Rice OsHSFA3 Gene Improves Drought Tolerance by Modulating Polyamine Biosynthesis Depending on Abscisic Acid and ROS Levels
Abstract
1. Introduction
2. Results
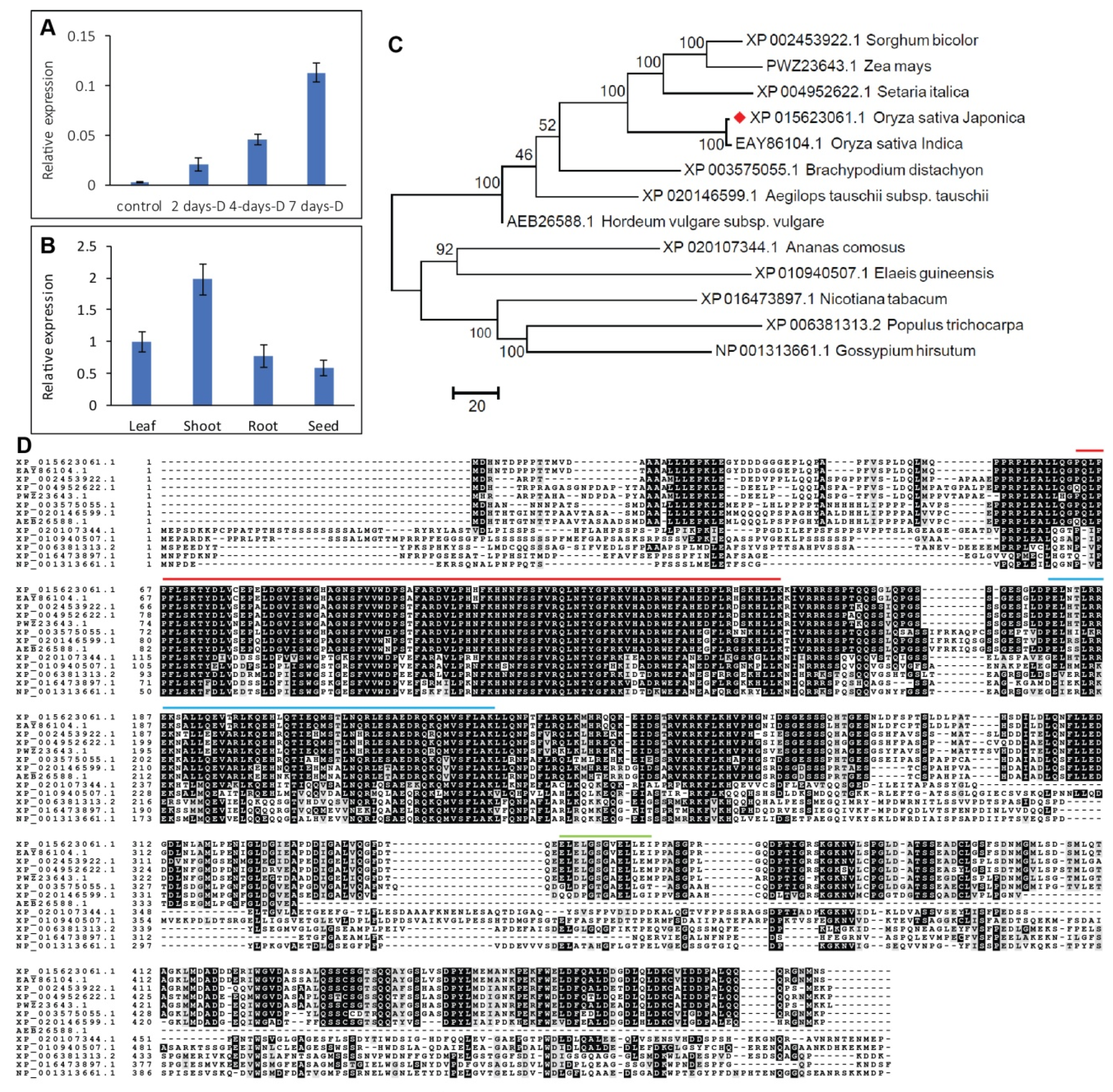
2.1. OsHSFA3 Is a Drought-Responsive Gene and Highly Accumulated in Shoot
2.2. Structural Features of OsHSFA3 and Evolutionary Relationship
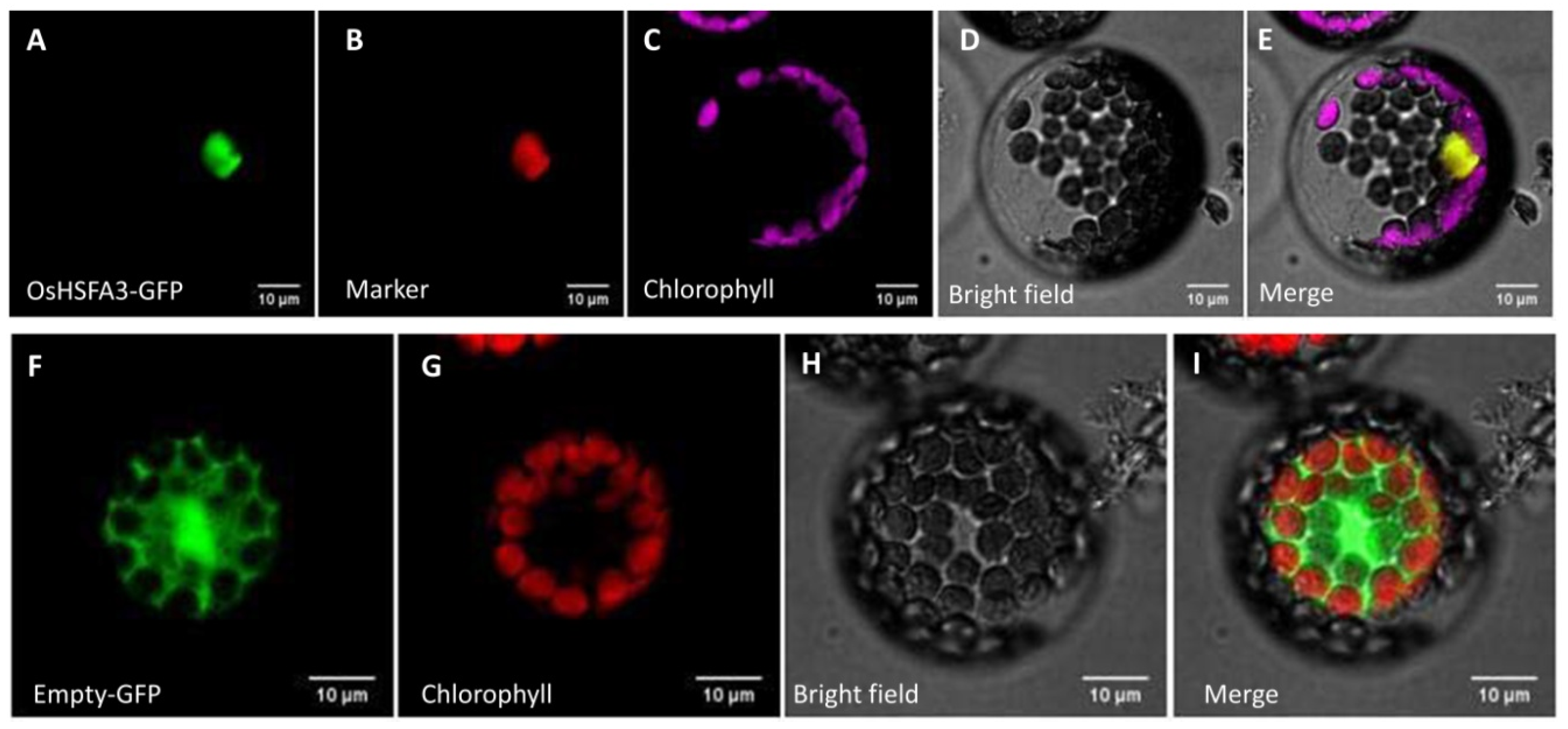
2.3. OsHSFA3 Localized in Nucleus
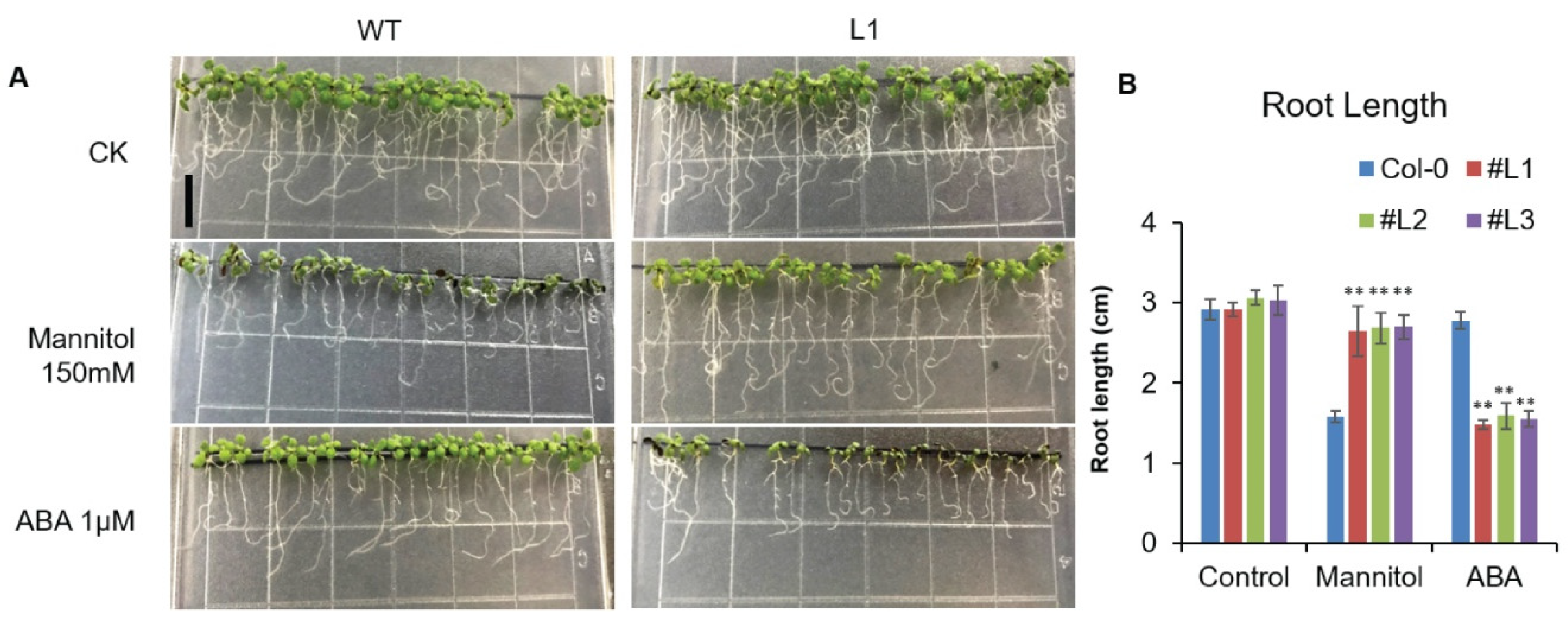
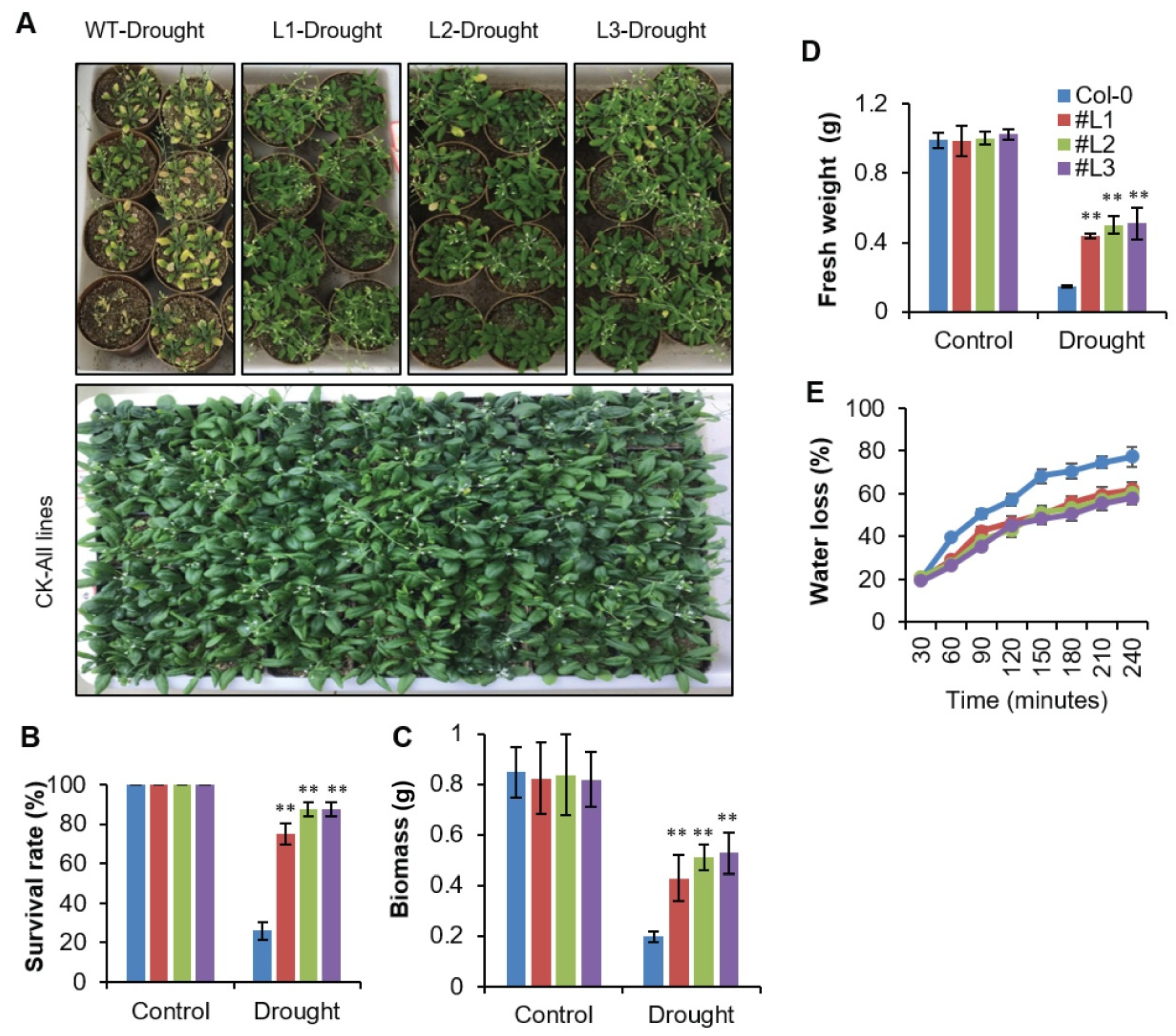
2.4. Overexpression of OsHSFA3 Increases Osmotic and Drought Stress Tolerance in Transgenic Arabidopsis
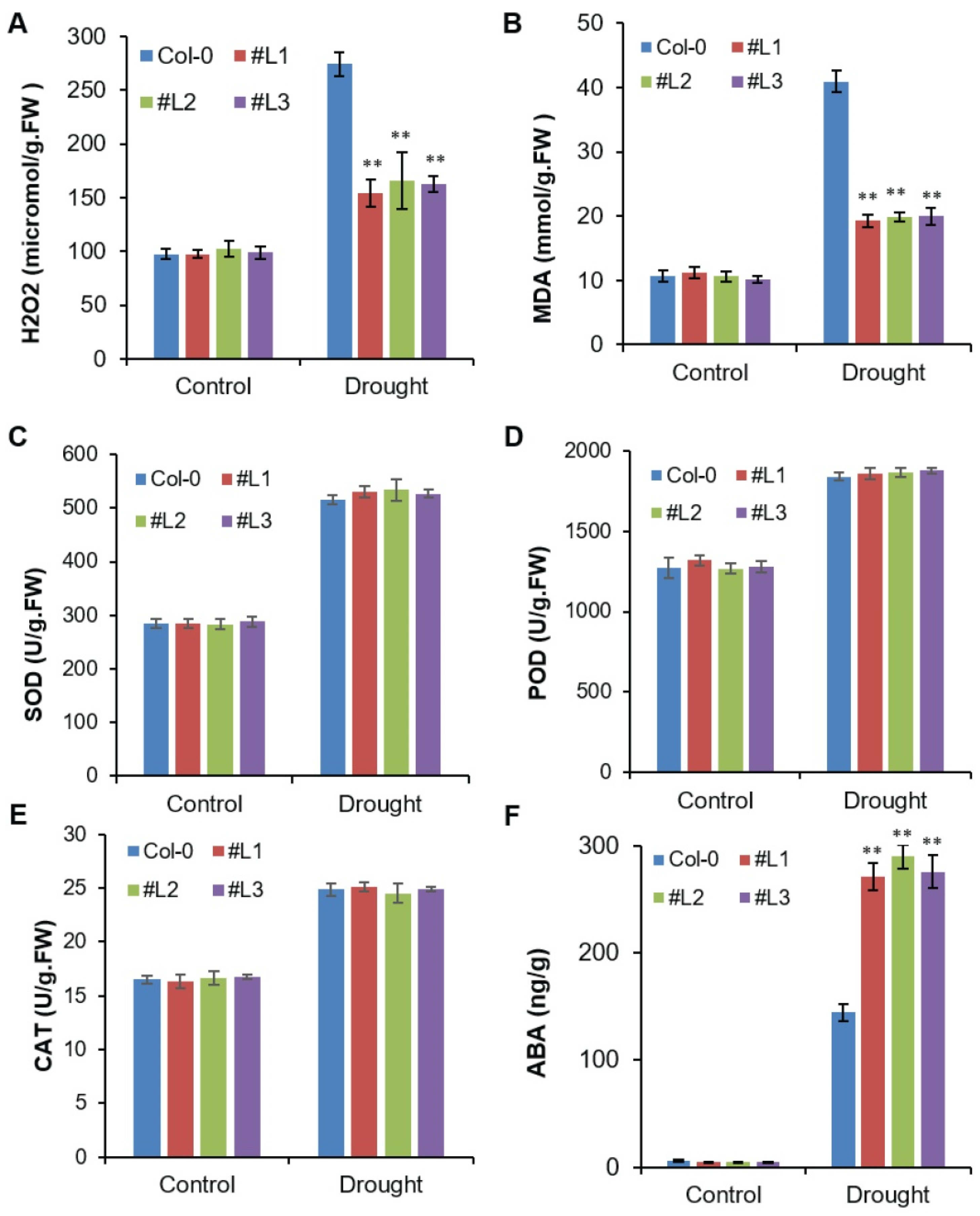
2.5. Overexpression of OsHSFA3 Increases Drought Tolerance by Decreasing H2O2 Accumulation and Increasing ABA Levels
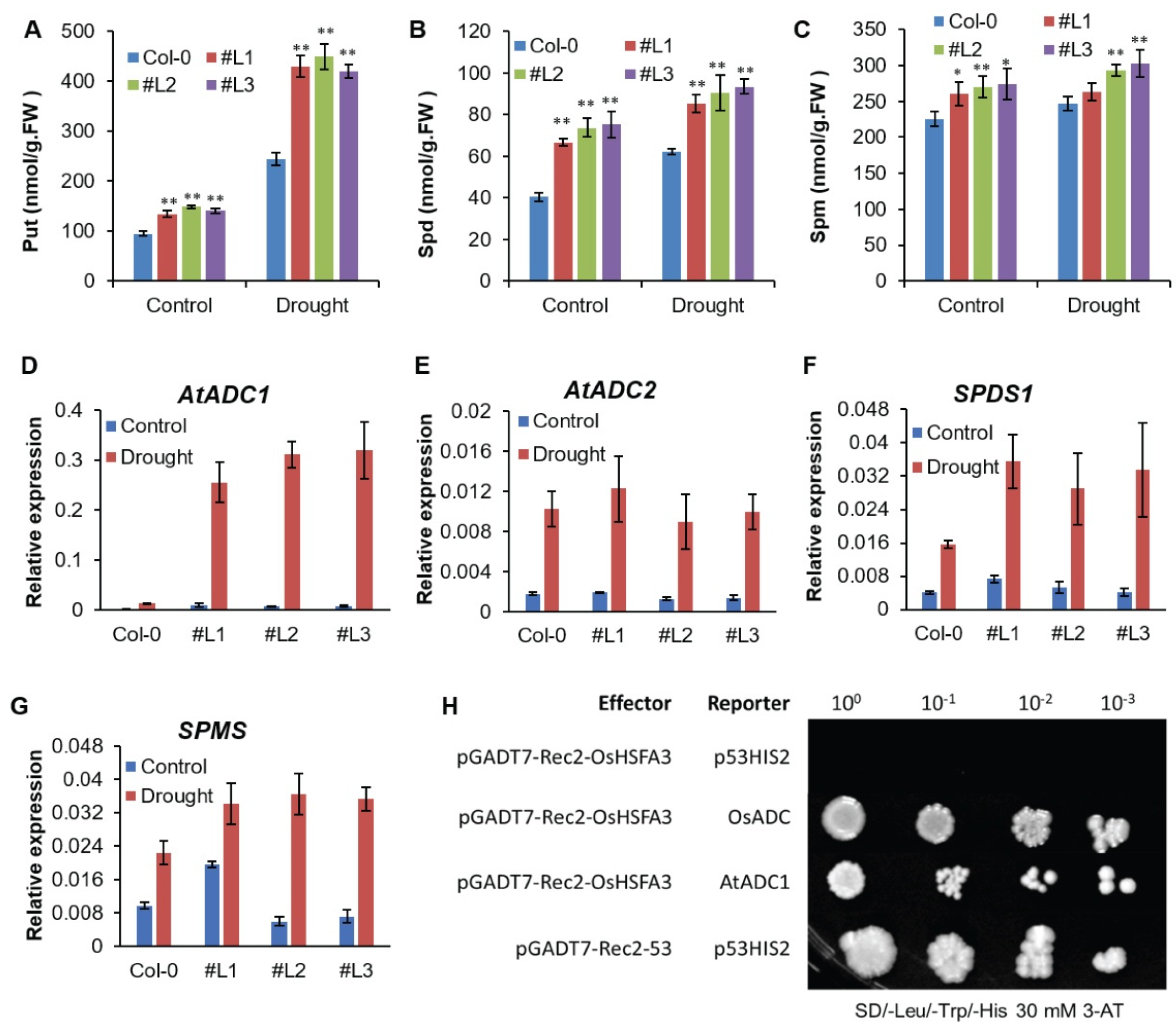
2.6. OsHSFA3 Regulates Polyamines Biosynthesis under Drought Stress
2.7. Expression of Polyamine Biosynthesis Genes is Induced under Drought Stress
2.8. OsHSFA3 Interacts with OsADC1 and AtADC1 Promoter
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions and Stress Treatments
4.2. Total RNA Isolation and qRT-PCR
4.3. Gene structure Prediction and Phylogenetic Analysis
4.4. Vector Construction and Transformation
4.5. Subcellular Localization
4.6. Measurement of ROS, MDA and Antioxidant Activities
4.7. Analysis of Polyamines and ABA
4.8. Yeast One Hybrid Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hussain, H.A.; Men, S.; Hussain, S.; Chen, Y.; Ali, S.; Zhang, S.; Zhang, K.; Li, Y.; Xu, Q.; Liao, C.; et al. Interactive effects of drought and heat stresses on morpho-physiological attributes, yield, nutrient uptake and oxidative status in maize hybrids. Sci. Rep. 2019, 9, 3890. [Google Scholar] [CrossRef]
- Zafar, S.A.; Zaidi, S.S.-e.-A.; Gaba, Y.; Singla-Pareek, S.L.; Dhankher, O.P.; Li, X.; Mansoor, S.; Pareek, A. Engineering Abiotic Stress Tolerance via CRISPR-Cas mediated genome editing. J. Exp. Bot. 2019, 71, 470–479. [Google Scholar] [CrossRef]
