Proteomic Analyses of Human Sperm Cells: Understanding the Role of Proteins and Molecular Pathways Affecting Male Reproductive Health
Abstract
:1. Introduction
2. Proteomics Overview
2.1. Process and Techniques
2.2. Proteomic Analysis Using LC-MS/MS
3. Proteomic Analysis of Human Spermatozoa
4. Clinical Implications of Sperm Proteomics
4.1. Protein Profiling in Male Infertility
4.1.1. Protein Profiling in Asthenozoospermia
4.1.2. Protein Profiling in Azoospermia
4.1.3. Protein Profiling in Oligoasthenozoospermia
4.1.4. Protein Profiling in Globozoospermia
4.2. Proteomic Profiling in Infertility-Related Conditions and Diseases
4.2.1. Protein Profiling in Varicocele
4.2.2. Protein Profiling in Testicular Cancer
4.2.3. Protein Profiling and Assisted Reproductive Technology
5. Sperm Proteomics in Deciphering Cellular Pathways Associated with Male Infertility
6. Conclusions
7. Future Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Plant, T.M.; Zeleznik, A.J. Knobil and Neill’s Physiology of Reproduction; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Zegers-Hochschild, F.; Adamson, G.D.; Dyer, S.; Racowsky, C.; de Mouzon, J.; Sokol, R.; Rienzi, L.; Sunde, A.; Schmidt, L.; Cooke, I.D.; et al. The International Glossary on Infertility and Fertility Care, 2017. Fertil. Steril. 2017, 108, 393–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inhorn, M.C.; Patrizio, P. Infertility around the globe: New thinking on gender, reproductive technologies and global movements in the 21st century. Hum. Reprod. Update 2015, 21, 411–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutstein, S.; Shah, I. DHS Comparative Reports 9. Infecundity, Infertility, and Childlessness in Developing Countries; World Health Organisation: Geneva, Switzerland, 2004. [Google Scholar]
- Agarwal, A.; Parekh, N.; Selvam, M.K.P.; Henkel, R.; Shah, R.; Homa, S.T.; Ramasamy, R.; Ko, E.; Tremellen, K.; Esteves, S.; et al. Male Oxidative Stress Infertility (MOSI): Proposed Terminology and Clinical Practice Guidelines for Management of Idiopathic Male Infertility. World J. Mens Health 2019, 37, 296–312. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. WHO Laboratory Manual for the Examination and Processing of Human Semen; WHO: Geneva, Switzerland, 2010. [Google Scholar]
- Wang, C.; Swerdloff, R.S. Limitations of semen analysis as a test of male fertility and anticipated needs from newer tests. Fertil. Steril. 2014, 102, 1502–1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, A.; Gupta, S.; Sharma, R. Oxidation–Reduction Potential Measurement in Ejaculated Semen Samples. In Andrological Evaluation of Male Infertility: A Laboratory Guide; Agarwal, A., Gupta, S., Sharma, R., Eds.; Springer International Publishing: Cham, Germany, 2016; pp. 165–170. [Google Scholar]
- Agarwal, A.; Bertolla, R.P.; Samanta, L. Sperm proteomics: Potential impact on male infertility treatment. Exp. Rev. Proteom. 2016, 13, 285–296. [Google Scholar] [CrossRef]
- Evenson, D.P. The Sperm Chromatin Structure Assay (SCSA®) and other sperm DNA fragmentation tests for evaluation of sperm nuclear DNA integrity as related to fertility. Anim. Reprod. Sci. 2016, 169, 56–75. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, A.; Sharma, R.; Roychoudhury, S.; Du Plessis, S.; Sabanegh, E. MiOXSYS: A novel method of measuring oxidation reduction potential in semen and seminal plasma. Fertil. Steril. 2016, 106, 566–573. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, A.; Gupta, S.; Sharma, R. Measurement of DNA fragmentation in spermatozoa by TUNEL assay using bench top flow cytometer. In Andrological Evaluation of Male Infertility; Springer: Berlin/Heidelberg, Germany, 2016; pp. 181–203. [Google Scholar]
- Jodar, M.; Soler-Ventura, A.; Oliva, R. Semen proteomics and male infertility. J. Proteom. 2017, 162, 125–134. [Google Scholar] [CrossRef]
- Mohanty, G.; Swain, N.; Samanta, L. Sperm Proteome:What Is on the Horizon? Reprod. Sci. 2015, 22, 638–653. [Google Scholar] [CrossRef]
- Codina, M.; Estanyol, J.M.; Fidalgo, M.J.; Ballescà, J.L.; Oliva, R. Advances in sperm proteomics: Best-practise methodology and clinical potential. Exp. Rev. Proteom. 2015, 12, 255–277. [Google Scholar] [CrossRef]
- Selvam, M.K.P.; Agarwal, A.; Pushparaj, P.N. A quantitative global proteomics approach to understanding the functional pathways dysregulated in the spermatozoa of asthenozoospermic testicular cancer patients. Andrology 2019, 7, 454–462. [Google Scholar]
- Selvam, M.K.P.; Agarwal, A.; Pushparaj, P.N. Altered Molecular Pathways in the Proteome of Cryopreserved Sperm in Testicular Cancer Patients before Treatment. Int. J. Mol. Sci. 2019, 20. [Google Scholar]
