Resveratrol as Inducer of Autophagy, Pro-Survival, and Anti-Inflammatory Stimuli in Cultured Human RPE Cells
Abstract
1. Introduction
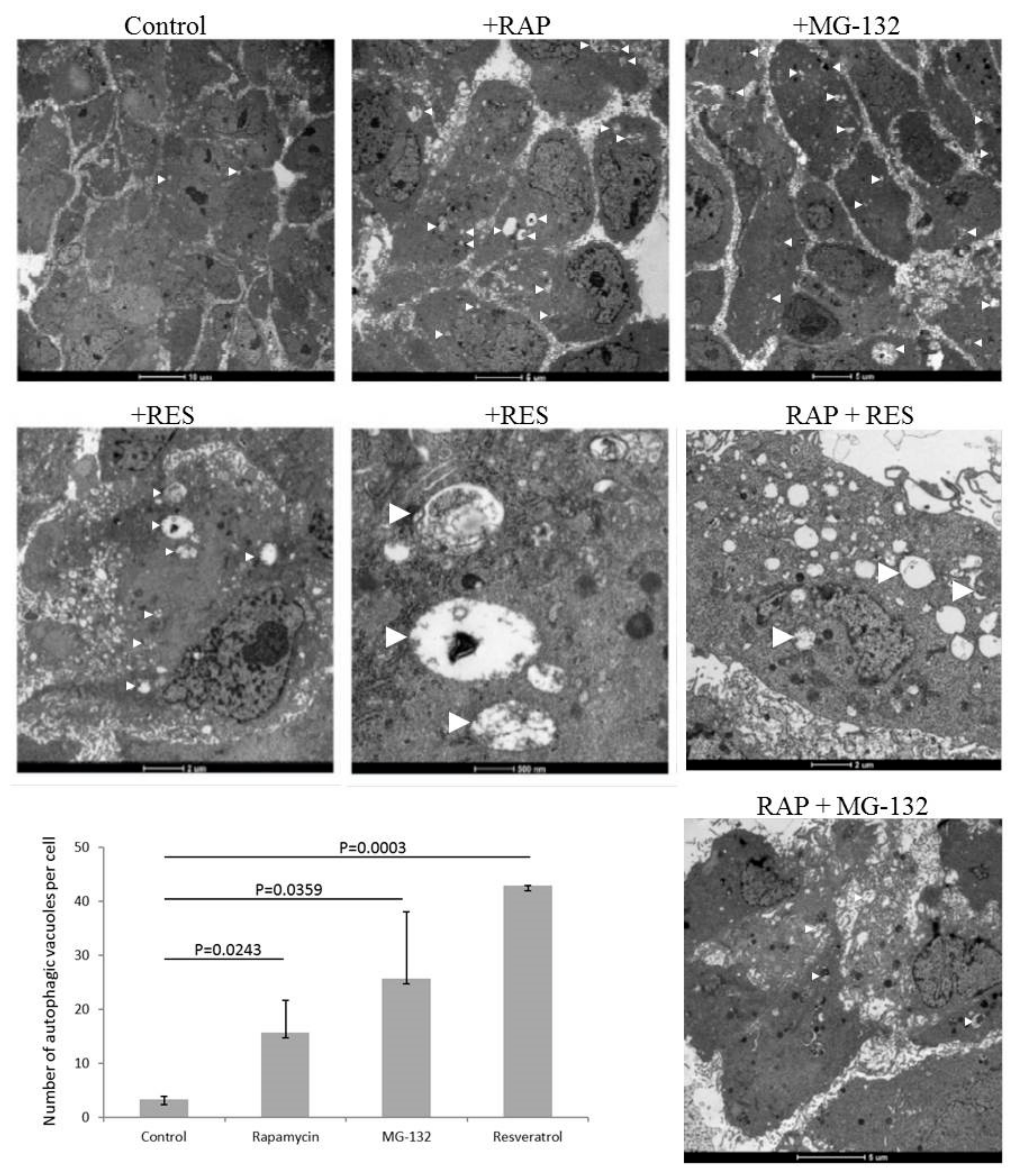
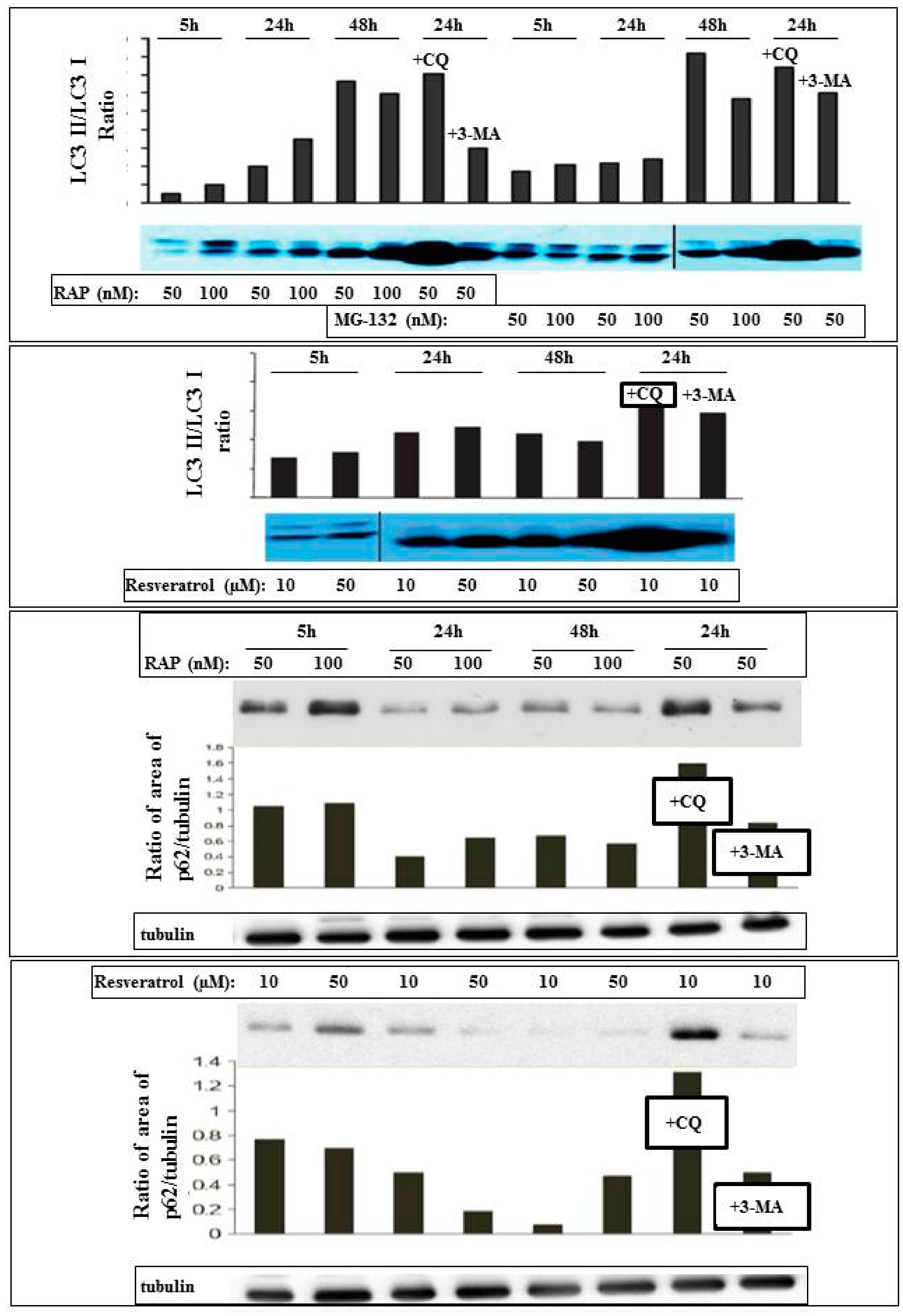
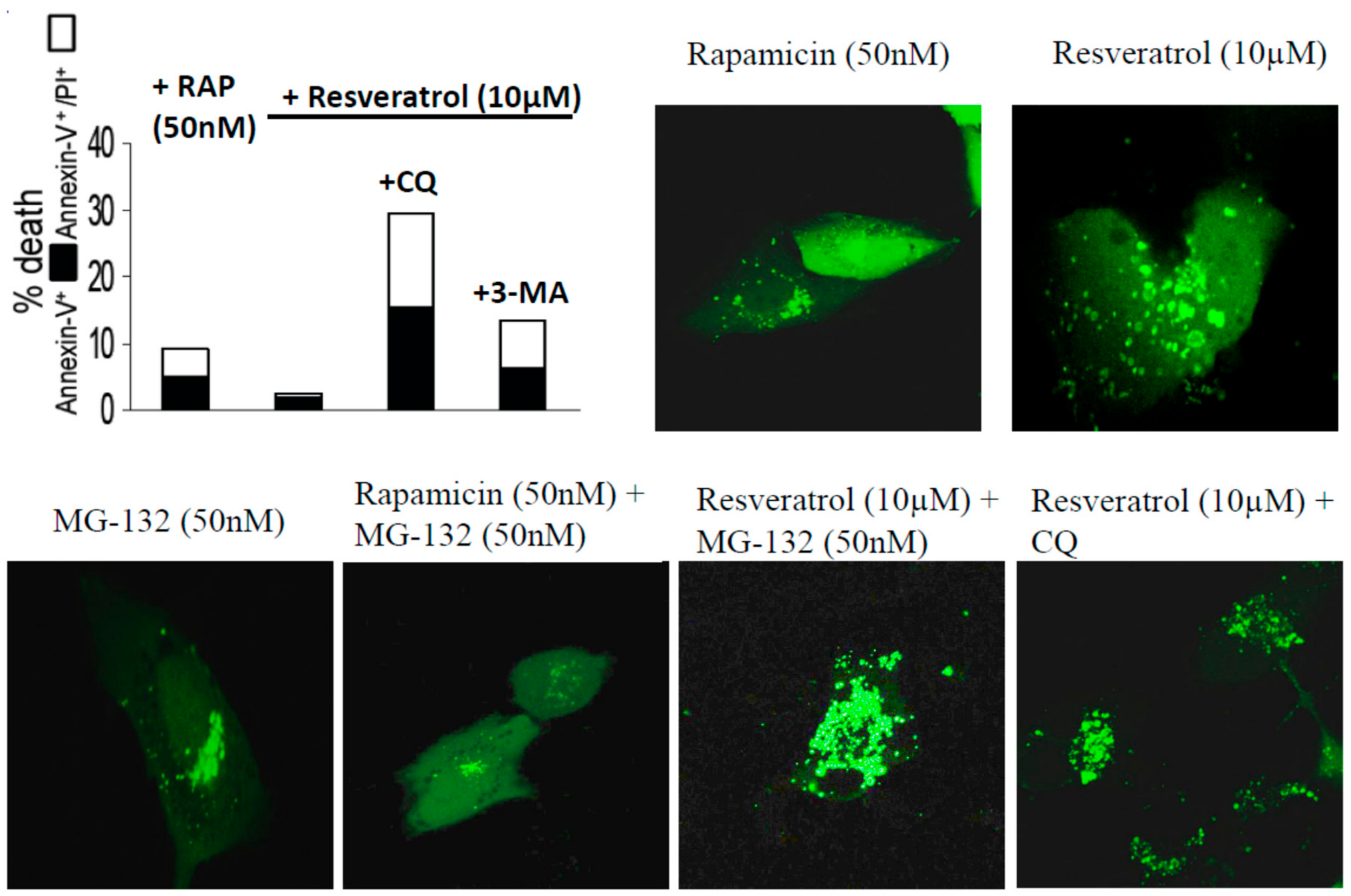
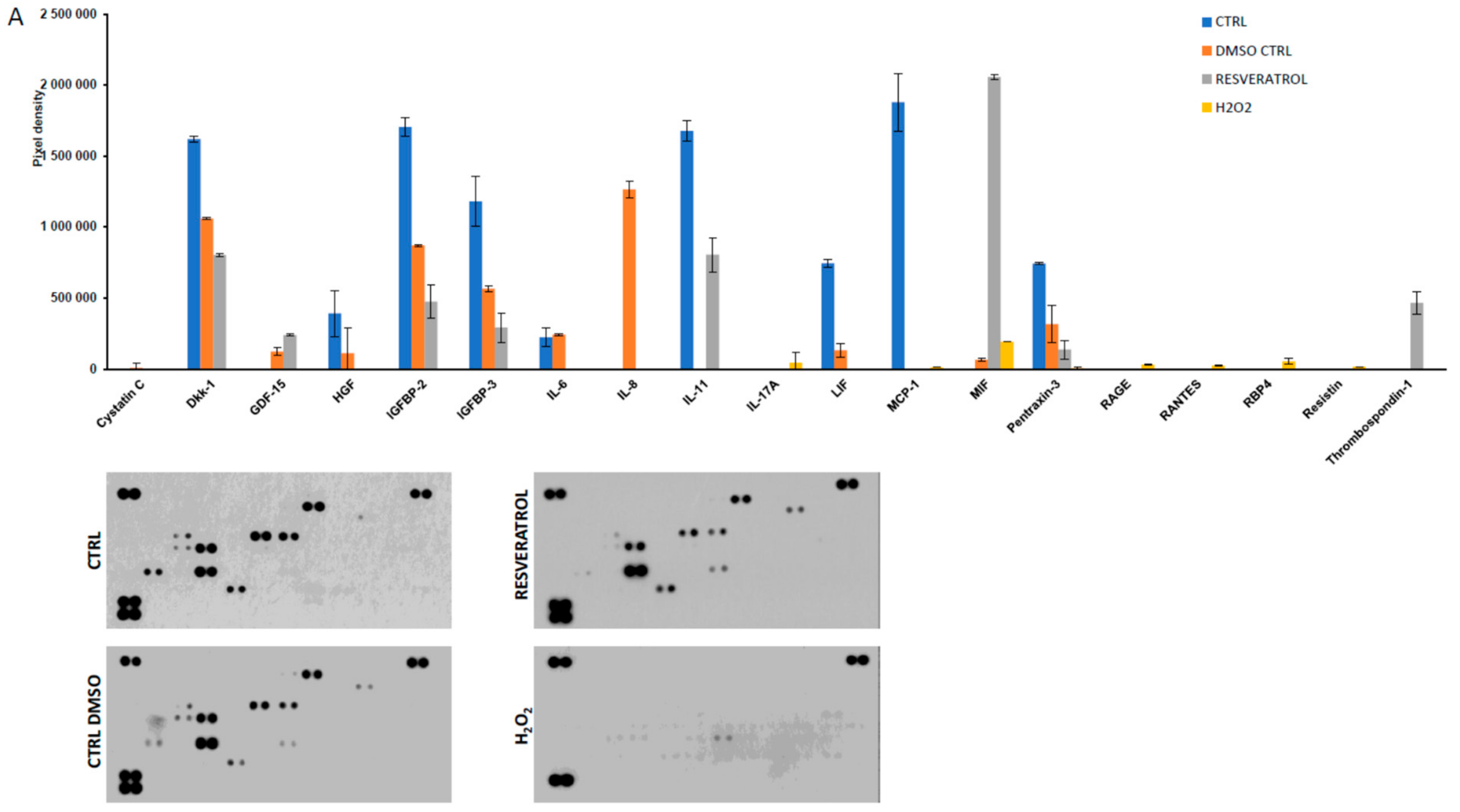
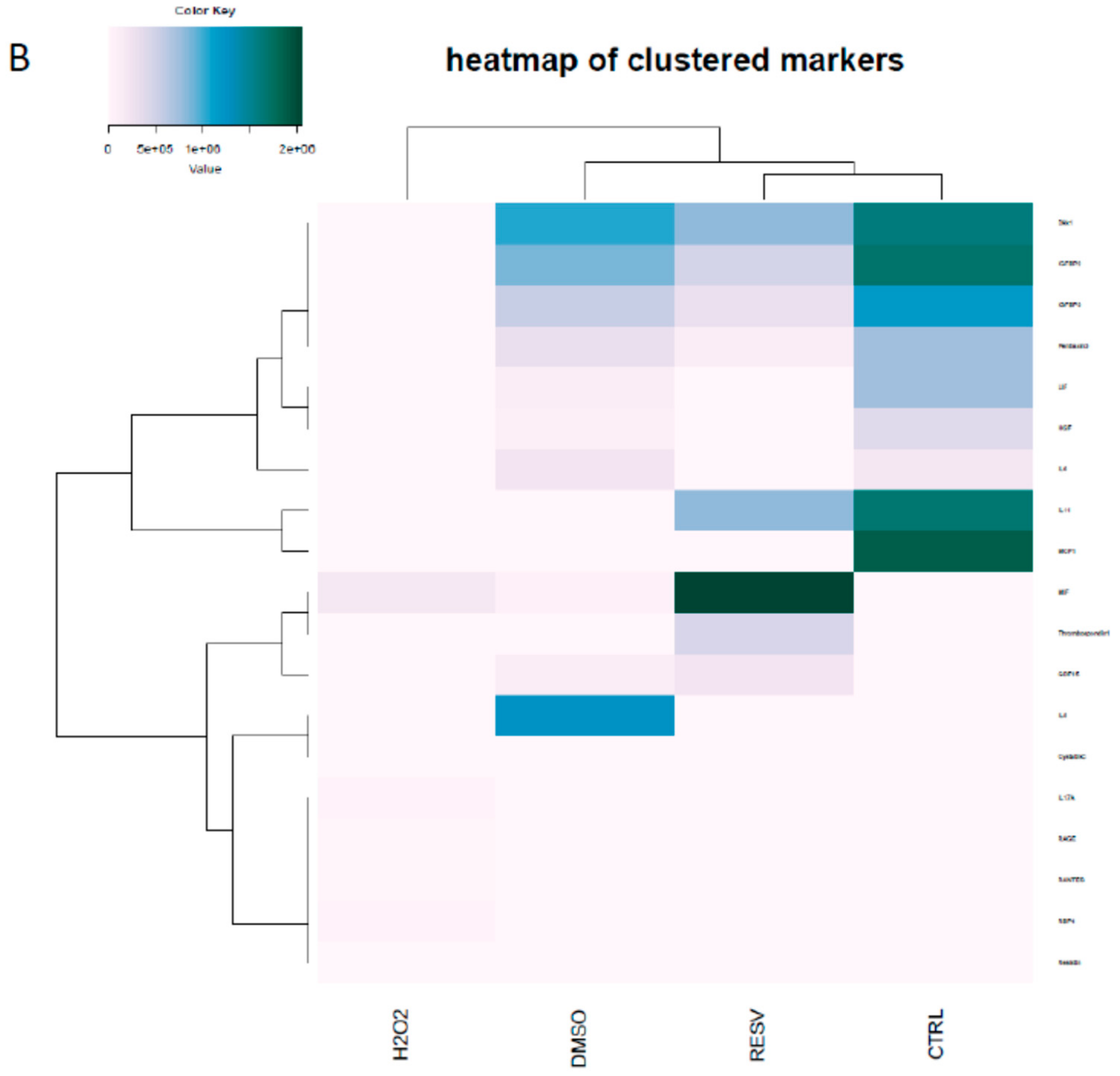
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatments
4.2. Assays of Cell Death and Autophagy
4.3. Transmission Electron Microscopy (TEM)
4.4. Antibodies and Immunoblotting
4.5. Detection of Secreted Cytokines by Protein Array
4.6. Statistical Analysis
4.7. Sample and Data Availability
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blasiak, J.; Piechota, M.; Pawlowska, E.; Szatkowska, M.; Sikora, E.; Kaarniranta, K. Cellular Senescence in Age-Related Macular Degeneration: Can Autophagy and DNA Damage Response Play a Role? Oxid. Med. Cell. Longev. 2017, 2017, 5293258. [Google Scholar] [CrossRef] [PubMed]
- Kaarniranta, K.; Tokarz, P.; Koskela, A.; Paterno, J.; Blasiak, J. Autophagy regulates death of retinal pigment epithelium cells in age-related macular degeneration. Cell Biol. Toxicol. 