NEAT1 is Required for the Expression of the Liver Cancer Stem Cell Marker CD44
Abstract
:1. Introduction
2. Results
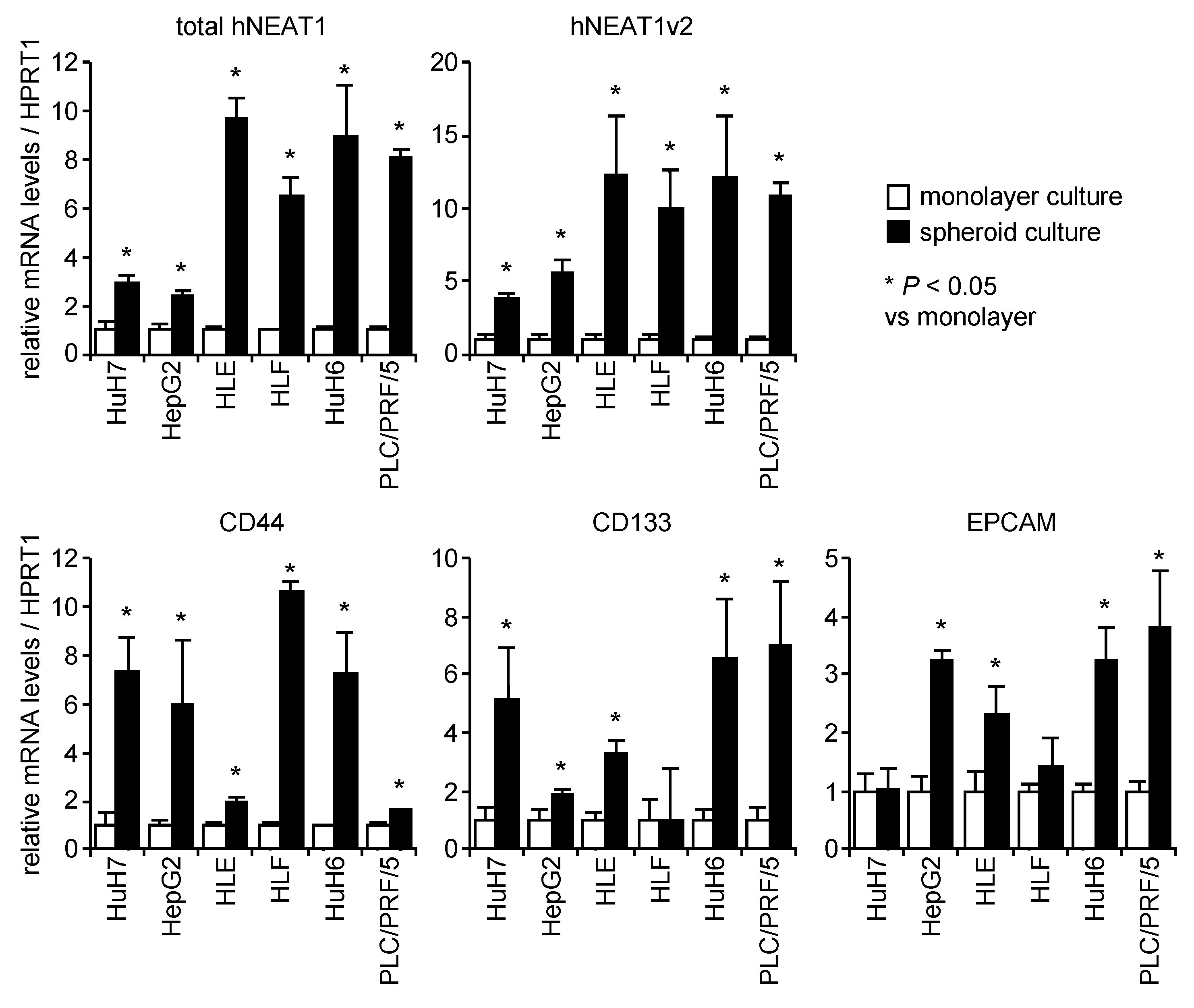
2.1. Increased Expression of NEAT1 in Spheroids
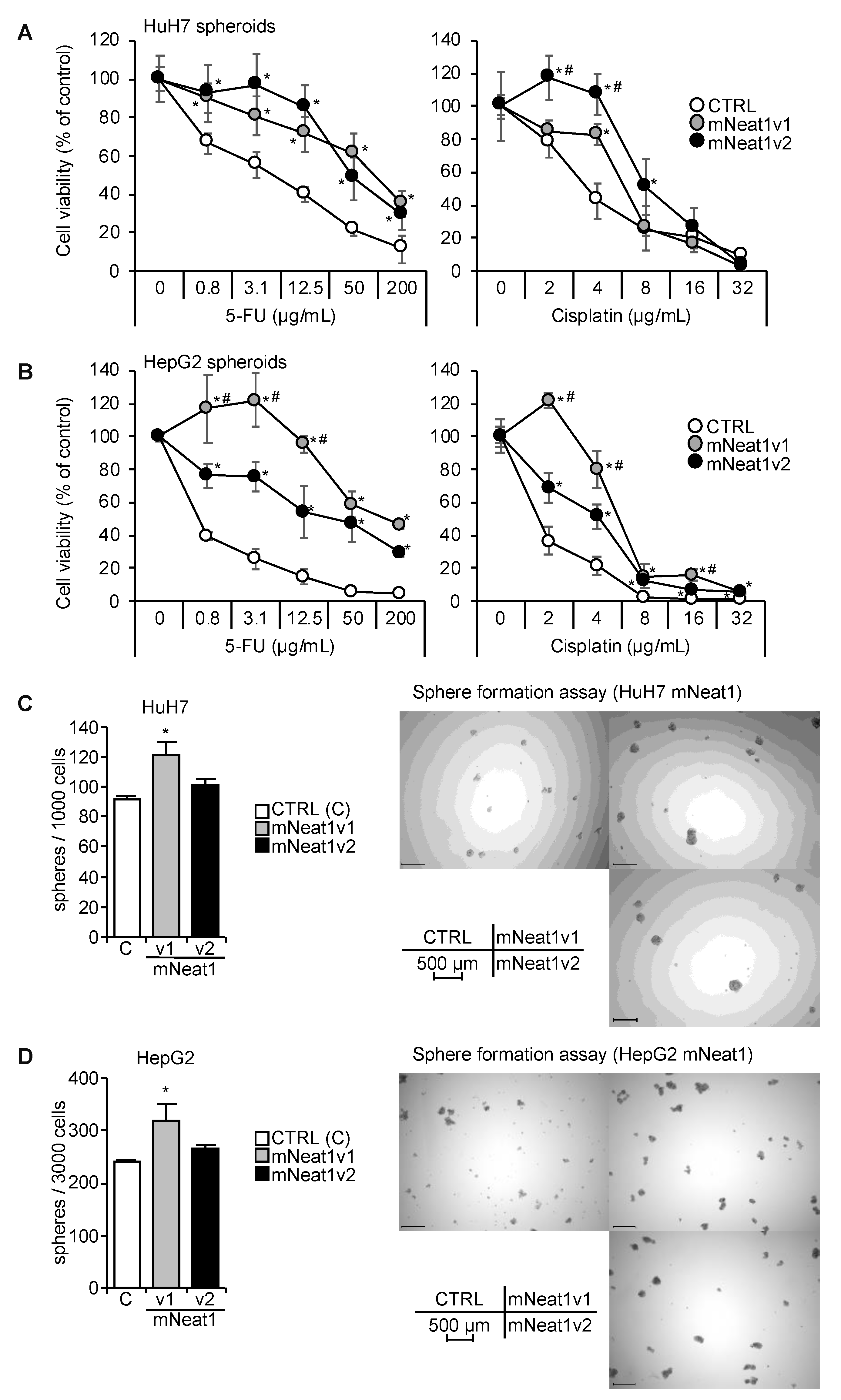
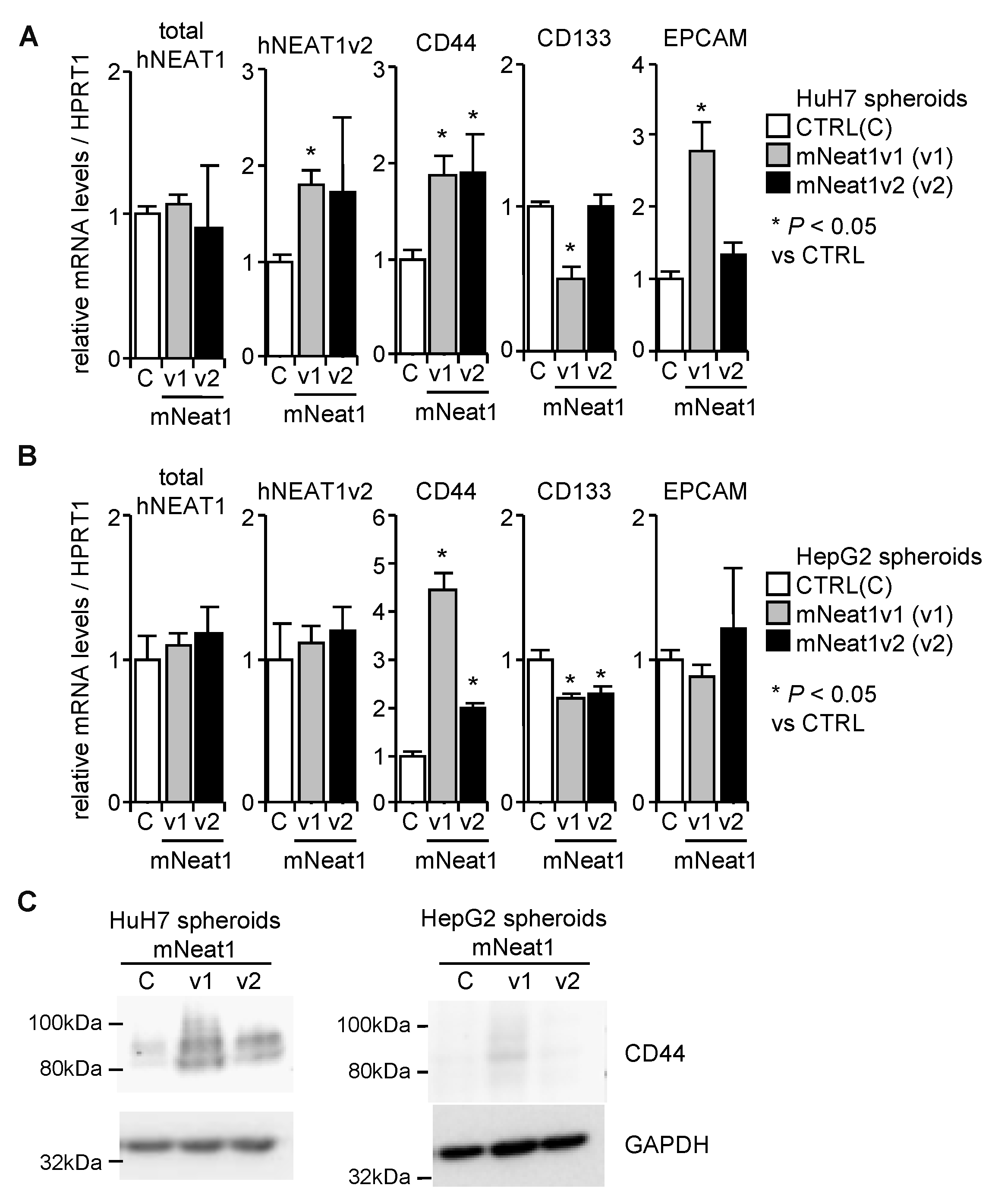
2.2. Increased Spheroid Formation and CD44 Expression by Mouse Neat1v1
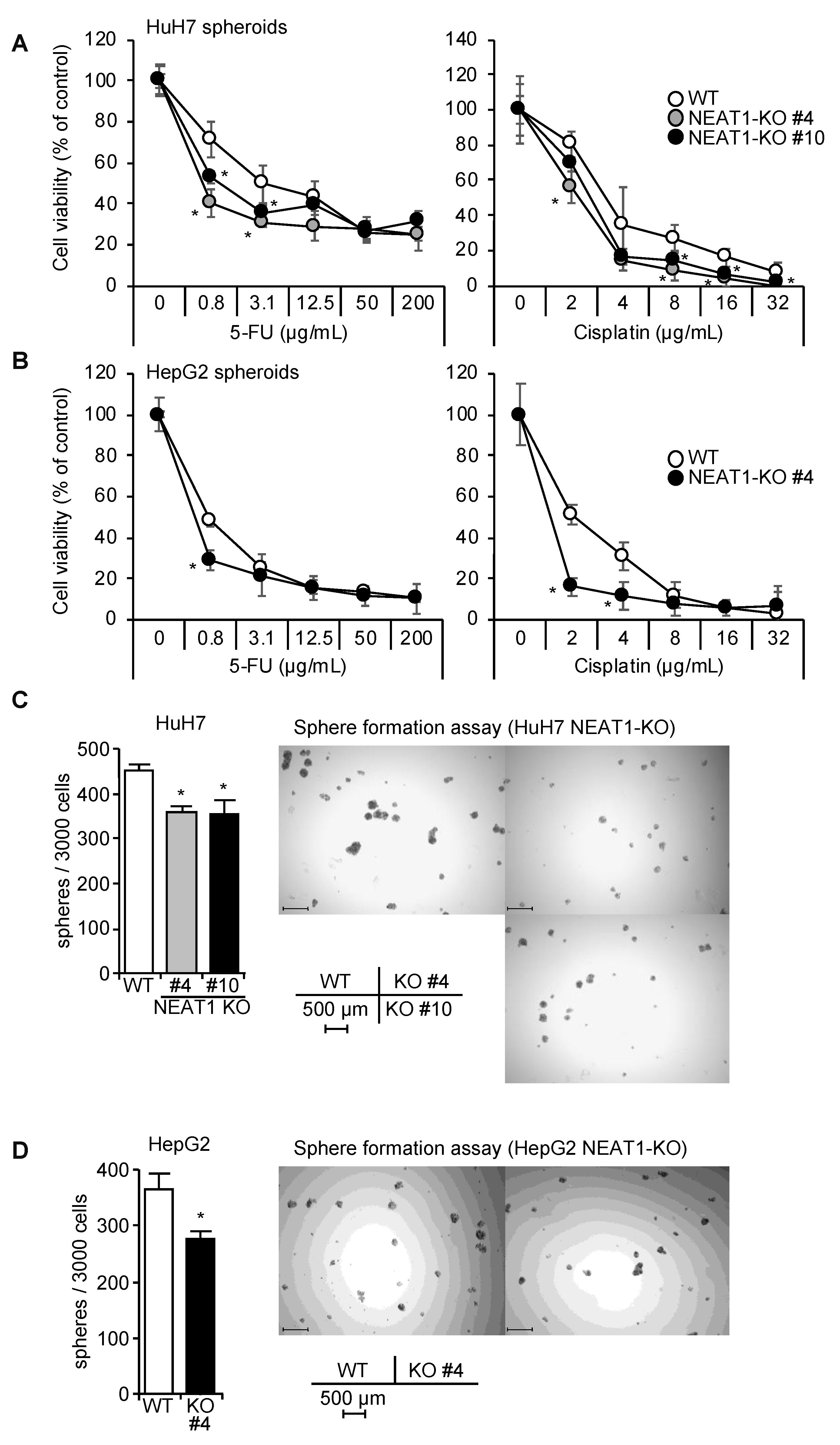
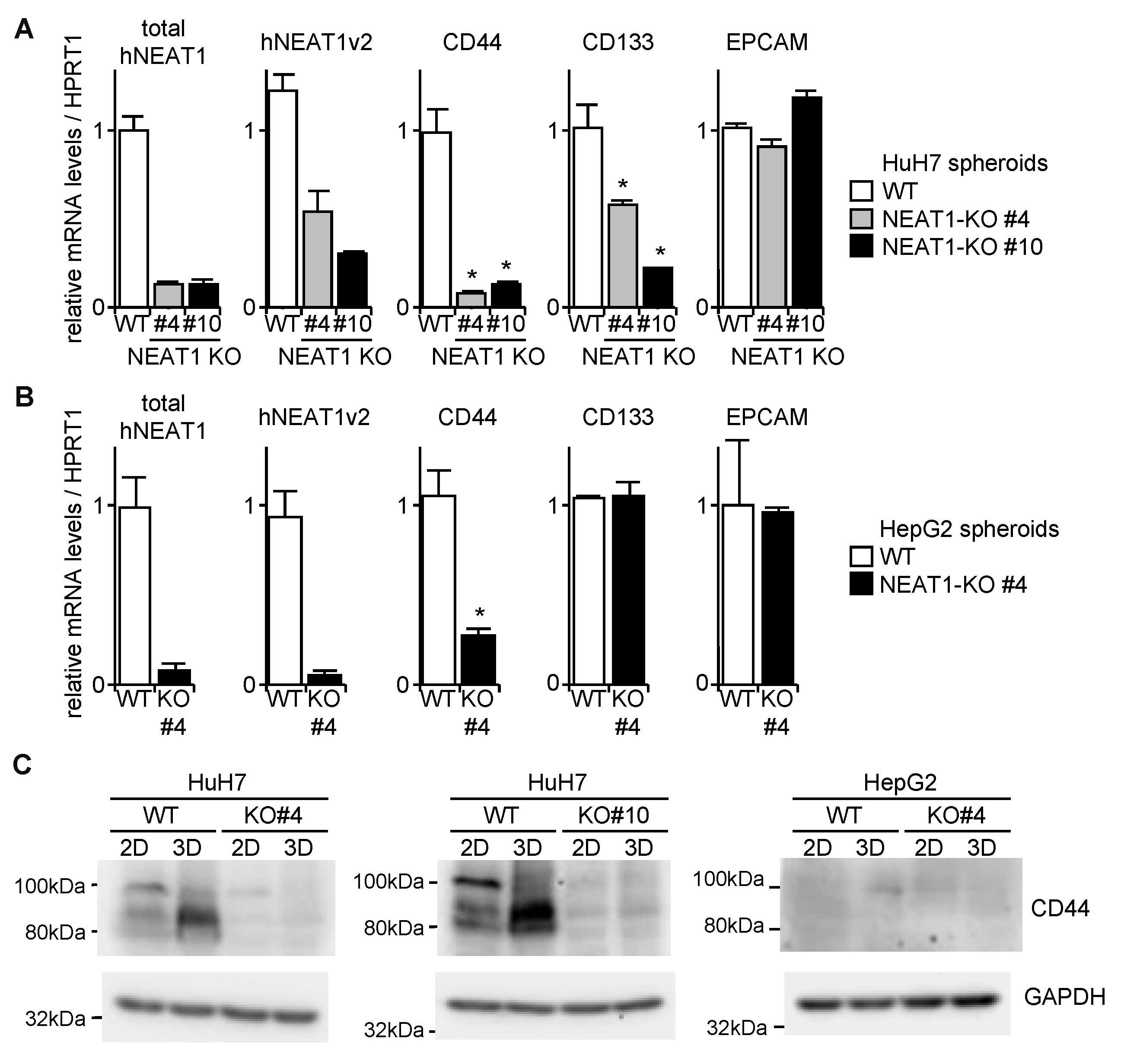
2.3. Decreased Spheroid Formation and CD44 Expression by NEAT1 Knockout
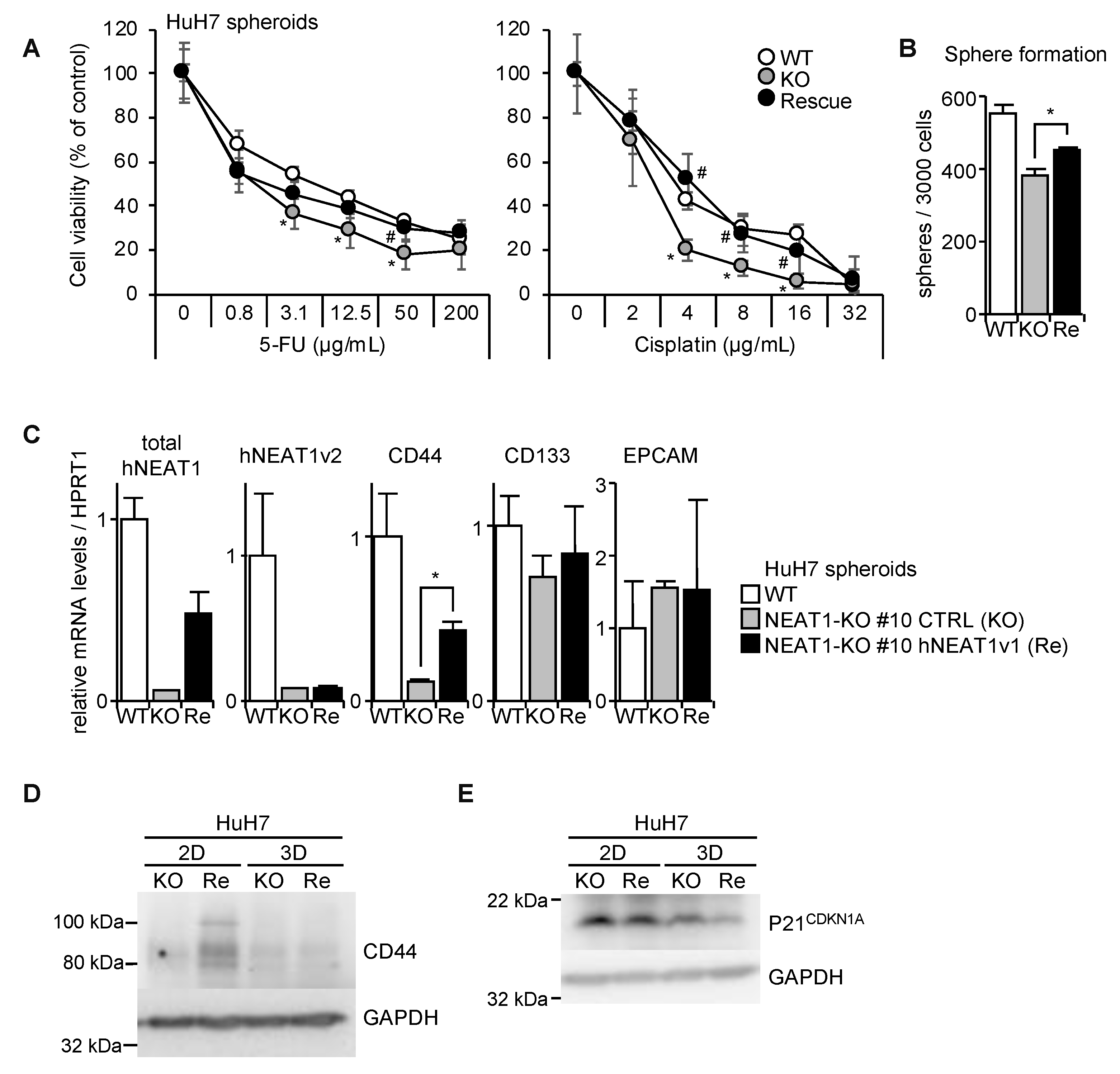
2.4. Increased Expression of p21CDKN1A in NEAT1-KO Cells
2.5. NEAT1v1-Induced CD44 Expression
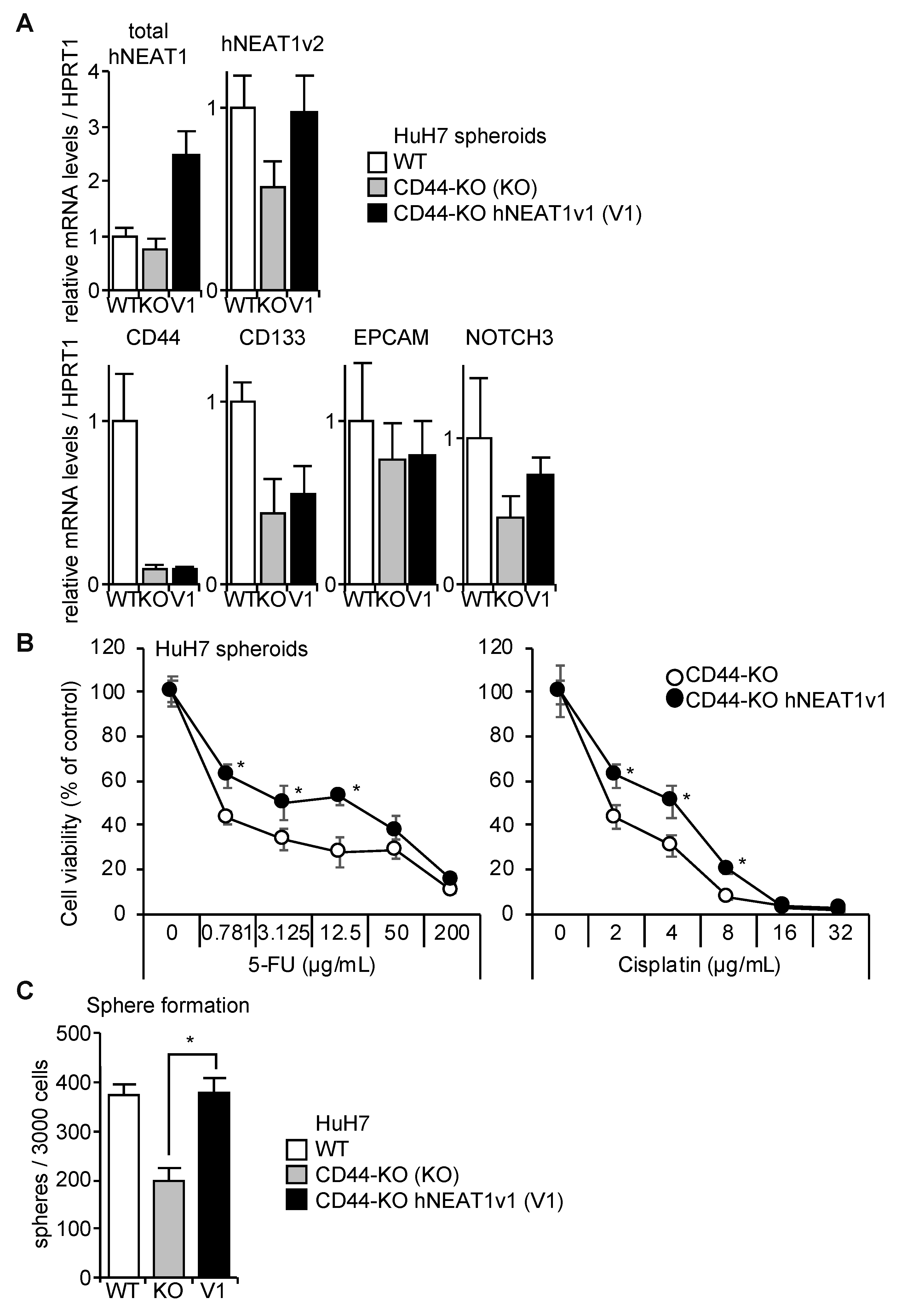
2.6. Rescue of CD44-KO Cells by NEAT1v1
2.7. Association of NEAT1v1 with Poor Prognosis in Patients with HCC
2.8. Association of NEAT1v1 with Poor Prognosis in Patients with HCC
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Reverse-Transcription Quantitative PCR (RT-qPCR)
4.3. Western Blot Analysis
4.4. Stable Transfection of NEAT1-Expressing Vectors
4.5. Knockout of the NEAT1 Gene
4.6. SiRNA Transfection
4.7. Cell viability Assay
4.8. Analysis of Human Samples
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-FU | 5-Fluorouracil |
| Blst | Blasticidin |
| CD44v | CD44 variant isoform |
| CD44s | CD44 standard isoform |
| CSC | Cancer stem cell |
| EPCAM | Epithelial adhesion molecule |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| HCC | Hepatocellular carcinoma |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 |
| KO | Knock out |
| lncRNA | Long noncoding RNA |
| miR | MicroRNA |
| NEAT1 | Nuclear enriched abundant transcript 1 |
