The SNP rs2298383 Reduces ADORA2A Gene Transcription and Positively Associates with Cytokine Production by Peripheral Blood Mononuclear Cells in Patients with Multiple Chemical Sensitivity
Abstract
1. Introduction
2. Results
2.1. Analysis of ADORA2A rs2298383 SNP Prevalence
2.2. Estimation of Odds Ratios of ADORA rs2298383 Variant Alleles
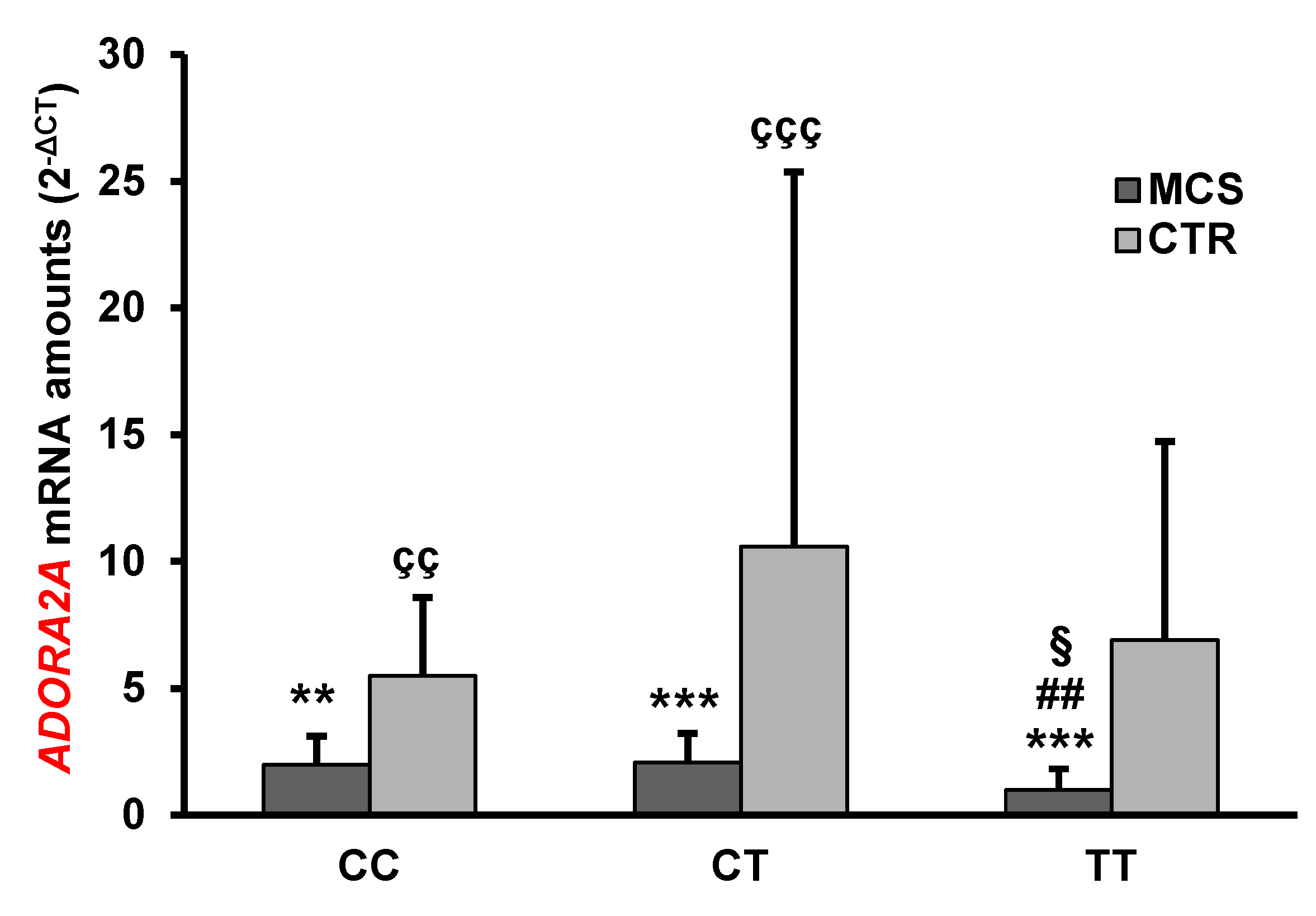
2.3. Analysis of ADORA2A Gene Expression
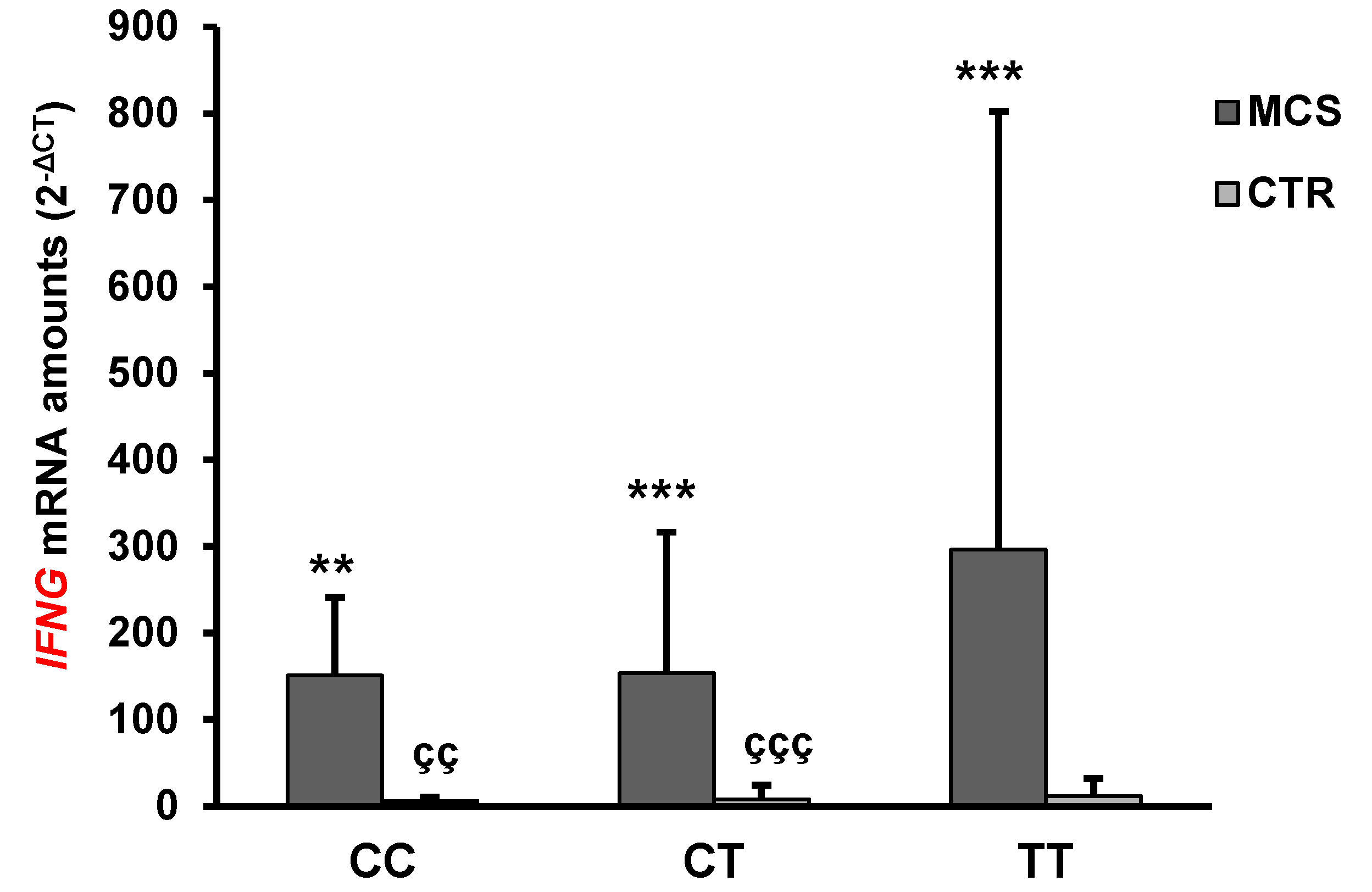
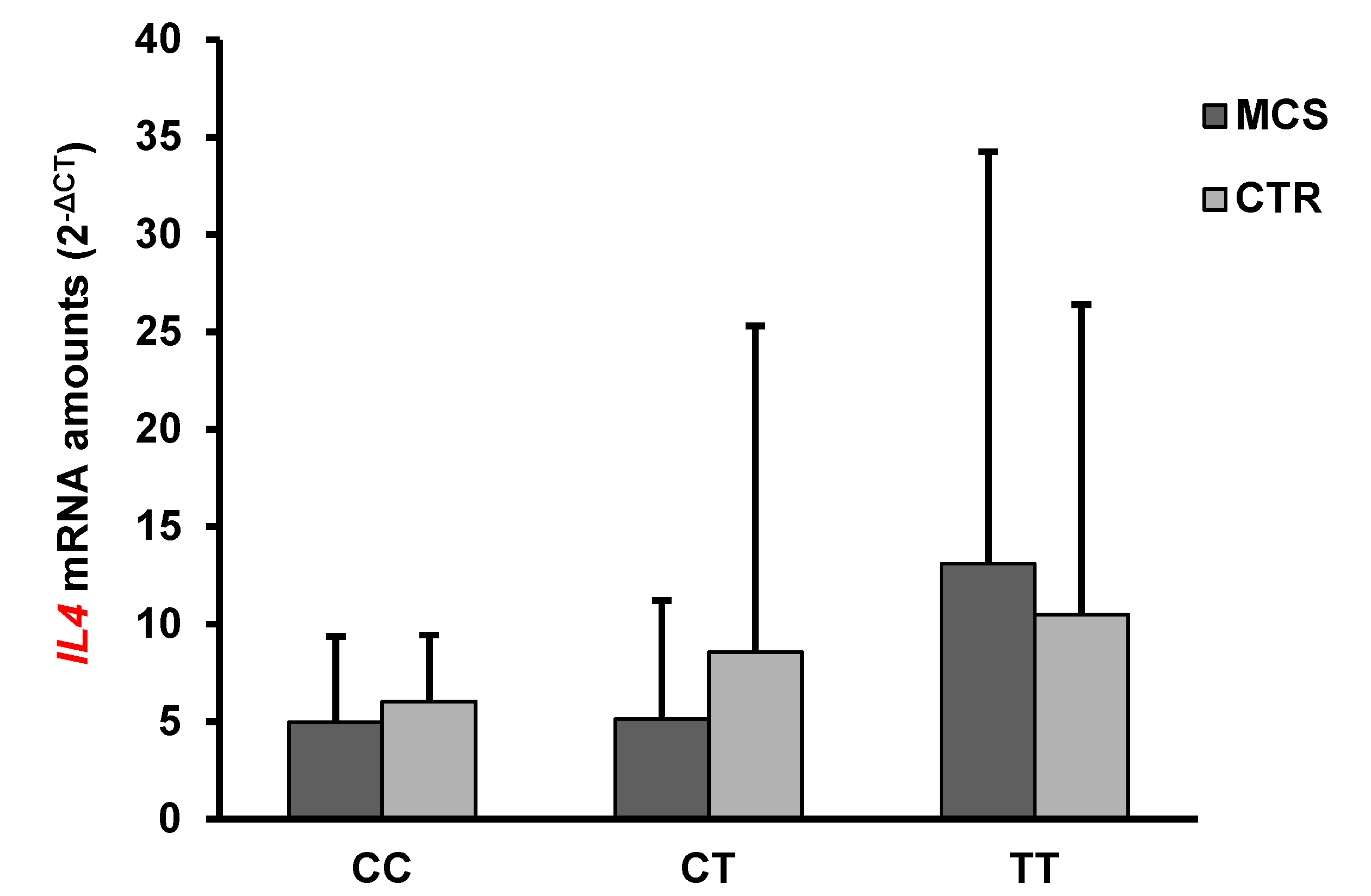
2.4. Analysis of IFN-γ and IL-4 mRNA and Protein Levels
2.5. Correlation Analysis
3. Discussion
4. Materials and Methods
4.1. Editorial Policies and Ethical Considerations
4.2. Study Cohorts
4.3. Isolation of Genomic DNA
4.4. Genotyping
4.5. Gene Expression Analysis
4.6. Assessment of IFN-γ and IL-4 Serum Concentrations
4.7. Statistical Analyses
Author Contributions
Funding
Conflicts of Interest
References
- Genuis, S.J. Sensitivity-related illness: The escalating pandemic of allergy, food intolerance and chemical sensitivity. Sci. Total Environ. 2010, 408, 6047–6061. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.S.; Prihoda, T.J. The Environmental Exposure and Sensitivity Inventory (EESI): A standardized approach for measuring chemical intolerances for research and clinical applications. Toxicol. Ind. Health 1999, 15, 370–385. [Google Scholar] [CrossRef] [PubMed]
- de Luca, C.; Raskovic, D.; Pacifico, V.; Thai, J.C.S.; Korkina, L. The search for reliable biomarkers of disease in multiple chemical sensitivity and other environmental intolerances. Int. J. Environ. Res. Public Health 2011, 8, 2770–2797. [Google Scholar] [CrossRef]
- Tuuminen, T.; Jääskeläinen, T.; Vaali, K.; Polo, O. Dampness and mold hypersensitivity syndrome and vaccination as risk factors for chronic fatigue syndrome. Autoimmun. Rev. 2019, 18, 107–108. [Google Scholar] [CrossRef] [PubMed]
- Shannon, M.; Woolf, A.; Goldman, R. Children’s environmental health: One year in a pediatric environmental health specialty unit. Ambul. Pediatr. 2003, 3, 53–56. [Google Scholar] [CrossRef]