- Saeed, Z.; Naveed, M.; Imran, M.; Bashir, M.A.; Sattar, A.; Mustafa, A.; Hussain, A.; Xu, M. Combined use of Enterobacter sp. MN17 and zeolite reverts the adverse effects of cadmium on growth, physiology and antioxidant activity of Brassica napus. PLoS ONE 2019, 14, e0213016. [Google Scholar] [CrossRef]
- Arshad, M.S.; Farooq, M.; Asch, F.; Krishna, J.S.; Prasad, P.V.; Siddique, K.H. Thermal stress impacts reproductive development and grain yield in rice. Plant Physiol. Biochem. 2017, 115, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Zafar, S.A.; Noor, M.A.; Waqas, M.A.; Wang, X.; Shaheen, T.; Raza, M. Temperature extremes in cotton production and mitigation strategies. In Past, Present and Future Trends in Cotton Breeding; Rahman, M.R., Zafar, Y., Eds.; IntechOpen: London, UK, 2018; pp. 65–91. [Google Scholar]
- Poli, Y.; Basava, R.K.; Panigrahy, M.; Vinukonda, V.P.; Dokula, N.R.; Voleti, S.R.; Desiraju, S.; Neelamraju, S. Characterization of a Nagina22 rice mutant for heat tolerance and mapping of yield traits. Rice 2013, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, N.; Audebert, A.; Bennett, M.J.; Bishopp, A.; de Oliveira, A.C.; Courtois, B.; Diedhiou, A.; Diévart, A.; Gantet, P.; Ghesquière, A. The roots of future rice harvests. Rice 2014, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought, high salt and cold responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Lakra, N.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. A unique bZIP transcription factor imparting multiple stress tolerance in Rice. Rice 2019, 12, 58. [Google Scholar] [CrossRef] [PubMed]
- Hussain, H.A.; Hussain, S.; Khaliq, A.; Ashraf, U.; Anjum, S.A.; Men, S.; Wang, L. Chilling and Drought Stresses in Crop Plants: Implications, Cross Talk, and Potential Management Opportunities. Front. Plant Sci. 2018, 9, 393. [Google Scholar] [CrossRef]
- Ribeiro, C.W.; Korbes, A.P.; Garighan, J.A.; Jardim-Messeder, D.; Carvalho, F.E.L.; Sousa, R.H.V.; Caverzan, A.; Teixeira, F.K.; Silveira, J.A.G.; Margis-Pinheiro, M. Rice peroxisomal ascorbate peroxidase knockdown affects ROS signaling and triggers early leaf senescence. Plant Sci. 2017, 263, 55–65. [Google Scholar] [CrossRef]