- Samanta, L.; Agarwal, A.; Swain, N.; Sharma, R.; Gopalan, B.; Esteves, S.C.; Durairajanayagam, D.; Sabanegh, E. Proteomic Signatures of Sperm Mitochondria in Varicocele: Clinical Use as Biomarkers of Varicocele Associated Infertility. J. Urol. 2018, 200, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Intasqui, P.; Agarwal, A.; Sharma, R.; Samanta, L.; Bertolla, R.P. Towards the identification of reliable sperm biomarkers for male infertility: A sperm proteomic approach. Andrologia 2018, 50, e12919. [Google Scholar] [CrossRef]
- Cao, X.; Cui, Y.; Zhang, X.; Lou, J.; Zhou, J.; Bei, H.; Wei, R. Proteomic profile of human spermatozoa in healthy and asthenozoospermic individuals. Reprod. Biol. Endocrinol. 2018, 16, 16. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Heredia, J.; Estanyol, J.M.; Ballesca, J.L.; Oliva, R. Proteomic identification of human sperm proteins. Proteomics 2006, 6, 4356–4369. [Google Scholar] [CrossRef]
- Gupta, S.; Ghulmiyyah, J.; Sharma, R.; Halabi, J.; Agarwal, A. Power of proteomics in linking oxidative stress and female infertility. BioMed Res. Int. 2014, 2014, 916212. [Google Scholar] [CrossRef] [Green Version]
- Hamada, A.; Sharma, R.; du Plessis, S.S.; Willard, B.; Yadav, S.P.; Sabanegh, E.; Agarwal, A. Two-dimensional differential in-gel electrophoresis-based proteomics of male gametes in relation to oxidative stress. Fertil. Steril. 2013, 99, 1216–1226. [Google Scholar] [CrossRef]
- Johnston, D.S.; Wooters, J.; Kopf, G.S.; Qiu, Y.; Roberts, K.P. Analysis of the Human Sperm Proteome. Ann. N. Y. Acad. Sci. 2005, 1061, 190–202. [Google Scholar] [CrossRef]
- Dias, T.R.; Agarwal, A.; Pushparaj, P.N.; Ahmad, G.; Sharma, R. New Insights on the Mechanisms Affecting Fertility in Men with Non-Seminoma Testicular Cancer before Cancer Therapy. World J. Mens Health 2018, 36. [Google Scholar] [CrossRef]
- Giacomini, E.; Ura, B.; Giolo, E.; Luppi, S.; Martinelli, M.; Garcia, R.C.; Ricci, G. Comparative analysis of the seminal plasma proteomes of oligoasthenozoospermic and normozoospermic men. Reprod. Biomed. Online 2015, 30, 522–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Liu, G.; Liu, J.; Zhu, P.; Wang, J.; Wang, Y.; Wang, W.; Li, N.; Wang, X.; Zhang, C.; et al. iTRAQ-based analysis of sperm proteome from normozoospermic men achieving the rescue-ICSI pregnancy after the IVF failure. Clin. Proteom. 2018, 15, 27. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ao, L.; Yan, Y.; Jiang, J.; Chen, B.; Duan, Y.; Shen, F.; Chen, J.; Inglis, B.; Ni, R.; et al. Differential motility parameters and identification of proteomic profiles of human sperm cryopreserved with cryostraw and cryovial. Clin. Proteom. 2019, 16, 24. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Guo, Y.; Song, N.; Fan, Y.; Li, K.; Teng, X.; Guo, Q.; Ding, Z. Proteomic pattern changes associated with obesity-induced asthenozoospermia. Andrology 2015, 3, 247–259. [Google Scholar] [CrossRef]
- Moscatelli, N.; Lunetti, P.; Braccia, C.; Armirotti, A.; Pisanello, F.; De Vittorio, M.; Zara, V.; Ferramosca, A. Comparative Proteomic Analysis of Proteins Involved in Bioenergetics Pathways Associated with Human Sperm Motility. Int. J. Mol. Sci. 2019, 20, 3000. [Google Scholar] [CrossRef] [Green Version]
- Milardi, D.; Grande, G.; Vincenzoni, F.; Castagnola, M.; Marana, R. Proteomics of human seminal plasma: Identification of biomarker candidates for fertility and infertility and the evolution of technology. Mol. Reprod. Dev. 2013, 80, 350–357. [Google Scholar] [CrossRef]
- Samanta, L.; Parida, R.; Dias, T.R.; Agarwal, A. The enigmatic seminal plasma: A proteomics insight from ejaculation to fertilization. Reprod. Biol. Endocrinol. 2018, 16, 41. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Wang, W.; Xu, Y.; Tang, M.; Fang, J.; Sun, H.; Sun, Y.; Gu, M.; Liu, Z.; Zhang, Z. Proteomic characteristics of human sperm cryopreservation. Proteomics 2014, 14, 298–310. [Google Scholar] [CrossRef]
- Amaral, A.; Paiva, C.; Attardo Parrinello, C.; Estanyol, J.M.; Ballescà, J.L.S.; Ramalho-Santos, J.O.; Oliva, R. Identification of proteins involved in human sperm motility using high-throughput differential proteomics. J. Proteom. Res. 2014, 13, 5670–5684. [Google Scholar] [CrossRef]
- Wang, X.M.; Xiang, Z.; Fu, Y.; Wu, H.L.; Zhu, W.B.; Fan, L.Q. Comparative Proteomics Reveal the Association between SPANX Proteins and Clinical Outcomes of Artificial Insemination with Donor Sperm. Sci. Rep. 2018, 8, 6850. [Google Scholar] [CrossRef] [Green Version]
