Dormancy-Associated MADS-Box (DAM) Genes Influence Chilling Requirement of Sweet Cherries and Co-Regulate Flower Development with SOC1 Gene
Abstract
1. Introduction
2. Results
2.1. Difference of Dormancy Status and Chilling Requirement between ‘Royal Lee’ and ‘Hongdeng’
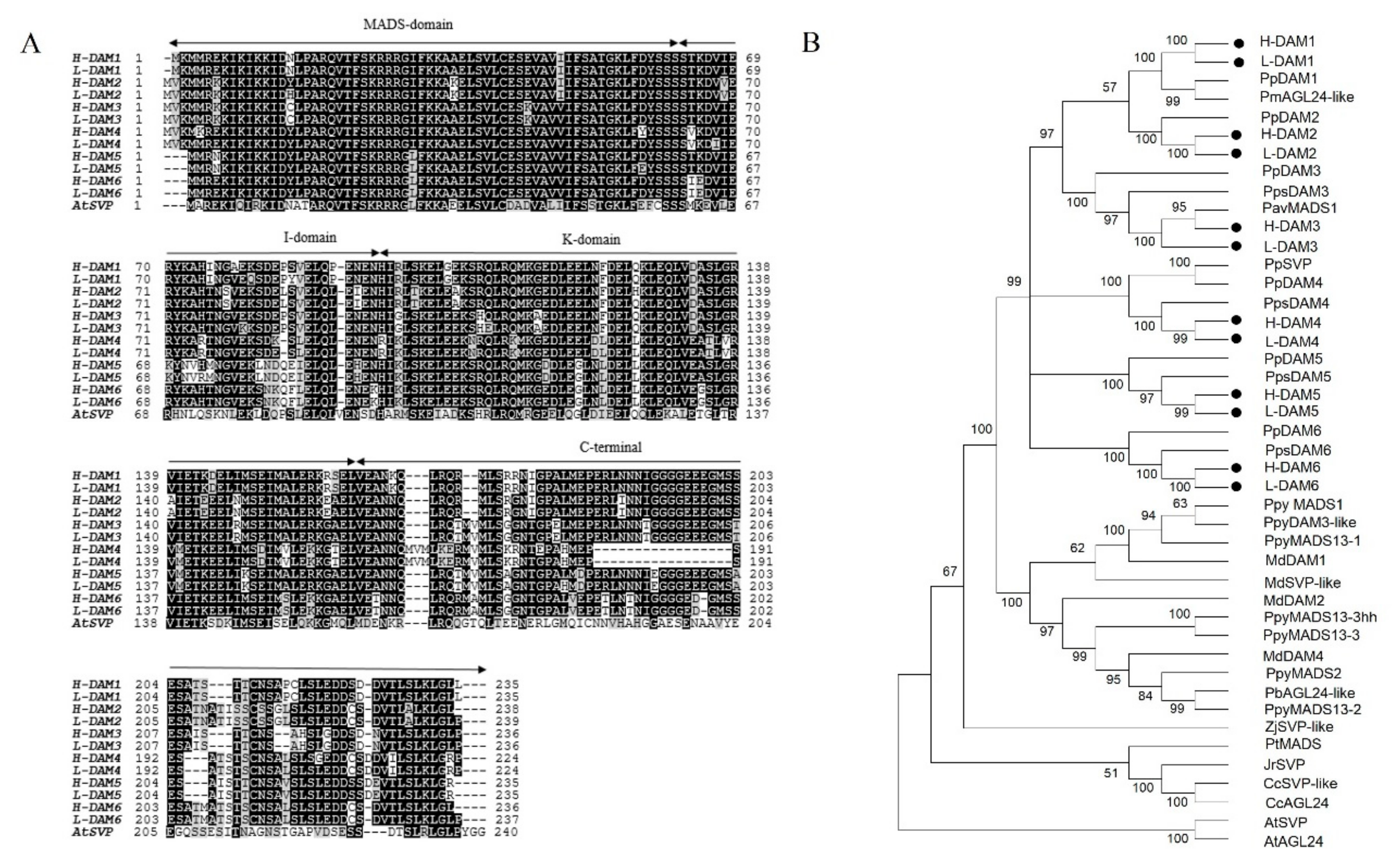
2.2. Identification and Phylogenetic Analysis of Six PavDAM Genes in Low- and High-Chill Cultivars
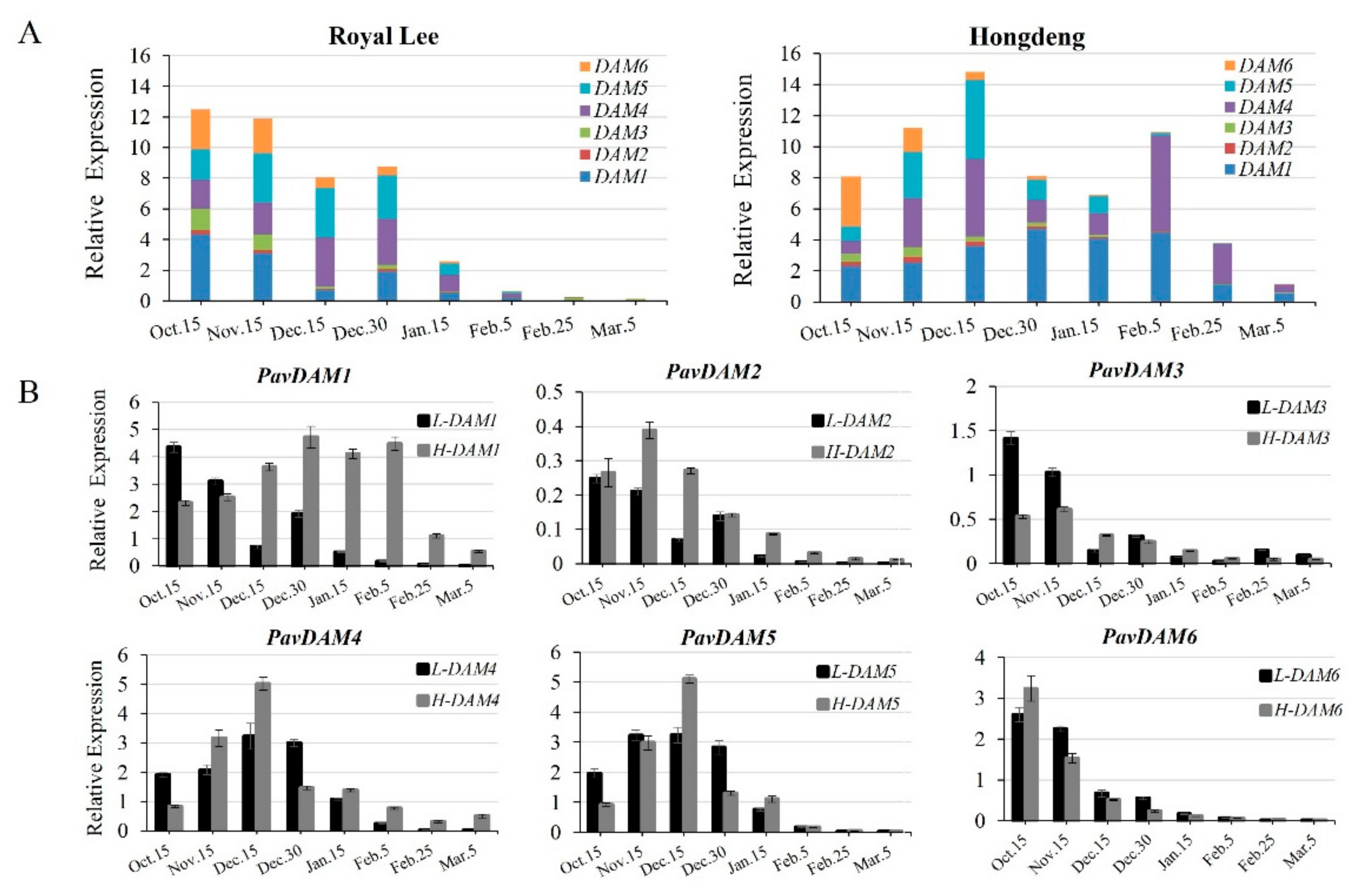
2.3. Expression Analysis of Six PavDAM Genes in Low- and High-Chill Cultivars
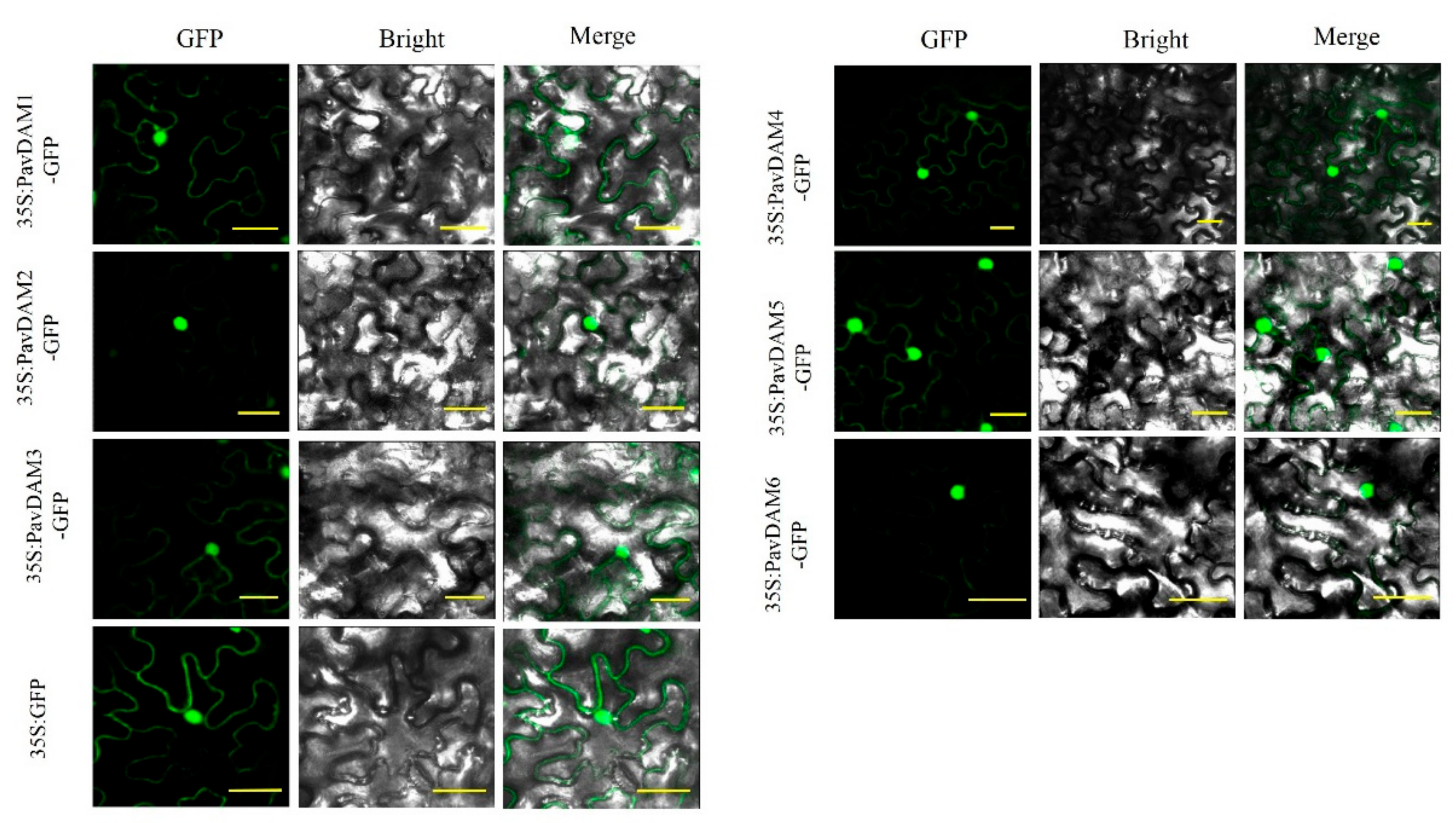
2.4. Subcellular Localization of Six PavDAMs
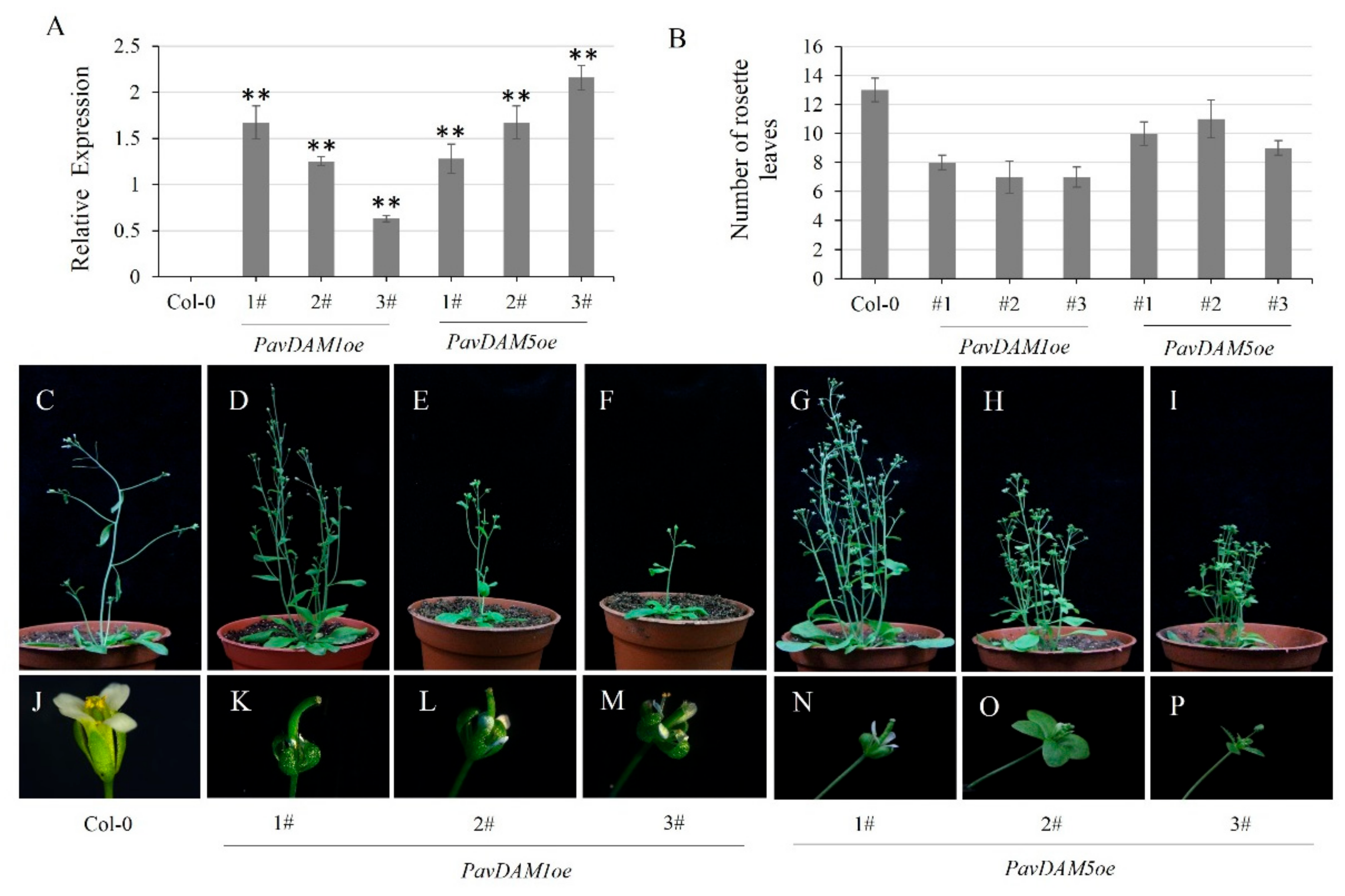
2.5. Ectopic Overexpression of PavDAM1/5 Affect Flower Development in Arabidopsis
2.6. Relative Expression of SOC1 in Arabidopsis and Sweet Cherries
2.7. Ectopic Overexpression of PavSOC1 in Arabidopsis
2.8. DAM Proteins Interact with SOC1 Protein In Vitro and In Vivo in Sweet Cherries
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Evaluation of Dormancy Status and Chilling Requirement for Bud Break
4.3. Characterization of Sweet Cherry DAM Sequences by Gene Cloning and Phylogenetic Analysis
4.4. Real-Time Quantitative RT-PCR Analysis
4.5. Subcellular Localization Assessment
4.6. Generation of Transgenic Arabidopsis
4.7. Bimolecular Fluorescence Complementation (BiFC) Assay
4.8. Yeast Two-Hybrid (Y2H) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
Abbreviations
| DAM | Dormancy-associated mads-box |
| SOC1 | Suppressor of overexpression of co1 |
| FT | Flowering locus T |
| CBF | Cold response genes C-repeat binding factors |
| AGL24 | Agamous-like 24 |
| SVP | Short vegetative phase |
References
- Lang, G.A.; Early, J.D.; Martin, G.C.; Darnell, R.L. Endo-, para- and ecodormancy: Physiological terminology and classification for dormancy research. Hortic. Sci. 1987, 22, 371–377. [Google Scholar]
