Analysis and Identification of Tumorigenic Targets of MicroRNA in Cancer Cells by Photoreactive Chemical Probes
Abstract
1. Introduction
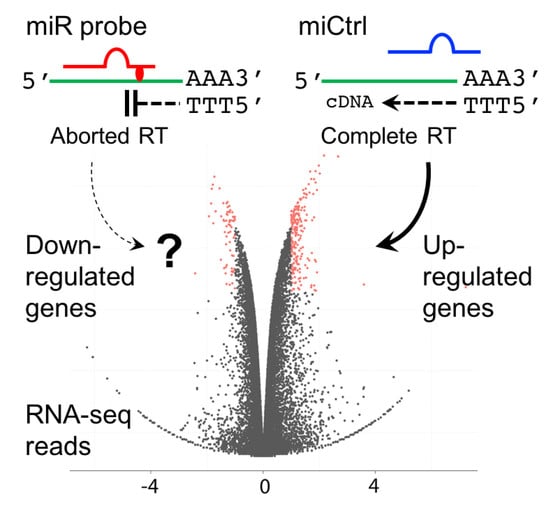
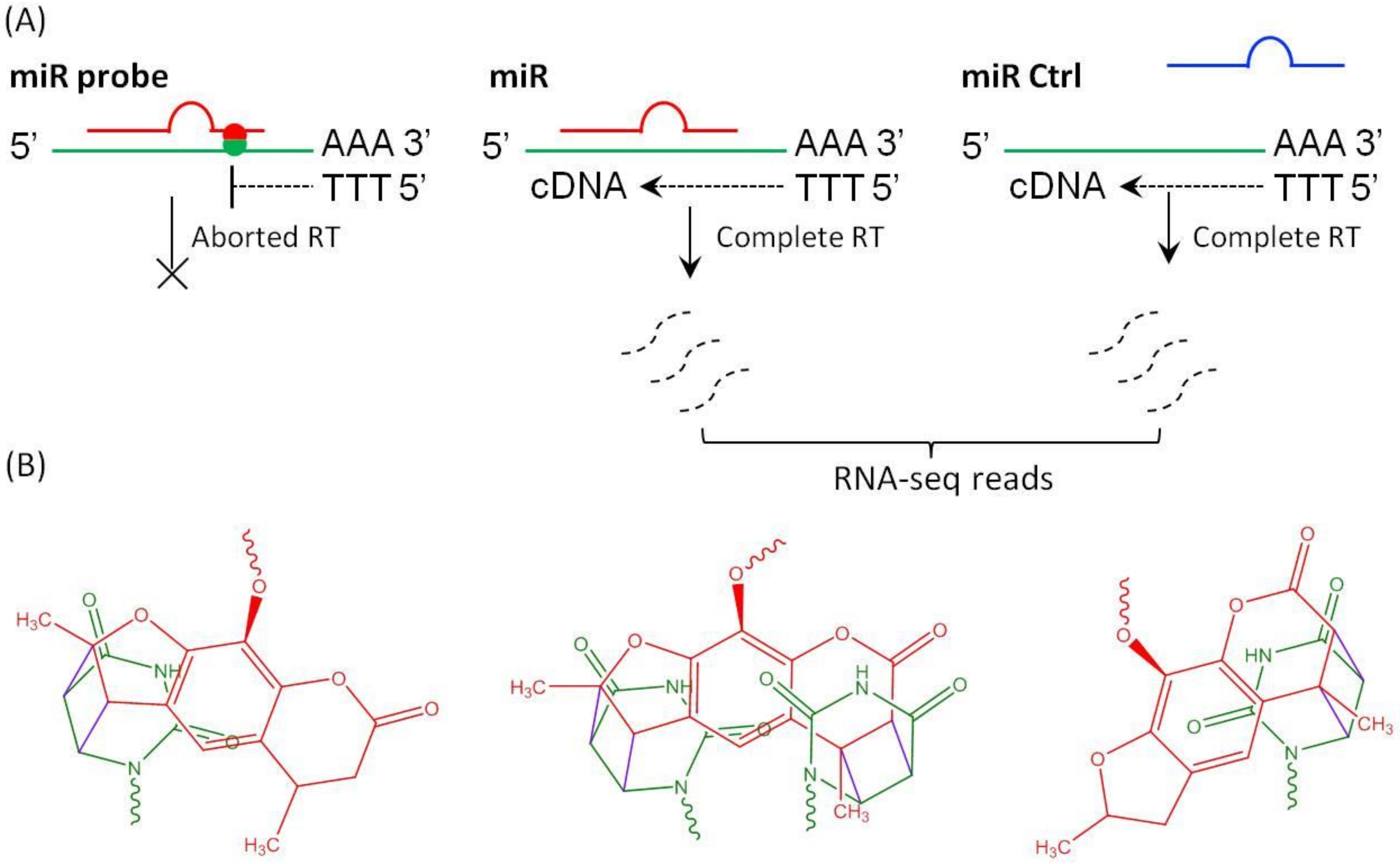
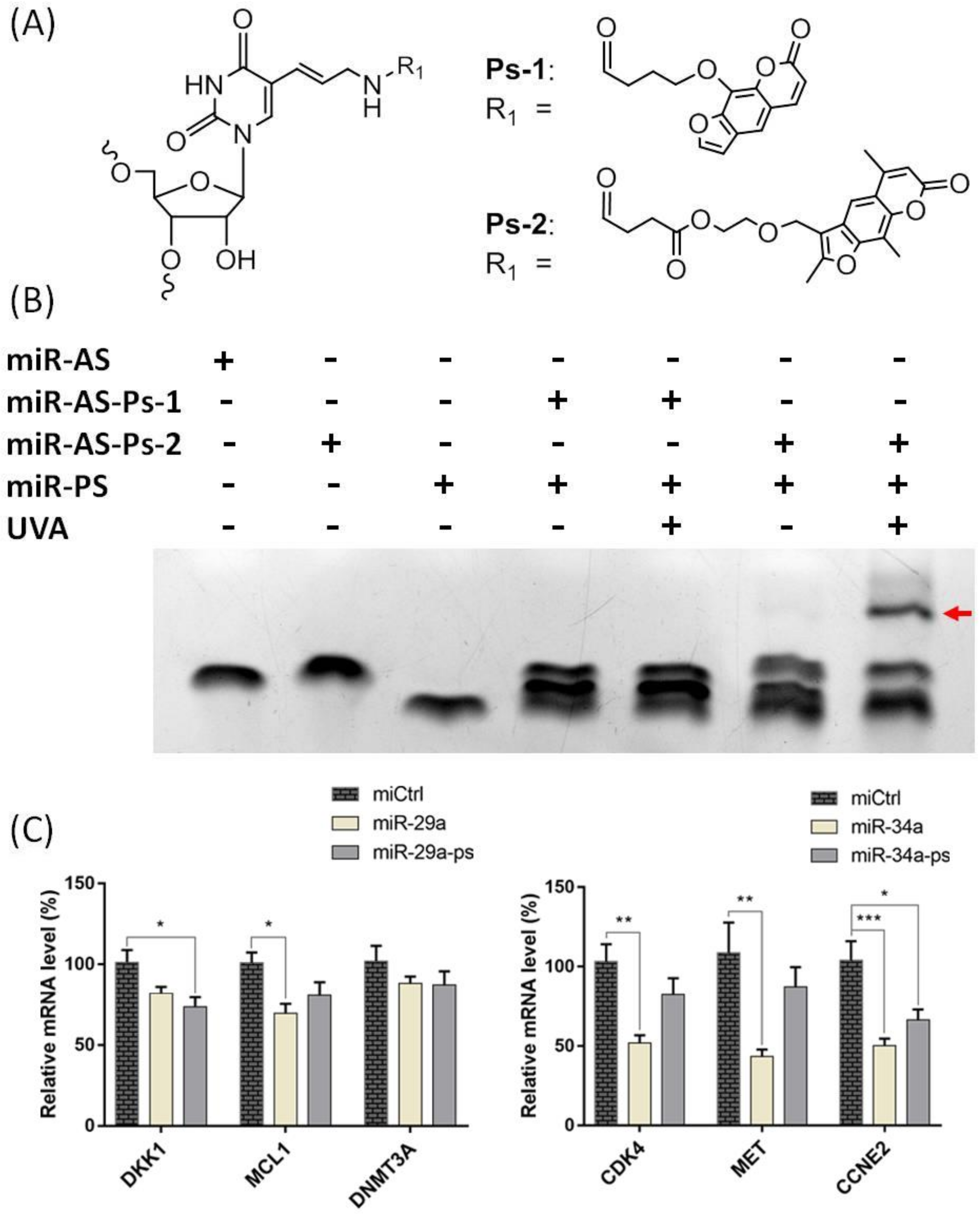
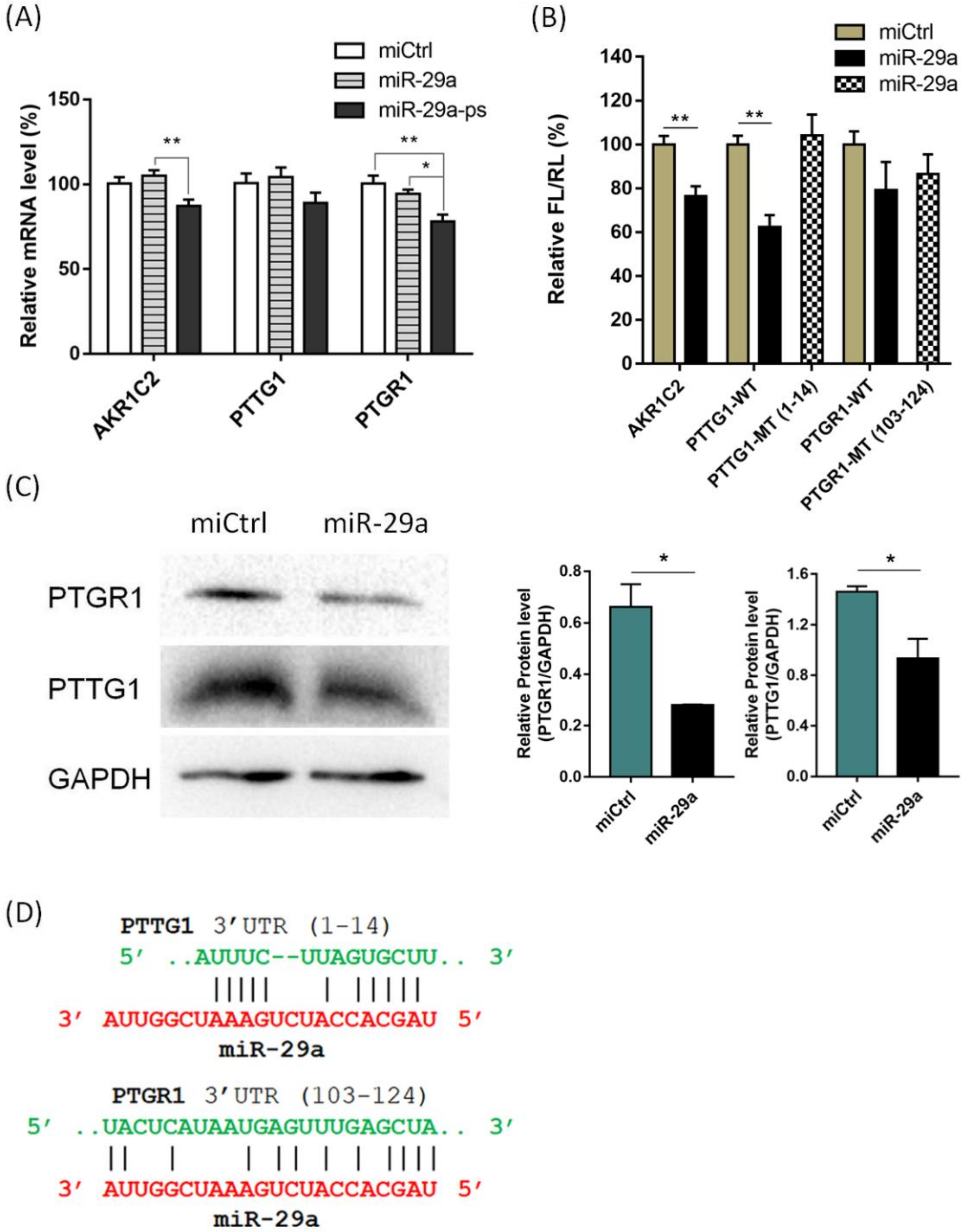
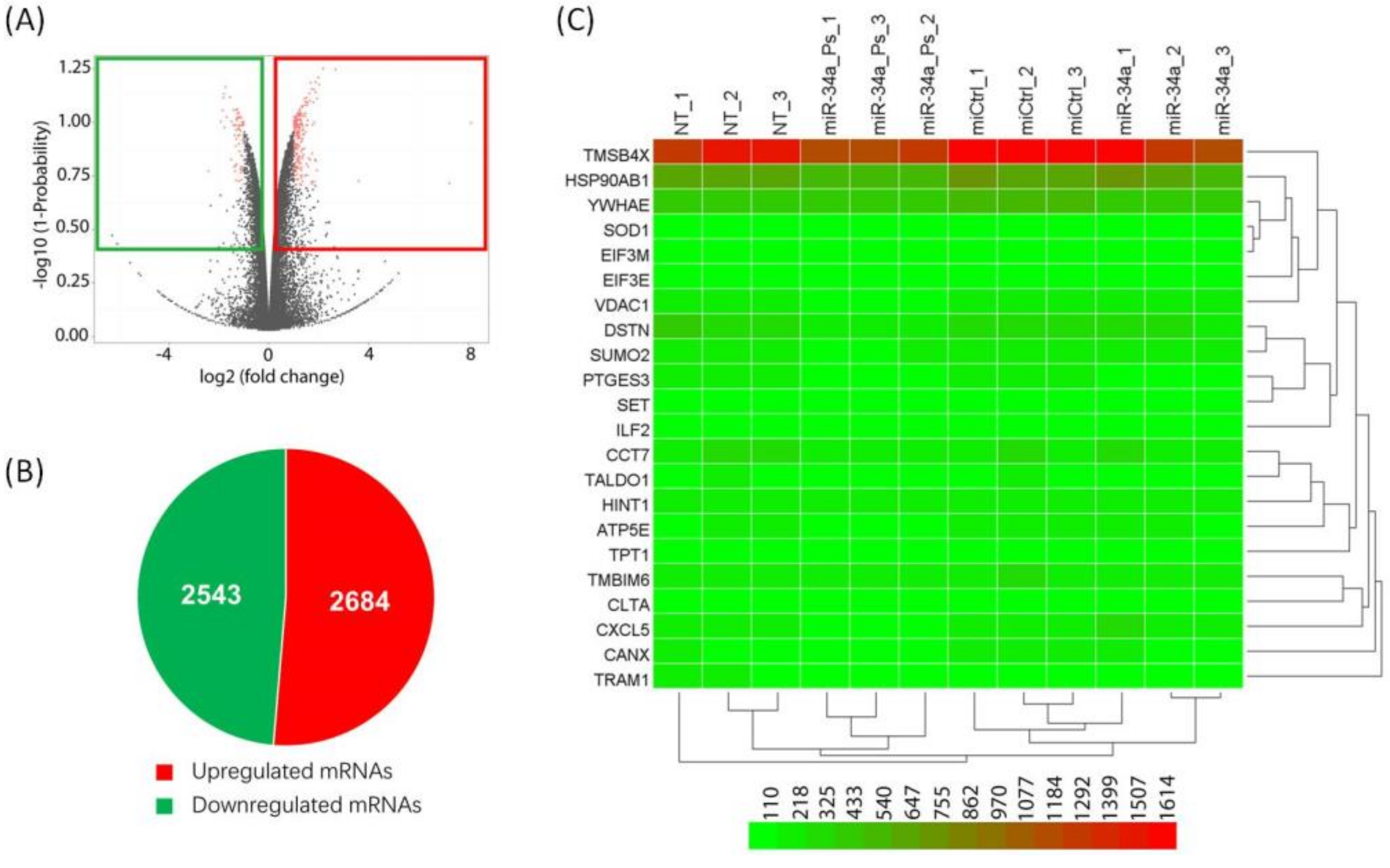
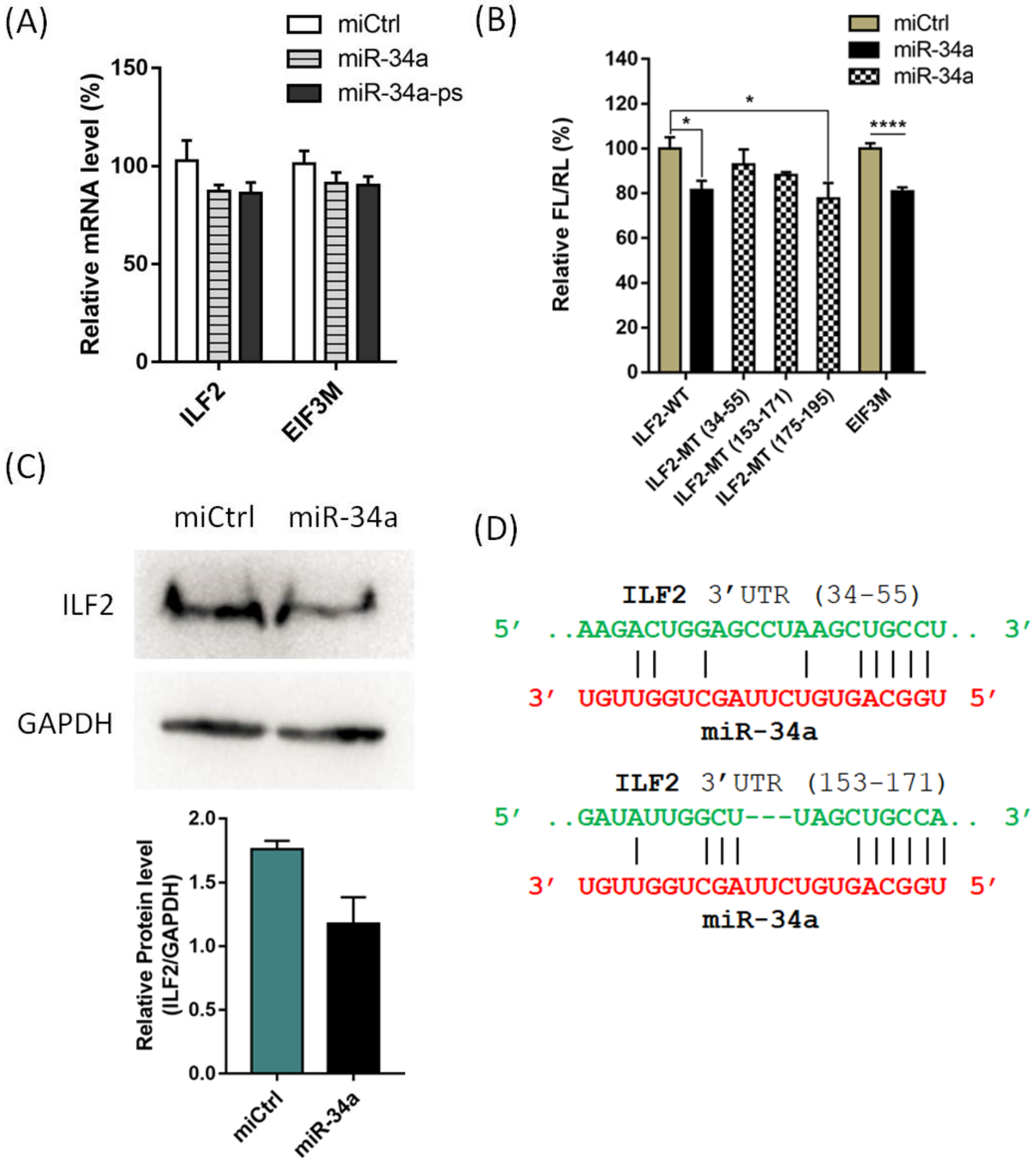
2. Results and Discussion
3. Materials and Methods
3.1. Preparation of Psoralen Modified miRNA Probes
3.2. MiR Probe-Mediated Crosslinking and RNA-Seq Analysis
3.3. Dual Luciferase Assay
3.4. Western Blotting Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef] [PubMed]
- Krek, A.; Grun, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Kiriakidou, M.; Nelson, P.T.; Kouranov, A.; Fitziev, P.; Bouyioukos, C.; Mourelatos, Z.; Hatzigeorgiou, A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004, 18, 1165–1178. [Google Scholar] [CrossRef]
- Kruger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Li, J.; Huang, L.; Xiao, X.; Chen, Y.; Wang, X.; Zhou, Z.; Zhang, C.; Zhang, Y. Photoclickable MicroRNA for the Intracellular Target Identification of MicroRNAs. J. Am. Chem. Soc. 2016, 138, 15943–15949. [Google Scholar] [CrossRef]
- Nakamoto, K.; Minami, K.; Akao, Y.; Ueno, Y. Labeling of target mRNAs using a photo-reactive microRNA probe. Chem. Commun. (Camb.) 2016, 52, 6720–6722. [Google Scholar] [CrossRef]
