Interactome and F-Actin Interaction Analysis of Dictyostelium discoideum Coronin A
Abstract
1. Introduction
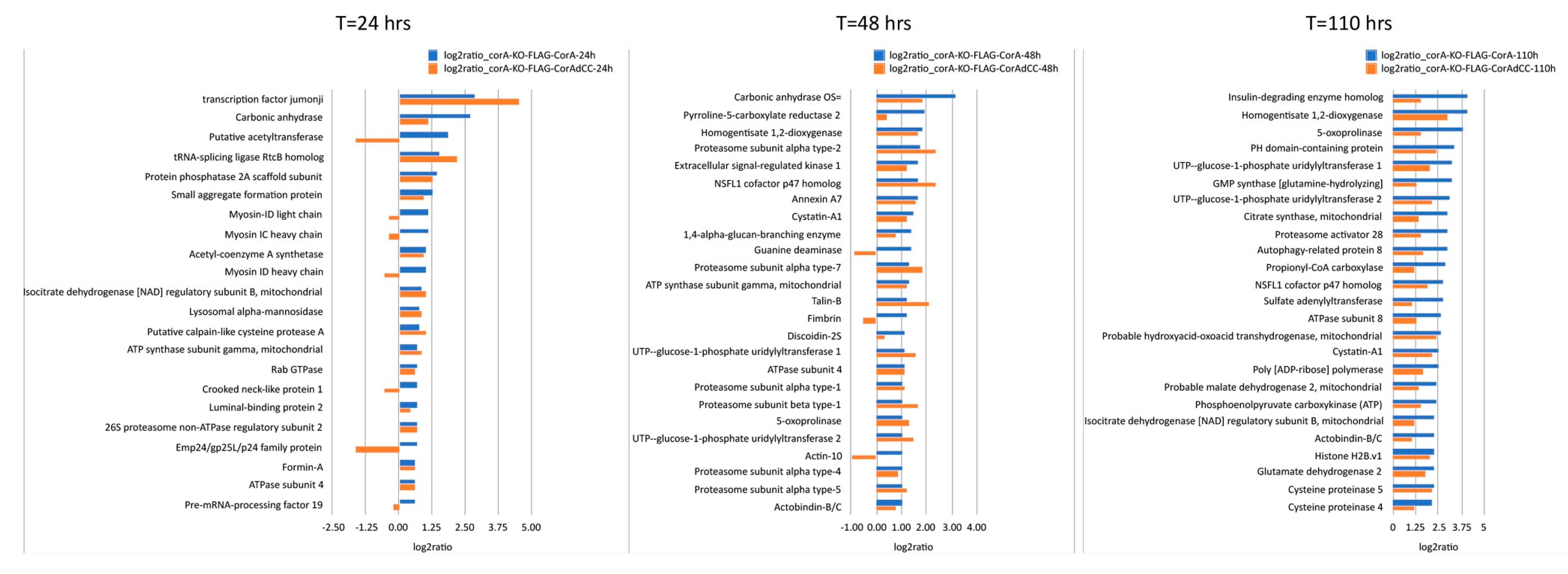
2. Results
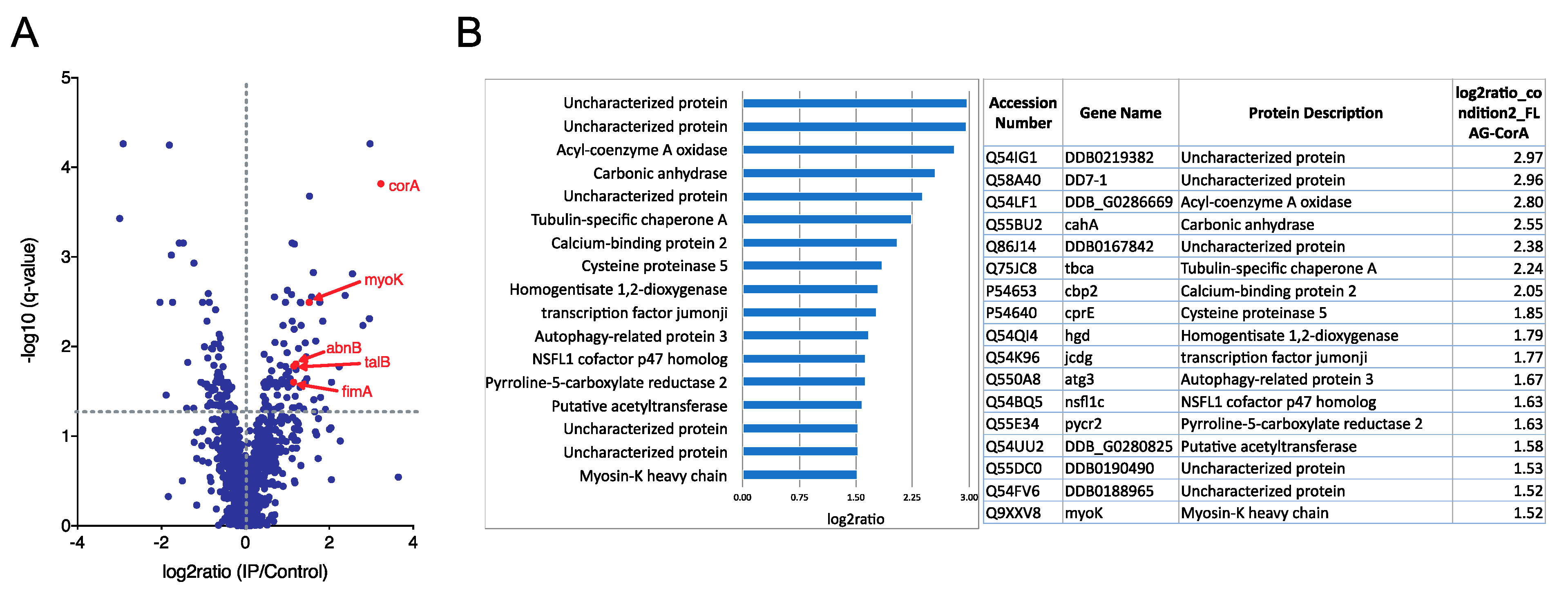
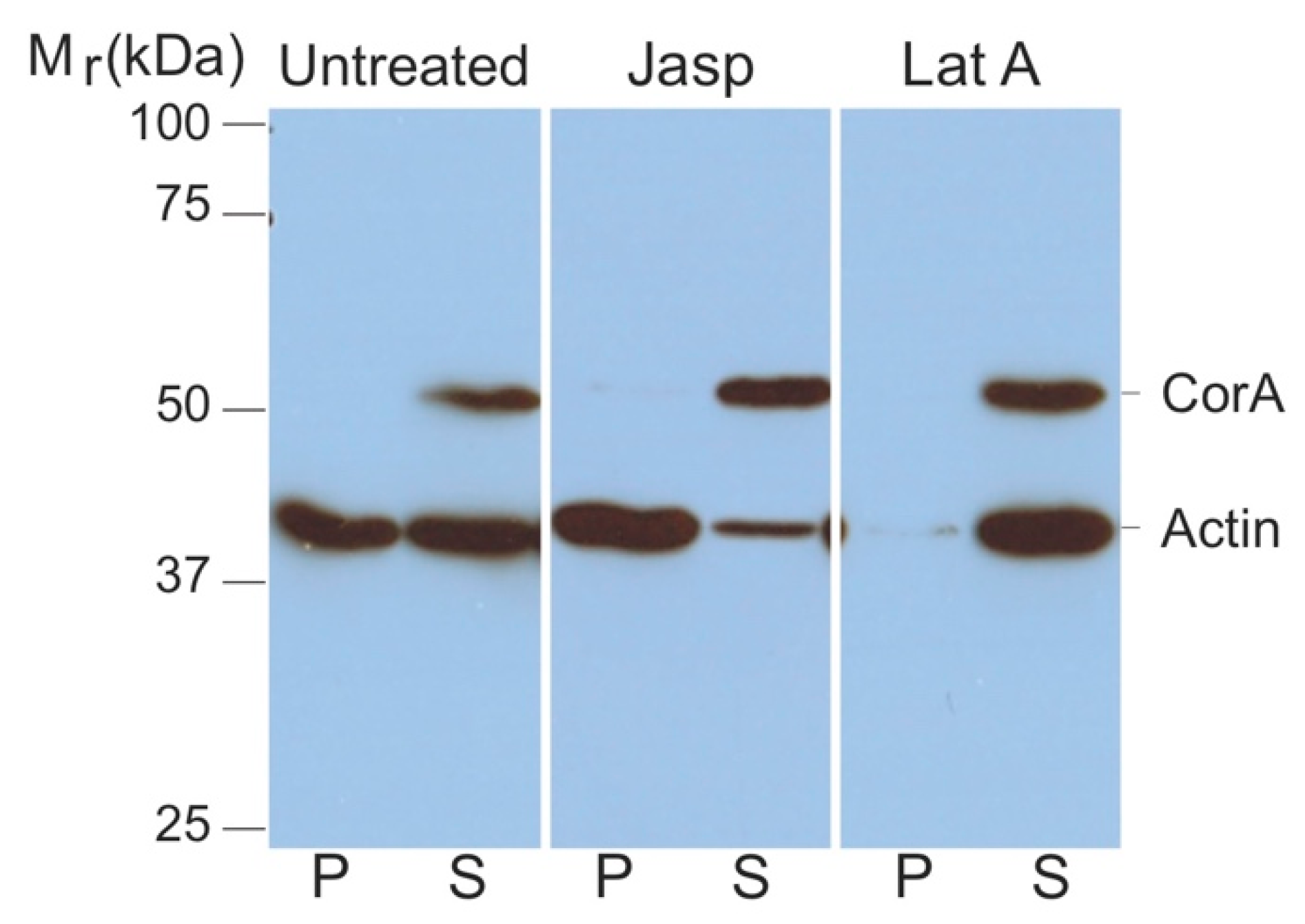
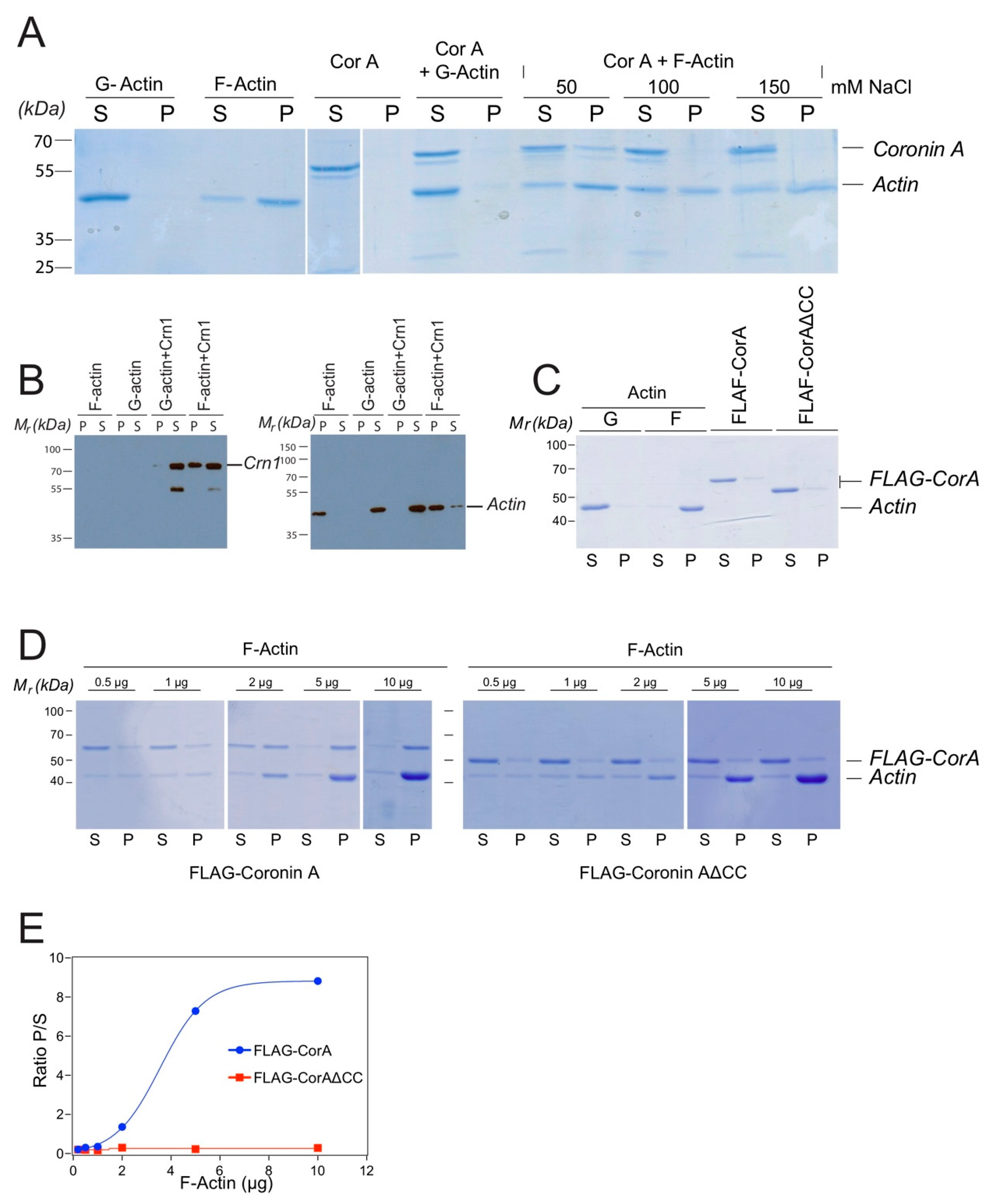
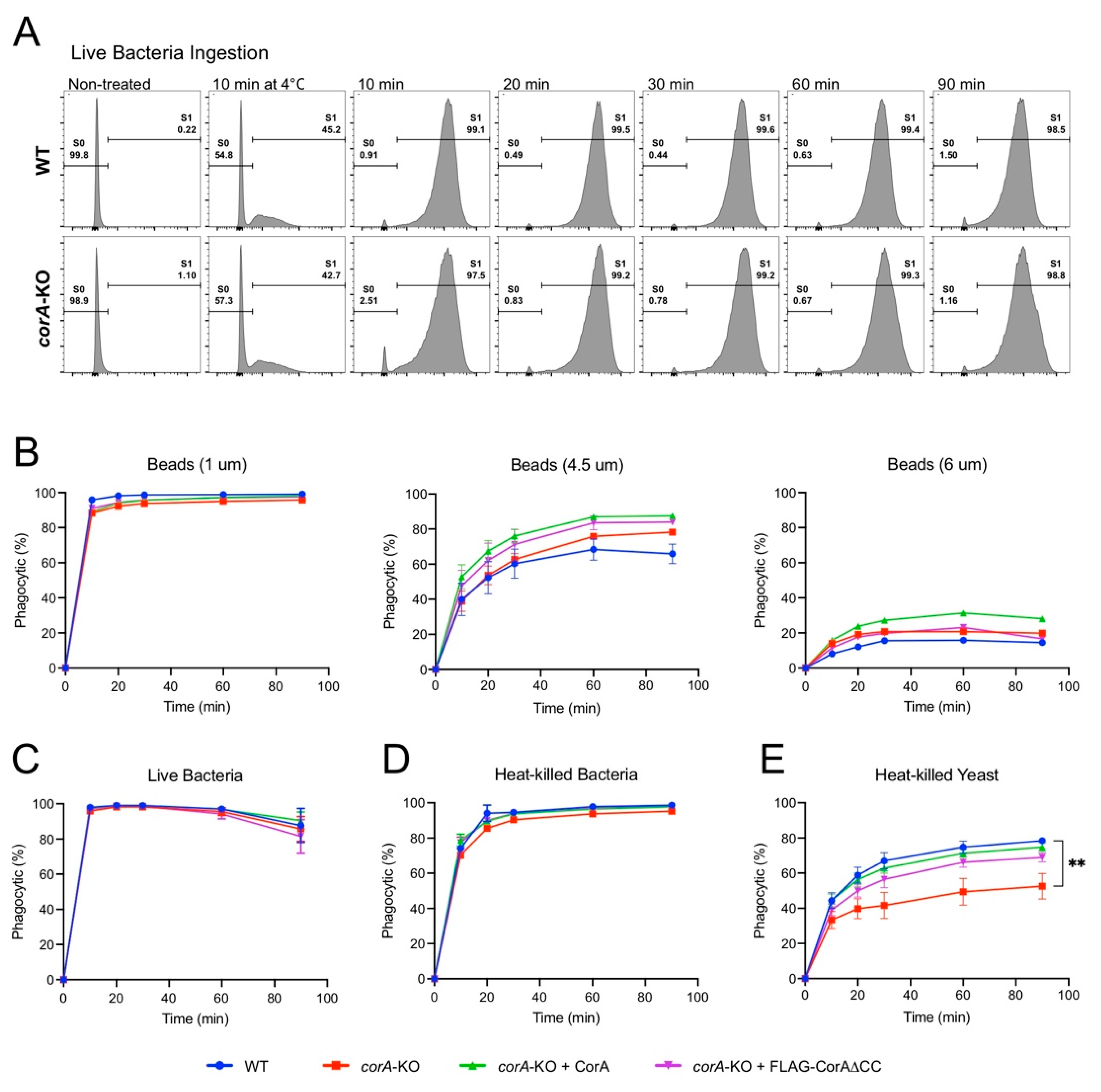
2.1. Coronin A-F-Actin Interaction in Vitro and within Cells
2.2. Coronin A is Required for the Phagocytosis of Yeast Particles, but not Bacteria and Inert Beads, Independent of the Coiled-Coil Domain
3. Discussion
4. Materials and Methods
4.1. Cells, Antibodies, and Growth Conditions
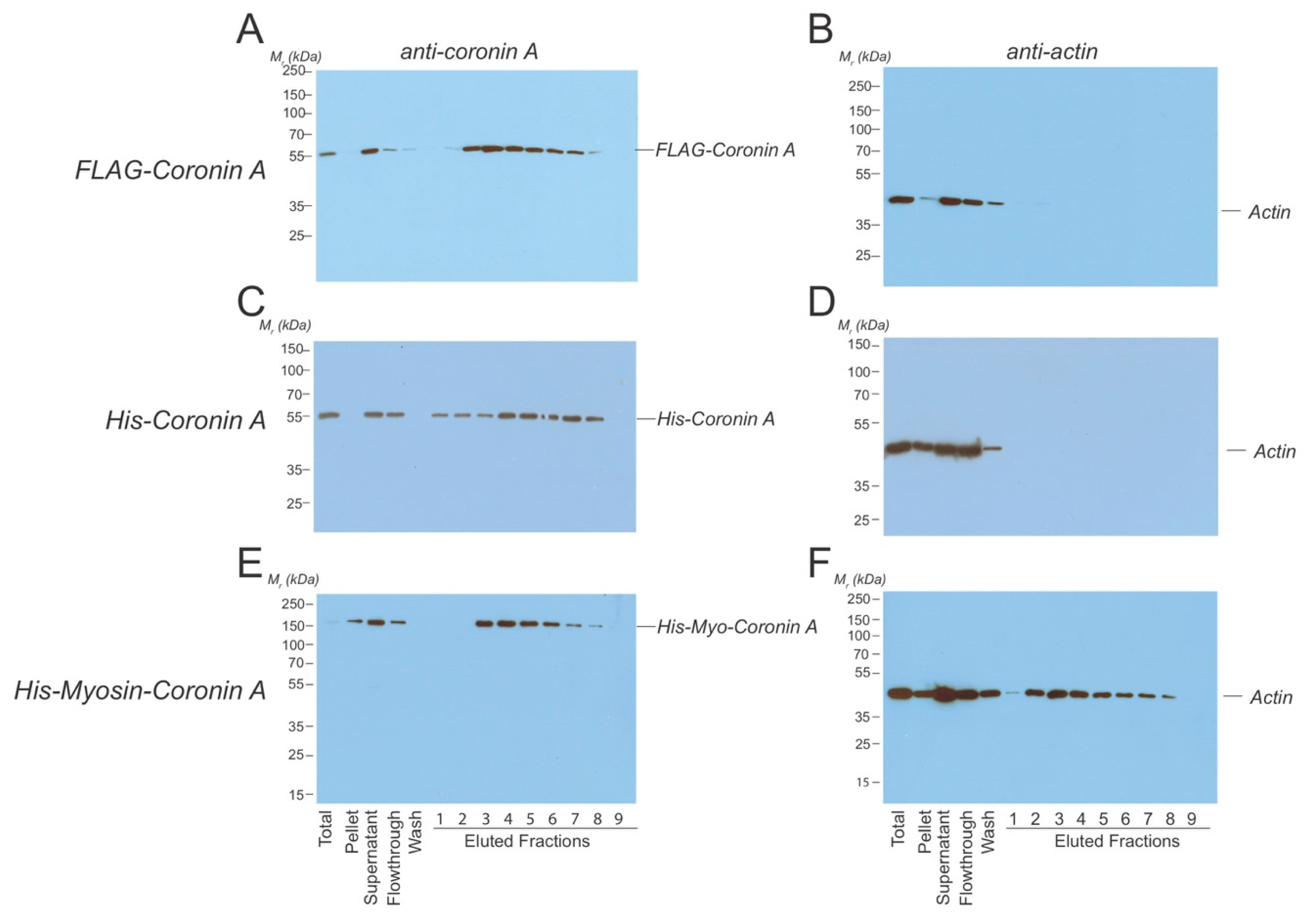
4.2. Anti-Flag Immunoprecipitation and Mass Spectrometry
4.3. Analysis of F-Actin and G-Actin from Cell Lysates
4.4. Protein Purifications
4.5. Coronin A-F-Actin co-Precipitation Analysis
4.6. Western Blotting
4.7. Phagocytosis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pieters, J.; Muller, P.; Jayachandran, R. On guard: Coronin proteins in innate and adaptive immunity. Nat. Rev. Immunol. 2013, 13, 510–518. [Google Scholar] [CrossRef] [PubMed]
- de Hostos, E.L. A brief history of the coronin family. Subcell. Biochem. 2008, 48, 31–40. [Google Scholar] [PubMed]
- Eckert, C.; Hammesfahr, B.; Kollmar, M. A holistic phylogeny of the coronin gene family reveals an ancient origin of the tandem-coronin, defines a new subfamily, and predicts protein function. BMC Evolut. Biol. 2011, 11, 268. [Google Scholar] [CrossRef]
- Goode, B.L.; Wong, J.J.; Butty, A.C.; Peter, M.; McCormack, A.L.; Yates, J.R.; Drubin, D.G.; Barnes, G. Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J. Cell Biol. 1999, 144, 83–98. [Google Scholar] [CrossRef]
- Tardieux, I.; Liu, X.; Poupel, O.; Parzy, D.; Dehoux, P.; Langsley, G. A Plasmodium falciparum novel gene encoding a coronin-like protein which associates with actin filaments. FEBS Lett. 1998, 441, 251–256. [Google Scholar] [CrossRef]
- Salamun, J.; Kallio, J.P.; Daher, W.; Soldati-Favre, D.; Kursula, I. Structure of Toxoplasma gondii coronin, an actin-binding protein that relocalizes to the posterior pole of invasive parasites and contributes to invasion and egress. FASEB J. 2014, 28, 4729–4747. [Google Scholar] [CrossRef]
- Okumura, M.; Kung, C.; Wong, S.; Rodgers, M.; Thomas, M.L. Definition of family of coronin-related proteins conserved between humans and mice: Close genetic linkage between coronin-2 and CD45-associated protein. DNA Cell Biol. 1998, 17, 779–787. [Google Scholar] [CrossRef]