- Mahmood, K.; Carew, J.G.; Hadley, P.; Battey, N.H. Chill unit models for the sweet cherry cvs Stella, Sunburst and Summit. J. Hortic. Sci. Biotech. 2010, 75, 602–606. [Google Scholar] [CrossRef]
- Alburquerque, N.; García-Montiel, F.; Carrillo, A.; Burgos, L. Chilling and heat requirements of sweet cherry cultivars and the relationship between altitude and the probability of satisfying the chill requirements. Environ. Exp. Bot. 2008, 64, 162–170. [Google Scholar] [CrossRef]
- Laube, J.; Sparks, T.H.; Estrella, N.; Hofler, J.; Ankerst, D.P.; Menzel, A. Chilling outweighs photoperiod in preventing precocious spring development. Global Change Biol. 2014, 20, 170–182. [Google Scholar] [CrossRef]
- Cook, N.C.; Calitz, F.J.; Allderman, L.A.; Steyn, W.J.; Louw, E.D. Diverse patterns in dormancy progression of apple buds under variable winter conditions. Sci. Hortic. 2017, 226, 307–315. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, L.; Ma, C.; Xu, W.; Liu, Z.; Zhang, C.; Matthew, W.D.; Wang, S.P. Impact of chilling accumulation and hydrogen cyanamide on floral organ development of sweet cherry in a warm region. J. Integr. Agric. 2016, 15, 2529–2538. [Google Scholar] [CrossRef]
- Liu, J.; Wang, J.; She, W.; Wang, L.; Luo, M.; Chen, Y.; Li, Y.; Wang, S.; Zhang, C. MADS-Box genes are involved in cultivar- and temperature-dependent formation of multi-pistil and polycarpy in Prunus avium L. J. Plant Growth Regu. 2019, 38, 1017–1027. [Google Scholar] [CrossRef]
- Bielenberg, D.G.; Wang, Y.; Li, Z.; Zhebentyayeva, T.; Fan, S.; Reighard, G.L.; Scorza, R.; Abbott, A.G. Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 2008, 4, 495–507. [Google Scholar] [CrossRef]
