Glioblastoma Cells Do Not Affect Axitinib-Dependent Senescence of HUVECs in a Transwell Coculture Model
Abstract
1. Introduction
2. Results
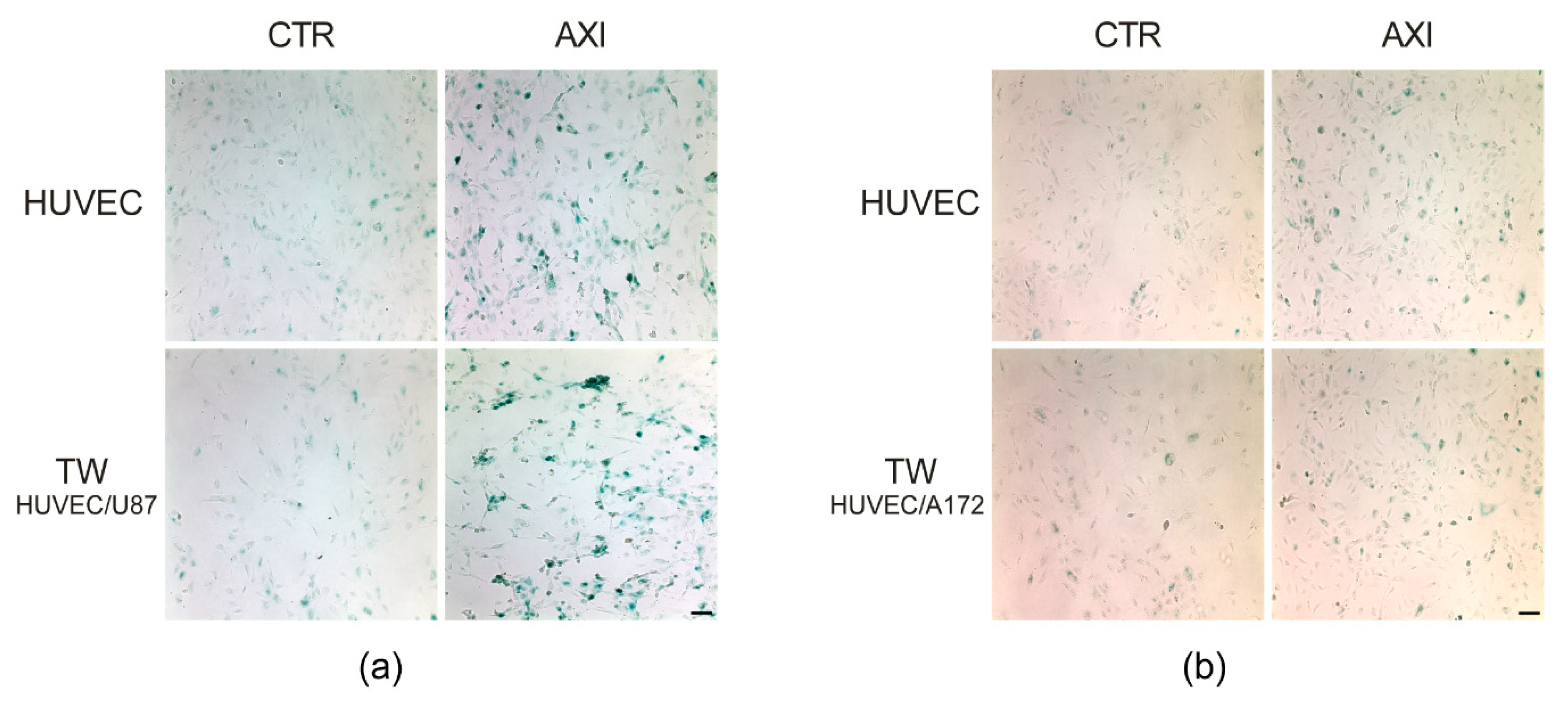
2.1. GBM Tumor Cells Do Not Affect Axitinib-Dependent Expression of SA-β-Galactosidase in HUVECs
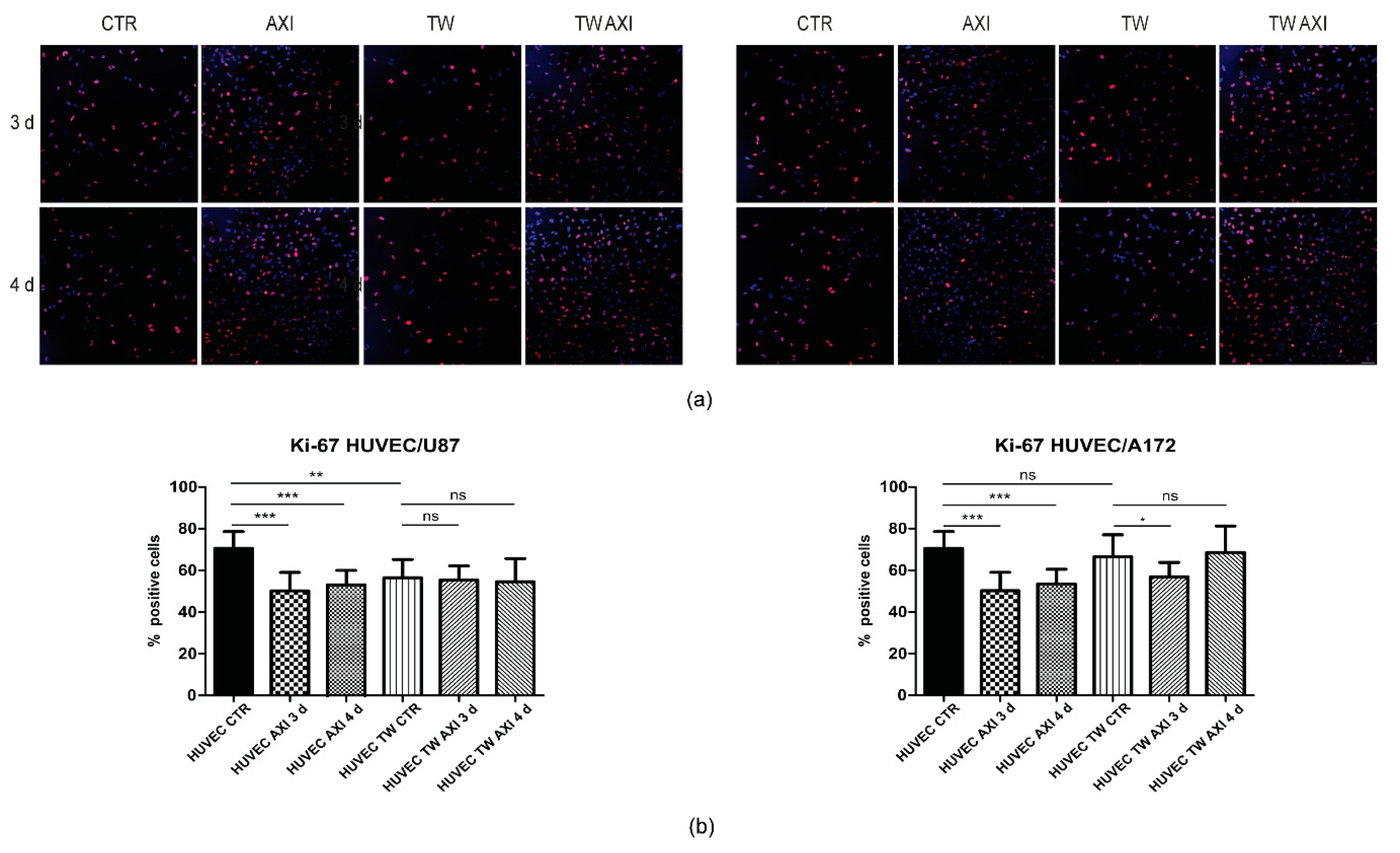
2.2. GBM Tumor Cells Do Not Affect Axitinib-Dependent Ki-67 Expression in HUVECs
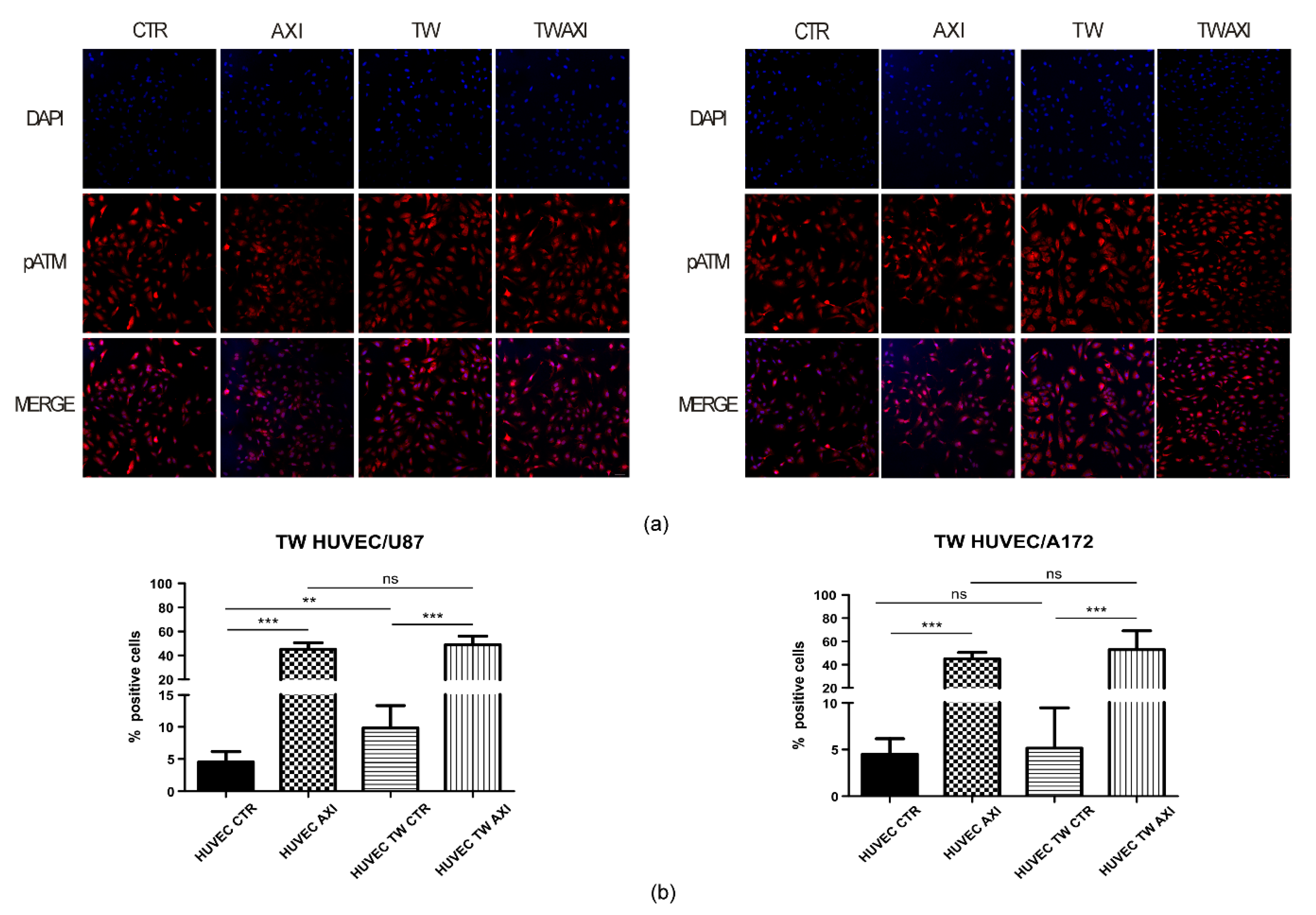
2.3. GBM Tumor Cells Do Not Affect Axitinib-Dependent Activation of ATM in HUVECs
2.4. GBM Tumor Cells Affect Axitinib-Dependent Gene Expression Profile of HUVECs
3. Discussion
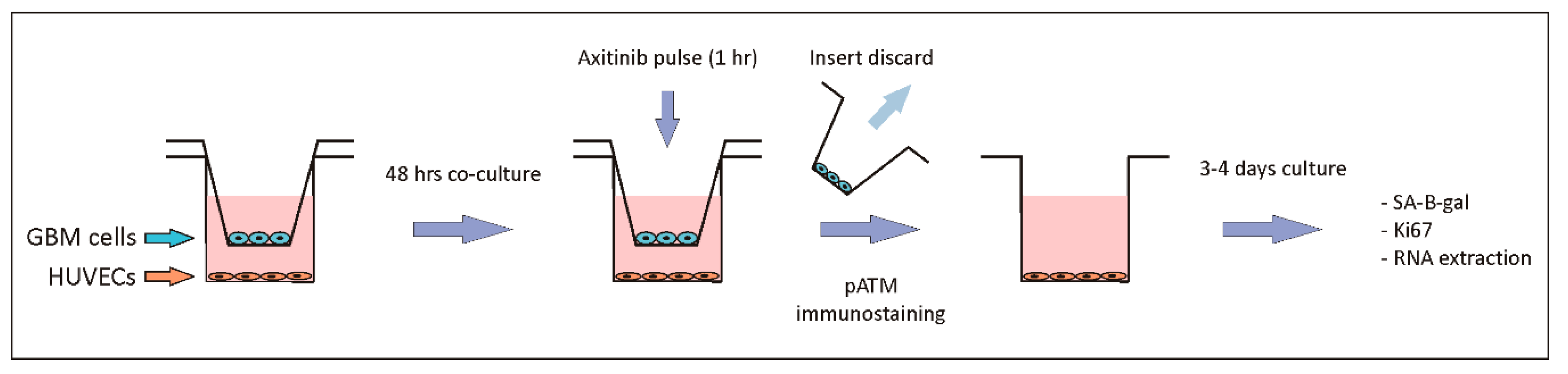
4. Materials and Methods
4.1. Cell Cultures and Chemicals
4.2. SA-β-Galactosidase Assay
4.3. Immunofluorescence
4.4. Reverse Transcription and Real-Time PCR Analyses
4.5. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| VEGFR | Vascular Endothelial Growth Factor Receptor |
| GBM | Glioblastoma |
| HUVECs | Human Umbilical Vein Endothelial Cells |
| ATM | Ataxia Telangectasia Mutated |
References
- Hu-Lowe, D.D.; Zou, H.Y.; Grazzini, M.L.; Hallin, M.E.; Wickman, G.R.; Amundson, K.; Chen, J.H.; Rewolinski, D.A.; Yamazaki, S.; Wu, E.Y.; et al. Nonclinical Antiangiogenesis and Antitumor Activities of Axitinib (AG-013736), an Oral, Potent, and Selective Inhibitor of Vascular Endothelial Growth Factor Receptor Tyrosine Kinases 1, 2, 3. Clin. Cancer Res. 2008, 14, 7272–7283. [Google Scholar] [CrossRef]
