A Link between Genetic Disorders and Cellular Impairment, Using Human Induced Pluripotent Stem Cells to Reveal the Functional Consequences of Copy Number Variations in the Central Nervous System—A Close Look at Chromosome 15
Abstract
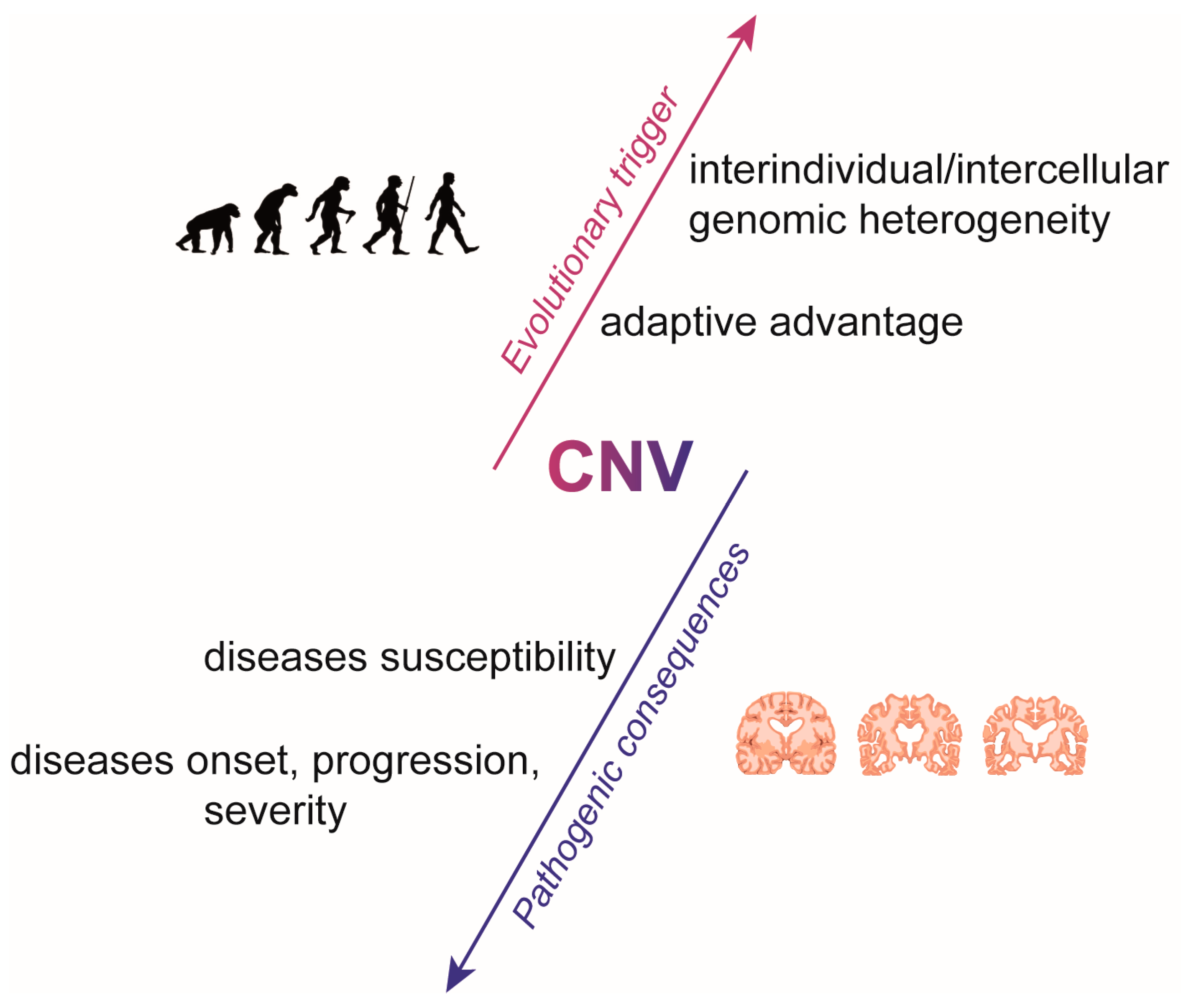
1. Introduction
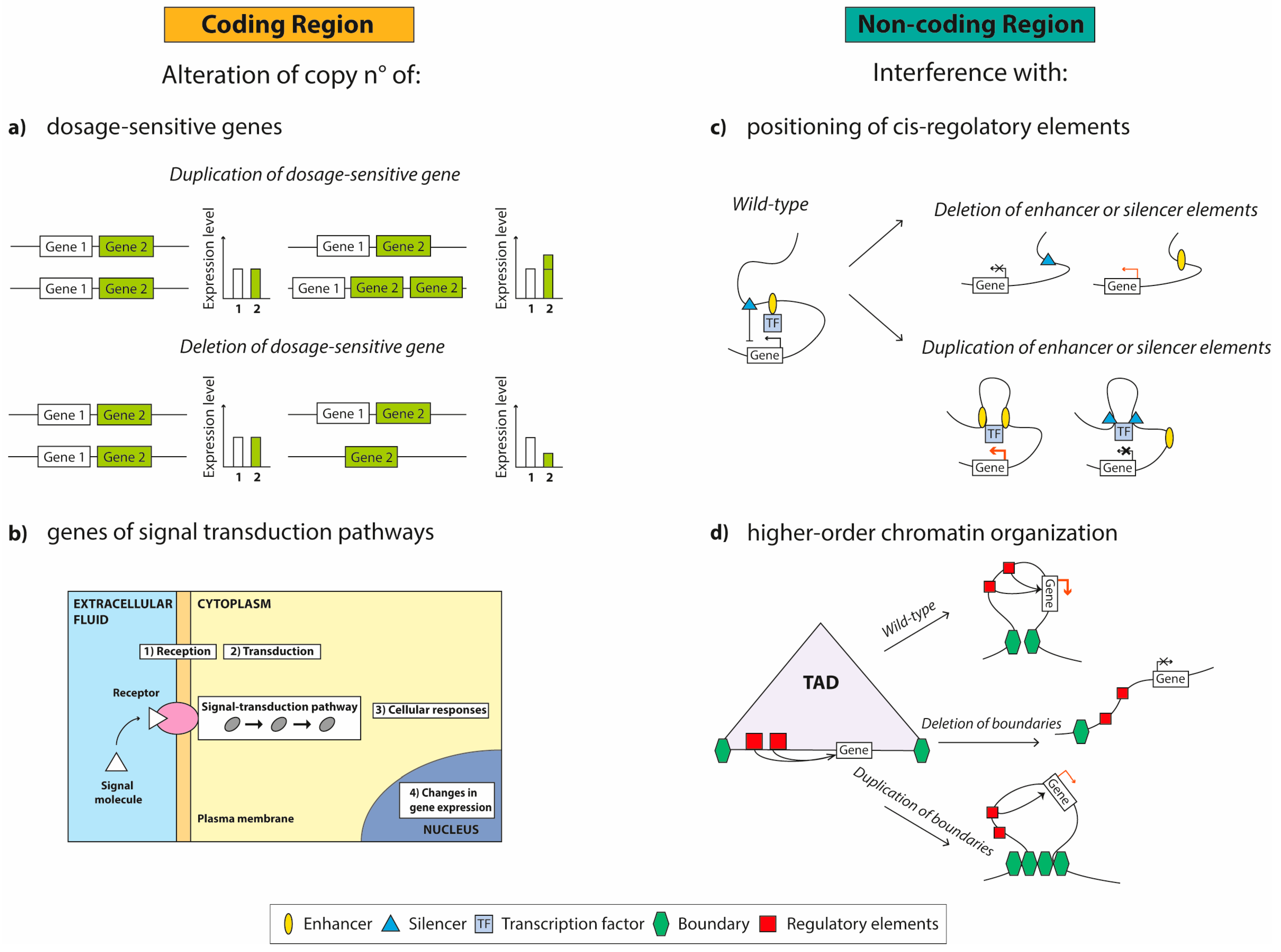
2. The Role of CNVs in Neuropsychiatric and Neurodevelopmental Diseases
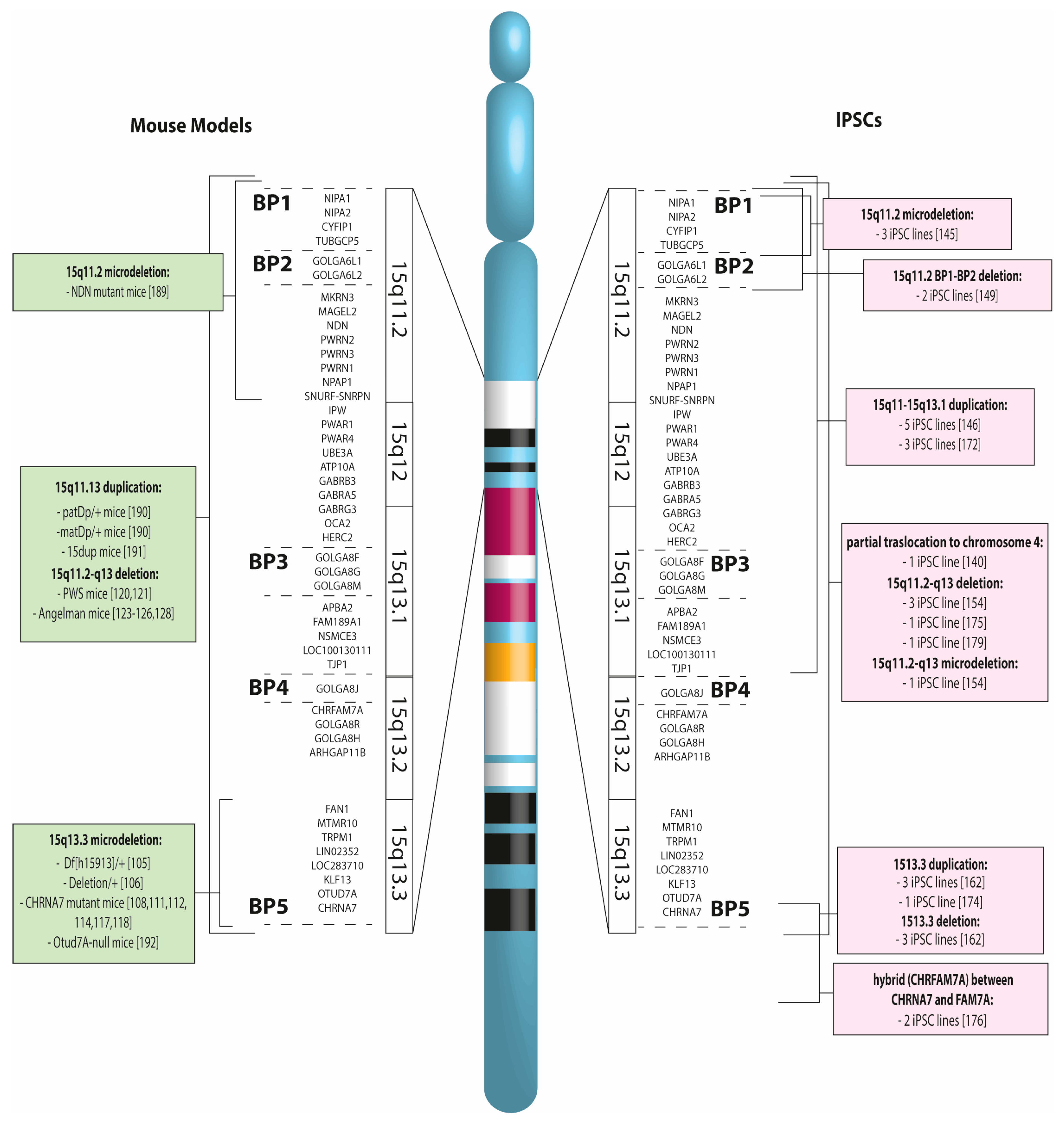
2.1. The BP1-BP2 Region, with an Approximate Length of 500 kb, Contains Four Non-Imprinted Genes
2.2. Within the BP2-BP3 Interval both Deletions and Duplications have been Mapped
2.3. CNVs between BP3 and BP4 are Rare
2.4. Within BP4 and BP5, There Is a Region Encompassing 6 Genes
3. CNV Mouse Models
4. Induced Pluripotent Stem Cells for Modeling CNV
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sebat, J.; Lakshmi, B.; Troge, J.; Alexander, J.; Young, J.; Lundin, P.; Månér, S.; Massa, H.; Walker, M.; Chi, M.; et al. Large-scale copy number polymorphism in the human genome. Science 2004, 305, 525–528. [Google Scholar] [CrossRef]
- Iafrate, A.J.; Feuk, L.; Rivera, M.N.; Listewnik, M.L.; Donahoe, P.K.; Qi, Y.; Scherer, S.W.; Lee, C. Detection of large-scale variation in the human genome. Nat. Genet. 2004, 36, 949–951. [Google Scholar] [CrossRef] [PubMed]
- Kearney, H.M.; Thorland, E.C.; Brown, K.K.; Quintero-Rivera, F.; South, S.T. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet. Med. 2011, 13, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, T.P.; Schroeder, J.P.; Gafford, G.M.; Warren, S.T.; Weinshenker, D.; Caspary, T.; Mulle, J.G. Unraveling the genetic architecture of copy number variants associated with schizophrenia and other neuropsychiatric disorders. J. Neurosci. Res. 2017, 95, 1144–1160. [Google Scholar] [CrossRef] [PubMed]
- Glessner, J.T.; Hakonarson, H. Common variants in polygenic schizophrenia. Genome Biol. 2009, 10, 236. [Google Scholar] [CrossRef] [PubMed]
- Sebat, J.; Lakshmi, B.; Malhotra, D.; Troge, J.; Lese-Martin, C.; Walsh, T.; Yamrom, B.; Yoon, S.; Krasnitz, A.; Kendall, J.; et al. Strong association of de novo copy number mutations with autism. Science 2007, 316, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Tammimies, K.; Li, D.; Rabkina, I.; Stamouli, S.; Becker, M.; Nicolaou, V.; Berggren, S.; Coco, C.; Falkmer, T.; Jonsson, U.; et al. Association between Copy Number Variation and Response to Social Skills Training in Autism Spectrum Disorder. Sci. Rep. 2019, 9, 9810. [Google Scholar] [CrossRef]
- Satterstrom, F.K.; Kosmicki, J.A.; Wang, J.; Breen, M.S.; De Rubeis, S.; An, J.Y.; Peng, M.; Collins, R.; Grove, J.; Klei, L.; et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2020, 180, 568–584. [Google Scholar] [CrossRef]
- Glessner, J.T.; Wang, K.; Cai, G.; Korvatska, O.; Kim, C.E.; Wood, S.; Zhang, H.; Estes, A.; Brune, C.W.; Bradfield, J.P.; et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 2009, 459, 569–573. [Google Scholar] [CrossRef]
- Hirabayashi, K.; Uehara, D.T.; Abe, H.; Ishii, A.; Moriyama, K.; Hirose, S.; Inazawa, J. Copy number variation analysis in 83 children with early-onset developmental and epileptic encephalopathy after targeted resequencing of a 109-epilepsy gene panel. J. Hum. Genet. 2019, 64, 1097–1106. [Google Scholar] [CrossRef]
- Girirajan, S.; Rosenfeld, J.A.; Coe, B.P.; Parikh, S.; Friedman, N.; Goldstein, A.; Filipink, R.A.; McConnell, J.S.; Angle, B.; Meschino, W.S.; et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl. J. Med. 2012, 367, 1321–1331. [Google Scholar] [CrossRef] [PubMed]
- Ou, Z.; Berg, J.S.; Yonath, H.; Enciso, V.B.; Miller, D.T.; Picker, J.; Lenzi, T.; Keegan, C.E.; Sutton, V.R.; Belmont, J.; et al. Microduplications of 22q11.2 are frequently inherited and are associated with variable phenotypes. Genet. Med. 2008, 10, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.; Pellecchia, G.; Thiruvahindrapuram, B.; D’Abate, L.; Merico, D.; Chan, A.; Zarrei, M.; Tammimies, K.; Walker, S.; Gazzellone, M.J.; et al. Indexing Effects of Copy Number Variation on Genes Involved in Developmental Delay. Sci. Rep. 2016, 6, 28663. [Google Scholar] [CrossRef] [PubMed]
- Zarrei, M.; Burton, C.L.; Engchuan, W.; Young, E.J.; Higginbotham, E.J.; MacDonald, J.R.; Trost, B.; Chan, A.J.S.; Walker, S.; Lamoureux, S.; et al. A large data resource of genomic copy number variation across neurodevelopmental disorders. NPJ Genom. Med. 2019, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, D.G.; Manolio, T.A.; Dimmock, D.P.; Rehm, H.L.; Shendure, J.; Abecasis, G.R.; Adams, D.R.; Altman, R.B.; Antonarakis, S.E.; Ashley, E.A.; et al. Guidelines for investigating causality of sequence variants in human disease. Nature 2014, 508, 469–476. [Google Scholar] [CrossRef]
- Zarrei, M.; MacDonald, J.R.; Merico, D.; Scherer, S.W. A copy number variation map of the human genome. Nat. Rev. Genet. 2015, 16, 172–183. [Google Scholar] [CrossRef]
- Myers, L.; Blyth, M.; Moradkhani, K.; Hranilović, D.; Polesie, S.; Isaksson, J.; Nordgren, A.; Bucan, M.; Vincent, M.; Bölte, S.; et al. Variable neurodevelopmental and morphological phenotypes of carriers with 12q12 duplications. Mol. Genet. Genomic Med. 2020, 8, e1013. [Google Scholar] [CrossRef]
- Zhang, S.Q.; Fleischer, J.; Al-Kateb, H.; Mito, Y.; Amarillo, I.; Shinawi, M. Intragenic CNTN4 copy number variants associated with a spectrum of neurobehavioral phenotypes. Eur. J. Med. Genet. 2019, 63, 103736. [Google Scholar] [CrossRef]
- Le, T.N.; Williams, S.R.; Alaimo, J.T.; Elsea, S.H. Genotype and phenotype correlation in 103 individuals with 2q37 deletion syndrome reveals incomplete penetrance and supports HDAC4 as the primary genetic contributor. Am. J. Med. Genet. A 2019, 179, 782–791. [Google Scholar] [CrossRef]
- Wang, X.B.; Cui, N.H.; Gao, J.J.; Qiu, X.P.; Zheng, F. SMN1 duplications contribute to sporadic amyotrophic lateral sclerosis susceptibility: Evidence from a meta-analysis. J. Neurol. Sci. 2014, 340, 63–68. [Google Scholar] [CrossRef]
- Morris, D.W.; Pearson, R.D.; Cormican, P.; Kenny, E.M.; O’Dushlaine, C.T.; Perreault, L.P.; Giannoulatou, E.; Tropea, D.; Maher, B.S.; Wormley, B.; et al. An inherited duplication at the gene p21 Protein-Activated Kinase 7 (PAK7) is a risk factor for psychosis. Hum. Mol. Genet. 2014, 23, 3316–3326. [Google Scholar] [CrossRef] [PubMed]
- Rohrback, S.; Siddoway, B.; Liu, C.S.; Chun, J. Genomic mosaicism in the developing and adult brain. Dev. Neurobiol. 2018, 78, 1026–1048. [Google Scholar] [CrossRef] [PubMed]
