Genome-Wide Identification, Evolution and Expression of the Complete Set of Cytoplasmic Ribosomal Protein Genes in Nile Tilapia
Abstract
1. Introduction
2. Results
2.1. Identification of RP Genes in Tilapia
2.2. Comparative Analysis of RP Genes in Chordates
2.3. Phylogenetic and Syntenic Analyses of RP Paralogous Genes in Tilapia
2.4. Tissue Distribution and Ontogeny Expression of RP Genes in Gonads of Tilapia by Transcriptomic Analysis
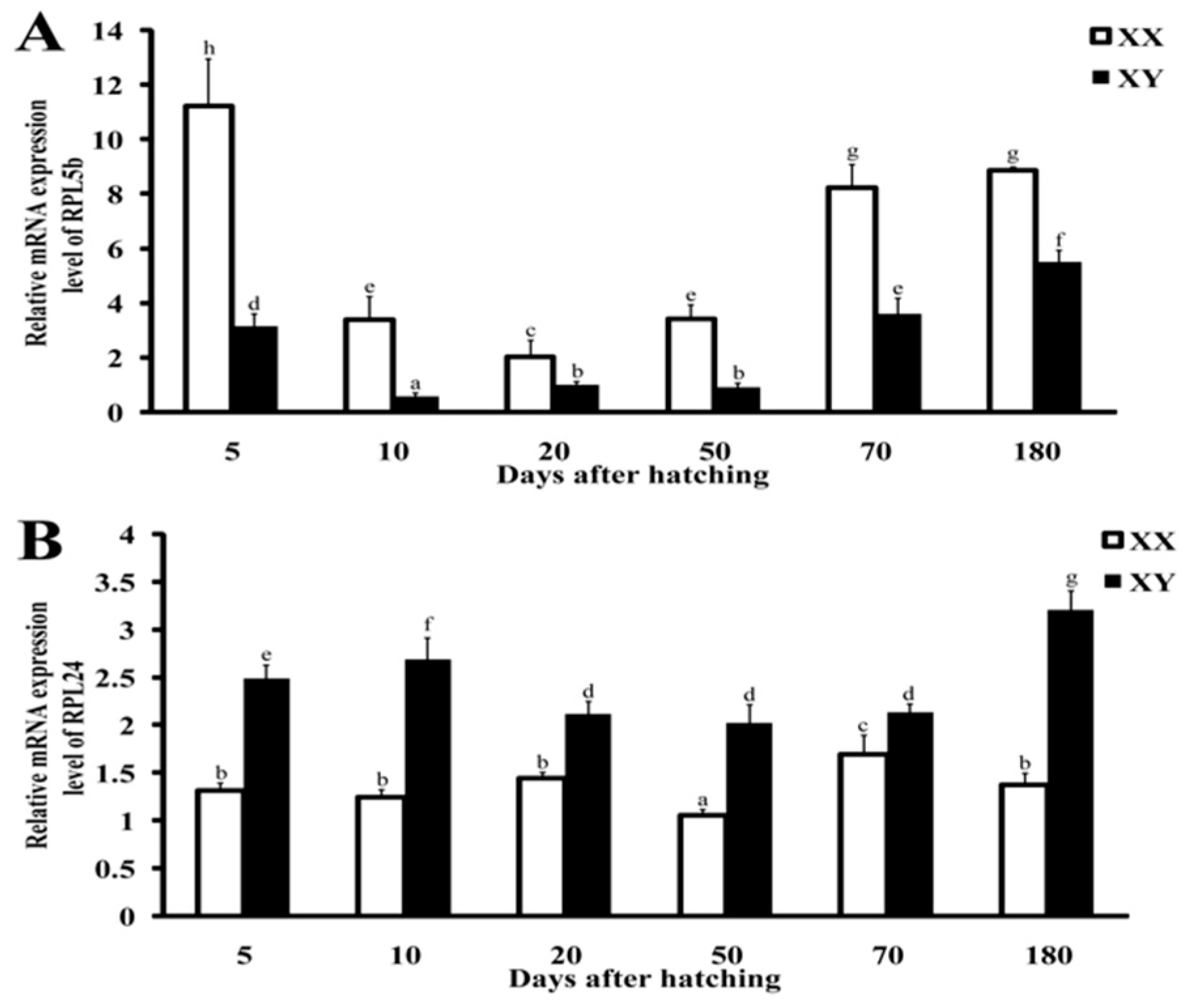
2.5. Validation by qRT-PCR and Immunohistochemistry
3. Discussion
3.1. Evolution of RP Genes in Chordates
3.2. Possible Roles of RP Genes in Different Tissues, Especially in Gonads
4. Materials and Methods
4.1. Animal Rearing
4.2. Identification of RP genes from different chordates
4.3. Phylogenetic and Syntenic Analyses
4.4. Expression Analyses of Tilapia RP Genes in Adult Tissues and Gonads at Different Developmental Stages
4.5. Validation of Differentially Expressed Genesby qRT-PCR and IHC
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| RP | ribosomal protein |
| WGD | whole genome duplication |
| 1R | first round of genome duplication |
| 2R | second round of genome duplication |
| 3R | third round of genome duplication |
| 4R | fourth round of genome duplication |
| qRT-PCR | quantitative real time polymerase chain reaction |
| IHC | Immunohistochemistry |
| ML | maximum-likelihood |
| NJ | neighbour-joining |
| dah | day after hatching |
| RPKM | reads per kb per million reads |
References
- McCann, K.L.; Baserga, S.J. Mysterious ribosome pathies. Sicence 2013, 341, 849–850. [Google Scholar] [CrossRef] [PubMed]
- Bolze, A.; Mahlaoui, N.; Byun, M.; Turner, B.; Trede, N.; Ellis, S.R.; Abhyankar, A.; Itan, Y.; Patin, E.; Brebner, S. Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science 2013, 340, 976–978. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Basu, A.; Biswas, A.; Poddar, D.; Andrews, J.; Barik, S.; Komar, A.A.; Mazumder, B. Insights into the mechanism of ribosomal incorporation of mammalian L13a protein during ribosome biogenesis. Mol. Cell Biol. 2013, 33, 2829–2842. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Gruppuso, P.A. Rapamycin inhibits liver growth during refeeding in rats via control of ribosomal protein translation but not cap-dependent translation initiation. J. Nutr. 2006, 136, 27–33. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kowalczyk, P.; Woszczynski, M.; Ostrowski, J. Increased expression of ribosomal protein S2 in liver tumors, posthepactomized livers, and proliferating hepatocytes in vitro. Acta. Biochim. Pol. 2002, 49, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Mathavan, S.; Lee, S.G.P.; Mak, A.; Miller, L.D.; Murthy, K.R.K.; Govindarajan, K.R.; Tong, Y.; Wu, Y.L.; Lam, S.H.; Yang, H.; et al. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 2005, 1, 260–276. [Google Scholar] [CrossRef]