| PTBP3 | Polypyrimidine tract-binding proteins 3 |
| RT-qPCR | Reverse-transcription quantitative PCR |
| SD | Standard deviation |
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Notake, T.; Kobayashi, A.; Shinkawa, H.; Kawahara, T.; Shimizu, A.; Yokoyama, T.; Hasegawa, K.; Kokudo, N.; Matsuyama, Y.; Makuuchi, M.; et al. Nomogram predicting long-term survival after the diagnosis of intrahepatic recurrence of hepatocellular carcinoma following an initial liver resection. Int. J. Clin. Oncol. 2017, 22, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Shindoh, J.; Makuuchi, M.; Matsuyama, Y.; Mise, Y.; Arita, J.; Sakamoto, Y.; Hasegawa, K.; Kokudo, N. Complete removal of the tumor-bearing portal territory decreases local tumor recurrence and improves disease-specific survival of patients with hepatocellular carcinoma. J. Hepatol. 2016, 64, 594–600. [Google Scholar] [CrossRef]
- Gao, W.; Chen, L.; Ma, Z.; Du, Z.; Zhao, Z.; Hu, Z.; Li, Q. Isolation and phenotypic characterization of colorectal cancer stem cells with organ-specific metastatic potential. Gastroenterology 2013, 145, 636–646. [Google Scholar] [CrossRef]
- Ayob, A.Z.; Ramasamy, T.S. liver stem cells as key drivers of tumour progression. J. Biomed. Sci. 2018, 25, 20. [Google Scholar] [CrossRef]
- Vlashi, E.; Lagadec, C.; Vergnes, L.; Matsutani, T.; Masui, K.; Poulou, M.; Popescu, R.; Della Donna, L.; Evers, P.; Dekmezian, C.; et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proc. Natl. Acad. Sci. USA 2011, 108, 16062–16067. [Google Scholar] [CrossRef] [Green Version]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.F.; Jackson, E.L.; Woolfenden, A.E.; Lawrence, S.; Babar, I.; Vogel, S.; Crowley, D.; Bronson, R.T.; Jacks, T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 2005, 121, 823–835. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Ji, J.; Budhu, A.; Forgues, M.; Yang, W.; Wang, H.Y.; Jia, H.; Ye, Q.; Qin, L.X.; Wauthier, E.; et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 2009, 136, 1012–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, S.; Chan, K.W.; Hu, L.; Lee, T.K.; Wo, J.Y.; Ng, I.O.; Zheng, B.J.; Guan, X.Y. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 2007, 132, 2542–2556. [Google Scholar] [CrossRef]
- Zhu, Z.; Hao, X.; Yan, M.; Yao, M.; Ge, C.; Gu, J.; Li, J. Cancer stem/progenitor cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int. J. Cancer. 2010, 126, 2067–2078. [Google Scholar] [CrossRef]
- Zöller, M. CD44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer 2011, 11, 254–267. [Google Scholar] [CrossRef]
- Yan, Y.; Zuo, X.; Wei, D. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem Cells Transl. Med. 2015, 4, 1033–1043. [Google Scholar] [CrossRef]
- Sakabe, T.; Azumi, J.; Umekita, Y.; Toriguchi, K.; Hatano, E.; Hirooka, Y.; Shiota, G. Prognostic relevance of miR-137 in patients with hepatocellular carcinoma. Liver Int. 2017, 37, 271–279. [Google Scholar] [CrossRef]
- Asai, R.; Tsuchiya, H.; Amisaki, M.; Makimoto, K.; Takenaga, A.; Sakabe, T.; Hoi, S.; Koyama, S.; Shiota, G. CD44 standard isoform is involved in maintenance of cancer stem cells of a hepatocellular carcinoma cell line. Cancer Med. 2019, 8, 773–782. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, S.; Naganuma, T.; Shioi, G.; Hirose, T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011, 193, 31–39. [Google Scholar] [CrossRef]