- Geller, J.I.; Fox, E.; Turpin, B.K.; Goldstein, S.L.; Liu, X.; Minard, C.G.; Kudgus, R.A.; Reid, J.M.; Berg, S.L.; Weigel, B.J. A study of axitinib, a VEGF receptor tyrosine kinase inhibitor, in children and adolescents with recurrent or refractory solid tumors: A Children’s Oncology Group phase 1 and pilot consortium trial (ADVL1315). Cancer 2018, 124, 4548–4555. [Google Scholar] [CrossRef] [PubMed]
- Grillo, F.; Florio, T.; Ferraù, F.; Kara, E.; Fanciulli, G.; Faggiano, A.; Colao, A.A.L. Emerging multitarget tyrosine kinase inhibitors in the treatment of neuroendocrine neoplasms. Endocrine-Related Cancer 2018, 25, R453–R466. [Google Scholar] [CrossRef] [PubMed]
- Rugo, H.S.; Stopeck, A.T.; Joy, A.A.; Chan, S.; Verma, S.; Lluch, A.; Liau, K.F.; Kim, S.; Bycott, P.; Rosbrook, B.; et al. Randomized, Placebo-Controlled, Double-Blind, Phase II Study of Axitinib Plus Docetaxel Versus Docetaxel Plus Placebo in Patients With Metastatic Breast Cancer. J. Clin. Oncol. 2011, 29, 2459–2465. [Google Scholar] [CrossRef] [PubMed]
- Fruehauf, J.P.; Lutzky, J.; McDermott, D.; Brown, C.K.; Meric, J.-B.; Rosbrook, B.; Shalinsky, D.R.; Liau, K.F.; Niethammer, A.G.; Kim, S.; et al. Multicenter, Phase II Study of Axitinib, a Selective Second-Generation Inhibitor of Vascular Endothelial Growth Factor Receptors 1, 2, and 3, in Patients with Metastatic Melanoma. Clin. Cancer Res. 2011, 17, 7462–7469. [Google Scholar] [CrossRef] [PubMed]