- Chan, K.T.; Creed, S.J.; Bear, J.E. Unraveling the enigma: Progress towards understanding the coronin family of actin regulators. Trends Cell Biol. 2011, 21, 481–488. [Google Scholar] [CrossRef]
- Pieters, J. Coronin 1 in innate immunity. Subcell. Biochem. 2008, 48, 116–123. [Google Scholar]
- Humphries, C.L.; Balcer, H.I.; D’Agostino, J.L.; Winsor, B.; Drubin, D.G.; Barnes, G.; Andrews, B.J.; Goode, B.L. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J. Cell Biol. 2002, 159, 993–1004. [Google Scholar] [CrossRef]
- Gandhi, M.; Jangi, M.; Goode, B.L. Functional surfaces on the actin-binding protein coronin revealed by systematic mutagenesis. J. Biol. Chem. 2010, 285, 34899–34908. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.L.; Needham, K.M.; May, J.R.; Nolen, B.J. Mechanism of a concentration-dependent switch between activation and inhibition of Arp2/3 complex by coronin. J. Biol. Chem. 2011, 286, 17039–17046. [Google Scholar] [CrossRef] [PubMed]
- Marchand, J.B.; Kaiser, D.A.; Pollard, T.D.; Higgs, H.N. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol. 2001, 3, 76–82. [Google Scholar] [CrossRef]
- Veltman, D.M.; Insall, R.H. WASP family proteins: Their evolution and its physiological implications. Mol. Biol. Cell 2010, 21, 2880–2893. [Google Scholar] [CrossRef] [PubMed]
- Heil-Chapdelaine, R.A.; Tran, N.K.; Cooper, J.A. The role of Saccharomyces cerevisiae coronin in the actin and microtubule cytoskeletons. Curr. Biol. 1998, 8, 1281–1284. [Google Scholar] [CrossRef]
- Sahasrabuddhe, A.A.; Nayak, R.C.; Gupta, C.M. Ancient Leishmania coronin (CRN12) is involved in microtubule remodeling during cytokinesis. J. Cell Sci. 2009, 122, 1691–1699. [Google Scholar] [CrossRef]
- Bane, K.S.; Lepper, S.; Kehrer, J.; Sattler, J.M.; Singer, M.; Reinig, M.; Klug, D.; Heiss, K.; Baum, J.; Mueller, A.-K.; et al. The Actin Filament-Binding Protein Coronin Regulates Motility in Plasmodium Sporozoites. PLoS Pathog. 2016, 12, e1005710. [Google Scholar] [CrossRef]
- Spoerl, Z.; Stumpf, M.; Noegel, A.A.; Hasse, A. Oligomerization, F-actin interaction, and membrane association of the ubiquitous mammalian coronin 3 are mediated by its carboxyl terminus. J. Biol. Chem. 2002, 277, 48858–48867. [Google Scholar] [CrossRef]