- Imamura, K.; Imamachi, N.; Akizuki, G.; Kumakura, M.; Kawaguchi, A.; Nagata, K.; Kato, A.; Kawaguchi, Y.; Sato, H.; Yoneda, M.; et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 2014, 53, 393–406. [Google Scholar] [CrossRef] [Green Version]
- Adriaens, C.; Standaert, L.; Barra, J.; Latil, M.; Verfaillie, A.; Kalev, P.; Boeckx, B.; Wijnhoven, P.W.; Radaelli, E.; Vermi, W.; et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016, 22, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Qu, S.; Wang, L.; Zhang, H.; Yang, Z.; Wang, J.; Dai, B.; Tao, K.; Shang, R.; Liu, Z.; et al. PTBP3 splicing factor promotes hepatocellular carcinoma by destroying the splicing balance of NEAT1 and pre-miR-612. Oncogene 2018, 37, 6399–6413. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, Z.; Liu, X.; Cheng, X.; Zhang, Y.; Han, X.; Zhang, Y.; Liu, S.; Yang, J.; Xu, B.; et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J. Clin. Investig. 2017, 127, 3421–3440. [Google Scholar] [CrossRef] [PubMed]
- Mello, S.S.; Sinow, C.; Raj, N.; Mazur, P.K.; Bieging-Rolett, K.; Broz, D.K.; Imam, J.F.C.; Vogel, H.; Wood, L.D.; Sage, J.; et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017, 31, 1095–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimoto, A.; Furuta, M.; Totoki, Y.; Tsunoda, T.; Kato, M.; Shiraishi, Y.; Tanaka, H.; Taniguchi, H.; Kawakami, Y.; Ueno, M.; et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 2016, 48, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Zhou, Y.; Zhai, B.; Liao, J.; Xu, W.; Zhang, R.; Li, J.; Zhang, Y.; Chen, L.; Qian, H.; et al. Sphere-forming cell subpopulations with cancer stem cell properties in human hepatoma cell lines. BMC Gastroenterol. 