- Mimida, N.; Saito, T.; Moriguchi, T.; Suzuki, A.; Komori, S.; Wada, M. Expression of DORMANCY-ASSOCIATED MADS-BOX (DAM)-like genes in apple. Biol. Plantarum 2015, 59, 237–244. [Google Scholar] [CrossRef]
- Porto, D.D.; da Silveira Falavigna, V.; Arenhart, R.A.; Perini, P.; Buffon, V.; Anzanello, R.; dos Santos, H.P.; Fialho, F.B.; de Oliveira, P.R.D.; Revers, L.F. Structural genomics and transcriptional characterization of the Dormancy-Associated MADS-box genes during bud dormancy progression in apple. Tree Genet. Genomes 2016, 12, 46. [Google Scholar] [CrossRef]
- Niu, Q.; Li, J.; Cai, D.; Qian, M.; Jia, H.; Bai, S.; Hussain, S.; Liu, G.; Teng, Y.; Zheng, X. Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 2015, 67, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Zhou, Y.; Ahmad, S.; Xu, Z.; Li, Y.; Yang, W.; Cheng, T.; Wang, J.; Zhang, Q. Comprehensive cloning of prunus mume dormancy associated MADS-Box genes and their response in flower bud development and dormancy. Front. Plant. Sci. 2018, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Bielenberg, D.G.; Wang, Y.; Fan, S.; Reighard, G.L.; Scorza, R.; Abbott, A.G. A deletion affecting several gene candidates is present in the Evergrowing peach mutant. J. Hered. 2004, 95, 436–444. [Google Scholar] [CrossRef]
- Yamane, H.; Ooka, T.; Jotatsu, H.; Hosaka, Y.; Sasaki, R.; Tao, R. Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. J. Exp. Bot. 2011, 62, 3481–3488. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, S.; Reighard, G.L.; Bielenberg, D.G. Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Mol. Biol. 2010, 73, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, R.; Yamane, H.; Ooka, T.; Jotatsu, H.; Kitamura, Y.; Akagi, T.; Tao, R. Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 2011, 157, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Yamane, H.; Tao, R. Functional characterization of japanese apricot (Prunus mume) DORMANCY-ASSOCIATED MADS-box1 (PmDAM1), a Paralog of PmDAM6, using Populus transformants. In Advances in Plant Dormancy; Springer: Berlin/Heidelberg, Germany, 2015; pp. 147–157. [Google Scholar]
- Wu, R.; Tomes, S.; Karunairetnam, S.; Tustin, S.D.; Hellens, R.P.; Allan, A.C.; Macknight, R.C.; Varkonyi-Gasic, E. SVP-like MADS-Box genes control dormancy and budbreak in apple. Front. Plant. Sci. 2017, 8, 477. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, Y.; Xin, D.; Chen, W.; Shao, X.; Wang, Y.; Guo, W. RNA-Seq-based transcriptome analysis of dormant flower buds of Chinese cherry (Prunus pseudocerasus). Gene 2015, 555, 362–376. [Google Scholar] [CrossRef]
- Zhao, K.; Zhou, Y.; Ahmad, S.; Yong, X.; Xie, X.; Han, Y.; Li, Y.; Sun, L.; Zhang, Q. PmCBFs synthetically affect PmDAM6 by alternative promoter binding and protein complexes towards the dormancy of bud for Prunus mume. Sci. Rep. 2018, 8, 4527. [Google Scholar] [CrossRef]
- Kitamura, Y.; Takeuchi, T.; Yamane, H.; Tao, R. Simultaneous down-regulation of DORMANCY-ASSOCIATED MADS-box 6 and SOC1 during dormancy release in Japanese apricot (Prunus mume) flower buds. J. Hortic. Sci. Biotech. 2016, 91, 476–482. [Google Scholar] [CrossRef]
- Lee, J.; Lee, I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef]
- Liu, C.; Chen, H.; Er, H.L.; Soo, H.M.; Kumar, P.P.; Han, J.H.; Liou, Y.C.; Yu, H. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 2008, 135, 1481–1491. [Google Scholar] [CrossRef] [PubMed]
- de Folter, S.; Immink, R.G.; Kieffer, M.; Parenicova, L.; Henz, S.R.; Weigel, D.; Busscher, M.; Kooiker, M.; Colombo, L.; Kater, M.M.; et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 2005, 17, 1424–1433. [Google Scholar] [CrossRef] [PubMed]
