonas, S. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS ONE 2011, 6, e25785. [Google Scholar] [CrossRef]
- Pellegrinet, L.; Rodilla, V.; Liu, Z.; Chen, S.; Koch, U.; Espinosa, L.; Kaestner, K.H.; Kopan, R.; Lewis, J.; Radtke, F. Dll1- and Dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 2011, 140, 1230–1240. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Koo, B.K.; Stange, D.E.; Sato, T.; Karthaus, W.; Farin, H.F.; Huch, M.; van Es, J.H.; Clevers, H. Controlling gene expression in primary Lgr5 organoid cultures. Nat. Methods 2011, 9, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Ohnishi, H.; Sugiura, Y.; Honda, K.; Suematsu, M.; Kawasaki, T.; Deguchi, T.; Fujii, T.; Orihashi, K.; Hippo, Y.; et al. Non-neuronal acetylcholine as an endogenous regulator of proliferation and differentiation of Lgr5-positive stem cells in mice. FEBS J. 2014, 281, 4672–4690. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Matano, M.; Nanki, K.; Sato, T. Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc. 2015, 10, 1474–1485. [Google Scholar] [CrossRef]
- Batlle, E.; Wilkinson, D.G. Molecular mechanisms of cell segmentation and boundary formation in development and tumorigenesis. Cold Spring Harbor Perspect. Biol. 2012, 4, a008227. [Google Scholar] [CrossRef]
- Pasquale, E.B. Eph receptor signalling casts a wide net on cell behaviour. Nat. Rev. Mol. Cell Biol. 2005, 6, 462–475. [Google Scholar] [CrossRef]
- Klein, R. Eph/ephrin signalling during development. Development 2012, 139, 4105–4109. [Google Scholar] [CrossRef]
- Lisabeth, E.M.; Falivelli, G.; Pasquale, E.B. Eph receptor signaling and ephrins. Cold Spring Harbor Perspect. Biol. 2013, 5, a009159. [Google Scholar] [CrossRef]
- Gale, N.W.; Holland, S.J.; Valenzuela, D.M.; Flenniken, A.; Pan, L.; Ryan, T.E.; Henkemeyer, M.; Strebhardt, K.; Hirai, H.; Wilkinson, D.G.; et al. Eph receptor and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron 1996, 17, 9–19. [Google Scholar] [CrossRef]
- Tanaka, M.; Kamo, T.; Ota, S.; Sugimura, H. Association of Dishevelled with Eph tyrosine kinase receptor and ephrin mediates cell repulsion. EMBO J. 2003, 22, 847–858. [Google Scholar] [CrossRef]
- Kida, Y.S.; Sato, T.; Miyasaka, K.Y.; Suto, A.; Ogura, T. Daam1 regulates the endocytosis of EphB during the convergent extension of the zebrafish notochord. Proc. Natl. Acad. Sci. USA 2007, 104, 6708–6713. [Google Scholar] [CrossRef]
- Batlle, E.; Bacani, J.; Begthel, H.; Jonkheer, S.; Gregorieff, A.; van de Born, M.; Malats, N.; Sancho, E.; Boon, E.; Pawson, T.; et al. EphB receptor activity suppresses colorectal cancer progression. Nature 2005, 435, 1126–1130. [Google Scholar] [CrossRef]
- Holmberg, J.; Frisén, J. Ephrins are not only unattractive. Trends Neurosci. 2002, 25, 239–243. [Google Scholar] [CrossRef]
- Palmer, A.; Klein, R. Multiple roles of ephrins in morphogenesis, neuronal networking, and brain function. Genes Dev. 2003, 17, 1429–1450. [Google Scholar] [CrossRef]
- Poliakov, A.; Cotrina, M.; Wilkinson, D.G. Diverse roles of eph receptors and ephrins in the regulation of cell migration and tissue assembly. Dev. Cell 2004, 7, 465–480. [Google Scholar] [CrossRef]
- Cowan, C.A.; Henkemeyer, M. Ephrins in reverse, park and drive. Trends Cell Biol. 2002, 12, 339–346. [Google Scholar] [CrossRef]
- Mo, J.-S.; Park, H.W.; Guan, K.-L. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014, 15, 642–656. [Google Scholar] [CrossRef]