2017, 33, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Mitter, S.K.; Qi, X.; Beli, E.; Rao, H.V.; Ding, J.; Ip, C.S.; Gu, H.; Akin, D.; Dunn, W.A., Jr.; et al. Oxidative stress-mediated NFkappaB phosphorylation upregulates p62/SQSTM1 and promotes retinal pigmented epithelial cell survival through increased autophagy. PLoS ONE 2017, 12, e0171940. [Google Scholar] [CrossRef]
- Mansoor, S.; Gupta, N.; Patil, A.J.; Estrago-Franco, M.F.; Ramirez, C.; Migon, R.; Sapkal, A.; Kuppermann, B.D.; Kenney, M.C. Inhibition of apoptosis in human retinal pigment epithelial cells treated with benzo(e)pyrene, a toxic component of cigarette smoke. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2601–2607. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, Z.; Yang, S.; Yin, T.; Zhang, Y.; Qin, Y.; Weinreb, R.N.; Sun, X. Tissue Distribution of trans-Resveratrol and Its Metabolites after Oral Administration in Human Eyes. J. Ophthalmol. 2017, 2017, 4052094. [Google Scholar] [CrossRef] [PubMed]
- Mitter, S.K.; Song, C.; Qi, X.; Mao, H.; Rao, H.; Akin, D.; Lewin, A.; Grant, M.; Dunn, W., Jr.; Ding, J.; et al. Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy 2014, 10, 1989–2005. [Google Scholar] [CrossRef]
- Sheu, S.J.; Chen, J.L.; Bee, Y.S.; Chen, Y.A.; Lin, S.H.; Shu, C.W. Differential autophagic effects of vital dyes in retinal pigment epithelial ARPE-19 and photoreceptor 661W cells. PLoS ONE 2017, 12, e0174736. [Google Scholar] [CrossRef]