- Jedrychowski, W.; Gałaś, A.; Whyatt, R.; Perera, F. The prenatal use of antibiotics and the development of allergic disease in one year-old infants. A preliminary study. Int. J. Occup. Med. Environ. Health 2006, 19, 70–76. [Google Scholar] [CrossRef]
- Ishibashi, M.; Tonori, H.; Miki, T.; Miyajima, E.; Kudo, Y.; Tsunoda, M.; Sakabe, K.; Aizawa, Y. Classification of patients complaining of sick house syndrome and/or multiple chemical sensitivity. Tohoku J. Exp. Med. 2007, 211, 223–233. [Google Scholar] [CrossRef]
- Pall, M.L. Elevated nitric oxide/peroxynitrite theory of multiple chemical sensitivity: Central role of N-methyl-D-aspartate receptors in the sensitivity mechanism. Environ. Health Perspect. 2003, 111, 1461–1464. [Google Scholar] [CrossRef]
- De Luca, C.; Scordo, M.G.; Cesareo, E.; Pastore, S.; Mariani, S.; Maiani, G.; Stancato, A.; Loreti, B.; Valacchi, G.; Lubrano, C.; et al. Biological definition of multiple chemical sensitivity from redox state and cytokine profiling and not from polymorphisms of xenobiotic-metabolizing enzymes. Toxicol. Appl. Pharmacol. 2010, 248, 285–292. [Google Scholar] [CrossRef]
- Dantoft, T.M.; Elberling, J.; Brix, S.; Szecsi, P.B.; Vesterhauge, S.; Skovbjerg, S. An elevated pro-inflammatory cytokine profile in multiple chemical sensitivity. Psychoneuroendocrinology 2014, 40, 140–150. [Google Scholar] [CrossRef]
- De Luca, C.; Thai, J.C.; Raskovic, D.; Cesareo, E.; Caccamo, D.; Trukhanov, A.; Korkina, L. Metabolic and genetic screening of electromagnetic hypersensitive subjects as a feasible tool for diagnostics and intervention. Mediators Inflamm. 2014, 2014, 924184. [Google Scholar] [CrossRef] [PubMed]
- De Luca, C.; Gugliandolo, A.; Calabrò, C.; Currò, M.; Ientile, R.; Raskovic, D.; Korkina, L.; Caccamo, D. Role of Polymorphisms of Inducible Nitric Oxide Synthase and Endothelial Nitric Oxide Synthase in Idiopathic Environmental Intolerances. Mediators Inflamm. 2015, 2015, 245308. [Google Scholar] [CrossRef] [PubMed]