- Ashraf, M. Inducing drought tolerance in plants: Recent advances. Biotechnol. Adv. 2010, 28, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Zafar, S.A.; Patil, S.B.; Uzair, M.; Fang, J.; Zhao, J.; Guo, T.; Yuan, S.; Uzair, M.; Luo, Q.; Shi, J.; et al. DEGENERATED PANICLE AND PARTIAL STERILITY 1 (DPS1) encodes a cystathionine beta-synthase domain containing protein required for anther cuticle and panicle development in rice. New Phytol. 2020, 225, 356–375. [Google Scholar] [CrossRef] [PubMed]
- Zafar, S.A.; Hameed, A.; Nawaz, M.A.; Wei, M.; Noor, M.A. Mechanisms and molecular approaches for heat tolerance in rice (Oryza sativa L.) under climate change scenario. J. Integr. Agric. 2018, 17, 726–738. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhou, L.; Liu, J.; Du, X.; Asad, M.A.; Huang, F.; Pan, G.; Cheng, F. Relationship of ROS accumulation and superoxide dismutase isozymes in developing anther with floret fertility of rice under heat stress. Plant Physiol. Biochem. 2018, 122, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Liang, W.; Yin, C.; Cui, X.; Zong, J.; Wang, X.; Hu, J.; Zhang, D. Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 2011, 23, 515–533. [Google Scholar] [CrossRef] [PubMed]
- Alcázar, R.; Bitrián, M.; Bartels, D.; Koncz, C.; Altabella, T.; Tiburcio, A.F. Polyamine metabolic canalization in response to drought stress in Arabidopsis and the resurrection plant Craterostigma plantagineum. Plant Signal. Behav. 2011, 6, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Xing, C.; Yao, Z.; Huang, X. PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol. J. 2017, 15, 1186–1203. [Google Scholar] [CrossRef]
- Sánchez-Rangel, D.; Chávez-Martínez, A.I.; Rodríguez-Hernández, A.A.; Maruri-López, I.; Urano, K.; Shinozaki, K.; Jiménez-Bremont, J.F. Simultaneous Silencing of Two Arginine Decarboxylase Genes Alters Development in Arabidopsis. Front. Plant Sci. 2016, 7, 300. [Google Scholar] [CrossRef]