- Ule, J.; Jensen, K.B.; Ruggiu, M.; Mele, A.; Ule, A.; Darnell, R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science 2003, 302, 1212–1215. [Google Scholar] [CrossRef] [PubMed]
- Ule, J.; Jensen, K.; Mele, A.; Darnell, R.B. CLIP: A method for identifying protein-RNA interaction sites in living cells. Methods 2005, 37, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Chi, S.W.; Zang, J.B.; Mele, A.; Darnell, R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009, 460, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M., Jr.; Jungkamp, A.C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Lianoglou, S.; Tuschl, T.; Betel, D. Genome-wide identification of miRNA targets by PAR-CLIP. Methods 2012, 58, 94–105. [Google Scholar] [CrossRef]
- Kudla, G.; Granneman, S.; Hahn, D.; Beggs, J.D.; Tollervey, D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 10010–10015. [Google Scholar] [CrossRef]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef]
- Song, Y.; Liu, K.J.; Wang, T.H. Elimination of ligation dependent artifacts in T4 RNA ligase to achieve high efficiency and low bias microRNA capture. PLoS ONE 2014, 9, e94619. [Google Scholar] [CrossRef]
- Baigude, H.; Li, Z.; Zhou, Y.; Rana, T.M. miR-TRAP: A benchtop chemical biology strategy to identify microRNA targets. Angew. Chem. Int. Ed. Engl. 2012, 51, 5880–5883. [Google Scholar] [CrossRef]