- Dutt, S.; Narla, A.; Lin, K.; Mullally, A.; Abayasekara, N.; Megerdichian, C.; Wilson, F.H.; Currie, T.; Khanna-Gupta, A.; Berliner, N. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood 2011, 117, 2567–2576. [Google Scholar] [CrossRef]
- Dionne, K.L.; Bergeron, D.; Landry-Voyer, A.M.; Bachand, F. The 40S ribosomal protein uS5 (RPS2) assembles into an extraribosomal complex with human ZNF277 that competes with the PRMT3-uS5 interaction. J. Biol. Chem. 2019, 294, 1944–1955. [Google Scholar] [CrossRef]
- Burwick, N.; Coats, S.A.; Nakamura, T.; Shimamura, A. Impaired ribosomal subunit association in Shwachman-Diamond syndrome. Blood 2012, 120, 5143–5152. [Google Scholar] [CrossRef]
- van Gijn, D.R.; Tucker, A.S.; Cobourne, M.T. Craniofacial development: Current concepts in the molecular basis of Treacher Collins syndrome. Br. J. Oral. Max. Surg. 2013, 51, 384–388. [Google Scholar] [CrossRef]
- Shenoy, N.; Kessel, R.; Bhagat, T.D.; Bhattacharyya, S.; Yu, Y.T.; Mcmahon, C.; Verma, A. Alterations in the ribosomal machinery in cancer and hematologic disorders. J. Hematol. Oncol. 2012, 5, 32. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.; Amsterdam, A.; Farrington, S.; Bronson, R.T.; Hopkins, N.; Lees, J.A. Many ribosomal protein mutations are associated with growth impairment and tumor predisposition in zebrafish. Dev. Dyn. 2009, 238, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Ba, Q.; Wang, Z.; Hao, M.; Li, X.; Hu, P.; Zhang, D.; Zhang, R.; Wang, H. Knockdown of ribosomal protein S7 causes developmental abnormalities via p53 dependent and independent pathways in zebrafish. Int. J. Bioche. Cell Biol. 2011, 43, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Wool, I.G.; Chan, Y.L.; Gluck, A. Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 1995, 73, 933–947. [Google Scholar] [CrossRef]
- Uechi, T.; Tanaka, T.; Kenmochi, N. A complete map of the human ribosomal protein genes: Assignment of 80 genes to the cytogenetic map and implications for human disorders. Genomics 2001, 72, 223–230. [Google Scholar] [CrossRef]
- Karsi, A.; Patterson, A.; Feng, J.; Liu, Z. Translational machinery of channel catfish: I. A transcriptomic approach to the analysis of 32 40S ribosomal protein genes and their expression. Gene 2002, 291, 177–186. [Google Scholar] [CrossRef]
- Patterson, A.; Karsi, A.; Feng, J.N.; Liu, Z.J. Translational machinery of channel catfish: II. Complementary DNA and expression of the complete set of 47 60S ribosomal proteins. Gene 2003, 305, 151–160. [Google Scholar] [CrossRef]
- Manchado, M.; Infante, C.; Asensio, E.; Canavate, J.P.; Douglas, S.E. Comparative sequence analysis of the complete set of 40S ribosomal proteins in the Senegalese sole (Soleasenegalensis Kaup) and Atlantic halibut (Hippoglossus hippoglossus L.) (Teleostei: Pleuronectiformes): Phylogeny and tissue- and development-specific expression. BMC Evol. Biol. 2007, 7, 107. [Google Scholar]