- Maruri-López, I.; Jiménez-Bremont, J.F. Hetero-and homodimerization of Arabidopsis thaliana arginine decarboxylase AtADC1 and AtADC2. Biochem. Biophys. Res. Commun. 2017, 484, 508–513. [Google Scholar] [CrossRef]
- Do, P.T.; Drechsel, O.; Heyer, A.G.; Hincha, D.K.; Zuther, E. Changes in free polyamine levels, expression of polyamine biosynthesis genes, and performance of rice cultivars under salt stress: A comparison with responses to drought. Front. Plant Sci. 2014, 5, 182. [Google Scholar] [CrossRef]
- Liu, T.; Wook Kim, D.; Niitsu, M.; Berberich, T.; Kusano, T. POLYAMINE OXIDASE 1 from rice (Oryza sativa) is a functional ortholog of Arabidopsis POLYAMINE OXIDASE 5. Plant Signal. Behav. 2014, 9, e29773. [Google Scholar] [CrossRef] [PubMed]
- Zafar, S.A.; Hussain, M.; Raza, M.; Ahmed, H.G.M.-D.; Rana, I.A.; Sadia, B.; Atif, R.M. Genome wide analysis of heat shock transcription factor (HSF) family in chickpea and its comparison with Arabidopsis. Plant Omics 2016, 9, 136–141. [Google Scholar] [CrossRef]
- Li, P.-S.; Yu, T.-F.; He, G.-H.; Chen, M.; Zhou, Y.-B.; Chai, S.-C.; Xu, Z.-S.; Ma, Y.-Z. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genom. 2014, 15, 1009. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Azeem, F.; Nawaz, M.A.; Acet, T.; Abbas, A.; Imran, Q.M.; Shah, K.H.; Rehman, H.M.; Chung, G.; Yang, S.H.; et al. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J. Plant Physiol. 2018, 226, 12–21. [Google Scholar] [CrossRef]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Zhang, J.; Zhang, H.; Zhang, Z.; Quan, R.; Zhou, S.; Huang, R. Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS ONE 2011, 6, e25216. [Google Scholar] [CrossRef]