2011, 11, 71. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.L.; Sun, Y.F.; Wang, B.L.; Shen, M.N.; Zhou, Y.; Chen, J.W.; Hu, B.; Gong, Z.J.; Zhang, X.; Cao, Y.; et al. Sphere-forming culture enriches liver cancer stem cells and reveals Stearoyl-CoA desaturase 1 as a potential therapeutic target. BMC Cancer 2019, 19, 760. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Schmidt, B.F.; Bruchez, M.P.; McManus, C.J. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 2018, 46, 3742–3752. [Google Scholar] [CrossRef]
- Valent, P.; Bonnet, D.; De Maria, R.; Lapidot, T.; Copland, M.; Melo, J.V.; Chomienne, C.; Ishikawa, F.; Schuringa, J.J.; Stassi, G.; et al. Cancer stem cell definitions and terminology: The devil is in the details. Nat. Rev. Cancer 2012, 12, 767–775. [Google Scholar] [CrossRef]
- Saygin, C.; Matei, D.; Majeti, R.; Reizes, O.; Lathia, J.D. Targeting Cancer Stemness in the Clinic: From Hype to Hope. Cell Stem Cell 2019, 24, 25–40. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.; Zou, Q.; Ge, R.; Shen, F.; Wang, Y. The critical role of CD133+CD44+/high tumor cells in hematogenous metastasis of liver cancers. Cell Res. 2012, 22, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003, 4, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wang, J.; Qian, J.; Kong, X.; Tang, J.; Wang, Y.; Chen, H.; Hong, J.; Zou, W.; Chen, Y.; et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014, 74, 6890–6902. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.M.; Lyu, Y.L.; Cai, L. NF-κB affects proliferation and invasiveness of breast cancer cells by regulating CD44 expression. PLoS ONE 2014, 9, e106966. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Xu, Y.; Hao, Q.; Wang, S.; Li, H.; Li, J.; Gao, Y.; Li, M.; Li, W.; Xue, X.; et al. FOXP3 suppresses breast cancer metastasis through downregulation of CD44. Int. J. Cancer. 2015, 137, 1279–1290. [Google Scholar] [CrossRef] [Green Version]
- Wei, B.; Sun, X.; Geng, Z.; Shi, M.; Chen, Z.; Chen, L.; Wang, Y.; Fu, X. Isoproterenol regulates CD44 expression in gastric cancer cells through STAT3/MicroRNA373 cascade. Biomaterials 2016, 105, 89–101. [Google Scholar] [CrossRef]
- Rimkus, T.K.; Carpenter, R.L.; Sirkisoon, S.; Zhu, D.; Pasche, B.C.; Chan, M.D.; Lesser, G.J.; Tatter, S.B.; Watabe, K.; Debinski, W.; et al. Truncated Glioma-Associated Oncogene Homolog 1 (tGLI1) Mediates Mesenchymal Glioblastoma via Transcriptional Activation of CD44. Cancer Res. 2018, 78, 2589–2600. [Google Scholar] [CrossRef] [Green Version]
- Fang, L.; Sun, J.; Pan, Z.; Song, Y.; Zhong, L.; Zhang, Y.; Liu, Y.; Zheng, X.; Huang, P. Long non-coding RNA NEAT1 promotes hepatocellular carcinoma cell proliferation through the regulation of miR-129-5p-VCP-IκB. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 313, G150–G156. [Google Scholar] [CrossRef]
- Wang, Z.; Zou, Q.; Song, M.; Chen, J. NEAT1 promotes cell proliferation and invasion in hepatocellular carcinoma by negative regulating miR-613 expression. Biomed. Pharmacother. 2017, 94, 612–618. [Google Scholar] [CrossRef]
