1. Introduction
Grape (
Vitis vinifera L.) is one of the most economically important fruit-tree crops in the world, with a global production of 74 million tons in 2017 (available online:
http://www.fao.org/faostat). Grape berries are widely used as table grapes and to produce wine, raisins, and juice [
1]. Grapes have a high nutritional value and provide health benefits [
2]. Berry firmness is one of the main factors affecting consumers’ acceptance [
3,
4,
5]. Therefore, it is an important trait in table grape breeding [
6,
7]. High berry firmness is associated with good shelf-life performance and less postharvest losses, which is especially important for year-round fruit marketability and shipping overseas [
8]. Therefore, breeding and propagating new cultivars with high firmness are very important for grape production.
Grapevines are woody perennials with a high degree of heterozygosity, and it generally takes 4 to 5 years for a seed to grow and develop into a fruit [
9,
10]. Traditional grape breeding to obtain a plant with all the desired features is time-consuming, laborious, and expensive [
11,
12,
13,
14]. With the development of modern molecular biology, plant breeders now use marker-assisted selection (MAS) for grape breeding. By screening molecular markers related to target traits, hybrid progenies can be selected at the early seedling stage; therefore, MAS can greatly shorten the breeding process and improve breeding efficiency [
15,
16].
Many important economic traits in grapes, such as yield, quality, and resistance, are quantitative traits; and their phenotypes are continuously distributed in the progeny [
17,
18,
19]. There is not a one-to-one correspondence between the phenotype and genotype. A quantitative trait locus (QTL) analysis based on linkage maps and phenotypic evaluation of segregating progenies has been widely used to investigate the genetic determinants of agronomic traits [
20].
A (high-density) genetic map is essential for QTL mapping [
21]. In the past ten years, a number of grape genetic maps have been constructed based on different mapping populations [
20,
22,
23,
24,
25,
26,
27,
28,
29,
30,
31,
32,
33]. Numerous QTLs for important economic traits, including resistance to downy mildew [
10,
16,
22,
34,
35,
36,
37,
38], powdery mildew [
34,
36,
39,
40,
41], anthracnose [
24], root-knot nematodes [
42], and grape phylloxera [
43,
44], as well as flower sex [
45,
46], berry color [
46,
47], seedlessness [
48], berry weight [
18,
32], soluble solid content [
32], acidity [
20], and muscat flavor [
49], have been identified.
Grape firmness, like most characteristics of agricultural interest, is a complex quantitative trait. Multiple QTLs for berry firmness have been identified in recent years. The first study for QTLs associated with berry firmness in table grapes found seven genomic regions on linkage groups (LGs) 1, 4, 5, 9, 10 (bottom region), 13, and 18, which individually explained up to 19.8% of the total phenotypic variance [
6]. A second QTL study identified firmness determinants distributed in LGs 8 and 18, which explained 27.6% of the phenotypic variance, with confidence intervals up to 10 cM [
50]. A third study using 98 F1 individuals from a
Vitis labruscana ×
V. vinifera cross reported two QTLs for firmness that were located on LGs 3 and 10, with the former being more stable [
18]. The identification of numerous polymorphic markers is an important prerequisite for constructing a genetic map [
51,
52]. The total number of markers in the LGs of maps generated in these previous studies is less than 500, and thus, the resolution of these genetic maps is low, which has limited the efficiency and accuracy of QTL mapping. Studies have demonstrated that the resolution of a genetic map can be significantly improved by increasing marker density [
53,
54]. Thus, it is necessary to construct a genetic linkage map using high-density molecular markers to improve the accuracy of QTL mapping for firmness-related traits in grapes.
Single-nucleotide polymorphisms (SNPs), as the third generation of molecular markers, have great advantages over other molecular markers in genomic and genetic studies. As SNPs are the most abundant heritable variations in genomes and can be detected and genotyped automatically and in a high-throughput manner, they have revolutionized high-quality genetic map construction [
55,
56]. Next-generation sequencing is a high-throughput, low-cost technology that has been used to identify a large number of SNPs for high-density genetic map construction in various crops, such as soybean [
57], cotton [
58], and pear [
59]. In recent years, several genetic maps for grapevine have been constructed on the basis of high-throughput sequencing technology; these maps have increased marker density and have led to the identification of some new QTLs [
10,
24,
28,
33,
46]. Nearly all of the genetic maps were constructed using reduced representation methods, such as restriction site-associated DNA sequencing and genotyping-by-sequencing, which are relatively cheap as they sample only a fraction of the genome, but they also produce incomplete data [
60]. With the development of high-throughput sequencing technologies and the availability of a reference genome for grapevines [
61], whole-genome resequencing (WGR) allows the identification of whole genome differences between individuals and large numbers of SNPs, and has become one of the most rapid and effective methods used in QTL mapping and breeding research [
62,
63].