- Serra, D.; Mayr, U.; Boni, A.; Lukonin, I.; Rempfler, M.; Challet-Meylan, L.; Stadler, M.B.; Strnad, P.; Papasaikas, P.; Vischi, D.; et al. Self-organization and symmetry breaking in intestinal organoid development. Nature 2019, 569, 66–72. [Google Scholar] [CrossRef]
- Driehuis, E.; Clevers, H. WNT signalling events near the cell membrane and their pharmacological targeting for the treatment of cancer. Br. J. Pharmacol. 2017, 174, 4547–4563. [Google Scholar] [CrossRef]
- Gregorieff, A.; Pinto, D.; Begthel, H.; Destrée, O.; Kielman, M.; Clevers, H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology 2005, 129, 626–638. [Google Scholar] [CrossRef]
- Farin, H.F.; Van Es, J.H.; Clevers, H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 2012, 143, 1518–1529.e7. [Google Scholar] [CrossRef]
- Durand, A.; Donahue, B.; Peignon, G.; Letoumeur, F.; Cagnard, N.; Slomianny, C.; Perret, C.; Shroyer, N.F.; Romagnolo, B. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc. Natl. Acad. Sci. USA 2012, 109, 8965–8970. [Google Scholar] [CrossRef]
- Kazanskaya, O.; Glinka, A.; del Barco Barrantes, I.; Stannek, P.; Niehrs, C.; Wu, W. R-Spondin2 is a secreted activator of Wnt/β-catenin signaling and is required for Xenopus myogenesis. Dev. Cell 2004, 7, 525–534. [Google Scholar] [CrossRef]
- Kamata, T.; Katsube, K.; Michikawa, M.; Yamada, M.; Takada, S.; Mizusawa, H. R-spondin, a novel gene with thrombospondin type 1 domain, was expressed in the dorsal neural tube and affected in Wnts mutants. Biochim. Biophys. Acta 2004, 1676, 51–62. [Google Scholar] [CrossRef]
- Kim, K.A.; Kakitani, M.; Zhao, J.; Oshima, T.; Tang, T.; Binnerts, M.; Liu, Y.; Boyle, B.; Park, E.; Emtage, P.; et al. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science 2005, 309, 1256–1259. [Google Scholar] [CrossRef]
- Carmon, K.S.; Gong, X.; Lin, Q.; Thomas, A.; Liu, Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 11452–11457. [Google Scholar] [CrossRef]
- Carmon, K.S.; Lin, Q.; Gong, X.; Thomas, A.; Liu, Q. LGR5 interacts and co-internalizes with Wnt receptors to modulate Wnt/β-catenin signaling. Mol. Cell Biol. 2012, 32, 2054–2064. [Google Scholar] [CrossRef]
- Glinka, A.; Dolde, C.; Kirsch, N.; Huang, Y.L.; Kazanskaya, O.; Ingelfinger, D.; Boutros, M.; Cruciat, C.M.; Niehrs, C. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signaling. EMBO Rep. 2011, 12, 1055–1061. [Google Scholar] [CrossRef]
- Ruffner, H.; Sprunger, J.; Charlat, O.; Leighton-Davies, J.; Grosshans, B.; Salathe, A.; Zietzling, S.; Beck, V.; Therier, M.; Isken, A.; et al. R-spondin potentiates Wnt/β-catenin signaling through orphan receptors LGR4 and LGR5. PLoS ONE 2012, 7, e40976. [Google Scholar] [CrossRef]
- Gong, X.; Carmon, K.S.; Lin, Q.; Thomas, A.; Yi, J.; Liu, Q. LGR6 is a high affinity receptor of R-spondins and potentially functions as a tumor suppressor. PLoS ONE 2012, 7, e37137. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; Clevers, H. Leucine-rich repeat-containing G-protein-coupled receptors as markers of adult stem cells. Gastroenterology 2010, 138, 1681–1696. [Google Scholar] [CrossRef]
- Barker, N.; Tan, S.; Clevers, H. Lgr proteins in epithelial stem cell biology. Development 2013, 140, 2484–2494. [Google Scholar] [CrossRef] [PubMed]