- Liu, X.; Liang, Y.; Song, R.; Yang, G.; Han, J.; Lan, Y.; Pan, S.; Zhu, M.; Liu, Y.; Wang, Y.; et al. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol. Cancer 2018, 17, 90. [Google Scholar] [CrossRef]
- Yu, H.M.; Wang, C.; Yuan, Z.; Chen, G.L.; Ye, T.; Yang, B.W. LncRNA NEAT1 promotes the tumorigenesis of colorectal cancer by sponging miR-193a-3p. Cell Prolif. 2019, 52, e12526. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, N. Downregulation of lncRNA NEAT1_2 radiosensitizes hepatocellular carcinoma cells through regulation of miR-101-3p/WEE1 axis. Cell Biol. Int. 2019, 43, 44–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Idogawa, M.; Ohashi, T.; Sasaki, Y.; Nakase, H.; Tokino, T. Long non-coding RNA NEAT1 is a transcriptional target of p53 and modulates p53-induced transactivation and tumor-suppressor function. Int. J. Cancer 2017, 140, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Huang, G.; Hua, X.; Li, Y.; Yan, H.; Che, X.; Tian, Z.; Liufu, H.; Huang, C.; Li, J.; et al. CD44s is a crucial ATG7 downstream regulator for stem-like property, invasion, and lung metastasis of human bladder cancer (BC) cells. Oncogene 2019, 38, 3301–3315. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Sun, D.; Li, W.; Shen, H.; Zhu, Y.; Li, C.; Chen, Y.; Lu, L.; Li, W.; Zhang, J.; et al. A c-Myc-MicroRNA functional feedback loop affects hepatocarcinogenesis. Hepatology 2013, 57, 2378–2389. [Google Scholar] [CrossRef] [PubMed]
- Idogawa, M.; Nakase, H.; Sasaki, Y.; Tokino, T. Prognostic Effect of Long Noncoding RNA NEAT1 Expression Depends on p53 Mutation Status in Cancer. J. Oncol. 2019, 2019, 4368068. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Harvey, A.R.; Hodgetts, S.I.; Fox, A.H. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA 2017, 23, 872–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, Y.T.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 106, 2525–2530. [Google Scholar] [CrossRef] [Green Version]