- Žilina, O.; Koltšina, M.; Raid, R.; Kurg, A.; Tõnisson, N.; Salumets, A. Somatic mosaicism for copy-neutral loss of heterozygosity and DNA copy number variations in the human genome. BMC Genomics 2015, 16, 703. [Google Scholar] [CrossRef]
- Iourov, I.Y.; Vorsanova, S.G.; Yurov, Y.B. Somatic cell genomics of brain disorders: A new opportunity to clarify genetic-environmental interactions. Cytogenet. Genome Res. 2013, 139, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Feuk, L.; Carson, A.R.; Scherer, S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006, 7, 85–97. [Google Scholar] [CrossRef] [PubMed]
- McConnell, M.J.; Lindberg, M.R.; Brennand, K.J.; Piper, J.C.; Voet, T.; Cowing-Zitron, C.; Shumilina, S.; Lasken, R.S.; Vermeesch, J.R.; Hall, I.M.; et al. Mosaic copy number variation in human neurons. Science 2013, 342, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Thean, L.F.; Low, Y.S.; Lo, M.; Teo, Y.Y.; Koh, W.P.; Yuan, J.M.; Chew, M.H.; Tang, C.L.; Cheah, P.Y. Genome-wide association study identified copy number variants associated with sporadic colorectal cancer risk. J. Med. Genet. 2018, 55, 181–188. [Google Scholar] [CrossRef]
- Xie, H.M.; Werner, P.; Stambolian, D.; Bailey-Wilson, J.E.; Hakonarson, H.; White, P.S.; Taylor, D.M.; Goldmuntz, E. Rare copy number variants in patients with congenital conotruncal heart defects. Birth Defects Res. 2017, 109, 271–295. [Google Scholar] [CrossRef]
- Lupski, J.R.; Stankiewicz, P. Genomic disorders: Molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005, 1, e49. [Google Scholar] [CrossRef]
- Gu, W.; Zhang, F.; Lupski, J.R. Mechanisms for human genomic rearrangements. Pathogenetics 2008, 1, 4. [Google Scholar] [CrossRef]
- Davis, A.J.; Chen, D.J. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2013, 2, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Gu, W.; Hurles, M.E.; Lupski, J.R. Copy number variation in human health, disease and evolution. Annu Rev. Genomics Hum. Genet. 2009, 10, 451–481. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.M.; McQuillan, M.A.; Seears, H.A.; Warren, J.A. Population differentiation at a regional scale in spadefoot toads: Contributions of distance and divergent selective environments. Curr. Zool. 2016, 62, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Verbitsky, M.; Westland, R.; Perez, A.; Kiryluk, K.; Liu, Q.; Krithivasan, P.; Mitrotti, A.; Fasel, D.A.; Batourina, E.; Sampson, M.G.; et al. Author Correction: The copy number variation landscape of congenital anomalies of the kidney and urinary tract. Nat. Genet. 2019, 51, 764. [Google Scholar] [CrossRef] [PubMed]
- Kun-Rodrigues, C.; Orme, T.; Carmona, S.; Hernandez, D.G.; Ross, O.A.; Eicher, J.D.; Shepherd, C.; Parkkinen, L.; Darwent, L.; Heckman, M.G.; et al. A comprehensive screening of copy number variability in dementia with Lewy bodies. Neurobiol. Aging 2019, 75, 221–223. [Google Scholar] [CrossRef] [PubMed]
- Kushima, I.; Aleksic, B.; Nakatochi, M.; Shimamura, T.; Okada, T.; Uno, Y.; Morikawa, M.; Ishizuka, K.; Shiino, T.; Kimura, H.; et al. Comparative Analyses of Copy-Number Variation in Autism Spectrum Disorder and Schizophrenia Reveal Etiological Overlap and Biological Insights. Cell Rep. 2018, 24, 2838–2856. [Google Scholar] [CrossRef]
- Di Gregorio, E.; Riberi, E.; Belligni, E.F.; Biamino, E.; Spielmann, M.; Ala, U.; Calcia, A.; Bagnasco, I.; Carli, D.; Gai, G.; et al. Copy number variants analysis in a cohort of isolated and syndromic developmental delay/intellectual disability reveals novel genomic disorders, position effects and candidate disease genes. Clin. Genet. 2017, 92, 415–422. [Google Scholar] [CrossRef]
- Weischenfeldt, J.; Symmons, O.; Spitz, F.; Korbel, J.O. Phenotypic impact of genomic structural variation: Insights from and for human disease. Nat. Rev. Genet. 2013, 14, 125–138. [Google Scholar] [CrossRef]
- Spielmann, M.; Lupiáñez, D.G.; Mundlos, S. Structural variation in the 3D genome. Nat. Rev. Genet. 2018, 19, 453–467. [Google Scholar] [CrossRef]
- Sudmant, P.H.; Kitzman, J.O.; Antonacci, F.; Alkan, C.; Malig, M.; Tsalenko, A.; Sampas, N.; Bruhn, L.; Shendure, J.; Eichler, E.E.; et al. Diversity of human copy number variation and multicopy genes. Science 2010, 330, 641–646. [Google Scholar] [CrossRef]
- Escaramís, G.; Docampo, E.; Rabionet, R. A decade of structural variants: Description, history and methods to detect structural variation. Brief. Funct Genomics 2015, 14, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Sanders, S.J.; He, X.; Willsey, A.J.; Ercan-Sencicek, A.G.; Samocha, K.E.; Cicek, A.E.; Murtha, M.T.; Bal, V.H.; Bishop, S.L.; Dong, S.; et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 2015, 87, 1215–1233. [Google Scholar] [CrossRef] [PubMed]
- Leppa, V.M.; Kravitz, S.N.; Martin, C.L.; Andrieux, J.; Le Caignec, C.; Martin-Coignard, D.; DyBuncio, C.; Sanders, S.J.; Lowe, J.K.; Cantor, R.M.; et al. Rare Inherited and De Novo CNVs Reveal Complex Contributions to ASD Risk in Multiplex Families. Am. J. Hum. Genet. 2016, 99, 540–554. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Delaby, E.; Merico, D.; Barbosa, M.; Merikangas, A.; Klei, L.; Thiruvahindrapuram, B.; Xu, X.; Ziman, R.; Wang, Z.; et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 2014, 94, 677–694. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.R.; Howrigan, D.P.; Merico, D.; Thiruvahindrapuram, B.; Wu, W.; Greer, D.S.; Antaki, D.; Shetty, A.; Holmans, P.A.; Pinto, D.; et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2017, 49, 27–35. [Google Scholar] [CrossRef]
- Green, E.K.; Rees, E.; Walters, J.T.; Smith, K.G.; Forty, L.; Grozeva, D.; Moran, J.L.; Sklar, P.; Ripke, S.; Chambert, K.D.; et al. Copy number variation in bipolar disorder. Mol. Psychiatry 2016, 21, 89–93. [Google Scholar] [CrossRef]
- Cai, X.; Evrony, G.D.; Lehmann, H.S.; Elhosary, P.C.; Mehta, B.K.; Poduri, A.; Walsh, C.A. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 2014, 8, 1280–1289. [Google Scholar] [CrossRef]
- Gole, J.; Gore, A.; Richards, A.; Chiu, Y.J.; Fung, H.L.; Bushman, D.; Chiang, H.I.; Chun, J.; Lo, Y.H.; Zhang, K. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat. Biotechnol. 2013, 31, 1126–1132. [Google Scholar] [CrossRef]
- Knouse, K.A.; Wu, J.; Amon, A. Assessment of megabase-scale somatic copy number variation using single-cell sequencing. Genome Res. 2016, 26, 376–384. [Google Scholar] [CrossRef]