- Dignass, A.U.; Stum, A. A peptide growth factors in the intestine. Eur. J. Gastroneterol. Hepatol. 2001, 13, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.Y.; Stange, D.E.; Page, M.E.; Buczacki, S.; Wabik, A.; Itami, S.; van de Wetering, M.; Poulsom, R.; Wright, N.A.; Trotter, M.W.B.; et al. Lrig1 controls intestinal stem cell homeostasis by negative regulation of ErbB signalling. Nat. Cell Biol. 2012, 14, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Gur, G.; Rubin, C.; Katz, M.; Amit, I.; Citri, A.; Nilsson, J.; Amariglio, N.; Henriksson, R.; Rechavi, G.; Hedman, H.; et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004, 23, 3270–3281. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.B.; Watt, F.M. Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence. Proc. Natl. Acad. Sci. USA 2006, 103, 11958–11963. [Google Scholar] [CrossRef] [PubMed]
- Laederich, M.B.; Funes-Duran, M.; Yen, L.; Ingalla, E.; Wu, X.; Carraway, K.L., 3rd; Sweeney, C. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J. Biol. Chem. 2004, 279, 47050–47056. [Google Scholar] [CrossRef]
- Bae, J.S.; Jeon, Y.; Kim, S.M.; Jang, J.Y.; Park, M.K.; Kim, I.-H.; Hwang, D.S.; Lim, D.-S.; Lee, H. Depletion of MOB1A/1B causes intestinal epithelial degeneration by suppressing Wnt activity and activating BMP/TGF-β signaling. Cell Death Dis. 2018, 9, 1083. [Google Scholar] [CrossRef]
- Zhang, J.; Ji, J.Y.; Overholtzer, M.; Smolen, G.A.; Wang, R.; Brugge, J.S.; Dyson, N.J.; Haber, D.A. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat. Cell Biol. 2009, 11, 1444–1450. [Google Scholar] [CrossRef]
- Gregorieff, A.; Liu, Y.; Inanlou, M.R.; Khomchuk, Y.; Wrana, J.L. Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature 2015, 526, 715–718. [Google Scholar] [CrossRef]
- Haramis, A.P.; Begthel, H.; van den Born, M.; van Es, J.; Jonkheer, S.; Offerhaus, G.J.; Clevers, H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 2004, 303, 1684–1686. [Google Scholar] [CrossRef]
- Batts, L.E.; Polk, D.B.; Dubois, R.N.; Kulessa, H. Bmp signaling is required for intestinal growth and morphogenesis. Dev. Dyn. 2006, 235, 1563–1570. [Google Scholar] [CrossRef]
- He, X.C.; Zhang, J.; Tong, W.G.; Tawfik, O.; Ross, J.; Scoville, D.H.; Tian, Q.; Zeng, X.; He, X.; Wiedemann, L.M.; et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 2004, 36, 1117–1121. [Google Scholar] [CrossRef]
- Aoki, R.; Shoshkes-Carmel, M.; Gao, N.; Shin, S.; May, C.L.; Golson, M.L.; Zahm, A.M.; Ray, M.; Wiser, C.L.; Wright, C.V.; et al. Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell Mol. Gastroenterol. Hepatol. 2016, 2, 175–188. [Google Scholar] [CrossRef]
- Sancho, E.; Batlle, E.; Clevers, H. Signaling pathways in intestinal development and cancer. Annu. Rev. Cell Dev. Biol. 2004, 20, 695–723. [Google Scholar] [CrossRef]
- Mou, H.; Vinarsky, V.; Tata, P.R.; Brazauskas, K.; Choi, S.H.; Crooke, A.K.; Zhang, B.; Solomon, G.M.; Tumer, B.; Bihler, H.; et al. Dual SMAD signaling inhibition enables long-term expansion of diverse epithelial basal cells. Cell Stem Cell 2016, 19, 217–231. [Google Scholar] [CrossRef]
- Sato, T.; Clevers, H. Growing self-organizing mini-guts from a single intestinal stem cell: Mechanism and applications. Science 2013, 340, 1190–1194. [Google Scholar] [CrossRef]
- Hansson, E.M.; Lendahl, U.; Chapman, G. Notch signaling in development and disease. Semin. Cancer Biol. 2004, 14, 320–328. [Google Scholar] [CrossRef]
- Kageyama, R.; Ohtsuka, T.; Kobayashi, T. The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development 2007, 134, 1243–1251. [Google Scholar] [CrossRef]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Fleming, R.J. Structural conservation of Notch receptors and ligands. Semin. Cell Dev. Biol. 1998, 9, 599–607. [Google Scholar] [CrossRef]