- Youvan, D.C.; Hearst, J.E. Sequencing psoralen photochemically reactive sites in Escherichia coli 16 S rRNA. Anal. Biochem. 1982, 119, 86–89. [Google Scholar] [CrossRef]
- Ericson, G.; Wollenzien, P. Use of reverse transcription to determine the exact locations of psoralen photochemical crosslinks in RNA. Anal. Biochem. 1988, 174, 215–223. [Google Scholar] [CrossRef]
- Teare, J.; Wollenzien, P. Specificity of site directed psoralen addition to RNA. Nucleic Acids Res. 1989, 17, 3359–3372. [Google Scholar] [CrossRef]
- Watkins, K.P.; Dungan, J.M.; Agabian, N. Identification of a small RNA that interacts with the 5′ splice site of the trypanosoma brucei spliced leader RNA in vivo. Cell 1994, 76, 171–182. [Google Scholar] [CrossRef]
- Wang, Z.; Rana, T.M. Chemical Conversion of a trans-Activation Responsive RNA-Binding Fragment of HIV-1 Tat Protein into a Site-Specific Crosslinking Agent. J. Am. Chem. Soc. 1995, 117, 5438–5444. [Google Scholar] [CrossRef]
- Ping, Y.H.; Rana, T.M. Mechanism of site-specific psoralen photoadducts formation in triplex DNA directed by psoralen-conjugated oligonucleotides. Biochemistry 2005, 44, 2501–2509. [Google Scholar] [CrossRef] [PubMed]