- Duerinck, J.; Du Four, S.; Vandervorst, F.; D’Haene, N.; Le Mercier, M.; Michotte, A.; Vanbinst, A.-M.; Everaert, H.; Salmon, I.; Bouttens, F.; et al. Randomized phase II study of axitinib versus physicians best alternative choice of therapy in patients with recurrent glioblastoma. J. Neuro-Oncology 2016, 128, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Bellesoeur, A.; Carton, E.; Alexandre, J.; Goldwasser, F.; Huillard, O. Axitinib in the treatment of renal cell carcinoma: Design, development, and place in therapy. Drug Des. Dev. Ther. 2017, 11, 2801–2811. [Google Scholar] [CrossRef]
- Morelli, M.B.; Amantini, C.; Nabissi, M.; Cardinali, C.; Santoni, M.; Bernardini, G.; Santoni, A.; Santoni, G. Axitinib induces senescence-associated cell death and necrosis in glioma cell lines: The proteasome inhibitor, bortezomib, potentiates axitinib-induced cytotoxicity in a p21(Waf/Cip1) dependent manner. Oncotarget 2016, 8, 3380–3395. [Google Scholar] [CrossRef]
- Mongiardi, M.P.; Radice, G.; Piras, M.; Stagni, V.; Pacioni, S.; Re, A.; Putti, S.; Ferrè, F.; Farsetti, A.; Pallini, R.; et al. Axitinib exposure triggers endothelial cells senescence through ROS accumulation and ATM activation. Oncogene 2019, 38, 5413–5424. [Google Scholar] [CrossRef]
- Mongiardi, M.P.; Merolle, M.; Fustaino, V.; Levi, A.; Falchetti, M. Gene expression profiling of hypoxic response in different models of senescent endothelial cells. Aging Clin. Exp. Res. 2019, 1–9. [Google Scholar] [CrossRef]