- Mo, J.; Zhang, M.; Marshall, B.; Smith, S.; Covar, J.; Atherton, S. Interplay of autophagy and apoptosis during murine cytomegalovirus infection of RPE cells. Mol. Vis. 2014, 20, 1161–1173. [Google Scholar]
- Petrovski, G.; Gurusamy, N.; Das, D.K. Resveratrol in cardiovascular health and disease. Ann. N. Y. Acad. Sci. 2011, 1215, 22–33. [Google Scholar] [CrossRef]
- Abu-Amero, K.K.; Kondkar, A.A.; Chalam, K.V. Resveratrol and Ophthalmic Diseases. Nutrients 2016, 8, 200. [Google Scholar] [CrossRef]
- Pintea, A.; Rugina, D.; Pop, R.; Bunea, A.; Socaciu, C.; Diehl, H.A. Antioxidant effect of trans-resveratrol in cultured human retinal pigment epithelial cells. J. Ocul. Pharmacol. Ther. 2011, 27, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.M.; Huang, C.H.; Li, H.J.; Hsiao, C.Y.; Su, C.C.; Lee, P.L.; Hung, C.F. Protective effects of resveratrol against UVA-induced damage in ARPE19 cells. Int. J. Mol. Sci. 2015, 16, 5789–5802. [Google Scholar] [CrossRef] [PubMed]
- Koskela, A.; Reinisalo, M.; Petrovski, G.; Sinha, D.; Olmiere, C.; Karjalainen, R.; Kaarniranta, K. Nutraceutical with Resveratrol and Omega-3 Fatty Acids Induces Autophagy in ARPE-19 Cells. Nutrients 2016, 8, 284. [Google Scholar] [CrossRef] [PubMed]