- Huang, W.; Ghisletti, S.; Saijo, K.; Gandhi, M.; Aouadi, M.; Tesz, G.J.; Zhang, D.X.; Yao, J.; Czech, M.P.; Goode, B.L.; et al. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature 2011, 470, 414–418. [Google Scholar] [CrossRef]
- Nakamura, T.; Takeuchi, K.; Muraoka, S.; Takezoe, H.; Takahashi, N.; Mori, N. A neurally enriched coronin-like protein, ClipinC, is a novel candidate for an actin cytoskeleton-cortical membrane-linking protein. J. Biol. Chem. 1999, 274, 13322–13327. [Google Scholar] [CrossRef]
- Chen, Y.; Ip, F.C.; Shi, L.; Zhang, Z.; Tang, H.; Ng, Y.P.; Ye, W.-C.; Fu, A.K.; Ip, N.Y. Coronin 6 regulates acetylcholine receptor clustering through modulating receptor anchorage to actin cytoskeleton. J. Neurosci. 2014, 34, 2413–2421. [Google Scholar] [CrossRef]
- Cai, L.; Marshall, T.W.; Uetrecht, A.C.; Schafer, D.A.; Bear, J.E. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell 2007, 128, 915–929. [Google Scholar] [CrossRef] [PubMed]
- Rybakin, V.; Stumpf, M.; Schulze, A.; Majoul, I.V.; Noegel, A.A.; Hasse, A. Coronin 7, the mammalian POD-1 homologue, localizes to the Golgi apparatus. FEBS Lett. 2004, 573, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, G.; Langen, H.; Naito, M.; Pieters, J. A coat protein on phagosomes involved in the intracellular survival of mycobacteria. Cell 1999, 97, 435–447. [Google Scholar] [CrossRef]
- Suzuki, K.; Nishihata, J.; Arai, Y.; Honma, N.; Yamamoto, K.; Irimura, T.; Toyoshima, S. Molecular cloning of a novel actin-binding protein, p57, with a WD repeat and a leucine zipper motif. FEBS Lett. 1995, 364, 283–288. [Google Scholar] [CrossRef]
- Jayachandran, R.; Liu, X.; BoseDasgupta, S.; Müller, P.; Zhang, C.L.; Moshous, D.; Studer, V.; Schneider, J.; Genoud, C.; Fossoud, C.; et al. Coronin 1 regulates cognition and behavior through modulation of cAMP/protein kinase A signaling. PLoS Biol. 2014, 12, e1001820. [Google Scholar] [CrossRef] [PubMed]
- Suo, D.; Park, J.; Harrington, A.W.; Zweifel, L.S.; Mihalas, S.; Deppmann, C.D. Coronin-1 is a neurotrophin endosomal effector that is required for developmental competition for survival. Nat. Neurosci. 2014, 17, 36–45. [Google Scholar] [CrossRef]
- Jayachandran, R.; Gumienny, A.; Bolinger, B.; Ruehl, S.; Lang, M.J.; Fucile, G.; Mazumder, S.; Tchang, V.; Woischnig, A.-K.; Stiess, M.; et al. Disruption of Coronin 1 Signaling in T Cells Promotes Allograft Tolerance while Maintaining Anti-Pathogen Immunity. Immunity 2019, 50, 152–165.e8. [Google Scholar] [CrossRef]