This study aimed to identify candidate regions and genes for berry firmness in grapevines to facilitate MAS. To this end, we used an F1 population consisting of 105 individuals to construct a high-resolution genetic map based on WGR. The F1 population was derived from a cross between Muscat Hamburg and Crimson Seedless (MH × CS), which have significantly different berry firmness.
3. Discussion
Berry firmness is one of the commercial qualities of grapes and an important standard in table grape breeding [
68]. The reliability of the berry firmness identification method is of great significance in grape QTL mapping research. For assessing the textural features of a grape, both sensory and instrumental analyses can be used [
18,
69,
70]. Traditionally, sensory tasting by trained judges is often subjective and more complicated when the number of samples is large or if the differences between samples are small [
71]. Texture profile analysis (TPA) is a well-developed and reliable method used for evaluating the textural characteristics of foods and fruits. The principle of this technique is to simulate the chewing movement of the human oral cavity, perform the compression process of the test sample twice, and output the texture parameters through the software. These parameters have been shown to be well-correlated with sensory evaluation of textural parameters [
72]. Giacosa et al. used a texture analyzer to measure the flesh firmness of the five reference table grape cultivars mentioned in the ampelographic descriptor for grapes (OIV code 235), and considered that soft, lightly soft, and firm levels were defined by 0.074–0.117, 0.121–0.158, and 0.205–0.391 N/mm, respectively [
73]. Conner et al. evaluated the berry firmness of 26 muscadine grape cultivars and showed that berry firmness of muscadine grapes ranged from very soft to firm, but still remained softer than
