The Control of Developmental Phase Transitions by microRNAs and Their Targets in Seed Plants
Abstract
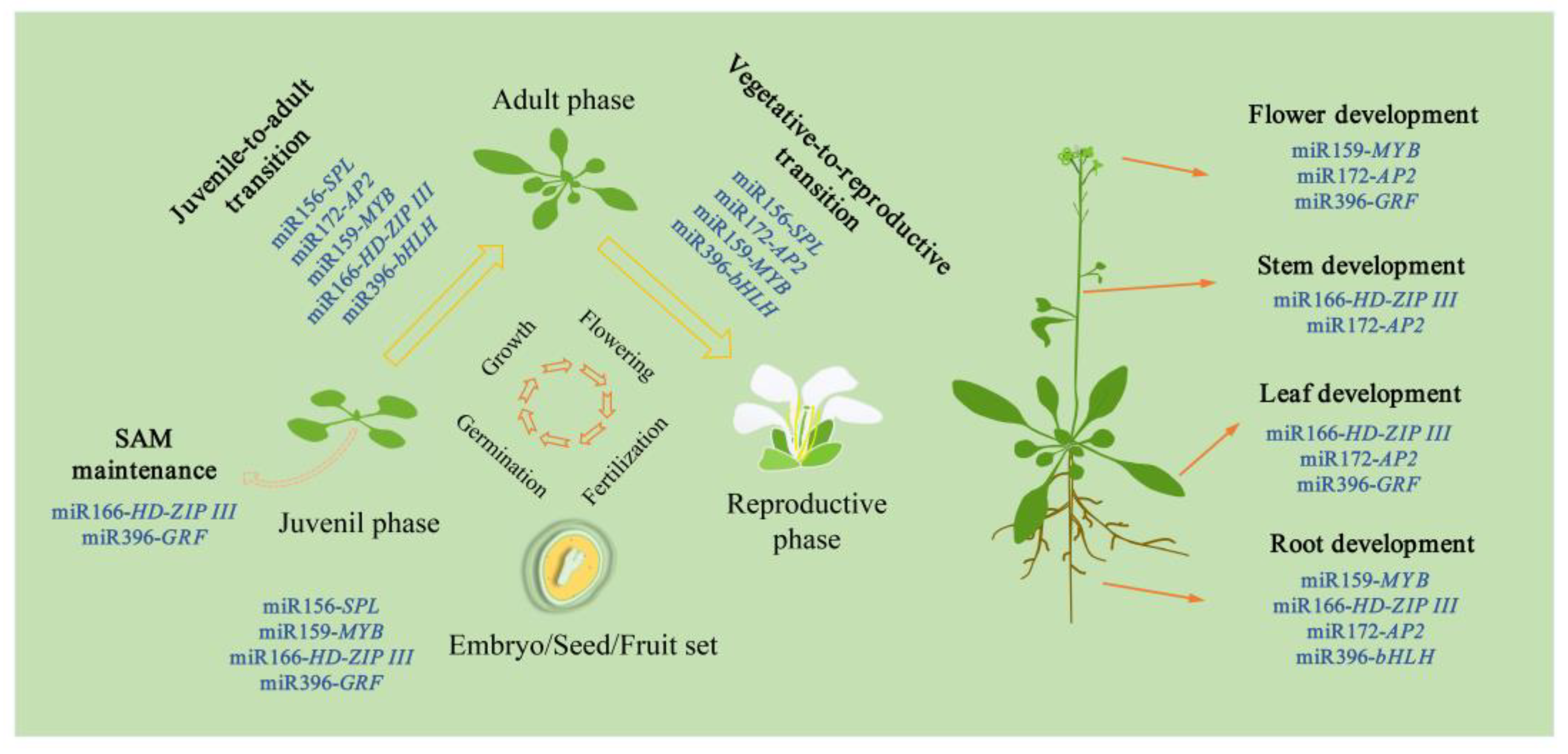
1. Introduction
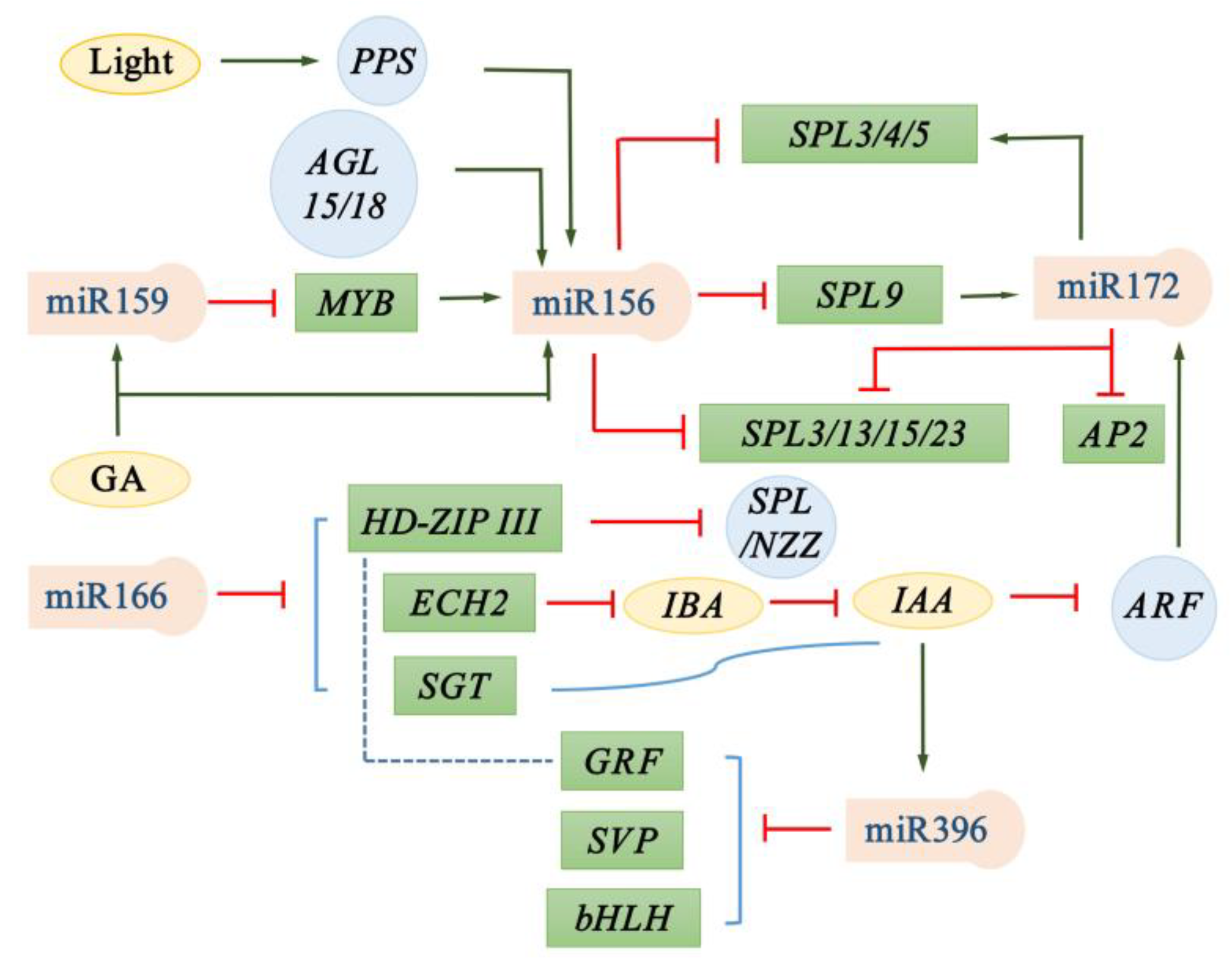
2. miR159
3. miR166
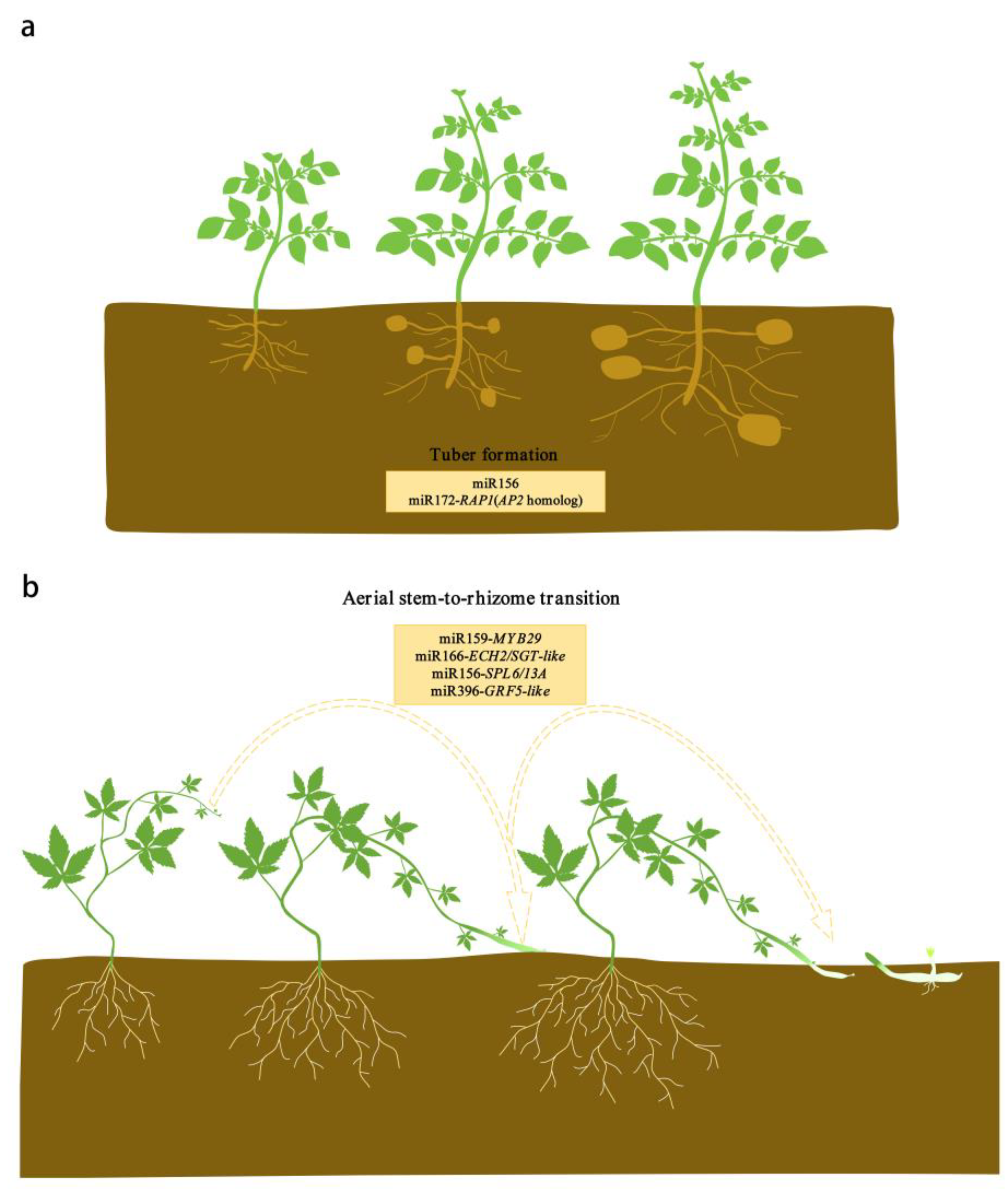
4. miR156
5. miR172
6. miR396
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| miRNAs | MicroRNAs |
| SAM | Shoot apical meristem |
| GA | Gibberellin |
| GSLs | Glucosinolates |
| HD-ZIP III | Homeodomain leucine zipper class III |
| PHB | Phabulosa |
| PHV | Phavoluta |
| REV | Revoluta |
| ATHB8 | Arabidopsis thaliana homeobox 8 |
| ATHB15 | Arabidopsis thaliana homeobox 15 |
| PCN | Popcorona |
| SPL/NZZ | Sporocyteless/Nozzle |
| ECH2 | Enoyl-CoA hydratase 2 |
| SGT | Scopoletin glucosyltransferase |
| SPL | Squamosa promoter binding protein like |
| SD | Short day |
| PPS | Peter pan syndrome |
| AP2 | Apetala2 |
| TOE1 | Target of eat 1 |
| SMZ | Schlafmutze |
| SNZ | Schnarchzapfen |
| LD | Long day |
| PHYB | Phytochrome B |
| RAP1 | Related to apetala2 1 |
| BEL5 | Bell5 |
| AuxREs | auxin response elements |
| ARF | Auxin response factors |
| GRF | Growth-regulating factor |
| bHLH | Basic helix-loop-helix |
| SVP | Short vegetative phase |
| FT | Flowering locus T |
References
- Huijser, P.; Schmid, M. The control of developmental phase transitions in plants. Development 2011, 138, 4117–4129. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Itoh, H.; Sentoku, N.; Kojima, M.; Sakakibara, H.; Izawa, T.; Itoh, J.; Nagato, Y. The COP1 ortholog PPS regulates the juvenile-adult and vegetative-reproductive phase changes in rice. Plant Cell 2011, 23, 2143–2154. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Guo, Z.; Li, L. Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev. Biol. 2013, 380, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Oh, T.J.; Wartell, R.M.; Cairney, J.; Pullman, G.S. Evidence for stage-specific modulation of specific microRNAs (miRNAs) and miRNA processing components in zygotic embryo and female gametophyte of loblolly pine (Pinus taeda). New Phytol. 2008, 179, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Li, S.; Yin, J.; Li, L.; Chen, X. Genome-wide analysis of differentially expressed genes relevant to rhizome formation in lotus root (Nelumbo nucifera Gaertn). PLoS ONE 2013, 8, e67116. [Google Scholar] [CrossRef] [PubMed]
- Bhogale, S.; Mahajan, A.S.; Natarajan, B.; Rajabhoj, M.; Thulasiram, H.V.; Banerjee, A.K. MicroRNA156: A potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 2014, 164, 1011–1027. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, S.; Han, X.; Ma, J.; Deng, W.; Wang, X.; Guo, H.; Xia, X. Integrated transcriptome and miRNA analysis uncovers molecular regulators of aerial stem-to-rhizome transition in the medical herb Gynostemma pentaphyllum. BMC Genomics 2019, 20, 865. [Google Scholar] [CrossRef]
- Aung, B.; Gruber, M.Y.; Amyot, L.; Omari, K.; Bertrand, A.; Hannoufa, A. MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol. J. 2015, 13, 779–790. [Google Scholar] [CrossRef]
- Ahsan, M.U.; Hayward, A.; Irihimovitch, V.; Fletcher, S.; Tanurdzic, M.; Pocock, A.; Beveridge, C.A.; Mitter, N. Juvenility and vegetative phase transition in tropical/subtropical tree crops. Front. Plant Sci. 2019, 10, 729. [Google Scholar] [CrossRef]