- Richer, S.; Patel, S.; Sockanathan, S.; Ulanski, L.J., 2nd; Miller, L.; Podella, C. Resveratrol based oral nutritional supplement produces long-term beneficial effects on structure and visual function in human patients. Nutrients 2014, 6, 4404–4420. [Google Scholar] [CrossRef]
- Lee, C.S.; Choi, E.Y.; Lee, S.C.; Koh, H.J.; Lee, J.H.; Chung, J.H. Resveratrol Inhibits Hypoxia-Induced Vascular Endothelial Growth Factor Expression and Pathological Neovascularization. Yonsei Med. J. 2015, 56, 1678–1685. [Google Scholar] [CrossRef]
- Fiorentini, D.; Zambonin, L.; Dalla Sega, F.V.; Hrelia, S. Polyphenols as Modulators of Aquaporin Family in Health and Disease. Oxid. Med. Cell. Longev. 2015, 196914. [Google Scholar] [CrossRef]
- Jang, M.; Cai, L.; Udeani, G.O.; Slowing, K.V.; Thomas, C.F.; Beecher, C.W.; Fong, H.H.; Farnsworth, N.R.; Kinghorn, A.D.; Mehta, R.G.; et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science 1997, 275, 218–220. [Google Scholar] [CrossRef]
- Bola, C.; Bartlett, H.; Eperjesi, F. Resveratrol and the eye: Activity and molecular mechanisms. Graefes Arch. Clin. Exp. Ophthalmol. 2014, 252, 699–713. [Google Scholar] [CrossRef]
- Subramani, M.; Ponnalagu, M.; Krishna, L.; Jeyabalan, N.; Chevour, P.; Sharma, A.; Jayadev, C.; Shetty, R.; Begum, N.; Archunan, G.; et al. Resveratrol reverses the adverse effects of bevacizumab on cultured ARPE-19 cells. Sci. Rep. 2017, 7, 12242. [Google Scholar] [CrossRef]