- Matsuoka, M.P.; Infante, C.; Reith, M.; Canavate, J.P.; Douglas, S.E.; Manchado, M. Translational machinery of Senegalese sole (Soleasenegalensis Kaup) and Atlantic halibut (Hippoglossus hippoglossus L.): Comparative sequence analysis of the complete set of 60s ribosomal proteins and their expression. Mar. Biotechnol. 2008, 10, 676–691. [Google Scholar] [CrossRef]
- Rahman, M.K.; Kim, B.; You, M. Molecular cloning, expression and impact of ribosomal protein S-27 silencing in Haemaphysalis longicornis (Acari: Ixodidae). Exp. Parasitol. 2019, 209, 107829. [Google Scholar] [CrossRef]
- Wade, J.L.; Tang, Y.P.; Peabody, C.P.; Tempelma, R.J. Enhanced gene expression in the forebrain of hatchling and juvenile male zebra finch. J. Neurobiol. 2005, 64, 224–248. [Google Scholar] [CrossRef]
- Tang, Y.P.; Wade, J. Sexually dimorphic expression of the genes encoding ribosomal proteins L17 and L37 in the song control nuclei of juvenile zebra finches. Brain Res. 2006, 1126, 102–108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tang, Y.P.; Wade, J. Sex- and age-related differences in ribosomal proteins L17 and L37, as well as androgen receptor protein, in the song control system of zebra finches. Neuroscience 2010, 171, 1131–1140. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Acharya, K.D.; Veney, S.L. Sexually dimorphic expression and estradiol mediated up-regulation of a sex-linked ribosomal gene, RPS6, in the zebra finch brain. Dev. Neurobiol. 2013, 73, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, Y.; Jiang, Y.; Lin, P.; Jia, X.; Zou, Z. Ribosomal protein L24 is differentially expressed in ovary and testis of the marine shrimp marsupenaeus japonicus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2007, 147, 466–474. [Google Scholar] [CrossRef]
- Zheng, W.L.; Xu, H.Y.; Lam, S.H.; Luo, H.; Karuturi, K.M.; Gong, Z.Y. Transcriptomic analyses of sexual dimorphism of the zebrafish liver and the effect of sex hormones. PLoS ONE 2013, 8, e53562. [Google Scholar] [CrossRef]
- Hafedh, Y.S.A. Effects of dietary protein on growth and body composition of Nile tilapia, Oreochromisniloticus L. Aquac. Res. 1999, 30, 385–393. [Google Scholar] [CrossRef]
- Brawand, D.; Wagner, C.E.; Li, Y.I.; Malinsky, M.; Keller, I.; Fan, S.; Simakov, O.; Ng, A.Y.; Lim, Z.W.; Bezault, E.; et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature 2014, 513, 375–381. [Google Scholar] [CrossRef]
- Conte, M.A.; Gammerdinger, W.J.; Bartie, K.L.; Penman, D.J.; Kocher, T.D. A high quality assembly of the Nile tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genomics 2017, 18, 341. [Google Scholar] [CrossRef]