- Tsai, Y.H.; VanDussen, K.L.; Sawey, E.T.; Wade, A.W.; Kasper, C.; Rakshit, S.; Bhatt, R.G.; Stoeck, A.; Mailland, I.; Crawford, H.C.; et al. ADAM10 regulates Notch function in intestinal stem cells of mice. Gastroenterology 2014, 147, 822–834. [Google Scholar] [CrossRef]
- Demitrack, E.S.; Samuelson, L.C. Notch regulation of gastrointestinal stem cells. J. Physiol. 2016, 594, 4791–4803. [Google Scholar] [CrossRef]
- Yang, Q.; Bermingham, N.A.; Finegold, M.J.; Zoghbi, H.Y. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 2001, 294, 2155–2158. [Google Scholar] [CrossRef]
- Shroyer, N.F.; Helmrath, M.A.; Wang, V.Y.; Antalffy, B.; Henning, S.J.; Zoghbi, H.Y. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology 2007, 132, 2478–2488. [Google Scholar] [CrossRef]
- van Es, J.H.; de Geest, N.; van de Born, M.; Clevers, H.; Hassan, B.A. Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitions. Nat. Commun. 2010, 1, 18. [Google Scholar] [CrossRef]
- Noah, T.K.; Shroyer, N.F. Notch in the intestine: Regulation of homeostasis and pathogenesis. Annu. Rev. Physiol. 2013, 75, 263–288. [Google Scholar] [CrossRef]
- VanDussen, K.L.; Samuelson, L.C. Mouse atonal homolog 1 directs intestinal progenitors to secretory cell rather than absorptive cell fate. Dev. Biol. 2010, 346, 215–223. [Google Scholar] [CrossRef]
- Ueo, T.; Imayoshi, I.; Kobayashi, T.; Ohtsuka, T.; Seno, H.; Nakase, H.; Chiba, T.; Kageyama, R. The role of Hes genes in intestinal development, homeostasis and tumor formation. Development 2012, 139, 1071–1082. [Google Scholar] [CrossRef]
- Stamataki, D.; Holder, M.; Hodgetts, C.; Jeffery, R.; Nye, E.; Spencer-Dene, B.; Winton, D.J.; Lewis, J. Delta1 expression, cell cycle exit, and commitment to a specific secretory fate coincide within a few hours in the mouse intestinal stem cell system. PLoS ONE 2011, 6, e24484. [Google Scholar] [CrossRef]
- Robinson, S.; Klobucar, K.; Pierre, C.C.; Ansari, A.; Zhenilo, S.; Prokhortchouk, E.; Daniel, J.M. Kaiso differentially regulates components of the Notch signaling pathway in intestinal cells. Cell Commun. Signal. 2017, 15, 24. [Google Scholar] [CrossRef]
- Jenny, M.; Uhl, C.; Roche, C.; Duluc, I.; Guillermin, V.; Guillemot, F.; Jensen, J.; Kedinger, M.; Gradwohl, G. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 2002, 21, 6338–6347. [Google Scholar] [CrossRef]
- Lee, C.S.; Perreault, N.; Brestelli, J.E.; Kaestner, K.H. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002, 16, 1488–1497. [Google Scholar] [CrossRef]
- Lopez-Diaz, L.; Jain, R.N.; Keeley, T.M.; VanDussen, K.L.; Brunkan, C.S.; Gumucio, D.L.; Samuelson, L.C. Intestinal Neurogenin 3 directs differentiation of a bipotential secretory progenitor to endocrine cell rather than goblet cell fate. Dev. Biol. 2007, 309, 298–305. [Google Scholar] [CrossRef]
- Shroyer, N.F.; Wallis, D.; Venken, K.J.; Bellen, H.J.; Zoghbi, H.Y. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005, 19, 2412–2417. [Google Scholar] [CrossRef]
- Noah, T.K.; Kazanjian, A.; Whitsett, J.; Shroyer, N.F. SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Exp. Cell Res. 2010, 316, 452–465. [Google Scholar] [CrossRef]
- Rothenberg, M.E.; Nusse, Y.; Kalisky, T.; Lee, J.J.; Dalerba, P.; Scheeren, F.; Lobo, N.; Kulkarni, S.; Sim, S.; Qian, D.; et al. Identification of a cKit+ colonic crypt base secretory cell that supports Lgr5+ stem cells in mice. Gastroenterology 2012, 152, 1195–1205. [Google Scholar] [CrossRef]