- Intasqui, P.; Camargo, M.; Del Giudice, P.T.; Spaine, D.M.; Carvalho, V.M.; Cardozo, K.H.M.; Cedenho, A.P.; Bertolla, R.P. Unraveling the sperm proteome and post-genomic pathways associated with sperm nuclear DNA fragmentation. J. Assist. Reprod. Genet. 2013, 30, 1187–1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amaral, A.; Castillo, J.; Estanyol, J.M.; Ballescà, J.L.; Ramalho-Santos, J.; Oliva, R. Human sperm tail proteome suggests new endogenous metabolic pathways. Mol. Cell. Proteom. 2013, 12, 330–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvam, M.K.P.; Agarwal, A.; Dias, T.R.; Martins, A.D.; Samanta, L. Presence of Round Cells Proteins do not Interfere with Identification of Human Sperm Proteins from Frozen Semen Samples by LC-MS/MS. Int. J. Mol. Sci. 2019, 20, 314. [Google Scholar] [CrossRef] [Green Version]
- Selvam, M.K.P.; Agarwal, A.; Dias, T.R.; Martins, A.D.; Baskaran, S.; Samanta, L. Molecular Pathways Associated with Sperm Biofunction Are Not Affected by the Presence of Round Cell and Leukocyte Proteins in Human Sperm Proteome. J. Proteome Res. 2018, 18, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Netherton, J.K.; Hetherington, L.; Ogle, R.A.; Velkov, T.; Baker, M.A. Proteomic analysis of good- and poor-quality human sperm demonstrates that several proteins are routinely aberrantly regulated. Biol. Reprod. 2018, 99, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Castillo, J.; Bogle, O.A.; Jodar, M.; Torabi, F.; Delgado-Duenas, D.; Estanyol, J.M.; Ballesca, J.L.; Miller, D.; Oliva, R. Proteomic Changes in Human Sperm During Sequential in vitro Capacitation and Acrosome Reaction. Front. Cell Dev. Biol. 2019, 7, 295. [Google Scholar] [CrossRef] [Green Version]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359. [Google Scholar] [CrossRef]
- Wang, G.; Wu, Y.; Zhou, T.; Guo, Y.; Zheng, B.; Wang, J.; Bi, Y.; Liu, F.; Zhou, Z.; Guo, X.; et al. Mapping of the N-Linked Glycoproteome of Human Spermatozoa. J. Proteome Res. 2013, 12, 5750–5759. [Google Scholar] [CrossRef]
- Glish, G.L.; Vachet, R.W. The basics of mass spectrometry in the twenty-first century. Nat. Rev. Drug Discov. 2003, 2, 140. [Google Scholar] [CrossRef]
- Oliva, R.; Martinez-Heredia, J.; Estanyol, J.M. Proteomics in the study of the sperm cell composition, differentiation and function. Syst. Biol. Reprod. Med. 2008, 54, 23–36. [Google Scholar] [CrossRef]
- Cadavid, J.A.; Alvarez, A.; Markert, U.R.; Cardona Maya, W. Differential protein expression in seminal plasma from fertile and infertile males. J. Hum. Reprod. Sci. 2014, 7, 206–211. [Google Scholar]
- Zhou, T.; Zhou, Z.-M.; Guo, X.-J. Bioinformatics for spermatogenesis: Annotation of male reproduction based on proteomics. Asian J. Androl. 2013, 15, 594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, N.; Montelione, G.T.; Gerstein, M. Ontologies for proteomics: Towards a systematic definition of structure and function that scales to the genome level. Curr. Opin. Chem. Biol. 2003, 7, 44–54. [Google Scholar] [CrossRef]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, A.; Durairajanayagam, D.; Halabi, J.; Peng, J.; Vazquez-Levin, M. Proteomics, oxidative stress and male infertility. Reprod. Biomed. Online 2014, 29, 32–58. [Google Scholar] [CrossRef] [Green Version]
- Jodar, M.; Selvaraju, S.; Sendler, E.; Diamond, M.P.; Krawetz, S.A. The presence, role and clinical use of spermatozoal RNAs. Hum. Reprod. Update 2013, 19, 604–624. [Google Scholar] [CrossRef]
- Baker, M.A.; Nixon, B.; Naumovski, N.; Aitken, R.J. Proteomic insights into the maturation and capacitation of mammalian spermatozoa. Syst. Biol. Reprod. Med. 2012, 58, 211–217. [Google Scholar] [CrossRef]
- Naaby-Hansen, S.; Flickinger, C.J.; Herr, J.C. Two-Dimensional Gel Electrophoretic Analysis of Vectorially Labeled Surface Proteins of Human Spermatozoa1. Biol. Reprod. 1997, 56, 771–787. [Google Scholar] [CrossRef] [Green Version]
- Shetty, J.; Diekman, A.B.; Jayes, F.C.; Sherman, N.E.; Naaby-Hansen, S.; Flickinger, C.J.; Herr, J.C. Differential extraction and enrichment of human sperm surface proteins in a proteome: Identification of immunocontraceptive candidates. Electrophoresis 2001, 22, 3053–3066. [Google Scholar] [CrossRef]
- Baker, M.A.; Reeves, G.; Hetherington, L.; Müller, J.; Baur, I.; Aitken, R.J. Identification of gene products present in Triton X-100 soluble and insoluble fractions of human spermatozoa lysates using LC-MS/MS analysis. Proteom. Clin. Appl. 2007, 1, 524–532. [Google Scholar] [CrossRef]
- Gilany, K.; Lakpour, N.; Vafakhah, M.; Sadeghi, M.R. The Profile of Human Sperm Proteome; A Mini-review. J. Reprod. Infertil. 2011, 12, 193–199. [Google Scholar] [PubMed]