- Tao, W.J.; Yuan, J.; Zhou, L.Y.; Sun, L.N.; Sun, Y.L.; Yang, S.J.; Li, M.H.; Zeng, S.; Huang, B.F.; Wang, D.S. Characterization of gonadal transcriptomes from Nile tilapia (Oreochromis niloticus) reveals differentially expressed genes. PLoS ONE 2013, 8, e63604. [Google Scholar] [CrossRef]
- Sun, L.N.; Jiang, X.L.; Xie, Q.P.; Yuan, J.; Huang, B.F.; Tao, W.J.; Zhou, L.Y.; Nagahama, Y.; Wang, D.S. Transdifferentiation of differentiated ovary into functional testis by long-term treatment of aromatase inhibitor in Nile tilapia. Endocrinology 2014, 155, 1476–1488. [Google Scholar] [CrossRef] [PubMed]
- Nomura, M.; Gourse, R.; Baughman, G. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 1984, 53, 75–117. [Google Scholar] [CrossRef] [PubMed]
- Barakat, A.; Szick-Miranda, K.; Chang, I.F.; Guyot, R.; Blanc, G.; Cooke, R.; Delseny, M.; Bailey-Serres, J. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 2001, 127, 398–415. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Das, S.; Moin, M.; Dutta, M.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Genome-wide identification and comprehensive expression profiling of ribosomal protein small subunit (rps) genes and their comparative analysis with the large subunit (rpl) genes in rice. Front. Plant Sci. 2017, 8, 1553. [Google Scholar] [CrossRef]
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.R.; Ceric, G.; Forslund, K.; Eddy, S.R.; Sonnhammer, E.L.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2008, 36, D281–D288. [Google Scholar] [CrossRef]
- Arragain, S.; Garcia-Serres, R.; Blondin, G.; Douki, T.; Clemancey, M.; Latour, J.M.; Forouhar, F.; Neely, H.; Montelione, G.T.; Hunt, J.F.; et al. Post-translational modification of ribosomal proteins structural and functional characterization of rimo from Thermotoga maritima, a radical s-adenosylmethionine methylthiotransferase. J. Biol. Chem. 2010, 285, 5792–5801. [Google Scholar] [CrossRef]
- Barthelemy, R.M.; Chenuil, A.; Blanquart, S.; Casanova, J.P.; Faure, E. Translational machinery of the chaetognath Spadella cephaloptera: A transcriptomic approach to the analysis of cytosolic ribosomal protein genes and their expression. BMC Evol. Biol. 2007, 7, 146–162. [Google Scholar] [CrossRef] [PubMed]
- Perina, D.; Cetkovic, H.; Harcet, M.; Premzl, M.; Lukic-Bilela, L.; Muller, W.E.; Gamulin, V. The complete set of ribosomal proteins from the marine sponge Suberites domuncula. Gene 2006, 366, 275–284. [Google Scholar] [CrossRef]
- Hoegg, S.; Brinkmann, H.; Taylor, J.S.; Meyer, A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J. Mol. Evol. 2004, 59, 190–203. [Google Scholar] [CrossRef]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noel, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014, 5, 3657. [Google Scholar] [CrossRef]