- Haraldsson, M.K.; Louis-Dit-Sully, C.A.; Lawson, B.R.; Sternik, G.; Santiago-Raber, M.L.; Gascoigne, N.R.; Theofilopoulos, A.N.; Kono, D.H. The lupus-related Lmb3 locus contains a disease-suppressing Coronin-1A gene mutation. Immunity 2008, 28, 40–51. [Google Scholar] [CrossRef]
- Vinet, A.F.; Fiedler, T.; Studer, V.; Froquet, R.; Dardel, A.; Cosson, P.; Pieters, J. Initiation of multicellular differentiation in Dictyostelium discoideum is regulated by coronin A. Mol. Biol. Cell 2014, 25, 688–701. [Google Scholar] [CrossRef]
- de Hostos, E.L.; Bradtke, B.; Lottspeich, F.; Guggenheim, R.; Gerisch, G. Coronin, an actin binding protein of Dictyostelium discoideum localized to cell surface projections, has sequence similarities to G protein beta subunits. EMBO J. 1991, 10, 4097–4104. [Google Scholar] [CrossRef] [PubMed]
- de Hostos, E.L.; Rehfuess, C.; Bradtke, B.; Waddell, D.R.; Albrecht, R.; Murphy, J.; Gerisch, G. Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J. Cell Biol. 1993, 120, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Gerisch, G.; Albrecht, R.; De Hostos, E.; Wallraff, E.; Heizer, C.; Kreitmeier, M.; Muller Taubenberger, A. Actin-associated proteins in motility and chemotaxis of Dictyostelium cells. Symp. Soc. Exp. Biol. 1993, 47, 297–315. [Google Scholar] [PubMed]
- Maniak, M.; Rauchenberger, R.; Albrecht, R.; Murphy, J.; Gerisch, G. Coronin involved in phagocytosis: Dynamics of particle-induced relocalization visualized by a green fluorescent protein Tag. Cell 1995, 83, 915–924. [Google Scholar] [CrossRef]
- Swaminathan, K.; Muller-Taubenberger, A.; Faix, J.; Rivero, F.; Noegel, A.A. A Cdc42- and Rac-interactive binding (CRIB) domain mediates functions of coronin. Proc. Natl. Acad. Sci. USA 2014, 111, E25–E33. [Google Scholar] [CrossRef] [PubMed]
- Shina, M.C.; Müller-Taubenberger, A.; Ünal, C.; Schleicher, M.; Steinert, M.; Eichinger, L.; Müller, R.; Blau-Wasser, R.; Glöckner, G.; Noegel, A.A. Redundant and unique roles of coronin proteins in Dictyostelium. Cell. Mol. Life Sci. 2011, 68, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Devreotes, P. Dictyostelium discoideum: A model system for cell-cell interactions in development. Science 1989, 245, 1054–1058. [Google Scholar] [CrossRef]