- Bibaki, E.; Tsitoura, E.; Vasarmidi, E.; Margaritopoulos, G.; Trachalaki, A.; Koutoulaki, C.; Georgopoulou, T.; Spandidos, D.A.; Tzanakis, N.; Antoniou, K.M. miR-185 and miR-29a are similarly expressed in the bronchoalveolar lavage cells in IPF and lung cancer but common targets DNMT1 and COL1A1 show disease specific patterns. Mol. Med. Rep. 2018, 17, 7105–7112. [Google Scholar] [CrossRef]
- Kamikawaji, K.; Seki, N.; Watanabe, M.; Mataki, H.; Kumamoto, T.; Takagi, K.; Mizuno, K.; Inoue, H. Regulation of LOXL2 and SERPINH1 by antitumor microRNA-29a in lung cancer with idiopathic pulmonary fibrosis. J. Hum. Genet. 2016, 61, 985–993. [Google Scholar] [CrossRef]
- Sherman, E.J.; Mitchell, D.C.; Garner, A.L. The RNA-binding protein SART3 promotes miR-34a biogenesis and G1 cell cycle arrest in lung cancer cells. J. Biol. Chem. 2019, 294, 17188–17196. [Google Scholar] [CrossRef]
- Cortez, M.A.; Valdecanas, D.; Niknam, S.; Peltier, H.J.; Diao, L.; Giri, U.; Komaki, R.; Calin, G.A.; Gomez, D.R.; Chang, J.Y.; et al. In Vivo Delivery of miR-34a Sensitizes Lung Tumors to Radiation Through RAD51 Regulation. Mol. Ther. Nucleic Acids 2015, 4, e270. [Google Scholar] [CrossRef]
- Candido, S.; Lupo, G.; Pennisi, M.; Basile, M.S.; Anfuso, C.D.; Petralia, M.C.; Gattuso, G.; Vivarelli, S.; Spandidos, D.A.; Libra, M.; et al. The analysis of miRNA expression profiling datasets reveals inverse microRNA patterns in glioblastoma and Alzheimer’s disease. Oncol. Rep. 2019, 42, 911–922. [Google Scholar] [CrossRef]