- Ghaleb, A.M.; McConnell, B.B.; Kaestner, K.H.; Yang, V.W. Altered intestinal epithelial homeostasis in mice with intestine-specific deletion of the Krüppel-like factor 4 gene. Dev. Biol. 2011, 349, 310–320. [Google Scholar] [CrossRef]
- Tian, H.; Biehs, B.; Chiu, C.; Siebel, C.W.; Wu, Y.; Costa, M.; de Sauvage, F.J.; Klein, O.D. Opposing activities of Notch and Wnt signaling regulate intestinal stem cell and gut homeostasis. Cell Rep. 2015, 11, 33–42. [Google Scholar] [CrossRef]
- Pitulescu, M.E.; Adams, R.H. Eph/ephrin molecules―a hub for signaling and endocytosis. Genes Dev. 2010, 24, 2480–2492. [Google Scholar] [CrossRef]
- Arvanitis, D.A.; Davy, A. Eph/ephrin signaling: Networks. Genes Dev. 2008, 22, 416–429. [Google Scholar] [CrossRef]
- Merlos-Suárez, A.; Barriga, F.M.; Jung, P.; Iglesias, M.; Céspedes, M.V.; Rossell, D.; Sevillano, M.; Hernando-Momblona, X.; da Silva-Diz, V.; Muñoz, P.; et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011, 8, 511–524. [Google Scholar] [CrossRef]
- Holmberg, J.; Genander, M.; Halford, M.M.; Annerén, C.; Sondell, M.; Chumley, M.J.; Silvany, R.E.; Henkemeyer, M.; Frisén, J. EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell 2006, 125, 1151–1163. [Google Scholar] [CrossRef]
- Takahashi, T.; Shiraishi, A.; Murata, Y. The coordinated activities of nAChR and Wnt signaling regulate intestinal stem cell function in mice. Int. J. Mol. Sci. 2018, 19, 738. [Google Scholar] [CrossRef]
- Wessler, I.; Kirkpatrick, C.J. Acetylcholine beyond neurons: The non-neuronal cholinergic system in humans. Br. J. Pharmacol. 2008, 154, 1558–1571. [Google Scholar] [CrossRef]
- Posadas, I.; Lópes-Hernández, B.; Ceňa, V. Nicotinic receptors in neurodegeneration. Curr. Neuropharmacol. 2013, 11, 298–314. [Google Scholar] [CrossRef]
- Cao, J.; Dwyer, J.B.; Mangold, J.E.; Wang, J.; Wei, J.; Leslie, F.M.; Li, M.D. Modulation of cell adhesion systems by prenatal nicotine exposure in limbic brain regions of adolescent female rats. Int. J. Neuropsychopharmacol. 2011, 14, 157–174. [Google Scholar] [CrossRef][Green Version]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- Gjorevski, N.; Lutolf, M.P. Synthesis and characterization of well-defined hydrogel matrices and their application to intestinal stem cell and organoid culture. Nature Protoc. 2017, 12, 2263–2274. [Google Scholar] [CrossRef]
- Capeling, M.M.; Czerwinski, M.; Huang, S.; Tsai, Y.-H.; Wu, A.; Nagy, M.S.; Juliar, B.; Sundaram, N.; Song, Y.; Han, W.M.; et al. Nonadhesive alginate hydrogels support growth of pluripotent stem cell-derived intestinal organoids. Stem Cell Rep. 2019, 12, 381–394. [Google Scholar] [CrossRef]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef]
- Camargo, F.D.; Gokhale, S.; Johnnidis, J.B.; Fu, D.; Bell, G.W.; Jaenisch, R.; Brummelkamp, T.R. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007, 17, 2054–2060. [Google Scholar] [CrossRef]
- Spradling, A.; Drummond-Barbosa, D.; Kai, T. Stem cells find their niche. Nature 2001, 414, 98–104. [Google Scholar] [CrossRef]
- Ootani, A.; Li, X.; Sangiorgi, E.; Ho, Q.T.; Ueno, H.; Toda, S.; Sugihara, H.; Fujimoto, K.; Weissman, I.L.; Capecchi, M.R.; et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 2009, 15, 701–706. [Google Scholar] [CrossRef]
- Kosinski, C.; Li, V.S.; Chan, A.S.; Zhang, J.; Ho, C.; Tsui, W.Y.; Chan, T.L.; Mifflin, R.C.; Powell, D.W.; Yuen, S.T.; et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc. Natl. Acad. Sci. USA 2007, 104, 15418–15423. [Google Scholar] [CrossRef]