- Guo, J.; Wu, J.; Ji, Q.; Wang, C.; Luo, L.; Yuan, Y.; Wang, Y.; Wang, J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genom. 2008, 35, 105–118. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Q.; Shou, H.-x. Identification and expression analysis of OsHsfs in rice. J. Zhejiang Univ. Sci. B 2009, 10, 291–300. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, L.; Zhang, M.; Zafar, S.A.; Fang, J.; Li, M.; Zhang, W.; Li, X. Arabidopsis E3 Ubiquitin Ligases PUB22 and PUB23 Negatively Regulate Drought Tolerance by Targeting ABA Receptor PYL9 for Degradation. Int. J. Mol. Sci. 2017, 18, 1841. [Google Scholar] [CrossRef]
- Dang, J.; Jiang, M.; Lin, F. ABA up-regulate the expression of OsHsf gene in leaves of rice plants. J. Nanjing Agric. Univ. 2010, 33, 11–15. [Google Scholar]
- Schmidt, R.; Schippers, J.H.; Welker, A.; Mieulet, D.; Guiderdoni, E.; Mueller-Roeber, B. Transcription factor OsHsfC1b regulates salt tolerance and development in Oryza sativa ssp. japonica. AoB Plants 2012, 2012, pls011. [Google Scholar] [CrossRef]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M.; Oda, K. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 2008, 227, 957–967. [Google Scholar] [CrossRef]
- Lv, Y.; Xu, L.; Dossa, K.; Zhou, K.; Zhu, M.; Xie, H.; Tang, S.; Yu, Y.; Guo, X.; Zhou, B. Identification of putative drought-responsive genes in rice using gene co-expression analysis. Bioinformation 2019, 15, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, H.; Khurana, N.; Agarwal, P.; Khurana, P. Heat shock factors in rice (Oryza sativa L.): Genome-wide expression analysis during reproductive development and abiotic stress. Mol. Genet. Genom. 2011, 286, 171. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Goher, M.; Iqbal, N. Heat stress-induced cell death, changes in antioxidants, lipid peroxidation, and protease activity in wheat leaves. J. Plant Growth Regul. 2012, 31, 283–291. [Google Scholar] [CrossRef]