- Blomme, T.; Vandepoele, K.; De Bodt, S.; Simillion, C.; Maere, S.; Van de Peer, Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006, 7, R43. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, C.; Sankoff, D. Evolutionary model for the statistical divergence of paralogous and orthologous gene pairs generated by whole genome duplication and speciation. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 15, 1579–1584. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.A.; Alvarez, C.E.; Sutcliffe, J.G. Evolutionarily distinct classes of S27 ribosomal proteins with differential mRNA expression in rat hypothalamus. J. Neurochem. 2000, 74, 2259–2267. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Yang, C.; Tao, W.J.; Wang, D.S. Genome-wide identification and transcriptome-based expression profiling of the Sox gene family in the Nile tilapia (Oreochromis niloticus). Int. J. Mol. Sci. 2016, 17, 270. [Google Scholar] [CrossRef] [PubMed]
- Thisse, C.; Degrave, A.; Kryukov, G.V.; Gladyshev, V.N.; Obrecht-Pflumio, S.; Krol, A.; Thisse, B.; Lescure, A. Spatial and temporal expression patterns of selenoprotein genes during embryogenesis in zebrafish. Gene Expr. Patterns 2003, 3, 525–532. [Google Scholar] [CrossRef]
- Fortier, S.; MacRae, T.; Sargeant, T.; Sauvageau, G. Regulation of mouse embryonic stem cell fate by ribosomal proteins. Exp. Hematol. 2013, 41, S29. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, W.J.; Liao, J.M.; Liao, P.; Lu, H. Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Biol. 2015, 7, 92–104. [Google Scholar] [CrossRef]
- Small, C.M.; Carney, G.E.; Mo, Q.X.; Vannucci, M.; Jones, A.G. A microarray analysis of sex- and gonad-biased gene expression in the zebrafish: Evidence for masculinization of the transcriptome. BMC Genom. 2019, 10, 579. [Google Scholar] [CrossRef]
- Bai, J.L.; Solberg, C.; Fernandes, J.M.O.; Johnston, I.A. Profiling of maternal and developmental-stage specific mRNA transcripts in Atlantic halibut Hippoglossus hippoglossus. Gene 2006, 386, 202–210. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, L.; Wang, Y.; Li, W.; He, L.; Jiang, H. Expression characteristics of two ubiquitin/ribosomal fusion protein genes in the developing testis, accessory gonad and ovary of chinese mitten crab, eriocheir sinensis. Mol. Biol. Rep. 2012, 39, 6683–6692. [Google Scholar] [CrossRef]
- Deschamps, S.; Morales, J.; Mazabraud, A.; Le, M.M.; Denis, H.; Brown, D.D. Two forms of elongation factor 1 alpha (EF-1 alpha and 42Sp50), present in oocytes, but absent in somatic cells of Xenopus laevis. J. Cell Biol. 1991, 114, 1109–1111. [Google Scholar] [CrossRef]
- Chambers, D.M.; Peters, J.; Abbott, C.M. The lethal mutation of the mouse wasted (wst) is a deletion that abolishes expression of a tissue-specific isoform of translation elongation factor 1α, encoded by the eEFla2 gene. Proc. Natl. Acad. Sci. USA 1998, 98, 4463–4468. [Google Scholar] [CrossRef]