- Wang, G.; Guo, Y.; Zhou, T.; Shi, X.; Yu, J.; Yang, Y.; Wu, Y.; Wang, J.; Liu, M.; Chen, X.; et al. In-depth proteomic analysis of the human sperm reveals complex protein compositions. J. Proteom. 2013, 79, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Nixon, B.; Mitchell, L.A.; Anderson, A.L.; Mclaughlin, E.A.; O’Bryan, M.K.; Aitken, R.J. Proteomic and functional analysis of human sperm detergent resistant membranes. J. Cell. Physiol. 2011, 226, 2651–2665. [Google Scholar] [CrossRef] [PubMed]
- De Mateo, S.; Castillo, J.; Estanyol, J.M.; Ballescà, J.L.; Oliva, R. Proteomic characterization of the human sperm nucleus. Proteomics 2011, 11, 2714–2726. [Google Scholar] [CrossRef]
- Baker, M.A.; Naumovski, N.; Hetherington, L.; Weinberg, A.; Velkov, T.; Aitken, R.J. Head and flagella subcompartmental proteomic analysis of human spermatozoa. Proteomics 2013, 13, 61–74. [Google Scholar] [CrossRef]
- De Mateo, S.; Martínez-Heredia, J.; Estanyol, J.M.; Domíguez-Fandos, D.; Vidal-Taboada, J.M.; Ballescà, J.L.; Oliva, R. Marked correlations in protein expression identified by proteomic analysis of human spermatozoa. Proteomics 2007, 7, 4264–4277. [Google Scholar] [CrossRef]
- Amaral, A.; Castillo, J.; Ramalho-Santos, J.; Oliva, R. The combined human sperm proteome: Cellular pathways and implications for basic and clinical science. Hum. Reprod. Update 2014, 20, 40–62. [Google Scholar] [CrossRef] [Green Version]
- Parte, P.P.; Rao, P.; Redij, S.; Lobo, V.; D’Souza, S.J.; Gajbhiye, R.; Kulkarni, V. Sperm phosphoproteome profiling by ultra performance liquid chromatography followed by data independent analysis (LC–MSE) reveals altered proteomic signatures in asthenozoospermia. J. Proteom. 2012, 75, 5861–5871. [Google Scholar] [CrossRef]
- Saraswat, M.; Joenväärä, S.; Jain, T.; Tomar, A.K.; Sinha, A.; Singh, S.; Yadav, S.; Renkonen, R. Human Spermatozoa Quantitative Proteomic Signature Classifies Normo- and Asthenozoospermia. Mol. Cell. Proteom. 2017, 16, 57–72. [Google Scholar] [CrossRef] [Green Version]
- Nowicka-Bauer, K.; Lepczynski, A.; Ozgo, M.; Kamieniczna, M.; Fraczek, M.; Stanski, L.; Olszewska, M.; Malcher, A.; Skrzypczak, W.; Kurpisz, M. Sperm mitochondrial dysfunction and oxidative stress as possible reasons for isolated asthenozoospermia. J. Physiol. Pharmacol. 2018, 69, 403–417. [Google Scholar]
- Sinha, A.; Singh, V.; Singh, S.; Yadav, S. Proteomic analyses reveal lower expression of TEX40 and ATP6V0A2 proteins related to calcium ion entry and acrosomal acidification in asthenozoospermic males. Life Sci. 2019, 218, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Siva, A.B.; Kameshwari, D.B.; Singh, V.; Pavani, K.; Sundaram, C.S.; Rangaraj, N.; Deenadayal, M.; Shivaji, S. Proteomics-based study on asthenozoospermia: Differential expression of proteasome alpha complex. MHR Basic Sci. Reprod. Med. 2010, 16, 452–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Heredia, J.; de Mateo, S.; Vidal-Taboada, J.M.; Ballescà, J.L.; Oliva, R. Identification of proteomic differences in asthenozoospermic sperm samples. Hum. Reprod. 2008, 23, 783–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Huo, R.; Wang, F.-Q.; Lin, M.; Zhou, Z.-M.; Sha, J.-H. Identification of several proteins involved in regulation of sperm motility by proteomic analysis. Fertil. Steril. 2007, 87, 436–438. [Google Scholar] [CrossRef] [PubMed]
- Hashemitabar, M.; Sabbagh, S.; Orazizadeh, M.; Ghadiri, A.; Bahmanzadeh, M. A proteomic analysis on human sperm tail: Comparison between normozoospermia and asthenozoospermia. J. Assist. Reprod. Genet. 2015, 32, 853–863. [Google Scholar] [CrossRef] [Green Version]
- Berookhim, B.M.; Schlegel, P.N. Azoospermia due to Spermatogenic Failure. Urol. Clin. N. Am. 2014, 41, 97–113. [Google Scholar] [CrossRef]
- Silber, S.J.; Nagy, Z.; Devroey, P.; Tournaye, H.; van Steirteghem, A.C. Distribution of spermatogenesis in the testicles of azoospermic men: The presence or absence of spermatids in the testes of men with germinal failure. Hum. Reprod. 1997, 12, 2422–2428. [Google Scholar] [CrossRef] [Green Version]
- Drabovich, A.P.; Dimitromanolakis, A.; Saraon, P.; Soosaipillai, A.; Batruch, I.; Mullen, B.; Jarvi, K.; Diamandis, E.P. Differential Diagnosis of Azoospermia with Proteomic Biomarkers ECM1 and TEX101 Quantified in Seminal Plasma. Sci. Transl. Med. 2013, 5, ra160–ra212. [Google Scholar] [CrossRef]
- Bieniek, J.M.; Drabovich, A.P.; Lo, K.C. Seminal biomarkers for the evaluation of male infertility. Asian J. Androl. 2016, 18, 426–433. [Google Scholar] [CrossRef]
- Alikhani, M.; Mirzaei, M.; Sabbaghian, M.; Parsamatin, P.; Karamzadeh, R.; Adib, S.; Sodeifi, N.; Gilani, M.A.S.; Zabet-Moghaddam, M.; Parker, L.; et al. Quantitative proteomic analysis of human testis reveals system-wide molecular and cellular pathways associated with non-obstructive azoospermia. J. Proteom. 2017, 162, 141–154. [Google Scholar] [CrossRef]