- Belpomme, D.; Campagnac, C.; Irigaray, P. Reliable disease biomarkers characterizing and identifying electrohypersensitivity and multiple chemical sensitivity as two etiopathogenic aspects of a unique pathological disorder. Rev. Environ Health 2015, 30, 251–271. [Google Scholar] [CrossRef]
- Gugliandolo, A.; Gangemi, C.; Calabrò, C.; Vecchio, M.; Di Mauro, D.; Renis, M.; Ientile, R.; Currò, M.; Caccamo, D. Assessment of glutathione peroxidase-1 polymorphisms, oxidative stress and DNA damage in sensitivity-related illnesses. Life Sci. 2016, 145, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Binkley, K.; King, N.; Poonai, N.; Seeman, P.; Ulpian, C.; Kennedy, J. Idiopathic environmental intolerance: Increased prevalence of panic disorder-associated cholecystokinin B receptor allele 7. J. Allergy Clin. Immunol. 2001, 149, 141–146. [Google Scholar] [CrossRef]
- McKeown-Eyssen, G.; Baines, C.; Cole, D.E.C.; Riley, N.; Tyndale, R.F.; Marshall, L.; Jazmaji, V. Case-control study of genotypes in multiple chemical sensitivity: CYP2D6, NAT1, NAT2, PON1, PON2 and MTHFR. Int. J. Epidemiol. 2004, 33, 971–978. [Google Scholar] [CrossRef]
- Schnakenberg, E.; Fabig, K.R.; Stanulla, M.; Strobl, N.; Lustig, M.; Fabig, N.; Schloot, W. A cross-sectional study of self-reported chemical-related sensitivity is associated with gene variants of drug-metabolizing enzymes. Environ. Heal. A Glob. Access Sci. Source 2007, 6, 6. [Google Scholar] [CrossRef]
- Wiesmüller, G.A.; Niggemann, H.; Weissbach, W.; Riley, F.; Maarouf, Z.; Dott, W.; Kunert, H.J.; Zerres, K.; Eggermann, T.; Blömeke, B. Sequence variations in subjects withself-reported multiple chemical sensitivity (sMCS): A case-control study. J. Toxicol. Environ. Health 2008, 71, 786–794. [Google Scholar] [CrossRef]
- Caccamo, D.; Cesareo, E.; Mariani, S.; Raskovic, D.; Ientile, R.; Currò, M.; Korkina, L.; De Luca, C. Xenobiotic sensor- and metabolism-related gene variants in environmental sensitivity-related illnesses: A survey on the Italian population. Oxid. Med. Cell. Longev. 2013, 2013, 831969. [Google Scholar] [CrossRef]