- He, J.; Xu, M.; Willmann, M.R.; McCormick, K.; Hu, T.; Yang, L.; Starker, C.G.; Voytas, D.F.; Meyers, B.C.; Poethig, R.S. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana. PLoS Genet. 2018, 14, e1007337. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, L.; Wu, G. Epigenetic regulation of juvenile-to-adult transition in plants. Front. Plant Sci. 2018, 9, 1048. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Bartel, D.P. Antiquity of microRNAs and their targets in land plants. Plant Cell 2005, 17, 1658–1673. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Bowman, J.L. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008, 13, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to line-14. Cell 1993, 75, 834–854. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef]
- Gai, Y.P.; Zhao, H.N.; Zhao, Y.N.; Zhu, B.S.; Yuan, S.S.; Li, S.; Guo, F.Y.; Ji, X.L. MiRNA-seq-based profiles of miRNAs in mulberry phloem sap provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease. Sci. Rep. 2018, 8, 812. [Google Scholar] [CrossRef]
- Nozawa, M.; Miura, S.; Nei, M. Origins and evolution of microRNA genes in plant species. Genome Biol. Evol. 2012, 4, 230–239. [Google Scholar] [CrossRef]
- Budak, H.; Akpinar, B.A. Plant miRNAs: Biogenesis, organization and origins. Funct. Integr. Genomics 2015, 15, 523–531. [Google Scholar] [CrossRef]
- Wang, J.; Mei, J.; Ren, G. Plant microRNAs: Biogenesis, Homeostasis, and Degradation. Front. Plant Sci. 2019, 10, 360. [Google Scholar] [CrossRef]
- Xie, Z.; Allen, E.; Fahlgren, N.; Calamar, A.; Givan, S.A.; Carrington, J.C. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005, 138, 2145–2154. [Google Scholar] [CrossRef]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Romero-Barrios, N.; Legascue, M.F.; Benhamed, M.; Ariel, F.; Crespi, M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018, 46, 2169–2184. [Google Scholar] [CrossRef] [PubMed]
- von Born, P.; Bernardo-Faura, M.; Rubio-Somoza, I. An artificial miRNA system reveals that relative contribution of translational inhibition to miRNA-mediated regulation depends on environmental and developmental factors in Arabidopsis thaliana. PLoS ONE 2018, 13, e0192984. [Google Scholar] [CrossRef] [PubMed]
- Couzigou, J.M.; Lauressergues, D.; Andre, O.; Gutjahr, C.; Guillotin, B.; Becard, G.; Combier, J.P. Positive gene regulation by a natural protective miRNA enables arbuscular mycorrhizal symbiosis. Cell Host Microbe. 2017, 21, 106–112. [Google Scholar] [CrossRef]
- Omidvar, V.; Mohorianu, I.; Dalmay, T.; Fellner, M. MicroRNA Regulation of Abiotic Stress Response in 7B-1 Male-Sterile Tomato Mutant. Plant Genome 2015, 8, 3. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Wang, Z.; Guo, X.; Wang, F.; Xia, X.J.; Zhou, J.; Shi, K.; Yu, J.Q.; Zhou, Y.H. Microarray and genetic analysis reveals that csa-miR159b plays a critical role in abscisic acid-mediated heat tolerance in grafted cucumber plants. Plant Cell Environ. 2016, 39, 1790–1804. [Google Scholar] [CrossRef]
- Millar, A.A.; Lohe, A.; Wong, G. Biology and Function of miR159 in Plants. Plants 2019, 8, 255. [Google Scholar] [CrossRef]
- Garcia, D. A miRacle in plant development: Role of microRNAs in cell differentiation and patterning. Semin. Cell Dev. Biol. 2008, 19, 586–595. [Google Scholar] [CrossRef]