- Herwig, R.; Knoll, C.; Planyavsky, M.; Pourbiabany, A.; Greilberger, J.; Bennett, K.L. Proteomic analysis of seminal plasma from infertile patients with oligoasthenoteratozoospermia due to oxidative stress and comparison with fertile volunteers. Fertil. Steril. 2013, 100, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, W.; Zhu, P.; Wang, J.; Wang, Y.; Wang, X.; Liu, J.; Li, N.; Lin, C.; Liu, F. In-depth quantitative proteome analysis of seminal plasma from men with oligoasthenozoospermia and normozoospermia. Reprod. Biomed. Online 2018, 37, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Dam, A.H.D.M.; Feenstra, I.; Westphal, J.R.; Ramos, L.; van Golde, R.J.T.; Kremer, J.A.M. Globozoospermia revisited. Hum. Reprod. Update 2006, 13, 63–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Braekeleer, M.; Nguyen, M.H.; Morel, F.; Perrin, A. Genetic aspects of monomorphic teratozoospermia: A review. J. Assist. Reprod. Genet. 2015, 32, 615–623. [Google Scholar] [CrossRef] [Green Version]
- Liao, T.T.; Xiang, Z.; Zhu, W.B.; Fan, L.Q. Proteome analysis of round-headed and normal spermatozoa by 2-D fluorescence difference gel electrophoresis and mass spectrometry. Asian J. Androl. 2009, 11, 683–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez Sedó, C.; Rawe, V.Y.; Chemes, H.E. Acrosomal biogenesis in human globozoospermia: Immunocytochemical, ultrastructural and proteomic studies. Hum. Reprod. 2012, 27, 1912–1921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kupis, Ł.; Dobroński, P.A.; Radziszewski, P. Varicocele as a source of male infertility—current treatment techniques. Cent. Eur. J. Urol. 2015, 68, 365–370. [Google Scholar] [CrossRef] [Green Version]
- Hosseinifar, H.; Gourabi, H.; Salekdeh, G.H.; Alikhani, M.; Mirshahvaladi, S.; Sabbaghian, M.; Modarresi, T.; Gilani, M.A.S. Study of Sperm Protein Profile in Men With and Without Varicocele Using Two-Dimensional Gel Electrophoresis. Urology 2013, 81, 293–300. [Google Scholar] [CrossRef]
- Chan, C.-C.; Sun, G.-H.; Shui, H.-A.; Wu, G.-J. Differential Spermatozoal Protein Expression Profiles in Men With Varicocele Compared to Control Subjects: Upregulation of Heat Shock Proteins 70 and 90 in Varicocele. Urology 2013, 81, 1379.e1–1379.e8. [Google Scholar] [CrossRef]
- Agarwal, A.; Sharma, R.; Durairajanayagam, D.; Ayaz, A.; Cui, Z.; Willard, B.; Gopalan, B.; Sabanegh, E. Major protein alterations in spermatozoa from infertile men with unilateral varicocele. Reprod. Biol. Endocrinol. 2015, 13, 8. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, A.; Sharma, R.; Durairajanayagam, D.; Cui, Z.; Ayaz, A.; Gupta, S.; Willard, B.; Gopalan, B.; Sabanegh, E. Differential proteomic profiling of spermatozoal proteins of infertile men with unilateral or bilateral varicocele. Urology 2015, 85, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Sharma, R.; Durairajanayagam, D.; Cui, Z.; Ayaz, A.; Gupta, S.; Willard, B.; Gopalan, B.; Sabanegh, E. Spermatozoa protein alterations in infertile men with bilateral varicocele. Asian J. Androl. 2016, 18, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Sharma, R.; Samanta, L.; Durairajanayagam, D.; Sabanegh, E. Proteomic signatures of infertile men with clinical varicocele and their validation studies reveal mitochondrial dysfunction leading to infertility. Asian J. Androl. 2016, 18, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Tvrda, E.; Sharma, R.; Gupta, S.; Ahmad, G.; Sabanegh, E.S. Spermatozoa protein profiles in cryobanked semen samples from testicular cancer patients before treatment. Fertil. Steril. 2015, 104, e260. [Google Scholar] [CrossRef]
- Dias, T.R.; Agarwal, A.; Pushparaj, P.N.; Ahmad, G.; Sharma, R. Reduced semen quality in patients with testicular cancer seminoma is associated with alterations in the expression of sperm proteins. Asian J. Androl. 2020, 22, 88. [Google Scholar]
- Sheehan, M.M.; Ramasamy, R.; Lamb, D.J. Molecular mechanisms involved in varicocele-associated infertility. J. Assist. Reprod. Genet. 2014, 31, 521–526. [Google Scholar] [CrossRef] [Green Version]
- Scieglinska, D.; Krawczyk, Z. Expression, function, and regulation of the testis-enriched heat shock HSPA2 gene in rodents and humans. Cell Stress Chaperones 2015, 20, 221–235. [Google Scholar] [CrossRef] [Green Version]
- Lima, S.B.; Cenedeze, M.A.; Bertolla, R.P.; Filho, P.A.H.; Oehninger, S.; Cedenho, A.P. Expression of the HSPA2 gene in ejaculated spermatozoa from adolescents with and without varicocele. Fertil. Steril. 2006, 86, 1659–1663. [Google Scholar] [CrossRef]