V. vinifera. It is recommended to use a texture analyzer rather than sensory evaluation of grape berry firmness, because instrument evaluation is controllable under laboratory conditions, is easier, and provides more accurate results [
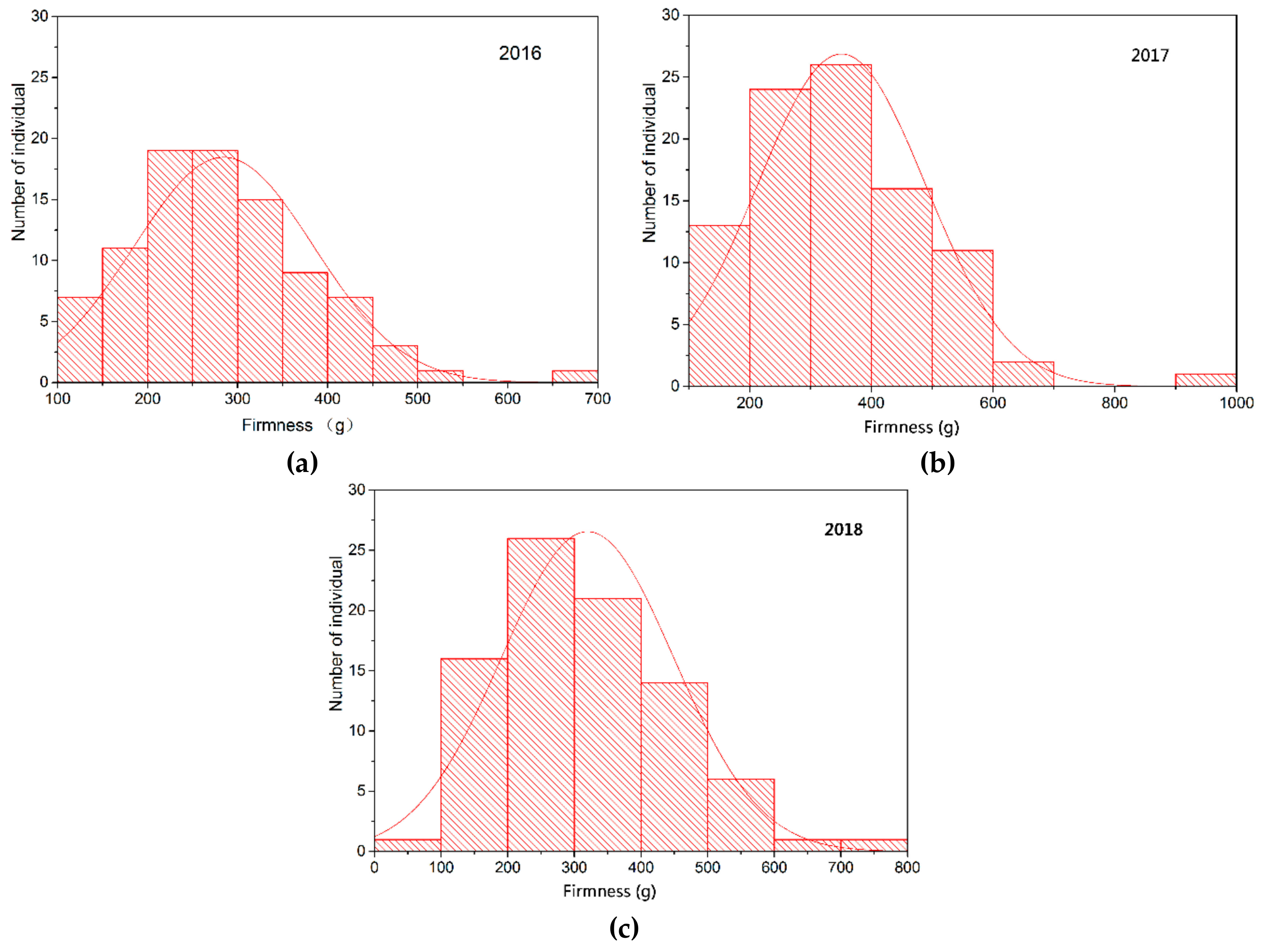
74]. In this study, berry firmness of parents and offspring was evaluated by texture profile analysis using a TA-XT2i texture analyzer. The firmness of CS was significantly higher than that of MH. Moreover, there were a certain number of “super-parent” individuals in the population, and berry firmness in the offspring showed continuous variation. The distribution was close to the normal distribution, indicating that this trait is quantitatively inherited and controlled by multiple genes, which is consistent with previous studies [
6,
50].
A reliable genetic map is essential for identifying QTLs of traits of interest and prediction of candidate genes [
75]; however, it is difficult to generate a hybrid population that is suitable for QTL mapping, such as recombinant inbred lines or F2 population in grapes and other fruit trees, which are all highly heterozygous, have long breeding cycles, and often have a smaller population size than those of annual crops [
59,
76]. In general, increasing the density of markers can increase the resolution of genetic maps, thereby improving the precision of QTL mapping [
54,
77]. Previous QTL studies of grape traits were mainly based on lower marker numbers (<1000) with relatively high QTL intervals, which affected the accuracy of QTLs and hard-to-locate candidate genes [
33,
78]. With the rapid development of sequencing technologies and bioinformatics, a large number of polymorphic molecular markers to meet the needs for high-density genetic map construction can be identified, and high-resolution linkage maps have been successfully used for QTL fine mapping in many crops, such as soybean, cotton, pear, and jujube [