- Achard, P.; Herr, A.; Baulcombe, D.C.; Harberd, N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development 2004, 131, 3357–3365. [Google Scholar] [CrossRef]
- Li, X.; Bian, H.; Song, D.; Ma, S.; Han, N.; Wang, J.; Zhu, M. Flowering time control in ornamental gloxinia (Sinningia speciosa) by manipulation of miR159 expression. Ann. Bot. 2013, 111, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, F.; Cao, H.; Peng, H.; Ni, Z.; Sun, Q.; Yao, Y. TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS ONE 2017, 7, e48445. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef]
- Alonso-Peral, M.M.; Li, J.; Li, Y.; Allen, R.S.; Schnippenkoetter, W.; Ohms, S.; White, R.G.; Millar, A.A. The MicroRNA159-Regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol. 2010, 154, 757–771. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Pandey, D.K.; Chaudhary, B. Target-mimicry based diminution of miRNA167 reinforced flowering-time phenotypes in tobacco via spatial-transcriptional biases of flowering associated miRNAs. Gene 2019, 682, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Li, W.F.; Zhang, S.G.; Han, S.Y.; Wu, T.; Zhang, J.H.; Qi, L.W. Regulation of LaMYB33 by miR159 during maintenance of embryogenic potential and somatic embryo maturation in Larix kaempferi (Lamb.) Carr. Plant Cell Tiss. Org. 2013, 113, 131–136. [Google Scholar] [CrossRef]
- Huang, D.; Koh, C.; Feurtado, J.A.; Tsang, E.W.; Cutler, A.J. MicroRNAs and their putative targets in Brassica napus seed maturation. BMC Genomics 2013, 14, 140. [Google Scholar] [CrossRef]
- Kitazumi, A.; Kawahara, Y.; Onda, T.S.; De Koeyer, D.; de los Reyes, B.G.; Cloutier, S. Implications of miR166 and miR159 induction to the basal response mechanisms of an andigena potato (Solanum tuberosum subsp. andigena) to salinity stress, predicted from network models in Arabidopsis. Genome 2015, 58, 13–24. [Google Scholar] [CrossRef]
- da Silva, E.; Silva, G.; Bidoia, D.; da Silva Azevedo, M.; de Jesus, F.; Pino, L.; Peres, L.; Carrera, E.; López-Díaz, I.; Nogueira, F. MicroRNA159-targeted SlGAMYB transcription factors are required for fruit set in tomato. Plant J. 2017, 92, 95–109. [Google Scholar] [CrossRef]
- Hu, Z.; Shen, X.; Xiang, X.; Cao, J. Evolution of MIR159/319 genes in Brassica campestris and their function in pollen development. Plant Mol. Biol. 2019, 101, 537–550. [Google Scholar] [CrossRef]
- Guo, C.; Xu, Y.; Shi, M.; Lai, Y.; Wu, X.; Wang, H.; Zhu, Z.; Poethig, R.S.; Wu, G. Repression of miR156 by miR159 regulates the timing of the juvenile-to-adult transition in Arabidopsis. Plant Cell 2017, 29, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Liu, Z.; Dai, X.; Xiang, F. Primary root growth in Arabidopsis thaliana is inhibited by the miR159 mediated repression of MYB33, MYB65 and MYB101. Plant Sci. 2017, 262, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Araki, R.; Hasumi, A.; Nishizawa, O.I.; Sasaki, K.; Kuwahara, A.; Sawada, Y.; Totoki, Y.; Toyoda, A.; Sakaki, Y.; Li, Y.; et al. Novel bioresources for studies of Brassica oleracea: Identification of a kale MYB transcription factor responsible for glucosinolate production. Plant Biotechnol. J. 2013, 11, 1017–1027. [Google Scholar] [CrossRef]
- Kim, Y.B.; Li, X.; Kim, S.J.; Kim, H.H.; Lee, J.; Kim, H.; Park, S.U. MYB transcription factors regulate glucosinolate biosynthesis in different organs of Chinese cabbage (Brassica rapa ssp. pekinensis). Molecules 2013, 18, 8682–8695. [Google Scholar] [CrossRef] [PubMed]