- Hafsi, S.; Candido, S.; Maestro, R.; Falzone, L.; Soua, Z.; Bonavida, B.; Spandidos, D.A.; Libra, M. Correlation between the overexpression of Yin Yang 1 and the expression levels of miRNAs in Burkitt’s lymphoma: A computational study. Oncol. Lett. 2016, 11, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Cimino, G.D.; Gamper, H.B.; Isaacs, S.T.; Hearst, J.E. Psoralens as photoactive probes of nucleic acid structure and function: Organic chemistry, photochemistry, and biochemistry. Annu. Rev. Biochem. 1985, 54, 1151–1193. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, P.; Gu, J. miR-29a modulates tumor necrosis factor-alpha-induced osteogenic inhibition by targeting Wnt antagonists. Dev. Growth Differ. 2015, 57, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Osaki, S.; Tazawa, H.; Hasei, J.; Yamakawa, Y.; Omori, T.; Sugiu, K.; Komatsubara, T.; Fujiwara, T.; Sasaki, T.; Kunisada, T.; et al. Ablation of MCL1 expression by virally induced microRNA-29 reverses chemoresistance in human osteosarcomas. Sci. Rep. 2016, 6, 28953. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, X.; Lim, L.P.; de Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef] [PubMed]
- Li, W.H.; Chang, L.; Xia, Y.X.; Wang, L.; Liu, Y.Y.; Wang, Y.H.; Jiang, Z.; Xiao, J.; Wang, Z.R. Knockdown of PTTG1 inhibits the growth and invasion of lung adenocarcinoma cells through regulation of TGFB1/SMAD3 signaling. Int. J. Immunopathol. Pharmacol. 2015, 28, 45–52. [Google Scholar] [CrossRef]