- Cui, X.; Lu, X.; Hiura, M.; Oda, M.; Miyazaki, W.; Katoh, T. Evaluation of Genetic Polymorphisms in Patients with Multiple Chemical Sensitivity. PLoS ONE 2013, 8, e73708. [Google Scholar] [CrossRef]
- Loria-Kohen, V.; Marcos-Pasero, H.; de la Iglesia, R.; Aguilar-Aguilar, E.; Espinosa-Salinas, I.; Herranz, J.; Ramírez de Molina, A.; Reglero, G. Multiple chemical sensitivity: Genotypic characterization, nutritional status and quality of life in 52 patients. Med. Clínica (English Ed.) 2017, 149, 141–146. [Google Scholar] [CrossRef]
- Ohta, A.; Sitkovsky, M. Extracellular adenosine-mediated modulation of regulatory T cells. Front Immunol. 2014, 5, 304. [Google Scholar] [CrossRef] [PubMed]
- Goñi, J.R.; de la Cruz, X.; Orozco, M. Triplex-forming oligonucleotide target sequences in the human genome. Nucleic Acids Res. 2004, 32, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Hohoff, C.; Mullings, E.L.; Heatherley, S.V.; Freitag, C.M.; Neumann, L.C.; Domschke, K.; Krakowitzky, P.; Rothermundt, M.; Keck, M.E.; Erhardt, A.; et al. Adenosine A2A receptor gene: Evidence for association of risk variants with panic disorder and anxious personality. J. Psychiatr. Res. 2010, 44, 930–937. [Google Scholar] [CrossRef]
- Guerrero, A. A2A Adenosine Receptor Agonists and their Potential Therapeutic Applications. An Update. Curr. Med. Chem. 2018, 25, 3597–3612. [Google Scholar] [CrossRef]
- Genuis, S.J.; Kyrillos, E. The chemical disruption of human metabolism. Toxicol. Mech. Methods. 2017, 27, 477–500. [Google Scholar] [CrossRef]
- Gostner, J.M.; Becker, K.; Fuchs, D.; Sucher, R. Redox regulation of the immune response. Redox Rep. 2013, 18, 88–94. [Google Scholar] [CrossRef]
- Lappas, C.M.; Sullivan, G.W.; Linden, J. Adenosine A 2A agonists in development for the treatment of inflammation. Expert Opin. Investig. Drugs 2005, 14, 797–806. [Google Scholar] [CrossRef]
- Pei, H.; Linden, J. Adenosine influences myeloid cells to inhibit aeroallergen sensitization. Am. J. Physiol. Cell. Mol. Physiol. 2016, 310, L985–L992. [Google Scholar] [CrossRef]