- Hsu, D.R.; Economides, A.N.; Wang, X.; Eimon, P.M.; Harland, R.M. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol. Cell 1998, 1, 673–683. [Google Scholar] [CrossRef]
- Lin, G.; Zhang, X.; Ren, J.; Pang, Z.; Wang, C.; Xu, N.; Xi, R. Integrin signaling is required for maintenance and proliferation of intestinal stem cells in Drosophila. Dev. Biol. 2013, 377, 177–187. [Google Scholar] [CrossRef]
- Schultz, K.M.; Kyburz, K.A.; Anseth, K.S. Measuring dynamic cell-material interactions and remodeling during 3D human mesenchymal stem cell migration in hydrogels. Proc. Natl. Acad. Sci. USA 2015, 112, E3757–E3764. [Google Scholar] [CrossRef]
- Panek, M.; Grabacka, M.; Pierzchalska, M. The formation of intestinal organoids in a hanging drop culture. Cytotechnology 2018, 70, 1085–1095. [Google Scholar] [CrossRef]
- Jee, J.H.; Lee, D.H.; Ko, J.; Hahn, S.; Jeong, S.Y.; Kim, H.K.; Park, E.; Choi, S.Y.; Jeong, S.; Lee, J.W.; et al. Development of collagen-based 3D matrix for gastrointestinal tract-derived organoid culture. Stem Cells Int. 2019, 2019, 8472712. [Google Scholar] [CrossRef]
- Meran, L.; Baulies, A.; Li, V.S.W. Intestinal stem cell niche: The extracellular matrix and cellular components. Stem Cells Int. 2017, 2017, 7970385. [Google Scholar] [CrossRef]
- Balbi, V.; Ciarletta, P. Morpho-elasticity of intestinal villi. J. R. Soc. Interface 2013, 10, 20130109. [Google Scholar] [CrossRef]
- Mathur, D.; Bost, A.; Driver, I.; Ohlstein, B. A transient niche regulates the specification of Drosophila intestinal stem cells. Science 2010, 327, 210–213. [Google Scholar] [CrossRef]
- Ryan, J.F.; Pang, K.; Schnitzler, C.E.; Nguyen, A.-D.; Moreland, R.T.; Simmons, D.K.; Koch, B.J.; Francis, W.R.; Havlak, P.; NISC Comparative Sequencing Program; et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 2013, 342, 1242592. [Google Scholar] [CrossRef]
- Srivastava, M.; Begovic, E.; Chapman, J.; Putnam, N.H.; Hellsten, U.; Kawashima, T.; Kuo, A.; Mitros, T.; Salamov, A.; Carpenter, M.L.; et al. The Trichoplax genome and the nature of placozoans. Nature 2008, 454, 955–960. [Google Scholar] [CrossRef]
- Peterson, C.P.; Reddien, P.W. Wnt signaling and the polarity of the primary body axis. Cell 2009, 139, 1056–1068. [Google Scholar] [CrossRef]
- Holstein, T.W.; Watanabe, H.; Ozbek, S. Signaling pathways and axis formation in the lower metazoa. Curr. Top. Dev. Biol. 2011, 97, 137–177. [Google Scholar]
- Snijder, B.; Sacher, R.; Rämö, P.; Damm, E.M.; Liberali, P.; Pelkmans, L. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature 2009, 461, 520–523. [Google Scholar] [CrossRef]
- Battich, N.; Stoeger, T.; Pelkmans, L. Control of transcript variability in single mammalian cells. Cell 2015, 163, 1596–1610. [Google Scholar] [CrossRef]
- Loewer, A.; Lahav, G. We are all individuals: Causes and consequences of non-genetic heterogeneity in mammalian cells. Curr. Opin. Genet. Dev. 2011, 21, 753–758. [Google Scholar] [CrossRef]
- Wennekamp, S.; Mesecke, S.; Médélec, F.; Hiiragi, T. A self-organization framework for symmetry breaking in the mammalian embryo. Nat. Rev. Mol. Cell Biol. 2013, 14, 452–459. [Google Scholar] [CrossRef]
- Ayyaz, A.; Kumar, S.; Sangiorgi, B.; Ghoshal, B.; Gosio, J.; Ouladan, S.; Fink, M.; Barutcu, S.; Trcka, D.; Shen, J.; et al. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 2019, 569, 121–125. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).