- Li, H.; Yin, C.; Zhang, B.; Sun, Y.; Shi, L.; Liu, N.; Liang, S.; Lu, S.; Liu, Y.; Zhang, J.; et al. PTTG1 promotes migration and invasion of human non-small cell lung cancer cells and is modulated by miR-186. Carcinogenesis 2013, 34, 2145–2155. [Google Scholar] [CrossRef]
- Sanchez-Rodriguez, R.; Torres-Mena, J.E.; De-la-Luz-Cruz, M.; Bernal-Ramos, G.A.; Villa-Trevino, S.; Chagoya-Hazas, V.; Landero-Lopez, L.; Garcia-Roman, R.; Rouimi, P.; Del-Pozo-Yauner, L.; et al. Increased expression of prostaglandin reductase 1 in hepatocellular carcinomas from clinical cases and experimental tumors in rats. Int. J. Biochem. Cell Biol. 2014, 53, 186–194. [Google Scholar] [CrossRef]
- Sanchez-Rodriguez, R.; Torres-Mena, J.E.; Quintanar-Jurado, V.; Chagoya-Hazas, V.; Rojas Del Castillo, E.; Del Pozo Yauner, L.; Villa-Trevino, S.; Perez-Carreon, J.I. Ptgr1 expression is regulated by NRF2 in rat hepatocarcinogenesis and promotes cell proliferation and resistance to oxidative stress. Free Radic. Biol. Med. 2017, 102, 87–99. [Google Scholar] [CrossRef]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Slabakova, E.; Culig, Z.; Remsik, J.; Soucek, K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017, 8, e3100. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Ivan, C.; Valdecanas, D.; Wang, X.; Peltier, H.J.; Ye, Y.; Araujo, L.; Carbone, D.P.; Shilo, K.; Giri, D.K.; et al. PDL1 Regulation by p53 via miR-34. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef]