- Nagineni, C.N.; Raju, R.; Nagineni, K.K.; Kommineni, V.K.; Cherukuri, A.; Kutty, R.K.; Hooks, J.J.; Detrick, B. Resveratrol Suppresses Expression of VEGF by Human Retinal Pigment Epithelial Cells: Potential Nutraceutical for Age-related Macular Degeneration. Aging Dis. 2014, 5, 88–100. [Google Scholar] [CrossRef]
- Lancon, A.; Frazzi, R.; Latruffe, N. Anti-Oxidant, Anti-Inflammatory and Anti-Angiogenic Properties of Resveratrol in Ocular Diseases. Molecules 2016, 21, 304. [Google Scholar] [CrossRef] [PubMed]
- Cunha-Vaz, J.; Bernardes, R.; Lobo, C. Blood-retinal barrier. Eur. J. Ophthalmol. 2011, 21 (Suppl. 6), S3–S9. [Google Scholar] [CrossRef] [PubMed]
- Hyttinen, J.M.; Petrovski, G.; Salminen, A.; Kaarniranta, K. 5’-Adenosine monophosphate-activated protein kinase--mammalian target of rapamycin axis as therapeutic target for age-related macular degeneration. Rejuvenation Res. 2011, 14, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Andjelić, S.; Drašlar, K.; Lumi, X.; Yan, X.; Graw, J.; Facskó, A.; Hawlina, M.; Petrovski, G. Morphological and proliferative studies on ex vivo cultured human anterior lens epithelial cells–relevance to capsular opacification. Acta Ophthalmol. 2015, 93, e499–e506. [Google Scholar] [CrossRef]