- Kamran, M.; Malik, Z.; Parveen, A.; Huang, L.; Riaz, M.; Bashir, S.; Mustafa, A.; Abbasi, G.H.; Xue, B.; Ali, U. Ameliorative Effects of Biochar on Rapeseed (Brassica napus L.) Growth and Heavy Metal Immobilization in Soil Irrigated with Untreated Wastewater. J. Plant Growth Regul. 2019. [Google Scholar] [CrossRef]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.-H.; Lee, S.C. Function of ABA in Stomatal Defense against Biotic and Drought Stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 Enhances Drought and Salt Tolerance Through an ABA-Mediated Pathway in Arabidopsis thaliana. Front. Plant Sci. 2019, 9, 1979. [Google Scholar] [CrossRef]
- Kasukabe, Y.; He, L.; Nada, K.; Misawa, S.; Ihara, I.; Tachibana, S. Overexpression of Spermidine Synthase Enhances Tolerance to Multiple Environmental Stresses and Up-Regulates the Expression of Various Stress-Regulated Genes in Transgenic Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 712–722. [Google Scholar] [CrossRef]
- Li, J.; Chauve, L.; Phelps, G.; Brielmann, R.M.; Morimoto, R.I. E2F coregulates an essential HSF developmental program that is distinct from the heat-shock response. Genes Dev. 2016, 30, 2062–2075. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, H.; Khurana, N.; Agarwal, P.; Khurana, J.P.; Khurana, P. A seed preferential heat shock transcription factor from wheat provides abiotic stress tolerance and yield enhancement in transgenic Arabidopsis under heat stress environment. PLoS ONE 2013, 8, e79577. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xu, Z.-S.; Li, P.; Yang, L.; Wei, Y.; Chen, M.; Li, L.; Zhang, G.; Ma, Y. Overexpression of TaHSF3 in Transgenic Arabidopsis Enhances Tolerance to Extreme Temperatures. Plant Mol. Biol. Report. 2013, 31, 688–697. [Google Scholar] [CrossRef]
- Gu, L.; Ma, Q.; Zhang, C.; Wang, C.; Wei, H.; Wang, H.; Yu, S. The Cotton GhWRKY91 Transcription Factor Mediates Leaf Senescence and Responses to Drought Stress in Transgenic Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 1352. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.-L.; Zou, J.; Liu, C.-F.; Zhou, X.-Y.; Zhang, X.-W.; Luo, G.-Y.; Chen, X.-B. Over-expression of OsHsfA7 enhanced salt and drought tolerance in transgenic rice. BMB Rep. 2013, 46, 31. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Magwanga, R.O.; Kirungu, J.N.; Hu, Y.; Dong, Q.; Cai, X.; Zhou, Z.; Wang, X.; Zhang, Z.; Hou, Y. Overexpression of Cotton a DTX/MATE Gene Enhances Drought, Salt and Cold Stress Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 2019, 10, 299. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.; Pandey, M. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef]
- Li, H.-M.; Liu, S.-D.; Ge, C.-W.; Zhang, X.-M.; Zhang, S.-P.; Chen, J.; Shen, Q.; Ju, F.-Y.; Yang, Y.-F.; Li, Y.; et al. Analysis of Drought Tolerance and Associated Traits in Upland Cotton at the Seedling Stage. Int. J. Mol. Sci. 2019, 20, 3888. [Google Scholar] [CrossRef]
- Wang, N.-N.; Xu, S.-W.; Sun, Y.-L.; Liu, D.; Zhou, L.; Li, Y.; Li, X.-B. The cotton WRKY transcription factor (GhWRKY33) reduces transgenic Arabidopsis resistance to drought stress. Sci. Rep. 2019, 9, 724. [Google Scholar] [CrossRef]
- Wattoo, F.M.; Rana, R.M.; Fiaz, S.; Zafar, S.A.; Noor, M.A.; Hassan, H.M.; Bhatti, M.H.; Rehman, S.U.; Anis, G.B.; Amir, R.M. Identification of drought tolerant maize genotypes and seedling based morpho-physiological selection indices for crop improvement. Sains Malays. 2018, 47, 295–302. [Google Scholar]
- Noman, M.; Jameel, A.; Qiang, W.-D.; Ahmad, N.; Liu, W.-C.; Wang, F.-W.; Li, H.-Y. Overexpression of GmCAMTA12 Enhanced Drought Tolerance in Arabidopsis and Soybean. Int. J. Mol. Sci. 2019, 20, 4849. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Kim, M.C.; Kumar, D.; Park, B.; Cheong, M.S.; Choi, W.; Park, H.C.; Chun, H.J.; Park, H.J.; Lee, S.Y.; et al. AtPR5K2, a PR5-Like Receptor Kinase, Modulates Plant Responses to Drought Stress by Phosphorylating Protein Phosphatase 2Cs. Front. Plant Sci. 2019, 10, 1146. [Google Scholar] [CrossRef] [PubMed]