- DeMaria, M. Senescent cells: New target for an old treatment? Mol. Cell. Oncol. 2017, 4, e1299666. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Jiang, F.; Kalkanis, S.N.; Zhang, Z.G.; Zhang, X.; Zheng, X.; Mikkelsen, T.; Jiang, H.; Chopp, M. Decrease of endogenous vascular endothelial growth factor may not affect glioma cell proliferation and invasion. J. Exp. Ther. Oncol. 2007, 6, 219–229. [Google Scholar] [PubMed]
- Kuilman, T.; Michaloglou, C.; Mooi, W.J.; Peeper, D.S. The essence of senescence. Genes Dev. 2010, 24, 2463–2479. [Google Scholar] [CrossRef] [PubMed]
- Rodier, F.; Campisi, J. Four faces of cellular senescence. J. Cell Boil. 2011, 192, 547–556. [Google Scholar] [CrossRef]
- Kuilman, T.; Peeper, D.S. Senescence-messaging secretome: SMS-ing cellular stress. Nat. Rev. Cancer 2009, 9, 81–94. [Google Scholar] [CrossRef]
- Coppé, J.-P.; Desprez, P.-Y.; Krtolica, A.; Campisi, J. The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu. Rev. Pathol. Mech. Dis. 2010, 5, 99–118. [Google Scholar] [CrossRef]
- Acosta, J.C.; Banito, A.; Wuestefeld, T.; Georgilis, A.; Janich, P.; Morton, J.P.; Athineos, D.; Kang, T.-W.; Lasitschka, F.; Andrulis, M.; et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol. 2013, 15, 978–990. [Google Scholar] [CrossRef]
- Lee, S.; A Schmitt, C. The dynamic nature of senescence in cancer. Nat. Cell Biol. 2019, 21, 94–101. [Google Scholar] [CrossRef]
- Wiley, C.D.; Velarde, M.C.; Lecot, P.; Liu, S.; Sarnoski, E.A.; Freund, A.; Shirakawa, K.; Lim, H.W.; Davis, S.S.; Ramanathan, A.; et al. Mitochondrial Dysfunction Induces Senescence with a Distinct Secretory Phenotype. Cell Metab. 2015, 23, 303–314. [Google Scholar] [CrossRef]