- Biesecker, L.G.; Spinner, N.B. A genomic view of mosaicism and human disease. Nat. Rev. Genet. 2013, 14, 307–320. [Google Scholar] [CrossRef]
- Poduri, A.; Evrony, G.D.; Cai, X.; Elhosary, P.C.; Beroukhim, R.; Lehtinen, M.K.; Hills, L.B.; Heinzen, E.L.; Hill, A.; Hill, R.S.; et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron 2012, 74, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Huynh, M.; Silhavy, J.L.; Kim, S.; Dixon-Salazar, T.; Heiberg, A.; Scott, E.; Bafna, V.; Hill, K.J.; Collazo, A.; et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat. Genet. 2012, 44, 941–945. [Google Scholar] [CrossRef] [PubMed]
- Maynard, T.M.; Haskell, G.T.; Lieberman, J.A.; LaMantia, A.S. 22q11 DS: Genomic mechanisms and gene function in DiGeorge/velocardiofacial syndrome. Int. J. Dev. Neurosci. 2002, 20, 407–419. [Google Scholar] [CrossRef]
- Leana-Cox, J.; Pangkanon, S.; Eanet, K.R.; Curtin, M.S.; Wulfsberg, E.A. Familial DiGeorge/velocardiofacial syndrome with deletions of chromosome area 22q11.2: Report of five families with a review of the literature. Am. J. Med. Genet. 1996, 65, 309–316. [Google Scholar] [CrossRef]
- Cuscó, I.; Corominas, R.; Bayés, M.; Flores, R.; Rivera-Brugués, N.; Campuzano, V.; Pérez-Jurado, L.A. Copy number variation at the 7q11.23 segmental duplications is a susceptibility factor for the Williams-Beuren syndrome deletion. Genome Res. 2008, 18, 683–694. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Williams, S.R.; Girirajan, S.; Tegay, D.; Nowak, N.; Hatchwell, E.; Elsea, S.H. Array comparative genomic hybridisation of 52 subjects with a Smith-Magenis-like phenotype: Identification of dosage sensitive loci also associated with schizophrenia, autism and developmental delay. J. Med. Genet. 2010, 47, 223–229. [Google Scholar] [CrossRef]
- Kalsner, L.; Chamberlain, S.J. Prader-Willi, Angelman and 15q11-q13 Duplication Syndromes. Pediatr. Clin. North. Am. 2015, 62, 587–606. [Google Scholar] [CrossRef]
- Brunetti-Pierri, N.; Berg, J.S.; Scaglia, F.; Belmont, J.; Bacino, C.A.; Sahoo, T.; Lalani, S.R.; Graham, B.; Lee, B.; Shinawi, M.; et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat. Genet. 2008, 40, 1466–1471. [Google Scholar] [CrossRef]
- Niarchou, M.; Chawner, S.J.R.A.; Doherty, J.L.; Maillard, A.M.; Jacquemont, S.; Chung, W.K.; Green-Snyder, L.; Bernier, R.A.; Goin-Kochel, R.P.; Hanson, E.; et al. Correction: Psychiatric disorders in children with 16p11.2 deletion and duplication. Transl. Psychiatry 2019, 9, 107. [Google Scholar] [CrossRef]
- Okamoto, N.; Fujii, T.; Tanaka, J.; Saito, K.; Matsui, T.; Harada, N. A clinical study of patients with pericentromeric deletion and duplication within 16p12.2-p11.2. Am. J. Med. Genet. A 2014, 164, 213–219. [Google Scholar] [CrossRef]
- Nagamani, S.C.; Erez, A.; Bader, P.; Lalani, S.R.; Scott, D.A.; Scaglia, F.; Plon, S.E.; Tsai, C.H.; Reimschisel, T.; Roeder, E.; et al. Phenotypic manifestations of copy number variation in chromosome 16p13.11. Eur. J. Hum. Genet. 2011, 19, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Burnside, R.D. 22q11.21 Deletion Syndromes: A Review of Proximal, Central and Distal Deletions and Their Associated Features. Cytogenet Genome Res. 2015, 146, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Chen, H.I.; Liao, H.M.; Chen, Y.J.; Fang, J.S.; Lee, K.F.; Gau, S.S. Clinical and molecular characterization of three genomic rearrangements at chromosome 22q13.3 associated with autism spectrum disorder. Psychiatr. Genet. 2017, 27, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Cattanach, B.M.; Kirk, M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature 1985, 315, 496–498. [Google Scholar] [CrossRef] [PubMed]
- Tucci, V.; Isles, A.R.; Kelsey, G.; Ferguson-Smith, A.C.; Group, E.I. Genomic Imprinting and Physiological Processes in Mammals. Cell 2019, 176, 952–965. [Google Scholar] [CrossRef] [PubMed]
- Barlow, D.P.; Bartolomei, M.S. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol. 2014, 6. [Google Scholar] [CrossRef]
- Bajrami, E.; Spiroski, M. Genomic Imprinting. Open Access Maced. J. Med. Sci. 2016, 4, 181–184. [Google Scholar] [CrossRef]
- Burnside, R.D.; Pasion, R.; Mikhail, F.M.; Carroll, A.J.; Robin, N.H.; Youngs, E.L.; Gadi, I.K.; Keitges, E.; Jaswaney, V.L.; Papenhausen, P.R.; et al. Microdeletion/microduplication of proximal 15q11.2 between BP1 and BP2: A susceptibility region for neurological dysfunction including developmental and language delay. Hum. Genet. 2011, 130, 517–528. [Google Scholar] [CrossRef]
- Stefansson, H.; Rujescu, D.; Cichon, S.; Pietilainen, O.P.; Ingason, A.; Steinberg, S.; Fossdal, R.; Sigurdsson, E.; Sigmundsson, T.; Buizer-Voskamp, J.E.; et al. Large recurrent microdeletions associated with schizophrenia. Nature 2008, 455, 232–236. [Google Scholar] [CrossRef]
- Cox, D.M.; Butler, M.G. The 15q11.2 BP1-BP2 microdeletion syndrome: A review. Int. J. Mol. Sci. 2015, 16, 4068–4082. [Google Scholar] [CrossRef]
- van der Meer, D.; Sønderby, I.E.; Kaufmann, T.; Walters, G.B.; Abdellaoui, A.; Ames, D.; Amunts, K.; Andersson, M.; Armstrong, N.J.; Bernard, M.; et al. Association of Copy Number Variation of the 15q11.2 BP1-BP2 Region With Cortical and Subcortical Morphology and Cognition. JAMA Psychiatry 2019, 3779, 1–11. [Google Scholar] [CrossRef]
- Ulfarsson, M.O.; Walters, G.B.; Gustafsson, O.; Steinberg, S.; Silva, A.; Doyle, O.M.; Brammer, M.; Gudbjartsson, D.F.; Arnarsdottir, S.; Jonsdottir, G.A.; et al. 15q11.2 CNV affects cognitive, structural and functional correlates of dyslexia and dyscalculia. Transl. Psychiatry 2017, 7, e1109. [Google Scholar] [CrossRef] [PubMed]
- Weiner, K.S.; Zilles, K. The anatomical and functional specialization of the fusiform gyrus. Neuropsychologia 2016, 83, 48–62. [Google Scholar] [CrossRef] [PubMed]
- Kanwisher, N.; McDermott, J.; Chun, M.M. The fusiform face area: A module in human extrastriate cortex specialized for face perception. J. Neurosci. 1997, 17, 4302–4311. [Google Scholar] [CrossRef] [PubMed]
- Schenck, A.; Bardoni, B.; Langmann, C.; Harden, N.; Mandel, J.L.; Giangrande, A. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron 2003, 38, 887–898. [Google Scholar] [CrossRef]
- Adam, M.P.; Ardinger, H.H.; Pagon, R.A.; Wallace, S.E.; Bean, L.J.H.; Stephens, K.; Amemiya, A. GeneReviews® [Internet], 1993–2020. Available online: https://www.ncbi.nlm.nih.gov/pubmed/20301295 (accessed on 31 January 2020).