- Yeşilli, Ç.; Mungan, G.; Seçkiner, I.; Akduman, B.; Açikgöz, Ş.; Altan, K.; Mungan, A. Effect of varicocelectomy on sperm creatine kinase, HspA2 chaperone protein (creatine kinase-M type), LDH, LDH-X, and lipid peroxidation product levels in infertile men with varicocele. Urology 2005, 66, 610–615. [Google Scholar] [CrossRef]
- Redgrove, K.A.; Nixon, B.; Baker, M.A.; Hetherington, L.; Baker, G.; Liu, D.Y.; Aitken, R.J. The molecular chaperone HSPA2 plays a key role in regulating the expression of sperm surface receptors that mediate sperm-egg recognition. PLoS ONE 2012, 7, e50851. [Google Scholar] [CrossRef] [Green Version]
- Dix, D.J.; Allen, J.W.; Collins, B.W.; Mori, C.; Nakamura, N.; Poorman-Allen, P.; Goulding, E.H.; Eddy, E.M. Targeted gene disruption of Hsp70-2 results in failed meiosis, germ cell apoptosis, and male infertility. Proc. Natl. Acad. Sci. USA 1996, 93, 3264–3268. [Google Scholar] [CrossRef] [Green Version]
- Society, A.C. Available online: https://www.cancer.org/cancer/testicular-cancer/about/key-statistics.html (accessed on 20 February 2019).
- Rives, N.; Perdrix, A.; Hennebicq, S.; Saïas-Magnan, J.; Melin, M.C.; Berthaut, I.; Barthélémy, C.; Daudin, M.; Szerman, E.; Bresson, J.L. The semen quality of 1158 men with testicular cancer at the time of cryopreservation: Results of the French National CECOS Network. J. Androl. 2012, 33, 1394–1401. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, U.; Junker, H.; Krämer, F.; Balabanov, S.; Kleist, B.; Kammer, W.; Nordheim, A.; Walther, R. Comparative proteomic analysis of neoplastic and non-neoplastic germ cell tissue. Biol. Chem. 2006, 387, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Leman, E.S.; Magheli, A.; Yong, K.M.A.; Netto, G.; Hinz, S.; Getzenberg, R.H. Identification of nuclear structural protein alterations associated with seminomas. J. Cell. Biochem. 2009, 108, 1274–1279. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Hu, Z.; Qi, L.; Wang, J.; Zhou, T.; Guo, Y.; Zeng, Y.; Zheng, B.; Wu, Y.; Zhang, P.; et al. Scanning of novel cancer/testis proteins by human testis proteomic analysis. Proteomics 2013, 13, 1200–1210. [Google Scholar] [CrossRef] [PubMed]
- Milardi, D.; Grande, G.; Vincenzoni, F.; Pierconti, F.; Pontecorvi, A. Proteomics for the Identification of Biomarkers in Testicular Cancer-Review. Front. Endocrinol. 2019, 10, 462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, A.D.; Agarwal, A.; Baskaran, S.; Pushparaj, P.N.; Ahmad, G.; Selvam, M.K.P. Alterations of Spermatozoa Proteomic Profile in Men with Hodgkin’s Disease Prior to Cancer Therapy. World J. Mens Health 2019, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Hu, H.; Wang, Z.; Chen, X.; Yang, F.; Zhu, Z.; Fang, P.; Dai, J.; Wang, L.; Shi, H.; et al. Proteomic characteristics of spermatozoa in normozoospermic patients with infertility. J. Proteom. 2012, 75, 5426–5436. [Google Scholar] [CrossRef]
- Pixton, K.L.; Deeks, E.D.; Flesch, F.M.; Moseley, F.L.C.; Björndahl, L.; Ashton, P.R.; Barratt, C.L.R.; Brewis, I.A. Sperm proteome mapping of a patient who experienced failed fertilization at IVF reveals altered expression of at least 20 proteins compared with fertile donors: Case report. Hum. Reprod. 2004, 19, 1438–1447. [Google Scholar] [CrossRef] [Green Version]
- Frapsauce, C.; Pionneau, C.; Bouley, J.; Delarouziere, V.; Berthaut, I.; Ravel, C.; Antoine, J.-M.; Soubrier, F.; Mandelbaum, J. Proteomic identification of target proteins in normal but nonfertilizing sperm. Fertil. Steril. 2014, 102, 372–380. [Google Scholar] [CrossRef]
- Bracke, A.; Peeters, K.; Punjabi, U.; Hoogewijs, D.; Dewilde, S. A search for molecular mechanisms underlying male idiopathic infertility. Reprod. Biomed. Online 2018, 36, 327–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azpiazu, R.; Amaral, A.; Castillo, J.; Estanyol, J.M.; Guimerà, M.; Ballescà, J.L.; Balasch, J.; Oliva, R. High-throughput sperm differential proteomics suggests that epigenetic alterations contribute to failed assisted reproduction. Hum. Reprod. 2014, 29, 1225–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Légaré, C.; Droit, A.; Fournier, F.; Bourassa, S.; Force, A.; Cloutier, F.; Tremblay, R.; Sullivan, R. Investigation of Male Infertility Using Quantitative Comparative Proteomics. J. Proteome Res. 2014, 13, 5403–5414. [Google Scholar] [CrossRef] [PubMed]