- Childs, E.; Hohoff, C.; Deckert, J.; Xu, K.; Badner, J.; De Wit, H. Association between ADORA2A and DRD2 polymorphisms and caffeine-induced anxiety. Neuropsychopharmacology 2008, 33, 2791–2800. [Google Scholar] [CrossRef]
- Gugliandolo, A.; Gangemi, C.; Caccamo, D.; Currò, M.; Pandolfo, G.; Quattrone, D.; Crucitti, M.; Zoccali, R.A.; Bruno, A.; Muscatello, M.R.A. The RS685012 Polymorphism of ACCN2, the Human Ortholog of Murine Acid-Sensing Ion Channel (ASIC1) Gene, is Highly Represented in Patients with Panic Disorder. Neuromol. Med. 2016, 18, 91–98. [Google Scholar] [CrossRef] [PubMed]
- da Silva, R.A.; Ardais, A.P.; Gazal, M.; Lara, D.R.; de Mattos Souza, L.; Oliveira, S.; Jansen, K.; Kaster, M.P.; Bastos, C.R.; Ghisleni, G. Impact of genetic variations in ADORA2A gene on depression and symptoms: A cross-sectional population-based study. Purinergic Signal. 2018, 15, 37–44. [Google Scholar]
- Shinohara, M.; Saitoh, M.; Nishizawa, D.; Ikeda, K.; Hirose, S.; Takanashi, J.I.; Takita, J.; Kikuchi, K.; Kubota, M.; Yamanaka, G.; et al. ADORA2A polymorphism predisposes children to encephalopathy with febrile status epilepticus. Neurology 2013, 80, 1571–1576. [Google Scholar] [CrossRef] [PubMed]
- Lohr, C.; Grosche, A.; Reichenbach, A.; Hirnet, D. Purinergic neuron-glia interactions in sensory systems. Pflugers Arch. 2014, 466, 1859–1872. [Google Scholar] [CrossRef] [PubMed]
- Micarelli, A.; Cormano, A.; Caccamo, D.; Alessandrini, M. Olfactory-Related Quality of Life in Multiple Chemical Sensitivity: A Genetic-Acquired Factors Model. Int. J. Mol. Sci. 2019, 21, 156. [Google Scholar] [CrossRef]
- Hu, X.; Ivashkiv, L.B. Cross-regulation of Signaling and Immune Responses by IFN-γ and STAT1. Immunity 2009, 31, 539–550. [Google Scholar] [CrossRef]
- Krumina, A.; Chapenko, S.; Kenina, V.; Mihailova, M.; Logina, I.; Rasa, S.; Gintere, S.; Viksna, L.; Svirskis, S.; Murovska, M. The role of HHV-6 and HHV-7 infections in the development of fibromyalgia. J. Neurovirol. 2019, 25, 194–207. [Google Scholar] [CrossRef]
- Presti, R.M.; Popkin, D.L.; Connick, M.; Paetzold, S.; Virgin, H.W. 4th. Novel cell type-specific antiviral mechanism of interferon gamma action in macrophages. J. Exp. Med. 2001, 193, 483–496. [Google Scholar] [CrossRef]