- Mouhu, K.; Kurokura, T.; Koskela, E.A.; Albert, V.A.; Elomaa, P.; Hytonen, T. The Fragaria vesca homolog of suppressor of overexpression of constans1 represses flowering and promotes vegetative growth. Plant Cell 2013, 25, 3296–3310. [Google Scholar] [CrossRef]
- Weinberger, J.H. Chilling requirements of peach varieties. Proc. Am. Soc. Hortic. Sci. 1950, 56, 122–128. [Google Scholar]
- Horvath, D.P.; Sung, S.; Kim, D.; Chao, W.; Anderson, J. Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol. Biol. 2010, 73, 169–179. [Google Scholar] [CrossRef]
- Kumar, G.; Arya, P.; Gupta, K.; Randhawa, V.; Acharya, V.; Singh, A.K. Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malusx domestica). Sci. Rep. 2016, 6, 20695. [Google Scholar] [CrossRef]
- Wu, R.; Wang, T.; Warren, B.A.W.; Allan, A.C.; Macknight, R.C.; Varkonyi-Gasic, E. Kiwifruit SVP2 gene prevents premature budbreak during dormancy. J. Exp. Bot. 2017, 68, 1071–1082. [Google Scholar] [CrossRef]
- Wisniewski, M.; Norelli, J.; Artlip, T. Overexpression of a peach CBF gene in apple: A model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant. Sci. 2015, 6, 85. [Google Scholar] [CrossRef]
- Zheng, C.; Halaly, T.; Acheampong, A.K.; Takebayashi, Y.; Jikumaru, Y.; Kamiya, Y.; Or, E. Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism. J. Exp. Bot. 2015, 66, 1527–1542. [Google Scholar] [CrossRef]
- Wang, D.; Gao, Z.; Du, P.; Xiao, W.; Tan, Q.; Chen, X.; Li, L.; Gao, D. Expression of ABA metabolism-related genes suggests similarities and differences between seed dormancy and bud dormancy of peach (Prunus persica). Front. Plant. Sci. 2015, 6, 1248. [Google Scholar] [CrossRef] [PubMed]