- Eckhardt, K.; Troger, J.; Reissmann, J.; Katschinski, D.M.; Wagner, K.F.; Stengel, P.; Spielmann, P.; Stiehl, D.P.; Camenisch, G.; Wenger, R.H. Male germ cell expression of the pas domain kinase paskin and its novel target eukaryotic translation elongation factor eEF1A1. Cell Physiol. Biochem. 2007, 20, 227–240. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, D.; Tan, D.; Fan, Z.; Wei, Y.; Li, M.; Wang, D. Heterozygous mutation of eEF1A1b resulted in spermatogenesis arrest and infertility in male tilapia, Oreochromis niloticus. Sci. Rep. 2017, 7, 43733. [Google Scholar] [CrossRef]
- Yoshihama, M.; Uechi, T.; Asakawa, S.; Kawasaki, K.; Kato, S.; Higa, S.; Maeda, N.; Minoshima, S.; Tanaka, T.; Shimizu, N.; et al. The human ribosomal protein genes: Sequencing and comparative analysis of 73 genes. Genome Res. 2002, 12, 379–390. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Muffato, M.; Louis, A.; Poisnel, C.E.; RoestCrollius, H. Genomicus: A database and a browser to study gene synteny in modern and ancestral genomes. Bioinformatics 2010, 26, 1119–1121. [Google Scholar] [CrossRef]
- Yuan, J.; Tao, W.J.; Cheng, Y.Y.; Huang, B.F.; Wang, D.S. Genome-wide identification, phylogeny, and gonadal expression of fox genes in Nile tilapia, Oreochromis niloticus. Fish Physiol. Biochem. 2014, 40, 1239–1252. [Google Scholar] [CrossRef]
- Chen, C.j.; Chen, H.; He, Y.H.; Xia, R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018. [Google Scholar] [CrossRef]
- Shved, N.; Berishvili, G.; D’Cotta, H.; Baroiller, J.F.; Segner, H.; Eppler, E.; Reinecke, M. Ethinylestradiol differentially interferes with IGF-I in liver and extrahepatic sites during development of male and female bony fish. J. Endocrinol. 2007, 195, 513–523. [Google Scholar] [CrossRef]
- Li, M.H.; Sun, Y.L.; Zhao, J.; Shi, H.J.; Zeng, S.; Ye, K.; Jiang, D.N.; Zhou, L.Y.; Sun, L.N.; Tao, W.J.; et al. A tandem duplicate of anti-Mullerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile tilapia, Oreochromis niloticus. PLoS Genet. 2015, 11, e1005678. [Google Scholar] [CrossRef] [PubMed]
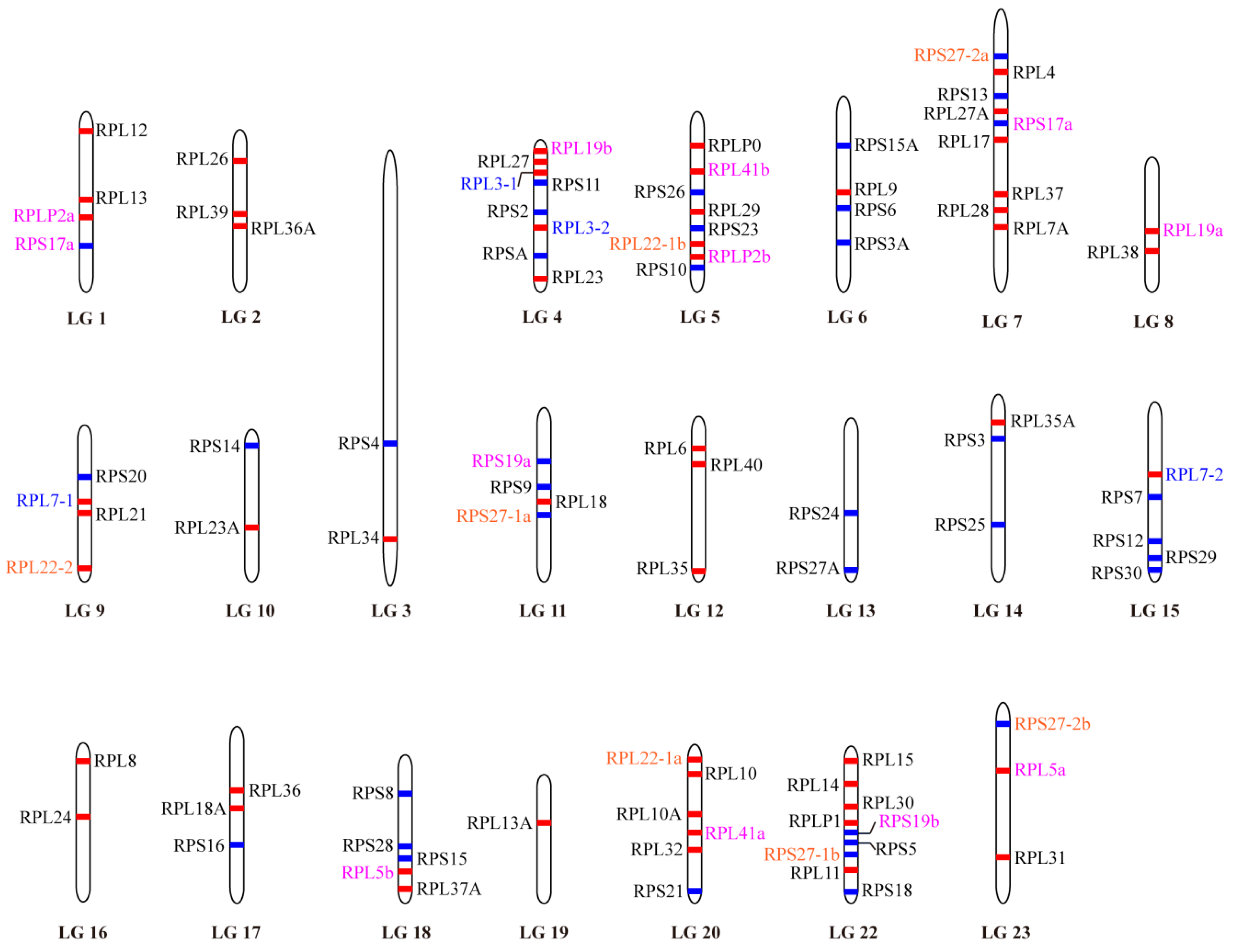


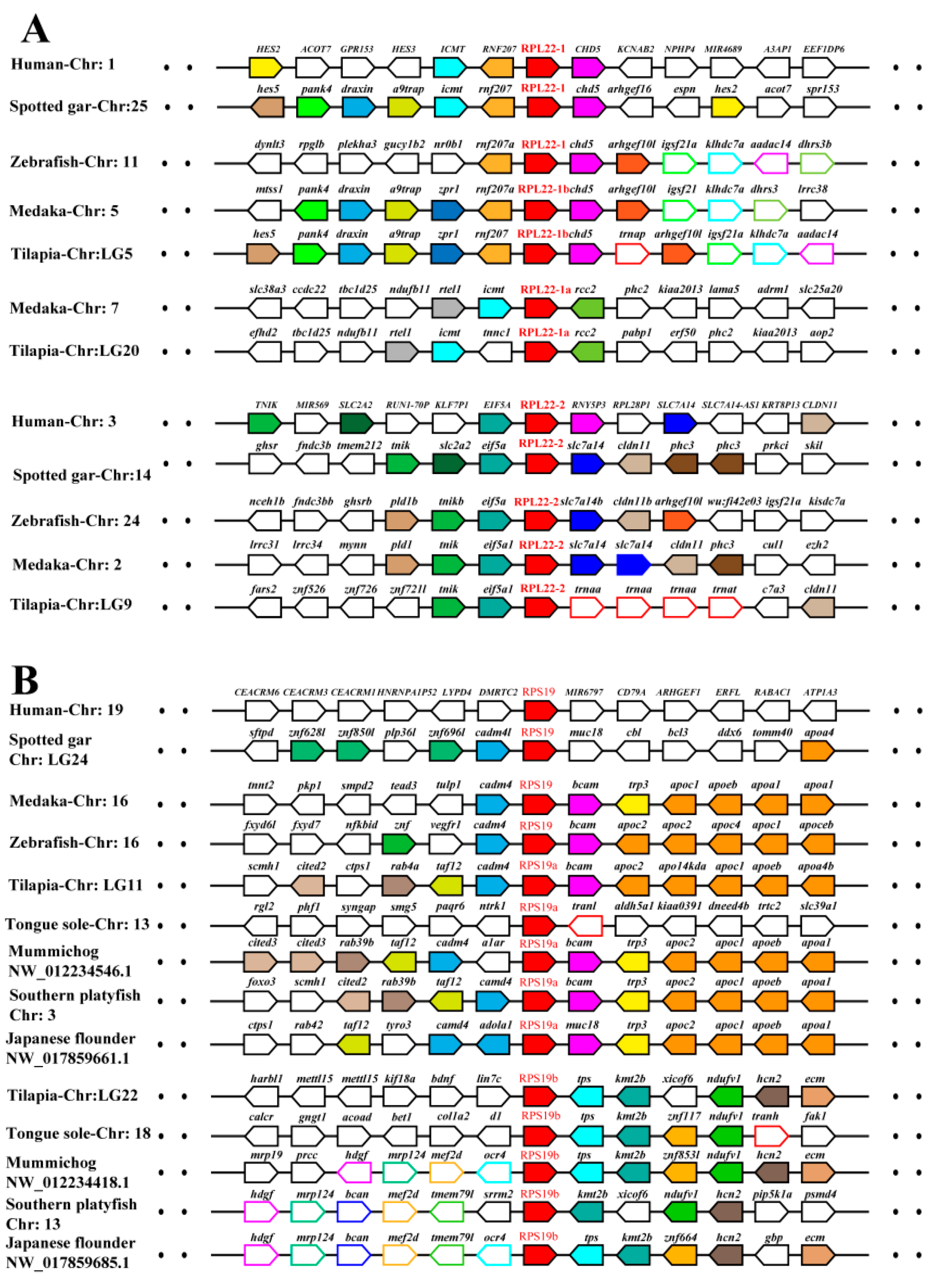
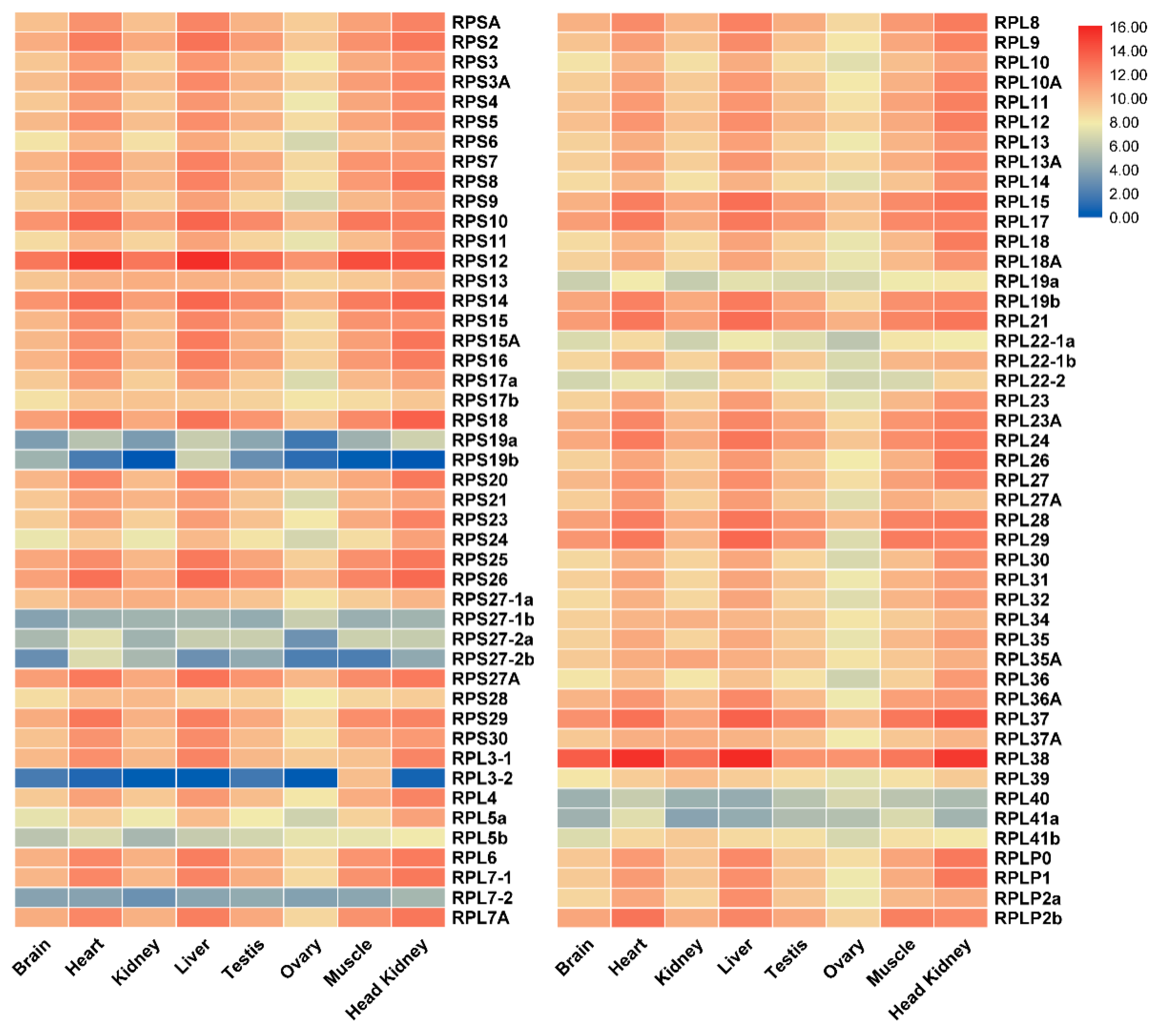
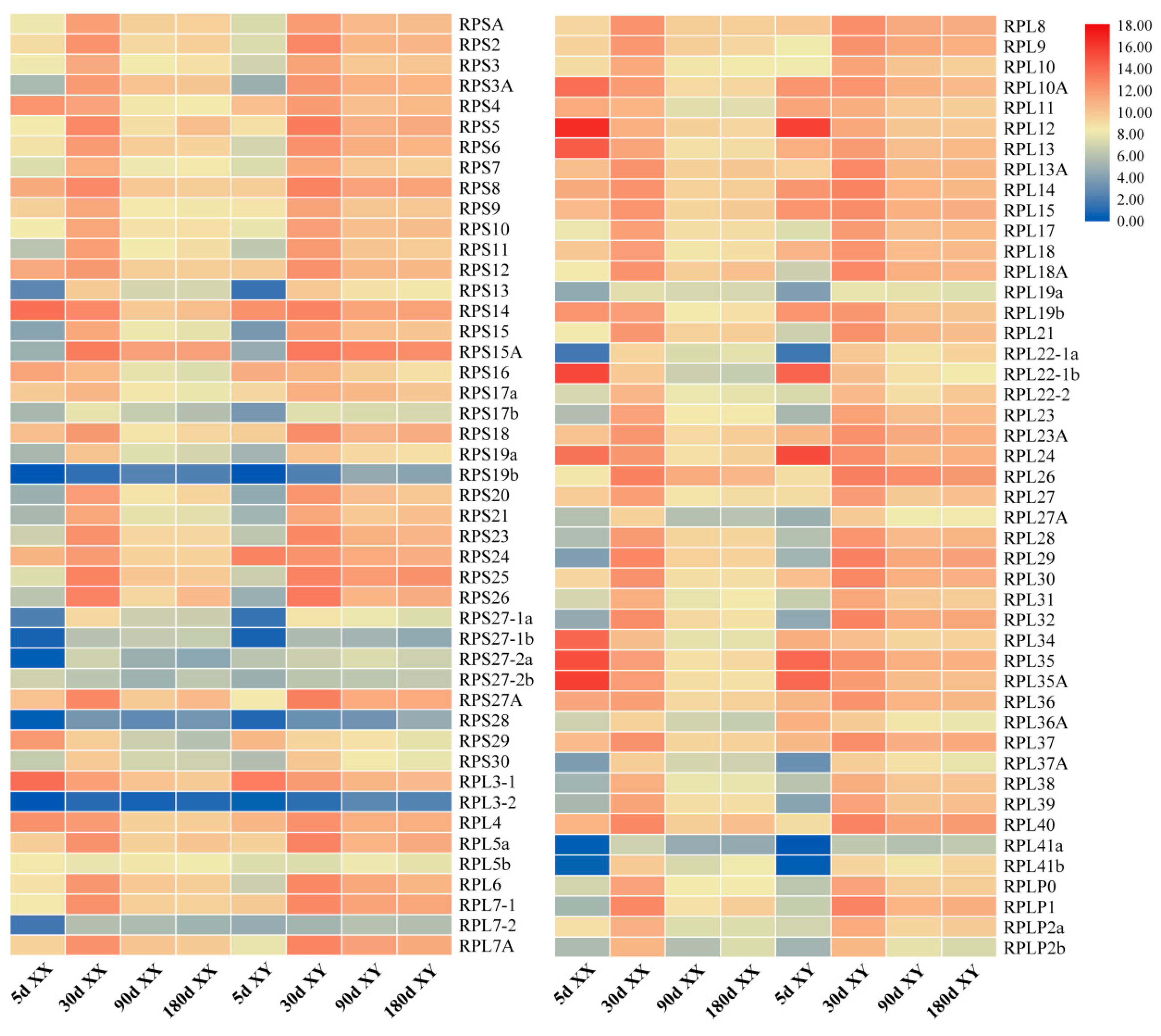



| Gene | Accession Number | Chromosome Location | Exon Count | Gene Length (bp) | Protein Length (aa) |
|---|---|---|---|---|---|
| RPSA | LOC100707344 | LG4 | 8 | 2836 | 307 |
| RPS2 | LOC100693457 | LG4 | 7 | 2113 | 279 |
| RPS3 | LOC100696630 | LG14 | 7 | 6031 | 245 |
| RPS3A | LOC100690034 | LG6 | 6 | 3940 | 266 |
| RPS4 | LOC100709648 | LG3 | 7 | 7165 | 263 |
| RPS5 | LOC100691205 | LG22 | 6 | 3187 | 203 |
| RPS6 | LOC100707655 | LG6 | 6 | 4440 | 249 |