- Burow, M.; Halkier, B.A.; Kliebenstein, D.J. Regulatory networks of glucosinolates shape Arabidopsis thaliana fitness. Curr. Opin. Plant Biol. 2010, 13, 347–352. [Google Scholar] [CrossRef]
- Moyers, B.T. Symphony of the Regulators: How do plants control complex responses to environmental signals? Plant Cell 2018, 30, 178–195. [Google Scholar] [CrossRef]
- Kim, J.; Jung, J.H.; Reyes, J.L.; Kim, Y.S.; Kim, S.Y.; Chung, K.S.; Kim, J.A.; Lee, M.; Lee, Y.; Kim, V.N.; et al. MicroRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 2005, 42, 84–94. [Google Scholar] [CrossRef]
- Jung, J.H.; Park, C.M. MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta 2007, 225, 1327–1338. [Google Scholar] [CrossRef]
- Tang, X.; Bian, S.; Tang, M.; Lu, Q.; Li, S.; Liu, X.; Tian, G.; Nguyen, V.; Tsang, E.W.T.; Wang, A.; et al. MicroRNA–mediated repression of the seed maturation program during vegetative development in Arabidopsis. PLoS Genet. 2012, 8, e1003091. [Google Scholar] [CrossRef]
- Li, Z.X.; Li, S.G.; Zhang, L.F.; Han, S.Y.; Li, W.F.; Xu, H.Y.; Yang, W.H.; Liu, Y.L.; Fan, Y.R.; Qi, L.W. Over-expression of miR166a inhibits cotyledon formation in somatic embryos and promotes lateral root development in seedlings of Larix leptolepis. Plant Cell Tiss. Org. 2016, 127, 461–473. [Google Scholar] [CrossRef]
- Williams, L.; Grigg, S.P.; Xie, M.; Christensen, S.; Fletcher, J.C. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 2005, 132, 3657–3668. [Google Scholar] [CrossRef] [PubMed]
- Boualem, A.; Laporte, P.; Jovanovic, M.; Laffont, C.; Plet, J.; Combier, J.-P.; Niebel, A.; Crespi, M.; Frugier, F. MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J. 2008, 54, 876–887. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Miura, E.; Robischon, M.; Martinez, C.; Groover, A. The Populus Class III HD ZIP transcription factor POPCORONA affects cell differentiation during secondary growth of woody stems. PLoS ONE 2011, 6, e17458. [Google Scholar] [CrossRef] [PubMed]
- Ragni, L.; Greb, T. Secondary growth as a determinant of plant shape and form. Semin. Cell Dev. Biol. 2018, 79, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Li, X.; Zhao, Y.; Zhang, M.; Wan, Y.; Cao, D.; Lu, S.; Lin, J. Genome-wide analysis reveals dynamic changes in expression of microRNAs during vascular cambium development in Chinese fir, Cunninghamia lanceolata. J. Exp. Bot. 2015, 66, 3041–3054. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gao, X.; Li, L.; Shi, X.; Zhang, J.; Shi, Z. Overexpression of Osta-siR2141 caused abnormal polarity establishment and retarded growth in rice. J. Exp. Bot. 2010, 61, 1885–1895. [Google Scholar] [CrossRef]
- Jian, C.; Han, R.; Chi, Q.; Wang, S.; Ma, M.; Liu, X.; Zhao, H.X. Virus-based microrna silencing and overexpressing in common wheat (Triticum aestivum L.). Front. Plant Sci. 2017, 8, 500. [Google Scholar] [CrossRef]
- Aydinoglu, F.; Lucas, S.J. Identification and expression profiles of putative leaf growth related microRNAs in maize (Zea mays L.) hybrid ADA313. Gene 2019, 690, 57–67. [Google Scholar] [CrossRef]
- Li, X.; Lian, H.; Zhao, Q.; He, Y. microRNA166 monitors SPOROCYTELESS/NOZZLE (SPL/NZZ) for building of the anther internal boundary. Plant Physiol. 2019, 181, 208–220. [Google Scholar] [CrossRef]