- Marini, C.; Cecconi, A.; Contini, E.; Pantaleo, M.; Metitieri, T.; Guarducci, S.; Giglio, S.; Guerrini, R.; Genuardi, M. Clinical and genetic study of a family with a paternally inherited 15q11-q13 duplication. Am. J. Med. Genet. A 2013, 161, 1459–1464. [Google Scholar] [CrossRef] [PubMed]
- Urraca, N.; Cleary, J.; Brewer, V.; Pivnick, E.K.; McVicar, K.; Thibert, R.L.; Schanen, N.C.; Esmer, C.; Lamport, D.; Reiter, L.T. The interstitial duplication 15q11.2-q13 syndrome includes autism, mild facial anomalies and a characteristic EEG signature. Autism. Res. 2013, 6, 268–279. [Google Scholar] [CrossRef]
- Rosenfeld, J.A.; Stephens, L.E.; Coppinger, J.; Ballif, B.C.; Hoo, J.J.; French, B.N.; Banks, V.C.; Smith, W.E.; Manchester, D.; Tsai, A.C.; et al. Deletions flanked by breakpoints 3 and 4 on 15q13 may contribute to abnormal phenotypes. Eur. J. Hum. Genet. 2011, 19, 547–554. [Google Scholar] [CrossRef]
- van Bon, B.W.; Mefford, H.C.; Menten, B.; Koolen, D.A.; Sharp, A.J.; Nillesen, W.M.; Innis, J.W.; de Ravel, T.J.; Mercer, C.L.; Fichera, M.; et al. Further delineation of the 15q13 microdeletion and duplication syndromes: A clinical spectrum varying from non-pathogenic to a severe outcome. J. Med. Genet. 2009, 46, 511–523. [Google Scholar] [CrossRef]
- Sharp, A.J.; Mefford, H.C.; Li, K.; Baker, C.; Skinner, C.; Stevenson, R.E.; Schroer, R.J.; Novara, F.; De Gregori, M.; Ciccone, R.; et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008, 40, 322–328. [Google Scholar] [CrossRef]
- Lupski, J.R. Genomic disorders: Structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998, 14, 417–422. [Google Scholar] [CrossRef]
- Stankiewicz, P.; Lupski, J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002, 18, 74–82. [Google Scholar] [CrossRef]
- Helbig, I.; Mefford, H.C.; Sharp, A.J.; Guipponi, M.; Fichera, M.; Franke, A.; Muhle, H.; de Kovel, C.; Baker, C.; von Spiczak, S.; et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat. Genet. 2009, 41, 160–162. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shachar, S.; Lanpher, B.; German, J.R.; Qasaymeh, M.; Potocki, L.; Nagamani, S.C.; Franco, L.M.; Malphrus, A.; Bottenfield, G.W.; Spence, J.E.; et al. Microdeletion 15q13.3: A locus with incomplete penetrance for autism, mental retardation and psychiatric disorders. J. Med. Genet. 2009, 46, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Lepichon, J.B.; Bittel, D.C.; Graf, W.D.; Yu, S. A 15q13.3 homozygous microdeletion associated with a severe neurodevelopmental disorder suggests putative functions of the TRPM1, CHRNA7 and other homozygously deleted genes. Am. J. Med. Genet. A 2010, 152A, 1300–1304. [Google Scholar] [CrossRef] [PubMed]
- Endris, V.; Hackmann, K.; Neuhann, T.M.; Grasshoff, U.; Bonin, M.; Haug, U.; Hahn, G.; Schallner, J.C.; Schröck, E.; Tinschert, S.; et al. Homozygous loss of CHRNA7 on chromosome 15q13.3 causes severe encephalopathy with seizures and hypotonia. Am. J. Med. Genet. A 2010, 152A, 2908–2911. [Google Scholar] [CrossRef]
- Spielmann, M.; Reichelt, G.; Hertzberg, C.; Trimborn, M.; Mundlos, S.; Horn, D.; Klopocki, E. Homozygous deletion of chromosome 15q13.3 including CHRNA7 causes severe mental retardation, seizures, muscular hypotonia and the loss of KLF13 and TRPM1 potentially cause macrocytosis and congenital retinal dysfunction in siblings. Eur. J. Med. Genet. 2011, 54, e441–e445. [Google Scholar] [CrossRef]
- Miller, D.T.; Shen, Y.; Weiss, L.A.; Korn, J.; Anselm, I.; Bridgemohan, C.; Cox, G.F.; Dickinson, H.; Gentile, J.; Harris, D.J.; et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J. Med. Genet. 2009, 46, 242–248. [Google Scholar] [CrossRef]
- Szafranski, P.; Schaaf, C.P.; Person, R.E.; Gibson, I.B.; Xia, Z.; Mahadevan, S.; Wiszniewska, J.; Bacino, C.A.; Lalani, S.; Potocki, L.; et al. Structures and molecular mechanisms for common 15q13.3 microduplications involving CHRNA7: Benign or pathological? Hum. Mutat. 2010, 31, 840–850. [Google Scholar] [CrossRef]
- Pettigrew, K.A.; Reeves, E.; Leavett, R.; Hayiou-Thomas, M.E.; Sharma, A.; Simpson, N.H.; Martinelli, A.; Thompson, P.; Hulme, C.; Snowling, M.J.; et al. Copy Number Variation Screen Identifies a Rare De Novo Deletion at Chromosome 15q13.1–13.3 in a Child with Language Impairment. PLoS ONE 2015, 10, e0134997. [Google Scholar] [CrossRef]
- Williams, N.M.; Franke, B.; Mick, E.; Anney, R.J.; Freitag, C.M.; Gill, M.; Thapar, A.; O’Donovan, M.C.; Owen, M.J.; Holmans, P.; et al. Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: The role of rare variants and duplications at 15q13.3. Am. J. Psychiatry 2012, 169, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Melchior, L.; Bertelsen, B.; Debes, N.M.; Groth, C.; Skov, L.; Mikkelsen, J.D.; Brøndum-Nielsen, K.; Tümer, Z. Microduplication of 15q13.3 and Xq21.31 in a family with Tourette syndrome and comorbidities. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2013, 162B, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Beal, J.C. Case report: Neuronal migration disorder associated with chromosome 15q13.3 duplication in a boy with autism and seizures. J. Child. Neurol. 2014, 29, 186–188. [Google Scholar] [CrossRef] [PubMed]
- Shinawi, M.; Schaaf, C.P.; Bhatt, S.S.; Xia, Z.; Patel, A.; Cheung, S.W.; Lanpher, B.; Nagl, S.; Herding, H.S.; Nevinny-Stickel, C.; et al. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat. Genet. 2009, 41, 1269–1271. [Google Scholar] [CrossRef] [PubMed]
- Gillentine, M.A.; Berry, L.N.; Goin-Kochel, R.P.; Ali, M.A.; Ge, J.; Guffey, D.; Rosenfeld, J.A.; Hannig, V.; Bader, P.; Proud, M.; et al. Erratum to: The Cognitive and Behavioral Phenotypes of Individuals with CHRNA7 Duplications. J. Autism. Dev. Disord. 2017, 47, 563. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.T.; Zanardo, É.; Dutra, R.L.; Piazzon, F.B.; Novo-Filho, G.M.; Montenegro, M.M.; Nascimento, A.M.; Rocha, M.; Madia, F.A.; Costa, T.V.; et al. Post-mortem cytogenomic investigations in patients with congenital malformations. Exp. Mol. Pathol. 2016, 101, 116–123. [Google Scholar] [CrossRef]
- Fischer, H.G.; Morawski, M.; Brückner, M.K.; Mittag, A.; Tarnok, A.; Arendt, T. Changes in neuronal DNA content variation in the human brain during aging. Aging Cell 2012, 11, 628–633. [Google Scholar] [CrossRef]
- Ye, T.; Lipska, B.K.; Tao, R.; Hyde, T.M.; Wang, L.; Li, C.; Choi, K.H.; Straub, R.E.; Kleinman, J.E.; Weinberger, D.R. Analysis of copy number variations in brain DNA from patients with schizophrenia and other psychiatric disorders. Biol. Psychiatry 2012, 72, 651–654. [Google Scholar] [CrossRef]
- Nomura, J.; Kannan, G.; Takumi, T. Rodent models of genetic and chromosomal variations in psychiatric disorders. Psychiatry Clin. Neurosci. 2017, 71, 508–517. [Google Scholar] [CrossRef]
- Arguello, P.A.; Markx, S.; Gogos, J.A.; Karayiorgou, M. Development of animal models for schizophrenia. Dis. Model. Mech. 2010, 3, 22–26. [Google Scholar] [CrossRef]
- Kazdoba, T.M.; Leach, P.T.; Crawley, J.N. Behavioral phenotypes of genetic mouse models of autism. Genes Brain Behav. 2016, 15, 7–26. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, H.G.; Kusek, G.K.; Yang, M.; Phoenix, J.L.; Bolivar, V.J.; Crawley, J.N. Autism-like behavioral phenotypes in BTBR T+tf/J mice. Genes Brain Behav. 2008, 7, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Tordjman, S.; Drapier, D.; Bonnot, O.; Graignic, R.; Fortes, S.; Cohen, D.; Millet, B.; Laurent, C.; Roubertoux, P.L. Animal models relevant to schizophrenia and autism: Validity and limitations. Behav. Genet. 2007, 37, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Fejgin, K.; Nielsen, J.; Birknow, M.R.; Bastlund, J.F.; Nielsen, V.; Lauridsen, J.B.; Stefansson, H.; Steinberg, S.; Sorensen, H.B.; Mortensen, T.E.; et al. A mouse model that recapitulates cardinal features of the 15q13.3 microdeletion syndrome including schizophrenia- and epilepsy-related alterations. Biol. Psychiatry 2014, 76, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Kogan, J.H.; Gross, A.K.; Featherstone, R.E.; Shin, R.; Chen, Q.; Heusner, C.L.; Adachi, M.; Lin, A.; Walton, N.M.; Miyoshi, S.; et al. Mouse Model of Chromosome 15q13.3 Microdeletion Syndrome Demonstrates Features Related to Autism Spectrum Disorder. J. Neurosci. 2015, 35, 16282–16294. [Google Scholar] [CrossRef] [PubMed]