- Radomska-Lesniewska, D.M.; Skopinski, P.; Balan, B.J.; Bialoszewska, A.; Jozwiak, J.; Rokicki, D.; Skopinska-Rozewska, E.; Borecka, A.; Hevelke, A. Angiomodulatory properties of Rhodiola spp. and other natural antioxidants. Cent. Eur. J. Immunol. 2015, 40, 249–262. [Google Scholar] [CrossRef]
- Kunchithapautham, K.; Coughlin, B.; Lemasters, J.J.; Rohrer, B. Differential effects of rapamycin on rods and cones during light-induced stress in albino mice. Investig. Ophthalmol. Vis. Sci. 2011, 52, 2967–2975. [Google Scholar] [CrossRef]
- Sinha, D.; Valapala, M.; Shang, P.; Hose, S.; Grebe, R.; Lutty, G.A.; Zigler, J.S., Jr.; Kaarniranta, K.; Handa, J.T. Lysosomes: Regulators of autophagy in the retinal pigmented epithelium. Exp. Eye Res. 2016, 144, 46–53. [Google Scholar] [CrossRef]
- Ye, H.; Chen, M.; Cao, F.; Huang, H.; Zhan, R.; Zheng, X. Chloroquine, an autophagy inhibitor, potentiates the radiosensitivity of glioma initiating cells by inhibiting autophagy and activating apoptosis. BMC Neurol. 2016, 16, 178. [Google Scholar] [CrossRef]
- Liang, X.; Tang, J.; Liang, Y.; Jin, R.; Cai, X. Suppression of autophagy by chloroquine sensitizes 5-fluorouracil-mediated cell death in gallbladder carcinoma cells. Cell Biosci. 2014, 4, 10. [Google Scholar] [CrossRef]