- Li, Z.X.; Fan, Y.R.; Dang, S.F.; Li, W.F.; Qi, L.W.; Han, S.Y. LaMIR166a-mediated auxin biosynthesis and signalling affect somatic embryogenesis in Larix leptolepis. Mol. Genet. Genomics 2018, 293, 1355–1363. [Google Scholar] [CrossRef]
- Strader, L.C.; Wheeler, D.L.; Christensen, S.E.; Berens, J.C.; Cohen, J.D.; Rampey, R.A.; Bartel, B. Multiple facets of Arabidopsis seedling development require indole-3-butyric acid–derived auxin. Plant Cell 2011, 23, 984–999. [Google Scholar] [CrossRef] [PubMed]
- Rayle, D.L.; Cleland, R.E. The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiol. 1992, 99, 1271–1274. [Google Scholar] [CrossRef] [PubMed]
- Katano, M.; Tsukaya, H.; Takahashi, K.; Hirano, T.; Kazama, Y.; Abe, T.; Ferjani, A. Suppressor screen and phenotype analyses revealed an emerging role of the monofunctional peroxisomal Enoyl-CoA Hydratase 2 in compensated cell enlargement. Front. Plant Sci. 2016, 7, 132. [Google Scholar] [CrossRef] [PubMed]
- Hino, F.; Okazaki, M.; Miura, Y. Effect of 2,4-Dichlorophenoxyacetic Acid on glucosylation of scopoletin to scopolin in tobacco tissue culture. Plant Physiol. 1982, 69, 810–813. [Google Scholar] [CrossRef]
- Gális, I.; Šimek, P.; van Onckelen, H.A.; Kakiuchi, Y.; Wabiko, H. Resistance of transgenic tobacco seedlings expressing the Agrobacterium tumefaciens C58-6b gene, to growth-inhibitory levels of cytokinin is associated with elevated IAA levels and activation of phenylpropanoid metabolism. Plant Cell Physiol. 2002, 43, 939–950. [Google Scholar] [CrossRef]
- Poethig, R.S. Vegetative phase change and shoot maturation in plants. Curr. Top Dev. Biol. 2013, 105, 125–152. [Google Scholar] [CrossRef]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E.; et al. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef]
- Guo, A.Y.; Zhu, Q.H.; Gu, X.; Ge, S.; Yang, J.; Luo, J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 2008, 418, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cardon, G.; Ho¨hmann, S.; Klein, J.; Nettesheim, K.; Saedler, H.; Huijser, P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene 1999, 237, 91–104. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, X.; Gu, S.; Hu, Z.; Xu, H.; Xu, C. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 2008, 407, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Cao, Y.; Yang, R.; Qi, T.; Hang, Y.; Lin, H.; Zhou, G.; Wang, Z.Y.; Fu, C. Switchgrass SBP-box transcription factors PvSPL1 and 2 function redundantly to initiate side tillers and affect biomass yield of energy crop. Biotechnol Biofuels 2016, 9, 101. [Google Scholar] [CrossRef]
- Li, X.Y.; Lin, E.P.; Huang, H.H.; Niu, M.Y.; Tong, Z.K.; Zhang, J.H. Molecular characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) gene family in Betula luminifera. Front. Plant Sci. 2018, 9, 608. [Google Scholar] [CrossRef]
- Xu, P.; Wu, F.; Ma, T.T.; Yan, Q.; Zong, X.F.; Li, J.; Zhao, Y.F.; Kanzana, G.; Zhang, J.Y. Analysis of Six Transcription Factor Families Explores Transcript Divergence of Cleistogamous and Chasmogamous Flowers in Cleistogenes songorica. Dna Cell Biol. 2020, 39, 2. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; Garcia, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.-J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef]