- O’Tuathaigh, C.M.; Waddington, J.L. Closing the translational gap between mutant mouse models and the clinical reality of psychotic illness. Neurosci. Biobehav. Rev. 2015, 58, 19–35. [Google Scholar] [CrossRef]
- Orr-Urtreger, A.; Göldner, F.M.; Saeki, M.; Lorenzo, I.; Goldberg, L.; De Biasi, M.; Dani, J.A.; Patrick, J.W.; Beaudet, A.L. Mice deficient in the alpha7 neuronal nicotinic acetylcholine receptor lack alpha-bungarotoxin binding sites and hippocampal fast nicotinic currents. J. Neurosci. 1997, 17, 9165–9171. [Google Scholar] [CrossRef]
- Stolerman, I.P.; Chamberlain, S.; Bizarro, L.; Fernandes, C.; Schalkwyk, L. The role of nicotinic receptor alpha 7 subunits in nicotine discrimination. Neuropharmacology 2004, 46, 363–371. [Google Scholar] [CrossRef]
- Naylor, C.; Quarta, D.; Fernandes, C.; Stolerman, I.P. Tolerance to nicotine in mice lacking alpha7 nicotinic receptors. Psychopharmacology 2005, 180, 558–563. [Google Scholar] [CrossRef]
- Fernandes, C.; Hoyle, E.; Dempster, E.; Schalkwyk, L.C.; Collier, D.A. Performance deficit of alpha7 nicotinic receptor knockout mice in a delayed matching-to-place task suggests a mild impairment of working/episodic-like memory. Genes Brain Behav. 2006, 5, 433–440. [Google Scholar] [CrossRef]
- Keller, J.J.; Keller, A.B.; Bowers, B.J.; Wehner, J.M. Performance of alpha7 nicotinic receptor null mutants is impaired in appetitive learning measured in a signaled nose poke task. Behav. Brain Res. 2005, 162, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Hoyle, E.; Genn, R.F.; Fernandes, C.; Stolerman, I.P. Impaired performance of alpha7 nicotinic receptor knockout mice in the five-choice serial reaction time task. Psychopharmacology 2006, 189, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Young, J.W.; Crawford, N.; Kelly, J.S.; Kerr, L.E.; Marston, H.M.; Spratt, C.; Finlayson, K.; Sharkey, J. Impaired attention is central to the cognitive deficits observed in alpha 7 deficient mice. Eur. Neuropsychopharmacol. 2007, 17, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Young, J.W.; Meves, J.M.; Tarantino, I.S.; Caldwell, S.; Geyer, M.A. Delayed procedural learning in α7-nicotinic acetylcholine receptor knockout mice. Genes Brain Behav. 2011, 10, 720–733. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.L.; Comalli, D.M.; De Biasi, M.; Woodruff-Pak, D.S. Trace eyeblink conditioning is impaired in α7 but not in β2 nicotinic acetylcholine receptor knockout mice. Front. Behav. Neurosci. 2010, 4, 166. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Chen, W.; Yang, H.; Xue, M.; Schaaf, C.P. Chrna7 deficient mice manifest no consistent neuropsychiatric and behavioral phenotypes. Sci. Rep. 2017, 7, 39941. [Google Scholar] [CrossRef]
- Morley, B.J.; Mervis, R.F. Dendritic spine alterations in the hippocampus and parietal cortex of alpha7 nicotinic acetylcholine receptor knockout mice. Neuroscience 2013, 233, 54–63. [Google Scholar] [CrossRef]
- Lozada, A.F.; Wang, X.; Gounko, N.V.; Massey, K.A.; Duan, J.; Liu, Z.; Berg, D.K. Glutamatergic synapse formation is promoted by α7-containing nicotinic acetylcholine receptors. J. Neurosci. 2012, 32, 7651–7661. [Google Scholar] [CrossRef]
- Cattanach, B.M.; Barr, J.A.; Evans, E.P.; Burtenshaw, M.; Beechey, C.V.; Leff, S.E.; Brannan, C.I.; Copeland, N.G.; Jenkins, N.A.; Jones, J. A candidate mouse model for Prader-Willi syndrome which shows an absence of Snrpn expression. Nat. Genet. 1992, 2, 270–274. [Google Scholar] [CrossRef]
- Gabriel, J.M.; Merchant, M.; Ohta, T.; Ji, Y.; Caldwell, R.G.; Ramsey, M.J.; Tucker, J.D.; Longnecker, R.; Nicholls, R.D. A transgene insertion creating a heritable chromosome deletion mouse model of Prader-Willi and angelman syndromes. Proc. Natl. Acad. Sci. USA 1999, 96, 9258–9263. [Google Scholar] [CrossRef]
- Davies, J.R.; Wilkinson, L.S.; Isles, A.R.; Humby, T. Prader-Willi syndrome imprinting centre deletion mice have impaired baseline and 5-HT2CR-mediated response inhibition. Hum. Mol. Genet. 2019, 28, 3013–3023. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.H.; Armstrong, D.; Albrecht, U.; Atkins, C.M.; Noebels, J.L.; Eichele, G.; Sweatt, J.D.; Beaudet, A.L. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron 1998, 21, 799–811. [Google Scholar] [CrossRef]
- Miura, K.; Kishino, T.; Li, E.; Webber, H.; Dikkes, P.; Holmes, G.L.; Wagstaff, J. Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiol. Dis. 2002, 9, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Cheron, G.; Servais, L.; Wagstaff, J.; Dan, B. Fast cerebellar oscillation associated with ataxia in a mouse model of Angelman syndrome. Neuroscience 2005, 130, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, K.A.; DuBose, A.J.; Futtner, C.R.; Elmore, M.D.; Brannan, C.I.; Resnick, J.L. A human imprinting centre demonstrates conserved acquisition but diverged maintenance of imprinting in a mouse model for Angelman syndrome imprinting defects. Hum. Mol. Genet. 2006, 15, 393–404. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Pan, Y.; Zhu, L.; Landa, L.; Yoo, J.; Spencer, C.; Lorenzo, I.; Brilliant, M.; Noebels, J.; Beaudet, A.L. Altered ultrasonic vocalization and impaired learning and memory in Angelman syndrome mouse model with a large maternal deletion from Ube3a to Gabrb3. PLoS ONE 2010, 5, e12278. [Google Scholar] [CrossRef]
- Lewis, M.W.; Vargas-Franco, D.; Morse, D.A.; Resnick, J.L. A mouse model of Angelman syndrome imprinting defects. Hum. Mol. Genet. 2019, 28, 220–229. [Google Scholar] [CrossRef]
- Chai, J.H.; Locke, D.P.; Ohta, T.; Greally, J.M.; Nicholls, R.D. Retrotransposed genes such as Frat3 in the mouse Chromosome 7C Prader-Willi syndrome region acquire the imprinted status of their insertion site. Mamm. Genome 2001, 12, 813–821. [Google Scholar] [CrossRef]
- Paulsen, M.; El-Maarri, O.; Engemann, S.; Strodicke, M.; Franck, O.; Davies, K.; Reinhardt, R.; Reik, W.; Walter, J. Sequence conservation and variability of imprinting in the Beckwith-Wiedemann syndrome gene cluster in human and mouse. Hum. Mol. Genet. 2000, 9, 1829–1841. [Google Scholar] [CrossRef][Green Version]
- Okamura, K.; Hagiwara-Takeuchi, Y.; Li, T.; Vu, T.H.; Hirai, M.; Hattori, M.; Sakaki, Y.; Hoffman, A.R.; Ito, T. Comparative genome analysis of the mouse imprinted gene impact and its nonimprinted human homolog IMPACT: Toward the structural basis for species-specific imprinting. Genome Res. 2000, 10, 1878–1889. [Google Scholar] [CrossRef]
- Morison, I.M.; Ramsay, J.P.; Spencer, H.G. A census of mammalian imprinting. Trends Genet. 2005, 21, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Blaydes, S.M.; Elmore, M.; Yang, T.; Brannan, C.I. Analysis of murine Snrpn and human SNRPN gene imprinting in transgenic mice. Mamm. Genome 1999, 10, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.K.; Levorse, J.; Tilghman, S.M. A human H19 transgene exhibits impaired paternal-specific imprint acquisition and maintenance in mice. Hum. Mol. Genet. 2002, 11, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Cavaille, J.; Buiting, K.; Kiefmann, M.; Lalande, M.; Brannan, C.I.; Horsthemke, B.; Bachellerie, J.P.; Brosius, J.; Huttenhofer, A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA 2000, 97, 14311–14316. [Google Scholar] [CrossRef]
- Davis, J.; Eyre, H.; Jacka, F.N.; Dodd, S.; Dean, O.; McEwen, S.; Debnath, M.; McGrath, J.; Maes, M.; Amminger, P.; et al. A review of vulnerability and risks for schizophrenia: Beyond the two hit hypothesis. Neurosci. Biobehav. Rev. 2016, 65, 185–194. [Google Scholar] [CrossRef]
- Pizzo, L.; Jensen, M.; Polyak, A.; Rosenfeld, J.A.; Mannik, K.; Krishnan, A.; McCready, E.; Pichon, O.; Le Caignec, C.; Van Dijck, A.; et al. Rare variants in the genetic background modulate cognitive and developmental phenotypes in individuals carrying disease-associated variants. Genet. Med. 2019, 21, 816–825. [Google Scholar] [CrossRef]
- Girirajan, S.; Rosenfeld, J.A.; Cooper, G.M.; Antonacci, F.; Siswara, P.; Itsara, A.; Vives, L.; Walsh, T.; McCarthy, S.E.; Baker, C.; et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat. Genet. 2010, 42, 203–209. [Google Scholar] [CrossRef]
- Pantelis, C.; Yücel, M.; Wood, S.J.; Velakoulis, D.; Sun, D.; Berger, G.; Stuart, G.W.; Yung, A.; Phillips, L.; McGorry, P.D. Structural brain imaging evidence for multiple pathological processes at different stages of brain development in schizophrenia. Schizophr. Bull. 2005, 31, 672–696. [Google Scholar] [CrossRef]