57,
58,
59,
76]. In a previous grape study, Wang et al. constructed the first high-density genetic map spanning a genetic distance of 1917.3 cM and with an average distance of 1.16 cM between markers [
31]. Sapkota et al. detected a major QTL for downy mildew resistance based on a high-resolution linkage map with 3825 markers in the cross-hybridization between Norton and Cabernet Sauvignon, which had 159 progenies, spanning a genetic distance of 2203.5 cM, and an average distance of 1.1 cM between markers [
10]. Based on the F1 population (91 offspring), Fu et al. constructed a high-density genetic map, which covered a total of 1665.31 cM in length, with an average of 1.81 cM between markers. The authors detected a major stable QTL for ripe rot resistance (
Cgr1) by using the constructed genetic map [
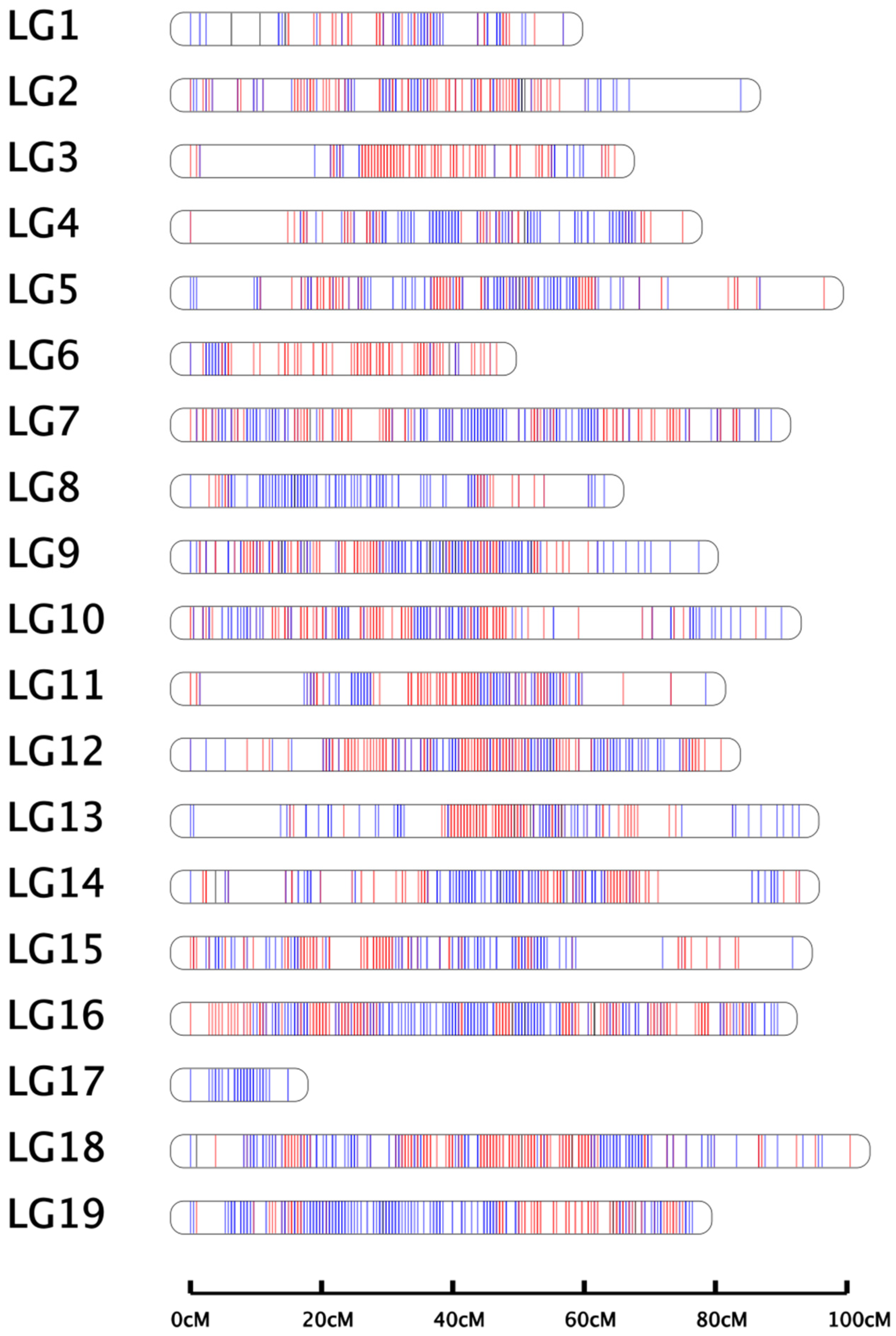
24]. In this study, a high-density linkage genetic map was constructed via the whole-genome resequencing method, and a total of 790.63 Gb clean data were generated. The effective sequencing depths of the female parent, the male parent, and their progeny were 44.93×, 47.05×, and 11.67–20.22×, respectively, which were sufficient to detect an appreciable number of recombination breakpoints. The genetic map contained 1662 bin markers (26,039 SNPs), with a total genetic length of 1460.38 cM and an average length of 0.88 cM for adjacent markers. The length of LGs ranged from 14.91 cM to 100.53 cM, with an average length of 76.86 cM. The average “Gap < 5 cM” percentage was 99.89%. The average marker distance ranged from 0.59 cM to 1.18 cM, and the map density was higher than other published maps [
20,
24,
26,
29]. The linkage map developed in this study possessed good quality and can therefore be used for QTL mapping.