- Nalbandian, A.; Llewellyn, K.J.; Nguyen, C.; Yazdi, P.G.; Kimonis, V.E. Rapamycin and chloroquine: The in vitro and in vivo effects of autophagy-modifying drugs show promising results in valosin containing protein multisystem proteinopathy. PLoS ONE 2015, 10, e0122888. [Google Scholar] [CrossRef]
- Szatmari-Toth, M.; Kristof, E.; Vereb, Z.; Akhtar, S.; Facsko, A.; Fesus, L.; Kauppinen, A.; Kaarniranta, K.; Petrovski, G. Clearance of autophagy-associated dying retinal pigment epithelial cells—A possible source for inflammation in age-related macular degeneration. Cell Death Dis. 2016, 7, e2367. [Google Scholar] [CrossRef] [PubMed]
- Turowski, P.; Adamson, P.; Sathia, J.; Zhang, J.J.; Moss, S.E.; Aylward, G.W.; Hayes, M.J.; Kanuga, N.; Greenwood, J. Basement membrane-dependent modification of phenotype and gene expression in human retinal pigment epithelial ARPE-19 cells. Investig. Ophthalmol. Vis. Sci. 2004, 45, 2786–2794. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed]
- Paglin, S.; Hollister, T.; Delohery, T.; Hackett, N.; McMahill, M.; Sphicas, E.; Domingo, D.; Yahalom, J. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 2001, 61, 439–444. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: https://www.R-project.org/.