- Bi, Y.; Shen, W.; Min, M.; Liu, Y. MicroRNA-7 functions as a tumor-suppressor gene by regulating ILF2 in pancreatic carcinoma. Int. J. Mol. Med. 2017, 39, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Sethupathy, P.; Corda, B.; Hatzigeorgiou, A.G. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA 2006, 12, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015, 43, D146–D152. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Zuo, Z.; Cai, G.; Kang, S.; Gao, X.; Li, T. miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009, 37, D105–D110. [Google Scholar] [CrossRef] [PubMed]

| Protocol | PCP-Seq | CLIP [11,12] | HITS-CLIP [13] | PAR-CLIP [14,15] | CLASH [16,17] |
|---|---|---|---|---|---|
| UV radiation | 360 nm | 254 nm | 254 nm | 360 nm | 254 nm |
| Photoactivatable nucleosides | ✓ | ✕ | ✕ | ✓ | ✕ |
| Total RNA isolation | ✓ | ✕ | ✕ | ✕ | ✕ |
| Partial RNase digestion | ✕ | ✓ | ✓ | ✓ | ✓ |
| Creation of recombinant protein | ✕ | ✕ | ✕ | ✕ | ✓ |
| Generation of stable cell line | ✕ | ✕ | ✕ | ✕ | ✓ |
| Recombinant protein purification | ✕ | ✕ | ✕ | ✕ | ✓ |
| Immunoprecipitation | ✕ | ✓ | ✓ | ✓ | ✕ |
| Phosphatase treatment | ✕ | ✓ | ✓ | ✓ | ✓ |