- Browning, G. Flower Bud Dormancy in Coffea Arabica L. I. Studies of gibberellin in flower buds and xylem sap and of abscisic acid in flower buds in relation to dormancy release. J. Hortic. Sci. 1973, 48, 29–41. [Google Scholar] [CrossRef]
- Wen, L.H.; Zhong, W.J.; Huo, X.M.; Zhuang, W.B.; Ni, Z.J.; Gao, Z.H. Expression analysis of ABA- and GA-related genes during four stages of bud dormancy in Japanese apricot (Prunus mume Sieb. et Zucc). J. Hortic. Sci. Biotech. 2016, 91, 362–369. [Google Scholar] [CrossRef]
- Zheng, C.; Kwame Acheampong, A.; Shi, Z.; Halaly, T.; Kamiya, Y.; Ophir, R.; Galbraith, D.W.; Or, E. Distinct gibberellin functions during and after grapevine bud dormancy release. J. Exp. Bot. 2018, 69, 1635–1648. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, C.; Huang, J.; Zhu, L.; Yu, X.; Li, J.; Lou, Y.; Xu, W.; Wang, S.; Ma, C. Hydrogen cyanamide improves endodormancy release and blooming associated with endogenous hormones in ‘Summit’ sweet cherry trees. New Zeal. J. Crop Hort. 2016, 45, 14–28. [Google Scholar] [CrossRef]
- Leseberg, C.H.; Li, A.; Kang, H.; Duvall, M.; Mao, L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene 2006, 378, 84–94. [Google Scholar] [CrossRef]
- Hartmann, U.; Höhmann, S.; Nettesheim, K.; Wisman, E.; Saedler, H.; Huijser, P. Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 2000, 21, 351–360. [Google Scholar] [CrossRef]
- Wu, R.; Wang, T.; Allan, A.C.; Macknight, R.C.; Varkonyi-Gasic, E. Overexpression of both AcSVP1 and AcSVP4 delays budbreak in kiwifruit A. chinensis var. deliciosa, but only AcSVP1 delays flowering in model plants. Environ. Exp. Bot. 2018, 153, 262–270. [Google Scholar]
- Wu, R.; Walton, E.; Richardson, A.C.; Wood, M.; Hellens, R.P.; Varkonyi-Gasic, E. Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J. Exp. Bot. 2012, 63, 797–807. [Google Scholar] [CrossRef]