Compared to research on traits such as disease resistance, stress resistance, sugar content, and color, fruit firmness has been rarely studied, and mostly in cherry, apple, peach, tomato, and melon. For example, a major fruit firmness QTL termed qP-FF4.1 was identified in three sweet cherry populations, the candidate genes were associated with cell wall modification and various hormone signaling pathways, and the extended protein gene, especially, was the most promising candidate gene [
64]. In apple, a QTL site termed Md-PG1, associated with apple hardness, which is regulated by ethylene, was identified based on an F1 mapping population generated from Fuji and Mondial Gala [
79]. Ogundiwin et al. used a hybrid of different varieties of peach varieties to construct a linkage map, and QTLs related to texture quality were located on LGs 1, 4, 5, 7, and 8, and encoded pectinase, pectin methylesterase, and endopolygalacturonase. In tomato, a firmness QTL with five distinct subpeaks was identified, and genes coding for ethylene response factor and pectin methylesterase were nominated as QTL candidate genes [
80]. Several QTLs for fruit flesh firmness have been identified in melon, and some candidate genes were speculated to be related to ethylene regulation, biosynthesis, and perception, and cell wall degradation [
81]. In grapes, ninety-eight individual progenies of a cross between 626-84 and Iku82 were used to identify QTLs for grape berry traits, including cracking, weight, firmness, harvest time, seed number, difficulty of breakdown, and soluble solids. Berry firmness was categorized as soft, medium, slightly firm, or very firm. The results of this study showed that the difficulty of breakdown and berry firmness were positively correlated (r = 0.542). Two loci related to berry firmness were located in LG3 and LG10. The LG3 locus was linked to the primer VMC2E7, the LG10 locus was close to the middle region of the primers VVIH01 and UDV073, and the LG3 locus was more stable. A genotyping-by-sequencing approach to analyze the fruit traits of 179 grape varieties, including berry color, firmness, and flavor, revealed two sites related to berry firmness, located on chromosomes 16 and 17 and encoding mainly proteins associated with calcium channels [
82]. Based on MH × Sugraone and Ruby Seedless × Moscatuel mapping populations, seven QTLs related to grape berry firmness, located on LGs 1, 4, 5, and 9 on chromosomes 10, 13, and 18, have been identified [
6]. Later, a Ruby Seedless × Sultanina mapping population was used to identify two QTLs for berry firmness on chromosomes 8 and 18. The QTL on chromosome 8 was located between the markers UDV125 and VMCNG2H2 and explained approximately 15.6% of the phenotypic variation, and the QTL on chromosome 18 was located between VVIN16 and VVCS1E103N17FM1 and explained approximately 12% of the phenotypic variation. This was the first report of a QTL for grape berry firmness that was stable in different seasons [