| 3′ or 5′ RNA linker ligation | ✕ | ✓ | ✓ | ✓ | ✓ |
| Radiolabeling of RNA segment | ✕ | ✓ | ✓ | ✓ | ✓ |
| Purification on SDS-PAGE | ✕ | ✓ | ✓ | ✓ | ✓ |
| RNA purification | ✕ | ✓ | ✓ | ✓ | ✓ |
| Reverse transcription | ✓ | ✓ | ✓ | ✓ | ✓ |
| PCR amplification | ✓ | ✓ | ✓ | ✓ | ✓ |
| High-throughput sequencing | ✓ | ✕ | ✓ | ✓ | ✓ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, Z.; Ganbold, T.; Baigude, H. Analysis and Identification of Tumorigenic Targets of MicroRNA in Cancer Cells by Photoreactive Chemical Probes. Int. J. Mol. Sci. 2020, 21, 1545. https://doi.org/10.3390/ijms21041545
Su Z, Ganbold T, Baigude H. Analysis and Identification of Tumorigenic Targets of MicroRNA in Cancer Cells by Photoreactive Chemical Probes. International Journal of Molecular Sciences. 2020; 21(4):1545. https://doi.org/10.3390/ijms21041545
Chicago/Turabian StyleSu, Zhiyu, Tsogzolmaa Ganbold, and Huricha Baigude. 2020. "Analysis and Identification of Tumorigenic Targets of MicroRNA in Cancer Cells by Photoreactive Chemical Probes" International Journal of Molecular Sciences 21, no. 4: 1545. https://doi.org/10.3390/ijms21041545
APA StyleSu, Z., Ganbold, T., & Baigude, H. (2020). Analysis and Identification of Tumorigenic Targets of MicroRNA in Cancer Cells by Photoreactive Chemical Probes. International Journal of Molecular Sciences, 21(4), 1545. https://doi.org/10.3390/ijms21041545