- Jaudal, M.; Monash, J.; Zhang, L.; Wen, J.; Mysore, K.S.; Macknight, R.; Putterill, J. Overexpression of Medicago SVP genes causes floral defects and delayed flowering in Arabidopsis but only affects floral development in Medicago. J. Exp. Bot. 2014, 65, 429–442. [Google Scholar] [CrossRef]
- Zhang, H.; Yan, H.; Zhang, D.; Yu, D. Ectopic expression of a soybean SVP-like gene in tobacco causes abnormal floral organs and shortens the vegetative phase. Plant Growth Regul. 2016, 80, 345–353. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, Y.; Yang, W.; Cheng, T.; Wang, J.; Zhang, Q. Isolation and functional characterization of SVP-like genes in Prunus mume. Sci. Hortic. 2017, 215, 91–101. [Google Scholar] [CrossRef]
- Wu, R.; Wang, T.; McGie, T.; Voogd, C.; Allan, A.C.; Hellens, R.P.; Varkonyi-Gasic, E. Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time. J. Exp. Bot. 2014, 65, 4985–4995. [Google Scholar] [CrossRef] [PubMed]
- Trainin, T.; Bar-Ya’akov, I.; Holland, D. ParSOC1, a MADS-box gene closely related to Arabidopsis AGL20/SOC1, is expressed in apricot leaves in a diurnal manner and is linked with chilling requirements for dormancy break. Tree Genet. Genomes 2013, 9, 753–766. [Google Scholar] [CrossRef]
- Trainin, T.; Bar-Ya’akov, I.; Holland, D. The genetic components involved in sensing chilling requirements in apricot. In Advances in Plant Dormancy; Anderson, J.V., Ed.; Springer: Berlin, Germany, 2015; pp. 159–168. [Google Scholar]
- Voogd, C.; Wang, T.; Varkonyi-Gasic, E. Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy. J. Exp. Bot. 2015, 66, 4699–4710. [Google Scholar] [CrossRef]