- Luo, L.; Li, W.; Miura, K.; Ashikari, M.; Kyozuka, J. Control of tiller growth of rice by OsSPL14 and Strigolactones, which work in two independent pathways. Plant Cell Physiol. 2012, 53, 1793–1801. [Google Scholar] [CrossRef]
- Liu, Q.; Shen, G.; Peng, K.; Huang, Z.; Tong, J.; Kabir, M.H.; Wang, J.; Zhang, J.; Qin, G.; Xiao, L. The alteration in the architecture of a T-DNA insertion rice mutantosmtd1is caused by up-regulation of MicroRNA156f. J. Integr. Plant Biol. 2015, 57, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Wu, C.; Xiong, L. Genomic Organization, Differential Expression, and Interaction of SQUAMOSA Promoter-Binding-Like Transcription Factors and microRNA156 in Rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zou, Z.; Zhang, J.; Zhang, Y.; Han, Q.; Hu, T.; Xu, X.; Liu, H.; Li, H.; Ye, Z. Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett. 2011, 585, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Sunkar, R.; Zhou, C.; Shen, H.; Zhang, J.Y.; Matts, J.; Wolf, J.; Mann, D.G.; Stewart, C.N., Jr.; Tang, Y.; et al. Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol. J. 2012, 10, 443–452. [Google Scholar] [CrossRef]
- Aung, B.; Gao, R.; Gruber, M.Y.; Yuan, Z.C.; Sumarah, M.; Hannoufa, A. MsmiR156 affects global gene expression and promotes root regenerative capacity and nitrogen fixation activity in alfalfa. Transgenic Res. 2017, 26, 541–557. [Google Scholar] [CrossRef]
- Wang, J.W.; Park, M.Y.; Wang, L.J.; Koo, Y.; Chen, X.Y.; Weigel, D.; Poethig, R.S. MiRNA control of vegetative phase change in trees. PLoS Genet. 2011, 7, e1002012. [Google Scholar] [CrossRef]
- Long, J.M.; Liu, C.Y.; Feng, M.Q.; Liu, Y.; Wu, X.M.; Guo, W.W. miR156-SPL modules regulate induction of somatic embryogenesis in citrus callus. J. Exp. Bot. 2018, 69, 2979–2993. [Google Scholar] [CrossRef]
- Chen, G.; Li, J.; Liu, Y.; Zhang, Q.; Gao, Y.; Fang, K.; Cao, Q.; Qin, L.; Xing, Y. Roles of the GA-mediated SPL Gene family and miR156 in the floral development of Chinese Chestnut (Castanea mollissima). Int. J. Mol. Sci. 2019, 20, 1577. [Google Scholar] [CrossRef]
- Xu, X.; Vreugdenhil, D.; Lammeren, A.A.M.V. Cell division and cell enlargement during potato tuber formation. J. Exp. Bot. 1998, 49, 573–582. [Google Scholar] [CrossRef]
- Serivichyaswat, P.; Ryu, H.S.; Kim, W.; Kim, S.; Chung, K.S.; Kim, J.J.; Ahn, J.H. Expression of the floral repressor miRNA156 is positively regulated by the agamous-like proteins AGL15 and AGL18. Mol. Cells 2015, 38, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2 -like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Bai, X.; Niu, L.-J.; Chai, X.; Chen, M.-S.; Xu, Z.-F. MiR172 regulates both vegetative and reproductive development in the perennial woody plant Jatropha curcas. Plant Cell Physiol. 2018, 59, 2549–2563. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Lu, W.; Lu, Z.; Ren, S.; Zhao, B.; Wang, L.; Teng, N.; Jin, B. Identification and analysis of microRNAs in the SAM and Leaves of Populus tomentosa. Forests 2019, 10, 130. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Lin, H.; Chuck, G.; Faris, J.D.; Dubcovsky, J. MicroRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 2017, 144, 1966–1975. [Google Scholar] [CrossRef]
- Li, X.Y.; Guo, F.; Ma, S.Y.; Zhu, M.Y.; Pan, W.H.; Bian, H.W. Regulation of flowering time via miR172-mediated APETALA2-like expression in ornamental gloxinia (Sinningia speciosa). J. Zhejiang Univ. Sci. B 2019, 20, 322–331. [Google Scholar] [CrossRef]