- Yang, J.; Cai, J.; Zhang, Y.; Wang, X.; Li, W.; Xu, J.; Li, F.; Guo, X.; Deng, K.; Zhong, M.; et al. Induced pluripotent stem cells can be used to model the genomic imprinting disorder Prader-Willi syndrome. J. Biol. Chem. 2010, 285, 40303–40311. [Google Scholar] [CrossRef]
- Lee, Y.; Mattai, A.; Long, R.; Rapoport, J.L.; Gogtay, N.; Addington, A.M. Microduplications disrupting the MYT1L gene (2p25.3) are associated with schizophrenia. Psychiatr. Genet. 2012, 22, 206–209. [Google Scholar] [CrossRef]
- Israel, M.A.; Yuan, S.H.; Bardy, C.; Reyna, S.M.; Mu, Y.; Herrera, C.; Hefferan, M.P.; Van Gorp, S.; Nazor, K.L.; Boscolo, F.S.; et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature 2012, 482, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Shimojima, K.; Inoue, T.; Imai, Y.; Arai, Y.; Komoike, Y.; Sugawara, M.; Fujita, T.; Ideguchi, H.; Yasumoto, S.; Kanno, H.; et al. Reduced PLP1 expression in induced pluripotent stem cells derived from a Pelizaeus-Merzbacher disease patient with a partial PLP1 duplication. J. Hum. Genet. 2012, 57, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Tucker, B.A.; Solivan-Timpe, F.; Roos, B.R.; Anfinson, K.R.; Robin, A.L.; Wiley, L.A.; Mullins, R.F.; Fingert, J.H. Duplication of TBK1 Stimulates Autophagy in iPSC-derived Retinal Cells from a Patient with Normal Tension Glaucoma. J. Stem Cell Res. Ther. 2014, 3, 161. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.J.; Nguyen, H.N.; Ursini, G.; Zhang, F.; Kim, N.S.; Wen, Z.; Makri, G.; Nauen, D.; Shin, J.H.; Park, Y.; et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell 2014, 15, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Germain, N.D.; Chen, P.F.; Plocik, A.M.; Glatt-Deeley, H.; Brown, J.; Fink, J.J.; Bolduc, K.A.; Robinson, T.M.; Levine, E.S.; Reiter, L.T.; et al. Gene expression analysis of human induced pluripotent stem cell-derived neurons carrying copy number variants of chromosome 15q11-q13.1. Mol. Autism. 2014, 5, 44. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Fujimoto, N.; Sasakawa, N.; Shirai, S.; Ohkame, T.; Sakuma, T.; Tanaka, M.; Amano, N.; Watanabe, A.; Sakurai, H.; et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports 2015, 4, 143–154. [Google Scholar] [CrossRef]
- Lee, I.S.; Carvalho, C.M.; Douvaras, P.; Ho, S.M.; Hartley, B.J.; Zuccherato, L.W.; Ladran, I.G.; Siegel, A.J.; McCarthy, S.; Malhotra, D.; et al. Characterization of molecular and cellular phenotypes associated with a heterozygous CNTNAP2 deletion using patient-derived hiPSC neural cells. NPJ Schizophr. 2015, 1. [Google Scholar] [CrossRef]
- Das, D.K.; Tapias, V.; D’Aiuto, L.; Chowdari, K.V.; Francis, L.; Zhi, Y.; Ghosh, B.A.; Surti, U.; Tischfield, J.; Sheldon, M.; et al. Genetic and morphological features of human iPSC-derived neurons with chromosome 15q11.2 (BP1-BP2) deletions. Mol. Neuropsychiatry 2015, 1, 116–123. [Google Scholar] [CrossRef]
- Zhao, D.; Lin, M.; Chen, J.; Pedrosa, E.; Hrabovsky, A.; Fourcade, H.M.; Zheng, D.; Lachman, H.M. MicroRNA Profiling of Neurons Generated Using Induced Pluripotent Stem Cells Derived from Patients with Schizophrenia and Schizoaffective Disorder and 22q11.2 Del. PLoS ONE 2015, 10, e0132387. [Google Scholar] [CrossRef]
- Kotini, A.G.; Chang, C.J.; Boussaad, I.; Delrow, J.J.; Dolezal, E.K.; Nagulapally, A.B.; Perna, F.; Fishbein, G.A.; Klimek, V.M.; Hawkins, R.D.; et al. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat. Biotechnol. 2015, 33, 646–655. [Google Scholar] [CrossRef]
- Maguire, J.A.; Lu, L.; Mills, J.A.; Sullivan, L.M.; Gagne, A.; Gadue, P.; French, D.L. Generation of Hermansky-Pudlak Syndrome Type 1 (HPS1) induced pluripotent stem cells (iPSCs). Stem Cell Res. 2016, 16, 233–235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schroter, F.; Sleegers, K.; Van Cauwenberghe, C.; Bohndorf, M.; Wruck, W.; Van Broeckhoven, C.; Adjaye, J. Lymphoblast-derived integration-free iPSC lines from a female and male Alzheimer’s disease patient expressing different copy numbers of a coding CNV in the Alzheimer risk gene CR1. Stem Cell Res. 2016, 17, 560–563. [Google Scholar] [CrossRef] [PubMed]
- Burnett, L.C.; LeDuc, C.A.; Sulsona, C.R.; Paull, D.; Eddiry, S.; Levy, B.; Salles, J.P.; Tauber, M.; Driscoll, D.J.; Egli, D.; et al. Induced pluripotent stem cells (iPSC) created from skin fibroblasts of patients with Prader-Willi syndrome (PWS) retain the molecular signature of PWS. Stem Cell Res. 2016, 17, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, M.; Akamatsu, W.; Okada, Y.; Ohnishi, T.; Balan, S.; Hisano, Y.; Iwayama, Y.; Toyota, T.; Matsumoto, T.; Itasaka, N.; et al. Analysis of induced pluripotent stem cells carrying 22q11.2 deletion. Transl. Psychiatry 2016, 6, e934. [Google Scholar] [CrossRef] [PubMed]
- Hunihan, L.; Brown, J.; Cacace, A.; Fernandes, A.; Weston, A. Generation of a clonal induced pluripotent stem cell (iPSC) line expressing the mutant MECP2 allele from a Rett Syndrome patient fibroblast line. Stem Cell Res. 2017, 20, 67–69. [Google Scholar] [CrossRef]
- Griscelli, F.; Oudrhiri, N.; Feraud, O.; Divers, D.; Portier, L.; Turhan, A.G.; Bennaceur Griscelli, A. Generation of induced pluripotent stem cell (iPSC) line from a patient with triple negative breast cancer with hereditary exon 17 deletion of BRCA1 gene. Stem Cell Res. 2017, 24, 135–138. [Google Scholar] [CrossRef]
- Gomez Limia, C.E.; Devalle, S.; Reis, M.; Sochacki, J.; Carneiro, M.; Madeiro da Costa, R.; D’Andrea, M.; Padilha, T.; Zalcberg, I.R.; Solza, C.; et al. Generation and characterization of a human induced pluripotent stem (iPS) cell line derived from an acute myeloid leukemia patient evolving from primary myelofibrosis carrying the CALR 52bp deletion and the ASXL1 p.R693X mutation. Stem Cell Res. 2017, 24, 16–20. [Google Scholar] [CrossRef]
- Heman-Ackah, S.M.; Manzano, R.; Hoozemans, J.J.M.; Scheper, W.; Flynn, R.; Haerty, W.; Cowley, S.A.; Bassett, A.R.; Wood, M.J.A. Alpha-synuclein induces the unfolded protein response in Parkinson’s disease SNCA triplication iPSC-derived neurons. Hum. Mol. Genet. 2017, 26, 4441–4450. [Google Scholar] [CrossRef]
- Omer, L.; Hudson, E.A.; Zheng, S.; Hoying, J.B.; Shan, Y.; Boyd, N.L. CRISPR Correction of a Homozygous Low-Density Lipoprotein Receptor Mutation in Familial Hypercholesterolemia Induced Pluripotent Stem Cells. Hepatol. Commun. 2017, 1, 886–898. [Google Scholar] [CrossRef]
- Deshpande, A.; Yadav, S.; Dao, D.Q.; Wu, Z.Y.; Hokanson, K.C.; Cahill, M.K.; Wiita, A.P.; Jan, Y.N.; Ullian, E.M.; Weiss, L.A. Cellular Phenotypes in Human iPSC-Derived Neurons from a Genetic Model of Autism Spectrum Disorder. Cell Rep. 2017, 21, 2678–2687. [Google Scholar] [CrossRef]
- Gillentine, M.A.; Yin, J.; Bajic, A.; Zhang, P.; Cummock, S.; Kim, J.J.; Schaaf, C.P. Functional Consequences of CHRNA7 Copy-Number Alterations in Induced Pluripotent Stem Cells and Neural Progenitor Cells. Am. J. Hum. Genet. 2017, 101, 874–887. [Google Scholar] [CrossRef] [PubMed]
- Spaltro, G.; Vigorelli, V.; Casalnuovo, F.; Spinelli, P.; Castiglioni, E.; Rovina, D.; Paganini, S.; Di Segni, M.; Nigro, P.; Gervasini, C.; et al. Derivation of the Duchenne muscular dystrophy patient-derived induced pluripotent stem cell line lacking DMD exons 49 and 50 (CCMi001DMD-A-3, 49, 50). Stem Cell Res. 2017, 25, 128–131. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Huang, L.; He, R.; Huang, W.; Wang, H.; Lai, X.; Zou, Z.; Sun, J.; Ke, Q.; Zheng, M.; et al. Modeling the Pathogenesis of Charcot-Marie-Tooth Disease Type 1A Using Patient-Specific iPSCs. Stem Cell Rep. 2018, 10, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Burnight, E.R.; Bohrer, L.R.; Giacalone, J.C.; Klaahsen, D.L.; Daggett, H.T.; East, J.S.; Madumba, R.A.; Worthington, K.S.; Mullins, R.F.; Stone, E.M.; et al. CRISPR-Cas9-Mediated Correction of the 1.02 kb Common Deletion in CLN3 in Induced Pluripotent Stem Cells from Patients with Batten Disease. Crispr. J. 2018, 1, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Meraviglia, V.; Benzoni, P.; Landi, S.; Murano, C.; Langione, M.; Motta, B.M.; Baratto, S.; Silipigni, R.; Di Segni, M.; Pramstaller, P.P.; et al. Generation of human induced pluripotent stem cells (EURACi001-A, EURACi002-A, EURACi003-A) from peripheral blood mononuclear cells of three patients carrying mutations in the CAV3 gene. Stem Cell Res. 2018, 27, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Gowran, A.; Spaltro, G.; Casalnuovo, F.; Vigorelli, V.; Spinelli, P.; Castiglioni, E.; Rovina, D.; Paganini, S.; Di Segni, M.; Gervasini, C.; et al. Generation of induced pluripotent stem cells from a Becker muscular dystrophy patient carrying a deletion of exons 45–55 of the dystrophin gene (CCMi002BMD-A-9 45–55). Stem Cell Res. 2018, 28, 21–24. [Google Scholar] [CrossRef]