- Hoare, M.; Ito, Y.; Kang, T.-W.; Weekes, M.P.; Matheson, N.J.; Patten, D.; Shetty, S.; Parry, A.J.; Menon, S.; Salama, R.; et al. NOTCH1 mediates a switch between two distinct secretomes during senescence. Nat. Cell Biol. 2016, 18, 979–992. [Google Scholar] [CrossRef]
- Belot, N.; Rorive, S.; Doyen, I.; Lefranc, F.; Bruyneel, E.; Dedecker, R.; Micik, S.; Brotchi, J.; Decaestecker, C.; Salmon, I.; et al. Molecular characterization of cell substratum attachments in human glial tumors relates to prognostic features. Glia 2001, 36, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Okuno, Y.; Nakamura-Ishizu, A.; Otsu, K.; Suda, T.; Kubota, Y. Pathological neoangiogenesis depends on oxidative stress regulation by ATM. Nat. Med. 2012, 18, 1208–1216. [Google Scholar] [CrossRef] [PubMed]
- Gems, D.; Partridge, L. Stress-Response Hormesis and Aging: “That which Does Not Kill Us Makes Us Stronger”. Cell Metab. 2008, 7, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Dimri, G.P.; Lee, X.; Basile, G.; Acosta, M.; Scott, G.; Roskelley, C.; Medrano, E.E.; Linskens, M.; Rubelj, I.; Pereira-Smith, O. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 9363–9367. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Primer Forward | Primer Reverse |
|---|---|---|
| TBP | TGCCCGAAACGCCGAATATAATC | TGGTTCGTGGCTCTCTTATCCTC |
| LMNB1 | GAAGAAGCAGCTGGAGTGGT | TTGGATGCTCTTGGGGTTCC |
| CDKN1/B | CCGCAACCAATGGATCTCCT | CAATATGGCGGTGGAAGGGA |