- Jia, H.; Jiu, S.; Zhang, C.; Wang, C.; Tariq, P.; Liu, Z.; Wang, B.; Cui, L.; Fang, J. Abscisic acid and sucrose regulate tomato and strawberry fruit ripening through the abscisic acid-stress-ripening transcription factor. Plant Biotechnol. J. 2016, 14, 2045–2065. [Google Scholar] [CrossRef]
- Jiu, S.; Wang, C.; Zheng, T.; Liu, Z.; Leng, X.; Pervaiz, T.; Lotfi, A.; Fang, J.; Wang, X. Characterization of VvPAL-like promoter from grapevine using transgenic tobacco plants. Funct. Integr. Genomic. 2016, 16, 595–617. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Gao, Z.; Li, H.; Jiu, S.; Qu, Y.; Wang, L.; Ma, C.; Xu, W.; Wang, S.; Zhang, C. Dormancy-Associated MADS-Box (DAM) Genes Influence Chilling Requirement of Sweet Cherries and Co-Regulate Flower Development with SOC1 Gene. Int. J. Mol. Sci. 2020, 21, 921. https://doi.org/10.3390/ijms21030921
Wang J, Gao Z, Li H, Jiu S, Qu Y, Wang L, Ma C, Xu W, Wang S, Zhang C. Dormancy-Associated MADS-Box (DAM) Genes Influence Chilling Requirement of Sweet Cherries and Co-Regulate Flower Development with SOC1 Gene. International Journal of Molecular Sciences. 2020; 21(3):921. https://doi.org/10.3390/ijms21030921
Chicago/Turabian StyleWang, Jiyuan, Zhen Gao, Hui Li, Songtao Jiu, Yueting Qu, Lei Wang, Chao Ma, Wenping Xu, Shiping Wang, and Caixi Zhang. 2020. "Dormancy-Associated MADS-Box (DAM) Genes Influence Chilling Requirement of Sweet Cherries and Co-Regulate Flower Development with SOC1 Gene" International Journal of Molecular Sciences 21, no. 3: 921. https://doi.org/10.3390/ijms21030921
APA StyleWang, J., Gao, Z., Li, H., Jiu, S., Qu, Y., Wang, L., Ma, C., Xu, W., Wang, S., & Zhang, C. (2020). Dormancy-Associated MADS-Box (DAM) Genes Influence Chilling Requirement of Sweet Cherries and Co-Regulate Flower Development with SOC1 Gene. International Journal of Molecular Sciences, 21(3), 921. https://doi.org/10.3390/ijms21030921