- Jansch, C.; Gunther, K.; Waider, J.; Ziegler, G.C.; Forero, A.; Kollert, S.; Svirin, E.; Puhringer, D.; Kwok, C.K.; Ullmann, R.; et al. Generation of a human induced pluripotent stem cell (iPSC) line from a 51-year-old female with attention-deficit/hyperactivity disorder (ADHD) carrying a duplication of SLC2A3. Stem Cell Res. 2018, 28, 136–140. [Google Scholar] [CrossRef]
- Eisen, B.; Ben Jehuda, R.; Cuttitta, A.J.; Mekies, L.N.; Reiter, I.; Ramchandren, S.; Arad, M.; Michele, D.E.; Binah, O. Generation of Duchenne muscular dystrophy patient-specific induced pluripotent stem cell line lacking exons 45–50 of the dystrophin gene (IITi001-A). Stem Cell Res. 2018, 29, 111–114. [Google Scholar] [CrossRef]
- Vincent, A.; Khetan, V.; Rishi, P.; Suganeswari, G.; Krishnakumar, S.; Krishnan, U.M.; Parameswaran, S. Generation of a human induced pluripotent stem cell line (VRFi001-A) from orbital adipose tissue of a bilateral retinoblastoma patient with heterozygous RB1 gene deletion. Stem Cell Res. 2018, 29, 42–45. [Google Scholar] [CrossRef]
- Arioka, Y.; Kushima, I.; Kubo, H.; Mori, D.; Ozaki, N. Induced pluripotent stem cells derived from a schizophrenia patient with ASTN2 deletion. Stem Cell Res. 2018, 30, 81–84. [Google Scholar] [CrossRef]
- Arioka, Y.; Kushima, I.; Mori, D.; Ozaki, N. Three lines of induced pluripotent stem cells derived from a 15q11.2-q13.1 duplication syndrome patient. Stem Cell Res. 2018, 31, 240–243. [Google Scholar] [CrossRef] [PubMed]
- Kathuria, A.; Nowosiad, P.; Jagasia, R.; Aigner, S.; Taylor, R.D.; Andreae, L.C.; Gatford, N.J.F.; Lucchesi, W.; Srivastava, D.P.; Price, J. Stem cell-derived neurons from autistic individuals with SHANK3 mutation show morphogenetic abnormalities during early development. Mol. Psychiatry 2018, 23, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Turco, E.M.; Vinci, E.; Altieri, F.; Ferrari, D.; Torres, B.; Goldoni, M.; Lamorte, G.; Tata, A.M.; Mazzoccoli, G.; Postorivo, D.; et al. Copy number variations in healthy subjects. Case study: iPSC line CSSi005-A (3544) production from an individual with variation in 15q13.3 chromosome duplicating gene CHRNA7. Stem Cell Res. 2018, 32, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Niki, T.; Imamura, K.; Enami, T.; Kinoshita, M.; Inoue, H. Establishment of human induced pluripotent stem cell line from a patient with Angelman syndrome carrying the deletion of maternal chromosome 15q11.2-q13. Stem Cell Res. 2019, 34, 101363. [Google Scholar] [CrossRef] [PubMed]
- Ihnatovych, I.; Nayak, T.K.; Ouf, A.; Sule, N.; Birkaya, B.; Chaves, L.; Auerbach, A.; Szigeti, K. iPSC model of CHRFAM7A effect on alpha7 nicotinic acetylcholine receptor function in the human context. Transl. Psychiatry 2019, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Piovani, G.; Lanzi, G.; Ferraro, R.M.; Masneri, S.; Barisani, C.; Savio, G.; Giliani, S.C. Generation of induced pluripotent stem cells (iPSCs) from patient with Cri du Chat Syndrome. Stem Cell Res. 2019, 35, 101393. [Google Scholar] [CrossRef]
- Valetdinova, K.R.; Maretina, M.A.; Kuranova, M.L.; Grigor’eva, E.V.; Minina, Y.M.; Kizilova, E.A.; Kiselev, A.V.; Medvedev, S.P.; Baranov, V.S.; Zakian, S.M. Generation of two spinal muscular atrophy (SMA) type I patient-derived induced pluripotent stem cell (iPSC) lines and two SMA type II patient-derived iPSC lines. Stem Cell Res. 2019, 34, 101376. [Google Scholar] [CrossRef]
- Soeda, S.; Saito, R.; Fujita, N.; Fukuta, K.; Taniura, H. Neuronal differentiation defects in induced pluripotent stem cells derived from a Prader-Willi syndrome patient. Neurosci. Lett. 2019, 703, 162–167. [Google Scholar] [CrossRef]
- Chen, G.; Jin, H.; Yu, Z.; Liu, Y.; Li, Z.; Navarengom, K.; Schwartzbeck, R.; Dmitrieva, N.; Cudrici, C.; Ferrante, E.A.; et al. Generation of human induced pluripotent stem cells from individuals with a homozygous CCR5Delta32 mutation. Stem Cell Res. 2019, 38, 101481. [Google Scholar] [CrossRef]
- Yang, X.; Zhou, T.; Zhang, H.; Li, Y.; Dong, R.; Liu, N.; Pan, G.; Liu, Y.; Gai, Z. Generation of an induced pluripotent stem cell line (SDQLCHi008-A) from a patient with ASD and DD carrying an 830kb de novo deletion at chr7q11.22 including the exon 1 of AUTS2 gene. Stem Cell Res. 2019, 40, 101557. [Google Scholar] [CrossRef]
- Rovina, D.; Castiglioni, E.; Farini, A.; Bellichi, M.; Gervasini, C.; Paganini, S.; Di Segni, M.; Santoro, R.; Torrente, Y.; Pompilio, G.; et al. Establishment of a Duchenne muscular dystrophy patient-derived induced pluripotent stem cell line carrying a deletion of exons 51–53 of the dystrophin gene (CCMi003-A). Stem Cell Res. 2019, 40, 101544. [Google Scholar] [CrossRef] [PubMed]
- Gridina, M.M.; Nikitina, T.V.; Pristyazhnyuk, I.E.; Kashevarova, A.A.; Lopatkina, M.E.; Vasilyev, S.A.; Nazarenko, L.P.; Lebedev, I.N.; Serov, O.L. Generation of the induced pluripotent stem cell line, ICAGi002-A, from unaffected carrier megabase scaled duplication involving the CNTN6 gene. Stem Cell Res. 2019, 40, 101556. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Ishikawa, M.; Fujimori, K.; Maeda, T.; Kushima, I.; Arioka, Y.; Mori, D.; Nakatake, Y.; Yamagata, B.; Nio, S.; et al. In Vitro Modeling of the Bipolar Disorder and Schizophrenia Using Patient-Derived Induced Pluripotent Stem Cells with Copy Number Variations of PCDH15 and RELN. eNeuro 2019, 6. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, Y.; Zhou, T.; Zhang, H.; Dong, R.; Li, Y.; Liu, N.; Gai, Z. An induced pluripotent stem cells line (SDQLCHi014-A) derived from urine cells of a patient with ASD and hyperactivity carrying a 303kb de novo deletion at chr3p26.1 implicating GRM7 gene. Stem Cell Res. 2019, 41, 101635. [Google Scholar] [CrossRef] [PubMed]
- Masneri, S.; Ferraro, R.M.; Lanzi, G.; Piovani, G.; Mori, L.; Barisani, C.; Moratto, D.; Plebani, A.; Badolato, R.; Soresina, A.; et al. Generation of induced Pluripotent Stem Cells (UNIBSi008-A, UNIBSi008-B, UNIBSi008-C) from an Ataxia-Telangiectasia (AT) patient carrying a novel homozygous deletion in ATM gene. Stem Cell Res. 2019, 41, 101596. [Google Scholar] [CrossRef] [PubMed]
- Zanon, A.; Riekschnitz, D.; von Troyer, M.; Volpato, C.; Picard, A.; Cantaloni, C.; Di Segni, M.; Silipigni, R.; Pramstaller, P.P.; Hicks, A.A.; et al. Generation of an induced pluripotent stem cell line (EURACi005-A) from a Parkinson’s disease patient carrying a homozygous exon 3 deletion in the PRKNgene. Stem Cell Res. 2019, 41, 101624. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, A.K.; Aghadi, M.; Ali, G.; Al-Khawaga, S.; Hussain, K.; Abdelalim, E.M. Generation of a human induced pluripotent stem cell line (QBRIi009-A) from a patient with a heterozygous deletion of FOXA2. Stem Cell Res. 2020, 42, 101705. [Google Scholar] [CrossRef]
- Gerard, M.; Hernandez, L.; Wevrick, R.; Stewart, C.L. Disruption of the mouse necdin gene results in early post-natal lethality. Nat. Genet. 1999, 23, 199–202. [Google Scholar] [CrossRef]
- Nakatani, J.; Tamada, K.; Hatanaka, F.; Ise, S.; Ohta, H.; Inoue, K.; Tomonaga, S.; Watanabe, Y.; Chung, Y.J.; Banerjee, R.; et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell 2009, 137, 1235–1246. [Google Scholar] [CrossRef]
- Sato, Y.; Okabe, S. Nano-scale analysis of synapse morphology in an autism mouse model with 15q11-13 copy number variation using focused ion beam milling and scanning electron microscopy. Microscopy 2019, 68, 122–132. [Google Scholar] [CrossRef]
- Yin, J.; Chen, W.; Chao, E.S.; Soriano, S.; Wang, L.; Wang, W.; Cummock, S.E.; Tao, H.; Pang, K.; Liu, Z.; et al. Otud7a Knockout Mice Recapitulate Many Neurological Features of 15q13.3 Microdeletion Syndrome. Am. J. Hum. Genet. 2018, 102, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, A.; Doerfler, W. Influence of in vitro manipulation on the stability of methylation patterns in the Snurf/Snrpn-imprinting region in mouse embryonic stem cells. Nucleic Acids Res. 2004, 32, 1566–1576. [Google Scholar] [CrossRef] [PubMed]