| RPS7 | LOC100704343 | LG15 | 7 | 6283 | 194 |
| RPS8 | LOC100693356 | LG18 | 6 | 4026 | 208 |
| RPS9 | LOC100694704 | LG11 | 5 | 2450 | 194 |
| RPS10 | LOC100708197 | LG5 | 6 | 4577 | 166 |
| RPS11 | LOC100696602 | LG4 | 5 | 2026 | 161 |
| RPS12 | LOC100534443 | LG15 | 5 | 2914 | 132 |
| RPS13 | LOC100696829 | LG7 | 6 | 2934 | 151 |
| RPS14 | LOC100696222 | LG10 | 5 | 2925 | 151 |
| RPS15 | LOC100695708 | LG18 | 4 | 2627 | 145 |
| RPS15A | LOC100698782 | LG6 | 5 | 5166 | 130 |
| RPS16 | LOC100703910 | LG17 | 6 | 3101 | 146 |
| RPS17a | LOC100691912 | LG7 | 5 | 7981 | 134 |
| RPS17b | LOC100699764 | LG1 | 5 | 5281 | 134 |
| RPS18 | LOC100699803 | LG22 | 6 | 3445 | 152 |
| RPS19a | LOC100689943 | LG11 | 6 | 6770 | 146 |
| RPS19b | LOC100690398 | LG22 | 5 | 7095 | 152 |
| RPS20 | LOC100696588 | LG9 | 4 | 1505 | 119 |
| RPS21 | LOC100701551 | LG20 | 6 | 4172 | 83 |
| RPS23 | LOC100690746 | LG5 | 4 | 2080 | 143 |
| RPS24 | LOC100700493 | LG13 | 5 | 4576 | 131 |
| RPS25 | LOC100690179 | LG14 | 5 | 2542 | 123 |
| RPS26 | LOC100700449 | LG5 | 4 | 1749 | 115 |
| RPS27-1a | LOC100705775 | LG11 | 4 | 5189 | 84 |
| RPS27-1b | LOC100696027 | LG22 | 4 | 2376 | 84 |
| RPS27-2a | LOC100691333 | LG7 | 4 | 2298 | 84 |
| RPS27-2b | LOC100690174 | LG23 | 4 | 2477 | 84 |
| RPS27A | LOC100697017 | LG13 | 6 | 2770 | 156 |
| RPS28 | LOC100696499 | LG18 | 4 | 2909 | 69 |
| RPS29 | LOC100703241 | LG15 | 3 | 1056 | 56 |
| RPS30 | LOC100704053 | LG15 | 5 | 1783 | 133 |
| RPL3-1 | LOC100711099 | LG4 | 9 | 4294 | 403 |
| RPL3-2 | LOC100694819 | LG4 | 10 | 3972 | 408 |
| RPL4 | LOC100694481 | LG7 | 10 | 3520 | 369 |
| RPL5a | LOC100708520 | LG23 | 8 | 6819 | 297 |
| RPL5b | LOC100700425 | LG18 | 8 | 3792 | 298 |
| RPL6 | LOC100699139 | LG12 | 6 | 6639 | 227 |
| RPL7-1 | LOC100702526 | LG9 | 7 | 4215 | 245 |
| RPL7-2 | LOC100700318 | LG15 | 7 | 4406 | 246 |
| RPL7A | LOC100693537 | LG7 | 8 | 3256 | 266 |
| RPL8 | LOC100706674 | LG16 | 6 | 3269 | 257 |
| RPL9 | LOC100703191 | LG6 | 8 | 5781 | 192 |
| RPL10 | LOC100697190 | LG20 | 6 | 3989 | 215 |
| RPL10A | LOC100702190 | LG20 | 6 | 1982 | 216 |
| RPL11 | LOC100695881 | LG22 | 6 | 3847 | 178 |
| RPL12 | LOC100695084 | LG1 | 6 | 3525 | 165 |
| RPL13 | LOC100712089 | LG1 | 6 | 4141 | 211 |
| RPL13A | LOC100708460 | LG19 | 7 | 3290 | 205 |
| RPL14 | LOC100534449 | LG22 | 6 | 3561 | 137 |
| RPL15 | LOC100697988 | LG22 | 4 | 2672 | 204 |
| RPL17 | LOC100707041 | LG7 | 7 | 4249 | 184 |
| RPL18 | LOC100534549 | LG11 | 7 | 5614 | 188 |
| RPL18A | LOC100706131 | LG17 | 5 | 3492 | 176 |
| RPL19a | LOC100706003 | LG8 | 6 | 2335 | 195 |
| RPL19b | LOC100709589 | LG4 | 6 | 3080 | 194 |
| RPL21 | LOC100711190 | LG9 | 6 | 2393 | 160 |
| RPL22-1a | LOC100699599 | LG20 | 4 | 3986 | 129 |
| RPL22-1b | LOC100710673 | LG5 | 4 | 3264 | 129 |
| RPL22-2 | LOC100699889 | LG9 | 4 | 2079 | 129 |