- Kimmel, A.R.; Firtel, R.A. cAMP signal transduction pathways regulating development of Dictyostelium discoideum. Curr. Opin. Genet. Dev. 1991, 1, 383–390. [Google Scholar] [CrossRef]
- Weijer, C.J. Morphogenetic cell movement in Dictyostelium. Semin. Cell Dev. Biol. 1999, 10, 609–619. [Google Scholar] [CrossRef]
- de Hostos, E.L. The coronin family of actin-associated proteins. Trends Cell Biol. 1999, 9, 345–350. [Google Scholar] [CrossRef]
- Uetrecht, A.C.; Bear, J.E. Coronins: The return of the crown. Trends Cell Biol. 2006, 16, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Shina, M.C.; Noegel, A.A. Invertebrate coronins. Subcell. Biochem. 2008, 48, 88–97. [Google Scholar] [PubMed]
- Mueller, P.; Quintana, A.; Griesemer, D.; Hoth, M.; Pieters, J. Disruption of the cortical actin cytoskeleton does not affect store operated Ca(2+) channels in human T-cells. FEBS Lett. 2007, 581, 3557–3562. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, Y.; Du, L.; Tang, D.D.; Gunst, S.J. Activation of the Arp2/3 complex by N-WASp is required for actin polymerization and contraction in smooth muscle. Am. J. Physiol. Cell Physiol. 2005, 288, C1145–C1160. [Google Scholar] [CrossRef]
- Mueller, P.; Liu, X.; Pieters, J. Migration and homeostasis of naive T cells depends on coronin 1-mediated prosurvival signals and not on coronin 1-dependent filamentous actin modulation. J. Immunol. 2011, 186, 4039–4050. [Google Scholar] [CrossRef] [PubMed]
- Fukui, Y.; Engler, S.; Inoue, S.; de Hostos, E.L. Architectural dynamics and gene replacement of coronin suggest its role in cytokinesis. Cell Motil. Cytoskeleton. 1999, 42, 204–217. [Google Scholar] [CrossRef]
- Shiow, L.R.; Roadcap, D.W.; Paris, K.; Watson, S.R.; Grigorova, I.L.; Lebet, T.; An, J.; Xu, Y.; Jenne, C.N.; Föger, N.; et al. The actin regulator coronin 1A is mutant in a thymic egress-deficient mouse strain and in a patient with severe combined immunodeficiency. Nat. Immunol. 2008, 9, 1307–1315. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, T.; Ndinyanka Fabrice, T.; Studer, V.; Vinet, A.; Faltova, L.; Sharp, T.; Pieters, J. Homodimerization of coronin A through the C-terminal coiled coil domain is essential for multicellular differentiation of Dictyostelium discoideum in preparation. Dictybase.org. (under review).