- McReynolds, S.; Dzieciatkowska, M.; Stevens, J.; Hansen, K.C.; Schoolcraft, W.B.; Katz-Jaffe, M.G. Toward the identification of a subset of unexplained infertility: A sperm proteomic approach. Fertil. Steril. 2014, 102, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Intasqui, P.; Antoniassi, M.P.; Camargo, M.; Nichi, M.; Carvalho, V.M.; Cardozo, K.H.; Zylbersztejn, D.S.; Bertolla, R.P. Differences in the seminal plasma proteome are associated with oxidative stress levels in men with normal semen parameters. Fertil. Steril. 2015, 104, 292–301. [Google Scholar] [CrossRef]
- Sharma, R.; Agarwal, A.; Mohanty, G.; Hamada, A.J.; Gopalan, B.; Willard, B.; Yadav, S.; du Plessis, S. Proteomic analysis of human spermatozoa proteins with oxidative stress. Reprod. Biol. Endocrinol. 2013, 11, 48. [Google Scholar] [CrossRef] [Green Version]
- Ayaz, A.; Agarwal, A.; Sharma, R.; Kothandaraman, N.; Cakar, Z.; Sikka, S. Proteomic analysis of sperm proteins in infertile men with high levels of reactive oxygen species. Andrologia 2018, 50, e13015. [Google Scholar] [CrossRef]
- Machesky, L.M.; Insall, R.H. Signaling to actin dynamics. J. Cell Biol. 1999, 146, 267–272. [Google Scholar] [CrossRef] [Green Version]
- Cui, Z.; Sharma, R.; Agarwal, A. Proteomic analysis of mature and immature ejaculated spermatozoa from fertile men. Asian J. Androl. 2016, 18, 735–746. [Google Scholar]
- Nixon, B.; Bromfield, E.G.; Dun, M.D.; Redgrove, K.A.; McLaughlin, E.A.; Aitken, R.J. The role of the molecular chaperone heat shock protein A2 (HSPA2) in regulating human sperm-egg recognition. Asian J. Androl. 2015, 17, 568–573. [Google Scholar] [CrossRef]
- Intasqui, P.; Camargo, M.; Antoniassi, M.P.; Cedenho, A.P.; Carvalho, V.M.; Cardozo, K.H.M.; Zylbersztejn, D.S.; Bertolla, R.P. Association between the seminal plasma proteome and sperm functional traits. Fertil. Steril. 2016, 105, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.W.; Manandhar, G.; Yi, Y.J.; Gupta, S.K.; Sutovsky, M.; Odhiambo, J.F.; Powell, M.D.; Miller, D.J.; Sutovsky, P. Sperm proteasomes degrade sperm receptor on the egg zona pellucida during mammalian fertilization. PLoS ONE 2011, 6, e17256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseinifar, H.; Sabbaghian, M.; Nasrabadi, D.; Modarresi, T.; Dizaj, A.V.; Gourabi, H.; Gilani, M.A. Study of the effect of varicocelectomy on sperm proteins expression in patients with varicocele and poor sperm quality by using two-dimensional gel electrophoresis. J. Assist. Reprod. Genet. 2014, 31, 725–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
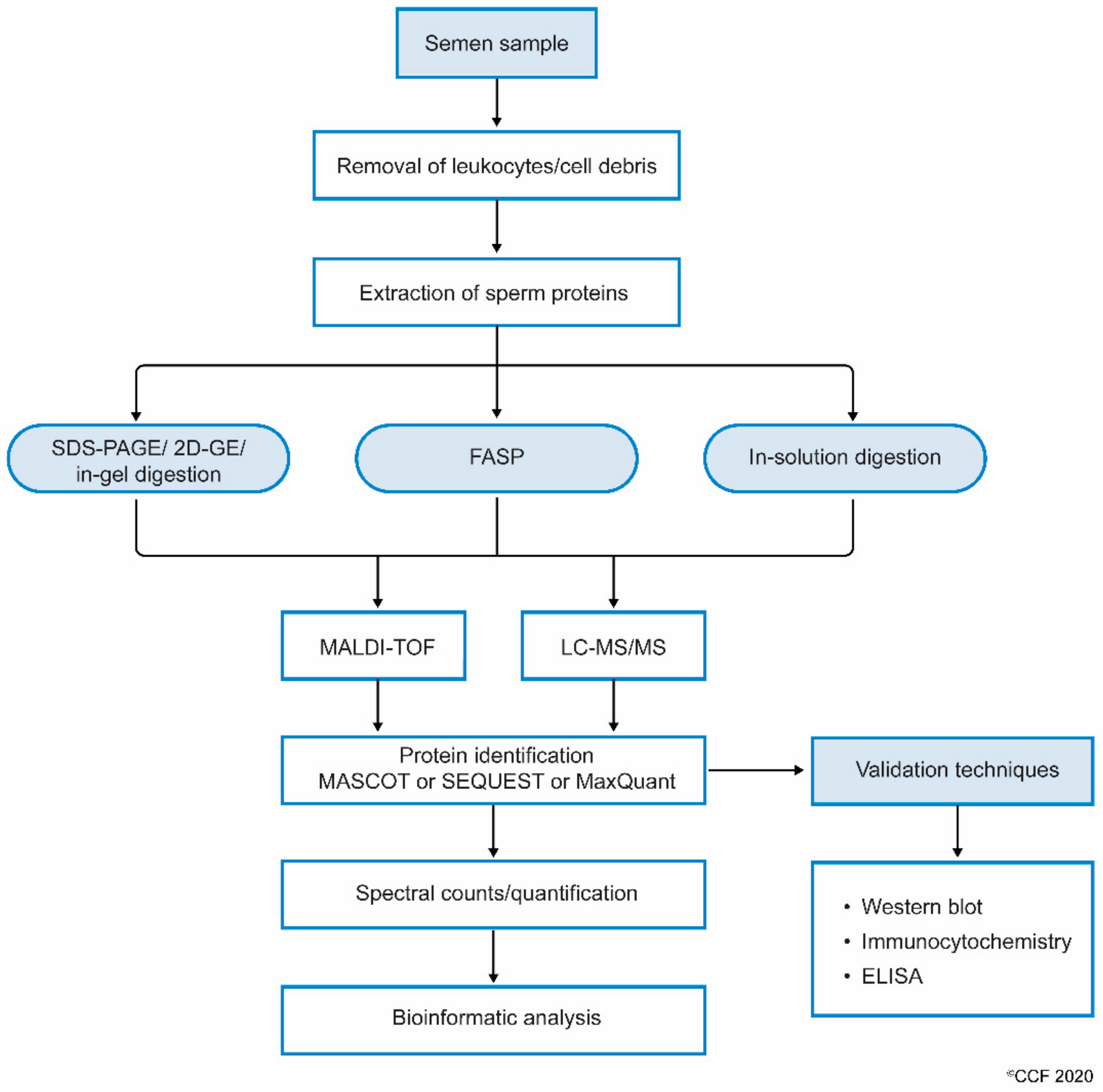
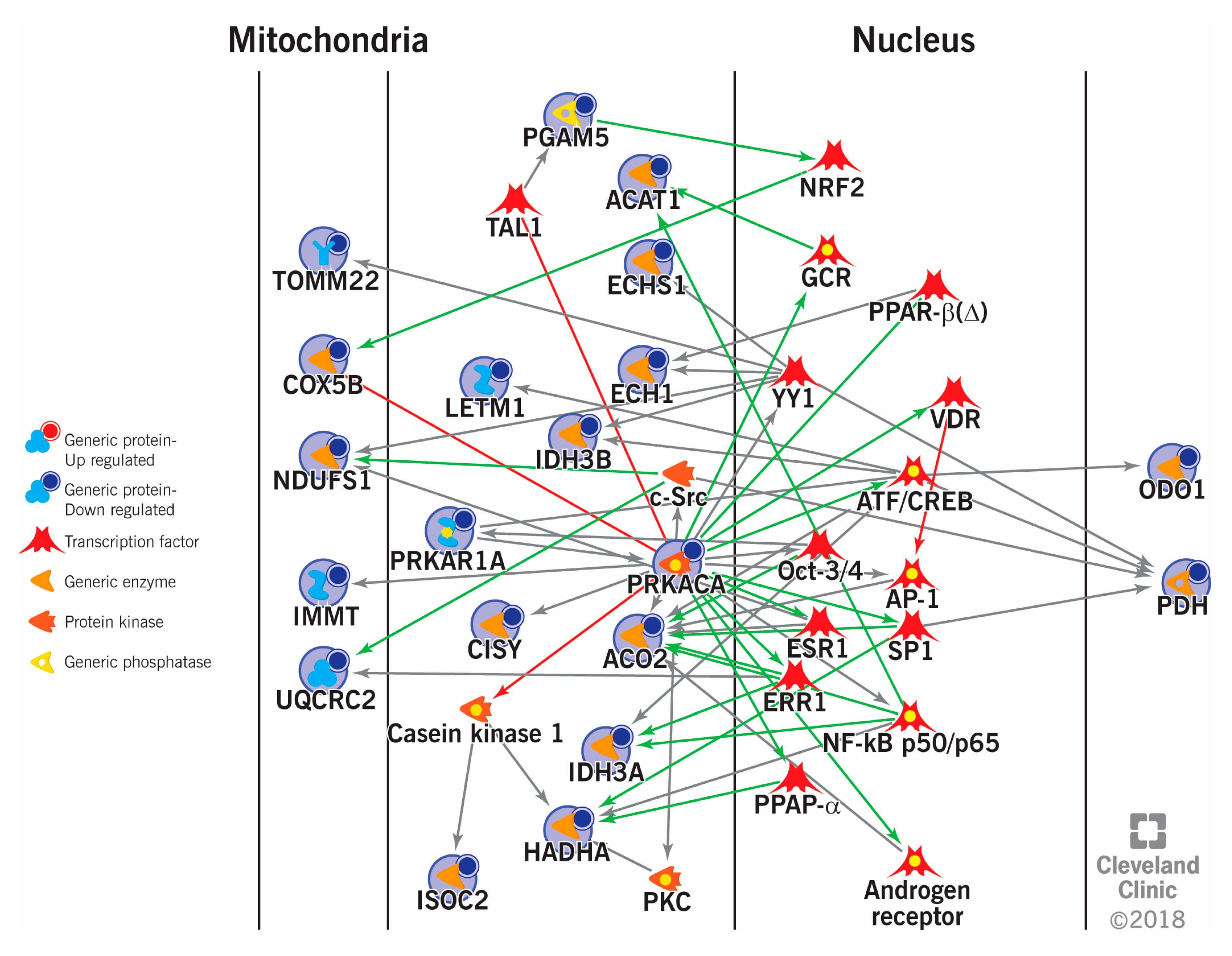
| Clinical Condition | Clinical Scenario | Exclusion/Filtering Criteria | Subjects Enrolled | Samples Used for Proteomics | Method | DEPs | Reference(s) |
|---|---|---|---|---|---|---|---|
| Varicocele | Oligozoospermic patients with varicocele | Systemic illnesses, cryptorchidism, orchitis, epididymitis, urethritis, testicular atrophy, or sexually transmitted diseases, including human immunodeficiency virus. Azoospermia and a sperm concentration <10 million sperm/mL. | 20 | 20 | 2D PAGE MALDI-TOF/TOF-MS | HSPA5, ATP5D, SOD1, ACPP, CLU, PARK7, KLK3, PIP, SEMG2, SEMG2pre | [83] |
| Unilateral varicocele | Systemic illnesses, cryptorchidism, orchitis, epididymitis, urethritis, testicular atrophy, or sexually transmitted diseases, including human immunodeficiency virus, Endtz-positive. Azoospermia and a sperm concentration <10 million sperm/mL. | 33 | Pooled sample (n = 5) | 1D PAGE LC-MS/MS | CABYR, AKAP, APOPA1, SEMG1, ACR, SPA17, RSPH1, RSPH9 DNAH17, DLD, GSTM3, TGM4, NPC23, ODF2GPR64, PSMA8, HIST1H2BA, PARK7 | [85] | |
| Unilateral and bilateral varicocele patients | Endtz-positive. Azoospermia and a sperm concentration <10 million sperm/mL. | Unilateral = 33, bilateral = 17 | Pooled sample (n = 5/each group) | 1D PAGE LC-MS/MS | GSTM3, SPANXB1, PARK7, PSMA8, DLD, SEMG1, SEMG2 | [86] | |
| Bilateral varicocele | Azoospermia and a sperm concentration <10 million sperm/mL. Smoker and abnormal body mass index | 17 | Pooled sample (n = 5) | 1D PAGE LC-MS/MS | ODF2, TEKT3, TCP11, TGM4, CLGN, TOM22, APOA1 | [87] | |
| Varicocele | Azoospermia and a sperm concentration <10 million sperm/mL. | 50 | Pooled sample (n = 10) | 1D PAGE LC-MS/MS | PKAR1A, AK7, CCT6B, HSPA2, ODF2 | [88] | |
| Varicocele | Endtz-positive and sperm concentration less than <10 million sperm/mL. Female factor infertility | 50 | Pooled sample (n = 10) | LC-MS/MS | LETM1, EFHC, MIC60, PGAM5, ISOC2, TOM22, NDFSU1, UQCRC2, COX5B, ATPase1A4, HSPA2, SPA17, APOA1 | [18] | |
| Testicular cancer | Testicular cancer | NA | 16 | 16 | 1D PAGE LC-MS/MS | PSA, PAcP, ZAG, SEMG 1 and 2, AKAP4, DNAH17 | [89] |
| Non-seminoma testicular cancer | NA | 15 | Pooled sample (n = 3) | 1D PAGE LC-MS/MS | NDUFS1, UQCRC2, ATP1A4, ANXA2, ATP1A2, ACR | [25] | |