| 5 h | 24 h | 48 h | ||||
|---|---|---|---|---|---|---|
| Rapamycin (RAP) | 50 nM | 100 nM | 50 nM | 100 nM | 50 nM | 100 nM |
| Rapamycin + CQ | 50 nM + 50 μM CQ | |||||
| Rapamycin + 3MA | 50 nM + 10 mM 3-MA | |||||
| MG-132 | 50 nM | 100 nM | 50 nM | 100 nM | 50 nM | 100 nM |
| MG-132 + Chloroquine | 50 nM + 50 μM CQ | |||||
| MG-132 + 3-MA | 50 nM + 10 mM 3-MA | |||||
| Rapamycin + MG-132 | 50 nM 50 nM | 100 nM 100 nM | 50 nM + 50 nM | 100 nM + 100 nM | ||
| Rapamycin + MG-132 + CQ. | RAP 50 nM + MG-132 50 nM + CQ 50 μ | |||||
| Rapamycin + MG-132 + 3-MA | RAP 50 nM + MG-132 50 nM + 10 mM 3-MA | |||||
| Resveratrol (RES) | 10 μM | 50 μM | 10 μM | 50 μM | 10 μM | 50 μM |
| Resveratrol + CQ | RES 10 μM + 50 μM CQ | |||||
| Resveratrol + 3-MA | RES 10 μM + 10 mM 3-MA | |||||
| Resveratrol + Rapamycin | 10 μM + 50 nM | 50 μM + 100 nM | 10 μM + 50nM | 50 μM + 100nM | 10 μM + 50 nM | 50 μM + 100 nM |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Josifovska, N.; Albert, R.; Nagymihály, R.; Lytvynchuk, L.; Moe, M.C.; Kaarniranta, K.; Veréb, Z.J.; Petrovski, G. Resveratrol as Inducer of Autophagy, Pro-Survival, and Anti-Inflammatory Stimuli in Cultured Human RPE Cells. Int. J. Mol. Sci. 2020, 21, 813. https://doi.org/10.3390/ijms21030813
Josifovska N, Albert R, Nagymihály R, Lytvynchuk L, Moe MC, Kaarniranta K, Veréb ZJ, Petrovski G. Resveratrol as Inducer of Autophagy, Pro-Survival, and Anti-Inflammatory Stimuli in Cultured Human RPE Cells. International Journal of Molecular Sciences. 2020; 21(3):813. https://doi.org/10.3390/ijms21030813
Chicago/Turabian StyleJosifovska, Natasha, Réka Albert, Richárd Nagymihály, Lyubomyr Lytvynchuk, Morten C. Moe, Kai Kaarniranta, Zoltán J. Veréb, and Goran Petrovski. 2020. "Resveratrol as Inducer of Autophagy, Pro-Survival, and Anti-Inflammatory Stimuli in Cultured Human RPE Cells" International Journal of Molecular Sciences 21, no. 3: 813. https://doi.org/10.3390/ijms21030813
APA StyleJosifovska, N., Albert, R., Nagymihály, R., Lytvynchuk, L., Moe, M. C., Kaarniranta, K., Veréb, Z. J., & Petrovski, G. (2020). Resveratrol as Inducer of Autophagy, Pro-Survival, and Anti-Inflammatory Stimuli in Cultured Human RPE Cells. International Journal of Molecular Sciences, 21(3), 813. https://doi.org/10.3390/ijms21030813