| RPL23 | LOC100701640 | LG4 | 5 | 4091 | 140 |
| RPL23A | LOC100705596 | LG10 | 5 | 2677 | 155 |
| RPL24 | LOC100707648 | LG16 | 6 | 3744 | 157 |
| RPL26 | LOC100708525 | LG2 | 4 | 3651 | 145 |
| RPL27 | LOC100711912 | LG4 | 5 | 3838 | 136 |
| RPL27A | LOC100702758 | LG7 | 5 | 6769 | 148 |
| RPL28 | LOC100704095 | LG7 | 5 | 2011 | 137 |
| RPL29 | LOC106098935 | LG5 | 4 | 3933 | 64 |
| RPL30 | LOC100710306 | LG22 | 5 | 3112 | 116 |
| RPL31 | LOC100696251 | LG23 | 5 | 3056 | 124 |
| RPL32 | LOC100534436 | LG20 | 4 | 3255 | 135 |
| RPL34 | LOC109195715 | LG3 | 5 | 2953 | 117 |
| RPL35 | LOC100711334 | LG12 | 4 | 2608 | 123 |
| RPL35A | LOC100711077 | LG14 | 5 | 4038 | 110 |
| RPL36 | LOC 100691103 | LG17 | 4 | 3201 | 105 |
| RPL36A | LOC100690460 | LG2 | 5 | 4870 | 106 |
| RPL37 | LOC100705081 | LG7 | 4 | 4669 | 97 |
| RPL37A | LOC100692490 | LG18 | 4 | 2432 | 92 |
| RPL38 | LOC100711498 | LG8 | 5 | 3295 | 70 |
| RPL39 | LOC100692256 | LG2 | 3 | 3583 | 51 |
| RPL40 | LOC100699676 | LG12 | 5 | 3438 | 128 |
| RPL41a | LOC100711481 | LG20 | 4 | 911 | 25 |
| RPL41b | LOC100699101 | LG5 | 4 | 1905 | 25 |
| RPLP0 | LOC100534569 | LG5 | 8 | 2532 | 315 |
| RPLP1 | LOC100692069 | LG22 | 4 | 2095 | 114 |
| RPLP2a | LOC100691806 | LG1 | 5 | 2962 | 114 |
| RPLP2b | LOC100697185 | LG5 | 5 | 1987 | 114 |
| 5 dah | 30 dah | 90 dah | 180 dah | |||||
|---|---|---|---|---|---|---|---|---|
| XX | XY | XX | XY | XX | XY | XX | XY | |
| Total | 409,026 | 229,345 | 299,234 | 372,252 | 45,903 | 133,854 | 50,678 | 131,097 |
| Average | 4446 | 2493 | 3253 | 4046 | 499 | 1455 | 551 | 1425 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuang, G.; Tao, W.; Zheng, S.; Wang, X.; Wang, D. Genome-Wide Identification, Evolution and Expression of the Complete Set of Cytoplasmic Ribosomal Protein Genes in Nile Tilapia. Int. J. Mol. Sci. 2020, 21, 1230. https://doi.org/10.3390/ijms21041230
Kuang G, Tao W, Zheng S, Wang X, Wang D. Genome-Wide Identification, Evolution and Expression of the Complete Set of Cytoplasmic Ribosomal Protein Genes in Nile Tilapia. International Journal of Molecular Sciences. 2020; 21(4):1230. https://doi.org/10.3390/ijms21041230
Chicago/Turabian StyleKuang, Gangqiao, Wenjing Tao, Shuqing Zheng, Xiaoshuang Wang, and Deshou Wang. 2020. "Genome-Wide Identification, Evolution and Expression of the Complete Set of Cytoplasmic Ribosomal Protein Genes in Nile Tilapia" International Journal of Molecular Sciences 21, no. 4: 1230. https://doi.org/10.3390/ijms21041230
APA StyleKuang, G., Tao, W., Zheng, S., Wang, X., & Wang, D. (2020). Genome-Wide Identification, Evolution and Expression of the Complete Set of Cytoplasmic Ribosomal Protein Genes in Nile Tilapia. International Journal of Molecular Sciences, 21(4), 1230. https://doi.org/10.3390/ijms21041230