- Rugg-Gunn, P.J.; Ferguson-Smith, A.C.; Pedersen, R.A. Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Hum. Mol. Genet. 2007, 16, R243–R251. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.P.; Thurston, A.; Mummery, C.; Ward-van Oostwaard, D.; Priddle, H.; Allegrucci, C.; Denning, C.; Young, L. Gene-specific vulnerability to imprinting variability in human embryonic stem cell lines. Genome Res. 2007, 17, 1731–1742. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, S.J.; Chen, P.F.; Ng, K.Y.; Bourgois-Rocha, F.; Lemtiri-Chlieh, F.; Levine, E.S.; Lalande, M. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc. Natl. Acad. Sci. USA 2010, 107, 17668–17673. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, G.E.; Schrode, N.; Flaherty, E.; Brennand, K.J. New considerations for hiPSC-based models of neuropsychiatric disorders. Mol. Psychiatry 2019, 24, 49–66. [Google Scholar] [CrossRef] [PubMed]
- Crawley, J.N. Translational animal models of autism and neurodevelopmental disorders. Dialogues Clin. Neurosci. 2012, 14, 293–305. [Google Scholar]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Marchetto, M.C.; Gage, F.H. Modeling brain disease in a dish: Really? Cell Stem Cell 2012, 10, 642–645. [Google Scholar] [CrossRef]
- Ernst, C. Proliferation and Differentiation Deficits are a Major Convergence Point for Neurodevelopmental Disorders. Trends Neurosci. 2016, 39, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Karaca, E.; Harel, T.; Pehlivan, D.; Jhangiani, S.N.; Gambin, T.; Coban Akdemir, Z.; Gonzaga-Jauregui, C.; Erdin, S.; Bayram, Y.; Campbell, I.M.; et al. Genes that Affect Brain Structure and Function Identified by Rare Variant Analyses of Mendelian Neurologic Disease. Neuron 2015, 88, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Alazami, A.M.; Patel, N.; Shamseldin, H.E.; Anazi, S.; Al-Dosari, M.S.; Alzahrani, F.; Hijazi, H.; Alshammari, M.; Aldahmesh, M.A.; Salih, M.A.; et al. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015, 10, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Najmabadi, H.; Hu, H.; Garshasbi, M.; Zemojtel, T.; Abedini, S.S.; Chen, W.; Hosseini, M.; Behjati, F.; Haas, S.; Jamali, P.; et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 2011, 478, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.S.; Gigek, C.O.; Rosenfeld, J.A.; Diallo, A.B.; Maussion, G.; Chen, G.G.; Vaillancourt, K.; Lopez, J.P.; Crapper, L.; Poujol, R.; et al. Molecular convergence of neurodevelopmental disorders. Am. J. Hum. Genet. 2014, 95, 490–508. [Google Scholar] [CrossRef] [PubMed]
| CNV-Locus | CNV-Type | Associated Human Genes | Associated Phenotypes | Subjects (n) | Reference |
|---|---|---|---|---|---|
| 15q11-q13 | partial translocation to chromosome 4 | MNKRN3, MAGEL2, Necdin, SNURF-SNRPN gene complex, SnoRNA gene cluster | PWS | 1 | [140] |
| 2p25.3 | deletion | MYT1L | DD, ID | 1 | [141] |
| 21q21.3 | duplication | APP | AD | 2 | [142] |
| Xq22.2 | partial duplication | PLP1 | PMD | 1 | [143] |
| 12q14.2 | duplication | TBK1 | NTG | 1 | [144] |
| 15q11.2 | microdeletion | CYFIP1, NIPA1, NIPA2, TUBGCP5 | SZ | 3 | [145] |
| 15q11-q13.1 | duplication | UBE3A, GABRB3, GABRG3, GABRA5, CYFIP1, NIPA1, NIPA2 | ASD | 5 | [146] |
| Xp21 | exon 44 deletion | DMD | DMD | 1 | [147] |
| 7q35 | exons 14-15 heterozygous deletion | CNTNAP2 | SZ | 1 | [148] |
| 15q11.2 | BP1-BP2 deletion | CYFIP1, NIPA1, NIPA2, TUBGCP5 | neurodevelopmental disorders | 2 | [149] |
| 22q11.2 | microdeletion | COMT, PRODH, TBX1, ZDHHC8, DGCR8 | SZ | 6 | [150] |
| 17q | deletion | EZH2 | MDS | 2 | [151] |
| 10q24.2 | homozygous duplication cΔ491-496 in exon 15 | HPS1 | HPS type 1 | 1 | [152] |
| 1q32.2 | CR1 CNV class 2; CR1-F/F | CR1 | AD | 1 | [153] |
| 1q32.2 | CR1 CNV class 3; CR1-F/S | CR1 | AD | 1 | [153] |
| 15q11.2-q13 | deletion | MNKRN3, MAGEL2, Necdin, SNURF-SNRPN gene complex, SnoRNA gene cluster | PWS | 3 | [154] |
| 15q11.2-q13 | microdeletion | SNOD109A, SNORD116, IPW | PWS | 1 | [154] |
| 22q11.2 | microdeletion | COMT, PRODH, TBX1, ZDHHC8, DGCR8 | SZ | 2 | [155] |
| Xq28 | deletion | MECP2 | RTT | 1 | [156] |
| 17q21.3 | exon 17 deletion | BRCA1 | Triple-negative breast cancer | 1 | [157] |
| 19p13.13 | deletion | CALR | AML | 1 | [158] |
| 4q22.1 | triplication | SNCA | PD | 1 | [159] |
| 19p13.2 | exon 4 homozygous deletion | LDLR | HoFH | 1 | [160] |
| 16p11.2 | deletion | region containing 29 genes | neurodevelopmental disorders | 3 | [161] |
| 16p11.2 | duplication | region containing 29 genes | neurodevelopmental disorders | 3 | [161] |
| 15q13.3 | heterozygous duplication | CHRNA7 | DD, ID, ADHD | 1 | [162] |
| 15q13.3 | heterozygous duplication | CHRNA7 | DD, ID, ADHD, ASD | 1 | [162] |
| 15q13.3 | heterozygous duplication | CHRNA7 | Healthy subject | 1 | [162] |
| 15q13.3 | heterozygous deletion | CHRNA7 | DD, ID, ASD | 2 | [162] |
| 15q13.3 | heterozygous deletion | CHRNA7 | DD, ID | 1 | [162] |
| Xp21 | exons 49–50 deletion | DMD | DMD | 1 | [163] |
| 17p12 | duplication | PMP22 | CMT1A | 2 | [164] |
| 16p12.1 | homozygous deletion spanning exons 7–8 | CLN3 | Batten disease | 1 | [165] |
| 16p12.1 | heterozygous deletion spanning exons 7–8 | CLN3 | Batten disease | 1 | [165] |
| 3p25.3 | heterozygous deletion cΔ184-192 | CAV3 | Caveolinopathy | 1 | [166] |
| Xp21 | exons 45–55 deletion | DMD | BMD | 1 | [167] |
| 12p13.31 | duplication | SLC2A3 | ADHD | 1 | [168] |
| Xp21 | exons 45–50 deletion | DMD | DMD | 1 | [169] |
| 13q14.1 | heterozygous deletion | RB1 | Retinoblastoma | 1 | [170] |
| 9q33.1 | exonic deletion | ASTN2 | SZ | 1 | [171] |
| 15q11.2-13.1 | duplication | MNKRN3, MAGEL2, Necdin, SNURF-SNRPN gene complex, SnoRNA gene cluster | 15q11.2-q13.1 duplication syndrome | 1 | [172] |
| 22q13 | microdeletion | SHANK3 | ASD | 2 | [173] |
| 15q13.3 | duplication | CHRNA7 | Healthy subject | 1 | [174] |
| 15q11.2-q13 | deletion | UBEA3 | AS | 1 | [175] |
| 15q13-14 | fusion gene | CHRFAM7A | AD | 2 | [176] |
| 5p14 | deletion | CTNND2 | CdCS | 1 | [177] |
| 5q13 | deletion | SMN1 | SMA | 2 | [178] |
| 15q11.2-q13 | deletion | MNKRN3, MAGEL2, Necdin, SNURF-SNRPN gene complex, SnoRNA gene cluster | PWS | 1 | [179] |
| 3p21.31 | homozygous deletion | CCR5 | Resistance to HIV infection | 3 | [180] |
| 7q11.22 | deletion | AUTS2 | DD, ASD | 1 | [181] |
| Xp21 | exons 51–53 deletion | DMD | DMD | 1 | [182] |
| 3p26.3 | microduplication | CNTN6 | DD, ID | 1 | [183] |
| 10q21.1 | deletion | PCDH15 | BD | 2 | [184] |
| 7q22.1 | deletion | RELN | SZ | 1 | [184] |
| 3p26.1 | deletion | GRM7 | ASD | 1 | [185] |
| 11q22.3 | homozygous deletion spanning exons 5–7 | ATM | AT | 1 | [186] |
| 6q26 | exon 3 homozygous deletion | PRKN | PD | 1 | [187] |
| 20p11.21 | deletion | FOXA2 | neurodevelopmental disorders | 1 | [188] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casamassa, A.; Ferrari, D.; Gelati, M.; Carella, M.; Vescovi, A.L.; Rosati, J. A Link between Genetic Disorders and Cellular Impairment, Using Human Induced Pluripotent Stem Cells to Reveal the Functional Consequences of Copy Number Variations in the Central Nervous System—A Close Look at Chromosome 15. Int. J. Mol. Sci. 2020, 21, 1860. https://doi.org/10.3390/ijms21051860
Casamassa A, Ferrari D, Gelati M, Carella M, Vescovi AL, Rosati J. A Link between Genetic Disorders and Cellular Impairment, Using Human Induced Pluripotent Stem Cells to Reveal the Functional Consequences of Copy Number Variations in the Central Nervous System—A Close Look at Chromosome 15. International Journal of Molecular Sciences. 2020; 21(5):1860. https://doi.org/10.3390/ijms21051860
Chicago/Turabian StyleCasamassa, Alessia, Daniela Ferrari, Maurizio Gelati, Massimo Carella, Angelo Luigi Vescovi, and Jessica Rosati. 2020. "A Link between Genetic Disorders and Cellular Impairment, Using Human Induced Pluripotent Stem Cells to Reveal the Functional Consequences of Copy Number Variations in the Central Nervous System—A Close Look at Chromosome 15" International Journal of Molecular Sciences 21, no. 5: 1860. https://doi.org/10.3390/ijms21051860
APA StyleCasamassa, A., Ferrari, D., Gelati, M., Carella, M., Vescovi, A. L., & Rosati, J. (2020). A Link between Genetic Disorders and Cellular Impairment, Using Human Induced Pluripotent Stem Cells to Reveal the Functional Consequences of Copy Number Variations in the Central Nervous System—A Close Look at Chromosome 15. International Journal of Molecular Sciences, 21(5), 1860. https://doi.org/10.3390/ijms21051860