- José Ripoll, J.; Bailey, L.J.; Mai, Q.A.; Wu, S.L.; Hon, C.T.; Chapman, E.J.; Ditta, G.S.; Estelle, M.; Yanofsky, M.F. microRNA regulation of fruit growth. Nature Plants 2015, 1, 15036. [Google Scholar] [CrossRef]
- Tripathi, R.K.; Bregitzer, P.; Singh, J. Genome-wide analysis of the SPL/miR156 module and its interaction with the AP2/miR172 unit in barley. Sci. Rep. 2018, 8, 7085. [Google Scholar] [CrossRef]
- Martin, A.; Adam, H.; Diaz-Mendoza, M.; Zurczak, M.; Gonzalez-Schain, N.D.; Suarez-Lopez, P. Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 2009, 136, 2873–2881. [Google Scholar] [CrossRef]
- Díaz-Manzano, F.; Cabrera, J.; Ripoll, J.; Del Olmo, I.; Andrés, M.; Silva, A.; Barcala, M.; Sánchez, M.; Ruíz-Ferrer, V.; de Almeida-Engler, J.; et al. A role for the gene regulatory module microRNA172/TARGET OF EARLY ACTIVATION TAGGED 1/FLOWERING LOCUS T (miRNA172/TOE1/FT) in the feeding sites induced by Meloidogyne javanica in Arabidopsis thaliana. New Phytol. 2018, 217, 813–827. [Google Scholar] [CrossRef]
- Yang, M.; Zhu, L.; Pan, C.; Xu, L.; Liu, Y.; Ke, W.; Yang, P. Transcriptomic analysis of the regulation of rhizome formation in temperate and tropical lotus (Nelumbo nucifera). Sci. Rep. 2015, 5, 13059. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Seo, P.J.; Kang, S.K.; Park, C.M. MiR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Mol. Biol. 2011, 76, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, S.; Jin, J.; Fu, D.; Yang, X.; Weng, X.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 15504–15509. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003, 36, 94–104. [Google Scholar] [CrossRef]
- Yang, C.Y.; Huang, Y.H.; Lin, C.P.; Lin, Y.Y.; Hsu, H.C.; Wang, C.N.; Liu, L.Y.; Shen, B.N.; Lin, S.S. MicroRNA396-targeted SHORT VEGETATIVE PHASE is required to repress flowering and is related to the development of abnormal flower symptoms by the phyllody symptoms1 effector. Plant Physiol. 2015, 168, 1702–1716. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Rodriguez, R.E.; Mecchia, M.A.; Palatnik, J.F. Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 2012, 8, e1002419. [Google Scholar] [CrossRef]
- Liu, D.; Song, Y.; Chen, Z.; Yu, D. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 2009, 136, 223–236. [Google Scholar] [CrossRef]
- Liu, W.; Zhou, Y.; Li, X.; Wang, X.; Dong, Y.; Wang, N.; Liu, X.; Chen, H.; Yao, N.; Cui, X.; et al. Tissue-specific regulation of Gma-miR396 family on coordinating development and low water availability responses. Front. Plant Sci. 2017, 8, 1112. [Google Scholar] [CrossRef]
- Szczygiel-Sommer, A.; Gaj, M.D. The miR396-GRF regulatory module controls the embryogenic response in Arabidopsis via an Auxin-related pathway. Int. J. Mol. Sci. 2019, 20, 5221. [Google Scholar] [CrossRef]
- Yang, F.; Liang, G.; Liu, D.; Yu, D. Arabidopsis miR396 mediates the development of leaves and flowers in transgenic tobacco. J. Plant Biol. 2009, 52, 475–481. [Google Scholar] [CrossRef]