| Normozoospermic and asthenozoosperic testicular cancer | NA | Normozoospermic testicular cancer = 20, | Pooled sample (n = 20) | 1D PAGE LC-MS/MS | CCT3, ATP5A1, UQCRC2, ATP1A4, MMP9 | [16] | |
| asthenozoosperic testicular cancer = 20 | Pooled sample (n = 11) | ||||||
| Normozoospermic and asthenozoosperic testicular cancer | NA | Normozoospermic testicular cancer = 20, | Pooled sample (n = 20) | 1D PAGE LC-MS/MS | NDUFS1, CD63 | [17] | |
| asthenozoosperic testicular cancer = 20 | Pooled sample (n = 11) | ||||||
| Testicular cancer seminoma | NA | 15 | Pooled sample (n = 3) | 1D PAGE LC-MS/MS | HSPA2, ATP1A4, UQCRC2, ACE | [90] | |
| Asthenozoospermia | Rapid motility (grade a) of 0–3% and progressive motility (grade a+b) of 5–20% | NA | 8 | 8 | 2-DE MALDI-TOF MS | IDH-α, ODF, SEMG1, ARHGDIB, GOT1, PGAM2, TPI1, CA2, GS10, MSS1 | [69] |
| Progressive motility <25% (grade a) or motile sperm <50% (grades a + b) | NA | 20 | 20 | 2D PAGE MS | ACTB, ANXA5, COX6B, H2A, PIP, PIPpre, S100A9, CLUpre, DLDpre, FHpre, HSPA2, IMPA1, MPST/ECH1pre, PSMB3, SEMG1pre, TEX12 | [68] | |
| Rapid linear progression <25% (Grade a) | Sexually transmitted diseases including human immunodeficiency virus (HIV), | 17 | Pooled sample (n = 5) | 2D PAGE MALDI MS/MS | TPIS, PSMA3, GKP2, HSPA2, OXCT1, TUBB2C, TEKT1 | [67] | |
| Progressive motility <10% | History of long term medication, varicocele and leukocytospermia. Hyperviscous and necrozoospermic samples, viability <70%. | 4 | Nano UPLC–MSE tandem mass spectrometry | GRP78, GAPDHS, HSP70-2, TUBA4A, TUBA3C, TUBA8, ODF1, AKAP3, AKAP4, ROPN1B, SPANXB, CLU, PIP, ATP5B, ALDOA, ARGDIA | [63] | ||
| Rapid progressive and slow progressive motility ≤30% | History of depression, diabetes, cancer, hypertension, hyperthyroidism, or sexually transmitted diseases. Exposed to environmental stress, including radiation or chemicals, smokers, and with abnormal body mass index. | 35 | 35 | 2-DE MALDI-TOF/TOF MS | UBB2B, ODF2, AKAP4, KRT1, CLU, COX6B, GAPDS, PHGPx, HSPA2, HSPA9, VDAC2, GSTMu3, ASRGL1, SPANXB | [70] | |
| Sperm motility <40% | Endtz-positive. | 10 | 10 | UPLC-MS | PLXNB2, POTEKP, NIN, PHF3, DYNLL1, PROCA1, FASCIN-3; LRRC37B, PLC | [64] | |
| Sperm motility <40% | Oligozoospermia, teratozoospermia or leukocytospermia | 4 | 4 | 2-DE MALDI-TOF MS | LFT, ATP5B, DJ-1, PARK7, ODF, TEKT1, AKAP4, ELSPBP1, PDHB, NDUS1, SUCLA2, SDHA | [65] | |
| progressive sperm motility ≤32% | HIV positive samples and sexually transmitted diseases. Samples contaminated with blood | 70 | Pooled sample (n = 5) | 2D-DIGE MALDI -TOF-MS | TEX40, ATP6V0A2, SERPINB9, PSA | [66] | |
| Globozoospermia | Round-headed acrosomeless sperm | NA | 1 | 1 | 2D DIGE MALDI-TOF/TOF MS/MS | SAMP1, ODF2, SPANXa/d, TUBA2, TPI1, PIP | [80] |
| Molecular Pathways | DEPs | Reference |
|---|---|---|
| Oxidative stress | HIST1H2BA, MDH2, TGM4, GPX4, GLUL, HSP90B1, HSPA5, ACE, HSPA2, RPS27A, MAP3K3 and APP, PRDX1, AKAP4 | [23,112,113] |
| Energy and metabolism and Mitochondrial dysfunction | PKAR1A, AK7, CCT6B, HSPA2, ODF2, DLD, ATP5D, NDUFS1, UQCRC2, COX5B, PDH, PHGPx, VDAC, COX6B, AKAP4 | [18,70,88,119] |
| Cytoskeleton integrity | ACTB, KRT1, ODF2, TEK1, TEK4, TEK5, TUBB2B, ACTB | [69,70,107] |
| Protein folding and degradation | HSPA2, CLU, PSMB4, PSMB5, PSMB6, PSMA3 | [36,90,107] |
| Spermatogenesis and sperm function | Importin, Exportin, HSP 70, AKAPs, HSPA2, | [115,116] |
| Sperm DNA damage | CRISPLD2, CRISPLD2, RARRES1 | [117] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agarwal, A.; Panner Selvam, M.K.; Baskaran, S. Proteomic Analyses of Human Sperm Cells: Understanding the Role of Proteins and Molecular Pathways Affecting Male Reproductive Health. Int. J. Mol. Sci. 2020, 21, 1621. https://doi.org/10.3390/ijms21051621
Agarwal A, Panner Selvam MK, Baskaran S. Proteomic Analyses of Human Sperm Cells: Understanding the Role of Proteins and Molecular Pathways Affecting Male Reproductive Health. International Journal of Molecular Sciences. 2020; 21(5):1621. https://doi.org/10.3390/ijms21051621
Chicago/Turabian StyleAgarwal, Ashok, Manesh Kumar Panner Selvam, and Saradha Baskaran. 2020. "Proteomic Analyses of Human Sperm Cells: Understanding the Role of Proteins and Molecular Pathways Affecting Male Reproductive Health" International Journal of Molecular Sciences 21, no. 5: 1621. https://doi.org/10.3390/ijms21051621
APA StyleAgarwal, A., Panner Selvam, M. K., & Baskaran, S. (2020). Proteomic Analyses of Human Sperm Cells: Understanding the Role of Proteins and Molecular Pathways Affecting Male Reproductive Health. International Journal of Molecular Sciences, 21(5), 1621. https://doi.org/10.3390/ijms21051621