- Di Meo, S.; Napolitano, G.; Venditti, P. Physiological and Pathological Role of ROS: Benefits and Limitations of Antioxidant Treatment. Int. J. Mol. Sci. 2019, 20, 4810. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Alhaithloul, H.A.S.; Parvin, K.; Bhuyan, M.H.M.B.; Tanveer, M.; Mohsin, S.M.; Nahar, K.; Soliman, M.H.; Mahmud, J.A.; Fujita, M. Polyamine Action under Metal/Metalloid Stress: Regulation of Biosynthesis, Metabolism, and Molecular Interactions. Int. J. Mol. Sci. 2019, 20, 3215. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, Y.; Shi, Y.; Zhang, Z.; Zou, Z.; Zhang, H.; Zhao, J. Effect of exogenous spermidine on polyamine content and metabolism in tomato exposed to salinity-alkalinity mixed stress. Plant Physiol. Biochem. 2012, 57, 200–209. [Google Scholar] [CrossRef]
- Okamoto, M.; Hanada, A.; Kamiya, Y.; Yamaguchi, S.; Nambara, E. Measurement of abscisic acid and gibberellins by gas chromatography/mass spectrometry. Methods Mol. Biol. 2009, 495, 53–60. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, M.-D.; Zhang, M.; Gao, D.-J.; Zhou, K.; Tang, S.-J.; Zhou, B.; Lv, Y.-M. Rice OsHSFA3 Gene Improves Drought Tolerance by Modulating Polyamine Biosynthesis Depending on Abscisic Acid and ROS Levels. Int. J. Mol. Sci. 2020, 21, 1857. https://doi.org/10.3390/ijms21051857
Zhu M-D, Zhang M, Gao D-J, Zhou K, Tang S-J, Zhou B, Lv Y-M. Rice OsHSFA3 Gene Improves Drought Tolerance by Modulating Polyamine Biosynthesis Depending on Abscisic Acid and ROS Levels. International Journal of Molecular Sciences. 2020; 21(5):1857. https://doi.org/10.3390/ijms21051857
Chicago/Turabian StyleZhu, Ming-Dong, Meng Zhang, Du-Juan Gao, Kun Zhou, Shan-Jun Tang, Bin Zhou, and Yan-Mei Lv. 2020. "Rice OsHSFA3 Gene Improves Drought Tolerance by Modulating Polyamine Biosynthesis Depending on Abscisic Acid and ROS Levels" International Journal of Molecular Sciences 21, no. 5: 1857. https://doi.org/10.3390/ijms21051857
APA StyleZhu, M.-D., Zhang, M., Gao, D.-J., Zhou, K., Tang, S.-J., Zhou, B., & Lv, Y.-M. (2020). Rice OsHSFA3 Gene Improves Drought Tolerance by Modulating Polyamine Biosynthesis Depending on Abscisic Acid and ROS Levels. International Journal of Molecular Sciences, 21(5), 1857. https://doi.org/10.3390/ijms21051857