- Baucher, M.; Moussawi, J.; Vandeputte, O.M.; Monteyne, D.; Mol, A.; Perez-Morga, D.; El Jaziri, M. A role for the miR396/GRF network in specification of organ type during flower development, as supported by ectopic expression of Populus trichocarpa miR396c in transgenic tobacco. Plant Biol. (Stuttg) 2013, 15, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Wang, J.; Ju, Z.; Liu, Q.; Li, S.; Tian, H.; Fu, D.; Zhu, H.; Luo, Y.; Zhu, B. Regulations on growth and development in tomato cotyledon, flower and fruit via destruction of miR396 with short tandem target mimic. Plant Sci. 2016, 247, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Guo, S.; Xu, Y.; Li, C.; Zhang, Z.; Zhang, D.; Xu, S.; Zhang, C.; Chong, K. OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 2014, 165, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Shi, X.; Ding, D.; Tang, J.; Zhao, X.; Niu, J. Investigation of miR396 and growth-regulating factor regulatory network in maize grain filling. Acta. Physiol. Plant 2015, 37, 28. [Google Scholar] [CrossRef]
- Diao, Z.; Yu, M.; Bu, S.; Duan, Y.; Zhang, L.; Wu, W. Functional characterization of OsmiR396a in rice (Oryza sativa L.). Plant Growth Regul. 2018, 85, 351–361. [Google Scholar] [CrossRef]
- McConnell, J.R.; Barton, M.K. Leaf polarity and meristem formation in Arabidopsis. Development 1998, 125, 2935–2942. [Google Scholar]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef]
- Mecchia, M.A.; Debernardi, J.M.; Rodriguez, R.E.; Schommer, C.; Palatnik, J.F. MicroRNA miR396 and RDR6 synergistically regulate leaf development. Mech. Dev. 2013, 130, 2–13. [Google Scholar] [CrossRef]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Lee, J.S.; Ahn, J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef]
- Bazin, J.; Khan, G.A.; Combier, J.P.; Bustos-Sanmamed, P.; Debernardi, J.M.; Rodriguez, R.; Sorin, C.; Palatnik, J.; Hartmann, C.; Crespi, M.; et al. MiR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 2013, 74, 920–934. [Google Scholar] [CrossRef]
- Bao, M.; Bian, H.; Zha, Y.; Li, F.; Sun, Y.; Bai, B.; Chen, Z.; Wang, J.; Zhu, M.; Han, N. MiR396a-Mediated basic helix-loop-helix transcription factor bHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant Cell Physiol. 2014, 55, 1343–1353. [Google Scholar] [CrossRef] [PubMed]
- Hou, N.; Cao, Y.; Li, F.; Yuan, W.; Bian, H.; Wang, J.; Zhu, M.; Han, N. Epigenetic regulation of miR396 expression by SWR1-C and the effect of miR396 on leaf growth and developmental phase transition in Arabidopsis. J. Exp. Bot. 2019, 70, 5217–5229. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Zhao, P.; Liu, S.; Yang, Q.; Guo, H. The Control of Developmental Phase Transitions by microRNAs and Their Targets in Seed Plants. Int. J. Mol. Sci. 2020, 21, 1971. https://doi.org/10.3390/ijms21061971
Ma J, Zhao P, Liu S, Yang Q, Guo H. The Control of Developmental Phase Transitions by microRNAs and Their Targets in Seed Plants. International Journal of Molecular Sciences. 2020; 21(6):1971. https://doi.org/10.3390/ijms21061971
Chicago/Turabian StyleMa, Jingyi, Pan Zhao, Shibiao Liu, Qi Yang, and Huihong Guo. 2020. "The Control of Developmental Phase Transitions by microRNAs and Their Targets in Seed Plants" International Journal of Molecular Sciences 21, no. 6: 1971. https://doi.org/10.3390/ijms21061971
APA StyleMa, J., Zhao, P., Liu, S., Yang, Q., & Guo, H. (2020). The Control of Developmental Phase Transitions by microRNAs and Their Targets in Seed Plants. International Journal of Molecular Sciences, 21(6), 1971. https://doi.org/10.3390/ijms21061971