- Joseph, J.M.; Fey, P.; Ramalingam, N.; Liu, X.I.; Rohlfs, M.; Noegel, A.A.; Müller-Taubenberger, A.; Schleicher, M. The actinome of Dictyostelium discoideum in comparison to actins and actin-related proteins from other organisms. PLoS ONE 2008, 3, e2654. [Google Scholar] [CrossRef][Green Version]
- Castellano, F.; Chavrier, P.; Caron, E. Actin dynamics during phagocytosis. Semin. Immunol. 2001, 13, 347–355. [Google Scholar] [CrossRef]
- May, R.C.; Machesky, L.M. Phagocytosis and the actin cytoskeleton. J. Cell Sci. 2001, 114, 1061–1077. [Google Scholar]
- Pan, M.; Xu, X.; Chen, Y.; Jin, T. Identification of a Chemoattractant G-Protein-Coupled Receptor for Folic Acid that Controls Both Chemotaxis and Phagocytosis. Dev. Cell 2016, 36, 428–439. [Google Scholar] [CrossRef]
- Vogel, G.; Thilo, L.; Schwarz, H.; Steinhart, R. Mechanism of phagocytosis in Dictyostelium discoideum: Phagocytosis is mediated by different recognition sites as disclosed by mutants with altered phagocytotic properties. J. Cell Biol. 1980, 86, 456–465. [Google Scholar] [CrossRef]
- Riyahi, T.Y.; Frese, F.; Steinert, M.; Omosigho, N.N.; Glöckner, G.; Eichinger, L.; Orabi, B.; Williams, R.S.B.; Noegel, A.A. RpkA, a highly conserved GPCR with a lipid kinase domain, has a role in phagocytosis and anti-bacterial defense. PLoS ONE 2011, 6, e27311. [Google Scholar] [CrossRef] [PubMed]
- Fukui, Y.; de Hostos, E.; Yumura, S.; Kitanishi-Yumura, T.; Inoué, S. Architectural dynamics of F-actin in eupodia suggests their role in invasive locomotion in Dictyostelium. Exp. Cell Res. 1999, 249, 33–45. [Google Scholar] [CrossRef]
- Tunnacliffe, E.; Corrigan, A.M.; Chubb, J.R. Promoter-mediated diversification of transcriptional bursting dynamics following gene duplication. Proc. Natl. Acad. Sci. USA 2018, 115, 8364–8369. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.T.; Roadcap, D.W.; Holoweckyj, N.; Bear, J.E. Coronin 1C harbours a second actin-binding site that confers co-operative binding to F-actin. Biochem. J. 2012, 444, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Berzat, A.; Hall, A. Cellular responses to extracellular guidance cues. EMBO J. 2010, 29, 2734–2745. [Google Scholar] [CrossRef]
- Hacker, U.; Albrecht, R.; Maniak, M. Fluid-phase uptake by macropinocytosis in Dictyostelium. J. Cell Sci. 1997, 110, 105–112. [Google Scholar]
- Nagasaki, A.; Hibi, M.; Asano, Y.; Uyeda, T.Q. Genetic approaches to dissect the mechanisms of two distinct pathways of cell cycle-coupled cytokinesis in Dictyostelium. Cell Struct. Funct. 2001, 26, 585–591. [Google Scholar] [CrossRef][Green Version]