- Austin, B.A.; Halford, W.; Silverman, R.H.; Williams, B.R.; Carr, D.J. OAS and PKR are not required for the antiviral effect of Ad:IFN-gamma against acute HSV-1 in primary trigeminal ganglia cultures. J. Interferon Cytokine Res. 2006, 26, 220–225. [Google Scholar] [CrossRef]
- Yang, W.C.; Hwang, Y.S.; Chen, Y.Y.; Liu, C.L.; Shen, C.N.; Hong, W.H.; Lo, S.M.; Shen, C.R. Interleukin-4 supports the suppressive immune responses elicited by regulatory T cells. Front. Immunol. 2017, 8, 1508. [Google Scholar] [CrossRef]
- Stefanello, N.; Spanevello, R.M.; Passamonti, S.; Porciúncula, L.; Bonan, C.D.; Olabiyi, A.A.; Teixeira da Rocha, J.B.; Assmann, C.E.; Morsch, V.M.; Schetinger, M.R.C. Coffee, caffeine, chlorogenic acid, and the purinergic system. Food Chem. Toxicol. 2019, 123, 298–313. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, M.C.; El-Sohemy, A.; Campos, H. Genetic polymorphism of the adenosine A2Areceptor is associated with habitual caffeine consumption. Am. J. Clin. Nutr. 2007, 86, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Arita, R.; Yanagi, Y.; Honda, N.; Maeda, S.; Maeda, K.; Kuchiba, A.; Yamaguchi, T.; Yanagihara, Y.; Suzuki, H.; Amano, S. Caffeine increases tear volume depending on polymorphisms within the adenosine A2A receptor gene and cytochrome P450 1A2. Ophthalmology 2012, 119, 972–978. [Google Scholar] [CrossRef]
- Wu, Y.M.; Luo, H.W.; Kou, H.; Wen, Y.X.; Shen, L.; Pei, L.G.; Zhou, J.; Zhang, Y.Z.; Wang, H. Prenatal caffeine exposure induced a lower level of fetal blood leptin mainly via placental mechanism. Toxicol. Appl. Pharmacol. 2015, 289, 109–116. [Google Scholar] [CrossRef] [PubMed]
| ADORA2A rs2298383 Genotype | MCS (n = 279) | Controls (n = 238) | p-Value |
|---|---|---|---|
| CC | 20.1% (56) | 20.2% (48) | 1 |
| CT | 24% (67) | 49.1% (117) | <0.00001 |
| TT | 55.9% (156) | 30.7% (73) | <0.00001 |
| Mutated allele T frequency | 0.679 | 0.4475 | - |
| Wild-type allele C frequency | 0.321 | 0.5525 | - |