- Derti, A.; Garrett-Engele, P.; Macisaac, K.D.; Stevens, R.C.; Sriram, S.; Chen, R.; Rohl, C.A.; Johnson, J.M.; Babak, T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012, 22, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [Green Version]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koyama, S.; Tsuchiya, H.; Amisaki, M.; Sakaguchi, H.; Honjo, S.; Fujiwara, Y.; Shiota, G. NEAT1 is Required for the Expression of the Liver Cancer Stem Cell Marker CD44. Int. J. Mol. Sci. 2020, 21, 1927. https://doi.org/10.3390/ijms21061927
Koyama S, Tsuchiya H, Amisaki M, Sakaguchi H, Honjo S, Fujiwara Y, Shiota G. NEAT1 is Required for the Expression of the Liver Cancer Stem Cell Marker CD44. International Journal of Molecular Sciences. 2020; 21(6):1927. https://doi.org/10.3390/ijms21061927
Chicago/Turabian StyleKoyama, Shigemi, Hiroyuki Tsuchiya, Masataka Amisaki, Hiromi Sakaguchi, Soichiro Honjo, Yoshiyuki Fujiwara, and Goshi Shiota. 2020. "NEAT1 is Required for the Expression of the Liver Cancer Stem Cell Marker CD44" International Journal of Molecular Sciences 21, no. 6: 1927. https://doi.org/10.3390/ijms21061927
APA StyleKoyama, S., Tsuchiya, H., Amisaki, M., Sakaguchi, H., Honjo, S., Fujiwara, Y., & Shiota, G. (2020). NEAT1 is Required for the Expression of the Liver Cancer Stem Cell Marker CD44. International Journal of Molecular Sciences, 21(6), 1927. https://doi.org/10.3390/ijms21061927