- Williams, T.D.; Peak-Chew, S.Y.; Paschke, P.; Kay, R.R. Akt and SGK protein kinases are required for efficient feeding by macropinocytosis. J. Cell Sci. 2019, 132, jcs224998. [Google Scholar] [CrossRef]
- Prassler, J.; Stocker, S.; Marriott, G.; Heidecker, M.; Kellermann, J.; Gerisch, G. Interaction of a Dictyostelium member of the plastin/fimbrin family with actin filaments and actin-myosin complexes. Mol. Biol. Cell 1997, 8, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Kollmar, M. Thirteen is enough: The myosins of Dictyostelium discoideum and their light chains. BMC Genomics 2006, 7, 183. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, F.; Rivero, F. Single and multiple CH (calponin homology) domain containing multidomain proteins in Dictyostelium discoideum: An inventory. Mol. Biol. Rep. 2010, 37, 2853–2862. [Google Scholar] [CrossRef]
- Lambooy, P.K.; Korn, E.D. Purification and characterization of actobindin, a new actin monomer-binding protein from Acanthamoeba castellanii. J. Biol. Chem. 1986, 261, 17150–17155. [Google Scholar] [PubMed]
- Howe, A.K. Regulation of actin-based cell migration by cAMP/PKA. Biochim. Biophys. Acta 2004, 1692, 159–174. [Google Scholar] [CrossRef]
- Mattila, P.K.; Batista, F.D.; Treanor, B. Dynamics of the actin cytoskeleton mediates receptor cross talk: An emerging concept in tuning receptor signaling. J. Cell Biol. 2016, 212, 267–280. [Google Scholar] [CrossRef]
- Ashworth, J.M.; Watts, D.J. Metabolism of the cellular slime mould Dictyostelium discoideum grown in axenic culture. Biochem. J. 1970, 119, 175–182. [Google Scholar] [CrossRef]
- Reubold, T.F.; Eschenburg, S.; Becker, A.; Kull, F.J.; Manstein, D.J. A structural model for actin-induced nucleotide release in myosin. Nat. Struct. Biol. 2003, 10, 826–830. [Google Scholar] [CrossRef]
- Fujita-Becker, S.; Reubold, T.F.; Holmes, K.C. The actin-binding cleft: Functional characterisation of myosin II with a strut mutation. J. Muscle Res. Cell Motil. 2006, 27, 115–123. [Google Scholar] [CrossRef]
- Mumberg, D.; Muller, R.; Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 1995, 156, 119–122. [Google Scholar] [CrossRef]
- Ahrne, E.; Glatter, T.; Vigano, C.; Schubert, C.; Nigg, E.A.; Schmidt, A. Evaluation and Improvement of Quantification Accuracy in Isobaric Mass Tag-Based Protein Quantification Experiments. J. Proteome Res. 2016, 15, 2537–2547. [Google Scholar] [CrossRef] [PubMed]