| CCL2 | AGTCTCTGCCGCCCTTCT | GTGACTGGGGCATTGATT |
| CX3CL1 | CCACCTTCTGCCATCTGACT | TTGACCCATTGCTCCTTCGG |
| BTG2 | GTCTTGATGCTGCTGCCATGATC | AATAAAGGACCTGGAGTTCCCTGTAG |
| COL1A2 | GGTGGGAACTTTGCTGCTCA | GCCTCTAGGTCCCATTAAGCC |
| ELN | GAGTTGGCATTTCCCCCGAA | CAAACTGGGCGGCTTTGG |
| PECAM 1 | GCAACACAGTCCAGATAGTCGT | GACCTCAAACTGGGCATCAT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merolle, M.; Mongiardi, M.P.; Piras, M.; Levi, A.; Falchetti, M.L. Glioblastoma Cells Do Not Affect Axitinib-Dependent Senescence of HUVECs in a Transwell Coculture Model. Int. J. Mol. Sci. 2020, 21, 1490. https://doi.org/10.3390/ijms21041490
Merolle M, Mongiardi MP, Piras M, Levi A, Falchetti ML. Glioblastoma Cells Do Not Affect Axitinib-Dependent Senescence of HUVECs in a Transwell Coculture Model. International Journal of Molecular Sciences. 2020; 21(4):1490. https://doi.org/10.3390/ijms21041490
Chicago/Turabian StyleMerolle, Matilde, Maria Patrizia Mongiardi, Maurizia Piras, Andrea Levi, and Maria Laura Falchetti. 2020. "Glioblastoma Cells Do Not Affect Axitinib-Dependent Senescence of HUVECs in a Transwell Coculture Model" International Journal of Molecular Sciences 21, no. 4: 1490. https://doi.org/10.3390/ijms21041490
APA StyleMerolle, M., Mongiardi, M. P., Piras, M., Levi, A., & Falchetti, M. L. (2020). Glioblastoma Cells Do Not Affect Axitinib-Dependent Senescence of HUVECs in a Transwell Coculture Model. International Journal of Molecular Sciences, 21(4), 1490. https://doi.org/10.3390/ijms21041490