| mRNA Levels (2−∆Ct) | MCS (n = 70) | Controls (n = 66) | p-Value |
|---|---|---|---|
| ADORA2A | 1.72 ± 1.15 *** | 7.66 ± 12.0 | <0.0001 |
| IFNG | 200.65 ± 315.05 *** | 14.9 ± 42.0 | <0.0001 |
| IL4 | 7.55 ± 12.72 | 8.8 ± 14.7 | n.s. |
| ADORA2A rs2298383 Genotype | IFN-γ (pg/mL) § | IL-4 (pg/mL) § |
|---|---|---|
| CC | 488.8 ± 364.7 | 5.5 ± 3.6 |
| CT | 515.7 ± 295.0 | 5.1 ± 3.7 |
| TT | 722.8 ± 461.1 | 6.7 ± 3.5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cannata, A.; De Luca, C.; Korkina, L.G.; Ferlazzo, N.; Ientile, R.; Currò, M.; Andolina, G.; Caccamo, D. The SNP rs2298383 Reduces ADORA2A Gene Transcription and Positively Associates with Cytokine Production by Peripheral Blood Mononuclear Cells in Patients with Multiple Chemical Sensitivity. Int. J. Mol. Sci. 2020, 21, 1858. https://doi.org/10.3390/ijms21051858
Cannata A, De Luca C, Korkina LG, Ferlazzo N, Ientile R, Currò M, Andolina G, Caccamo D. The SNP rs2298383 Reduces ADORA2A Gene Transcription and Positively Associates with Cytokine Production by Peripheral Blood Mononuclear Cells in Patients with Multiple Chemical Sensitivity. International Journal of Molecular Sciences. 2020; 21(5):1858. https://doi.org/10.3390/ijms21051858
Chicago/Turabian StyleCannata, Attilio, Chiara De Luca, Liudmila G. Korkina, Nadia Ferlazzo, Riccardo Ientile, Monica Currò, Giulia Andolina, and Daniela Caccamo. 2020. "The SNP rs2298383 Reduces ADORA2A Gene Transcription and Positively Associates with Cytokine Production by Peripheral Blood Mononuclear Cells in Patients with Multiple Chemical Sensitivity" International Journal of Molecular Sciences 21, no. 5: 1858. https://doi.org/10.3390/ijms21051858
APA StyleCannata, A., De Luca, C., Korkina, L. G., Ferlazzo, N., Ientile, R., Currò, M., Andolina, G., & Caccamo, D. (2020). The SNP rs2298383 Reduces ADORA2A Gene Transcription and Positively Associates with Cytokine Production by Peripheral Blood Mononuclear Cells in Patients with Multiple Chemical Sensitivity. International Journal of Molecular Sciences, 21(5), 1858. https://doi.org/10.3390/ijms21051858