- Vizcaino, J.A.; Csordas, A.; Del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef] [PubMed]
- Mueller, P.; Massner, J.; Jayachandran, R.; Combaluzier, B.; Albrecht, I.; Gatfield, J.; Blum, C.; Ceredig, R.; Rodewald, H.-R.; Rolink, A.G.; et al. Regulation of T cell survival through coronin-1-mediated generation of inositol-1,4,5-trisphosphate and calcium mobilization after T cell receptor triggering. Nat. Immunol. 2008, 9, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Eichinger, L.; Rivero, F. Dictyostelium discoideum protocols. In Methods in Molecular Biology; Clifton, N.J., Ed.; Humana Press: New York, NY, USA, 2006. [Google Scholar]
- Pardee, J.D.; Spudich, J.A. Purification of muscle actin. Methods Enzymol. 1982, 85, 164–181. [Google Scholar]
- Yahara, N.; Ueda, T.; Sato, K.; Nakano, A. Multiple roles of Arf1 GTPase in the yeast exocytic and endocytic pathways. Mol. Biol. Cell 2001, 12, 221–238. [Google Scholar] [CrossRef]
- Sattler, N.; Monroy, R.; Soldati, T. Quantitative analysis of phagocytosis and phagosome maturation. Methods Mol. Biol. 2013, 983, 383–402. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabrice, T.N.; Fiedler, T.; Studer, V.; Vinet, A.; Brogna, F.; Schmidt, A.; Pieters, J. Interactome and F-Actin Interaction Analysis of Dictyostelium discoideum Coronin A. Int. J. Mol. Sci. 2020, 21, 1469. https://doi.org/10.3390/ijms21041469
Fabrice TN, Fiedler T, Studer V, Vinet A, Brogna F, Schmidt A, Pieters J. Interactome and F-Actin Interaction Analysis of Dictyostelium discoideum Coronin A. International Journal of Molecular Sciences. 2020; 21(4):1469. https://doi.org/10.3390/ijms21041469
Chicago/Turabian StyleFabrice, Tohnyui Ndinyanka, Thomas Fiedler, Vera Studer, Adrien Vinet, Francesco Brogna, Alexander Schmidt, and Jean Pieters. 2020. "Interactome and F-Actin Interaction Analysis of Dictyostelium discoideum Coronin A" International Journal of Molecular Sciences 21, no. 4: 1469. https://doi.org/10.3390/ijms21041469
APA StyleFabrice, T. N., Fiedler, T., Studer, V., Vinet, A., Brogna, F., Schmidt, A., & Pieters, J. (2020). Interactome and F-Actin Interaction Analysis of Dictyostelium discoideum Coronin A. International Journal of Molecular Sciences, 21(4), 1469. https://doi.org/10.3390/ijms21041469