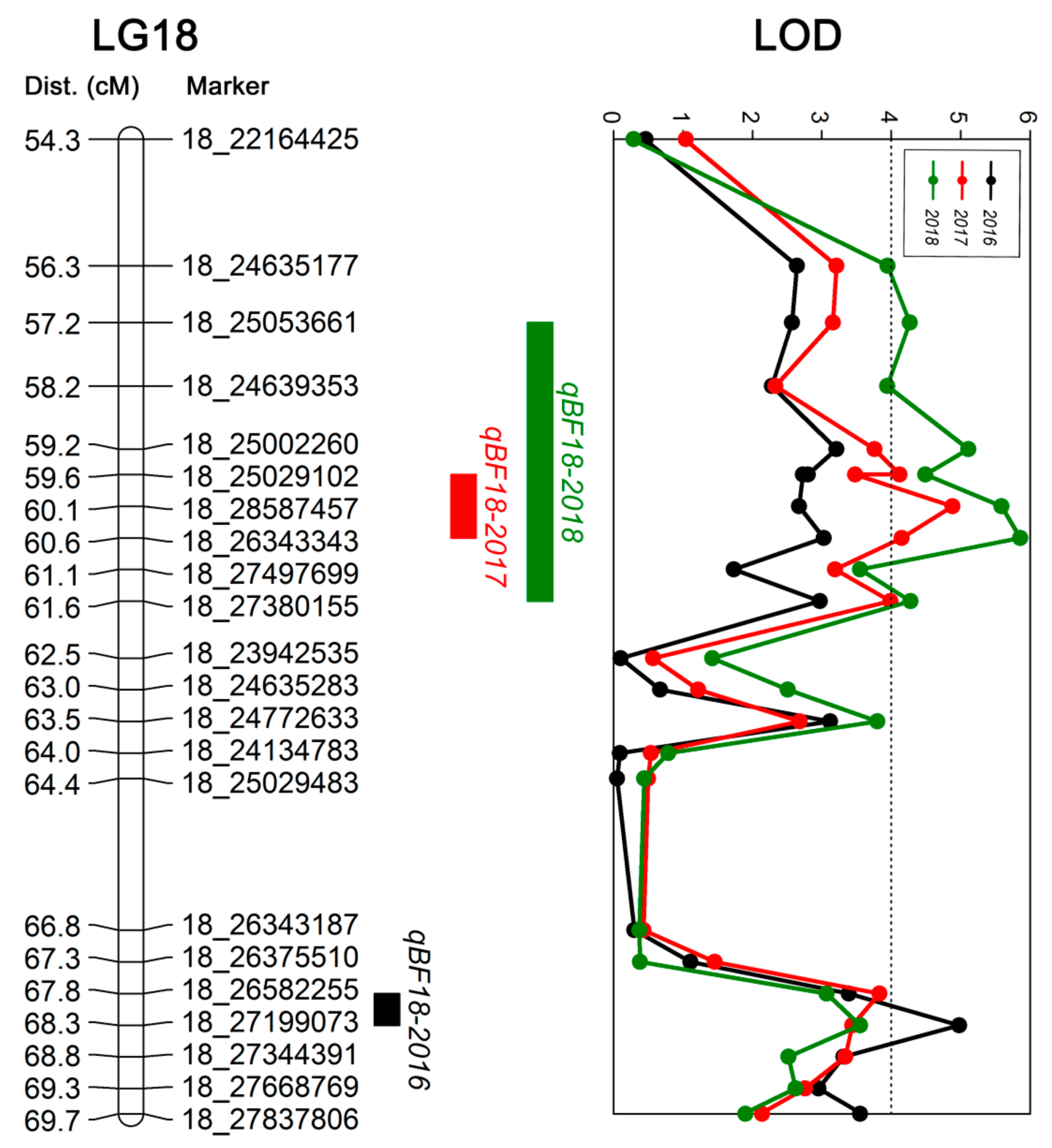
50]. In this study, we identified three QTLs related to berry firmness located on LG 18, which explained 21.5%–28.6% of the phenotypic variation. The genetic positions were 67.81–68.29 cM, 59.64–60.60 cM, and 57.23–61.56 cM, and corresponded to the physical positions 26.58–27.20 Mb, 25.03–28.59 Mb, and 24.64–28.59 Mb of chromosome 18, respectively. These QTLs are similar to those found by Carreño et al. [
6] and Correa et al. [
50] on LG18. The physical position of the SNPs did not coincide with genetic position in this study, which was likely attributed to errors in the alignment of reference genome sequence, as well as different microstructures on chromosomes [
10].
Softening was one of the main characteristics for fruit ripening and senescence. During fruit ripening and softening, cell wall components such as pectin, cellulose, and hemicellulose are degraded, and the microfibril filament structure of the cell wall is loosened and softened. Moreover, ripening leads to the disappearance of intercellular filaments, thinning of the cell wall, and cell dispersal, which results in destruction of the cell wall structure and fruit softening. Previous studies have shown that cell wall enzymes and plant hormones play an important role in this process [
64,
65,
66,
67]. Therefore, genes related to the metabolism of plant cell wall or various hormone signaling pathways were all considered as candidate genes for this study. Four candidate genes were obtained through functional annotation prediction. Glucanase is one of the key cell wall hydrolytic enzymes, which can promote the random hydrolysis of β-1,4-glycosidic bonds to cleave cellulose into smaller fragments and loosen the cell wall [
83]. Moreover, increased gene expression of glucanase has been associated with fruit ripening in bananas, pears, strawberries, and grapes [
84,
85,
86,
87,
88]. The plant hormone, abscisic acid (ABA), plays a crucial role in fruit ripening and responses to environmental stresses [
89]. The application of ABA during the grape growing period will soften the grape berry texture of Red Globe, Flame Seedless, and Crimson Seedless [
90,
91,
92], suggesting that ABA is a major regulator of grape berry ripening onset [
93,
94]. It is likely that the expression of ABA genes was associated with changes in berry firmness. Transcription factors, such as MADS-box and NAC, have also been reported to be involved in fruit ripening of bananas, tomatoes apples, grapes, etc. [
95,
96,
97,
98]. Previous studies highlight that berry softening occurs primarily during the veraison stage, the stage in which berry firmness decreases sharply in the soft flesh variety [
99].
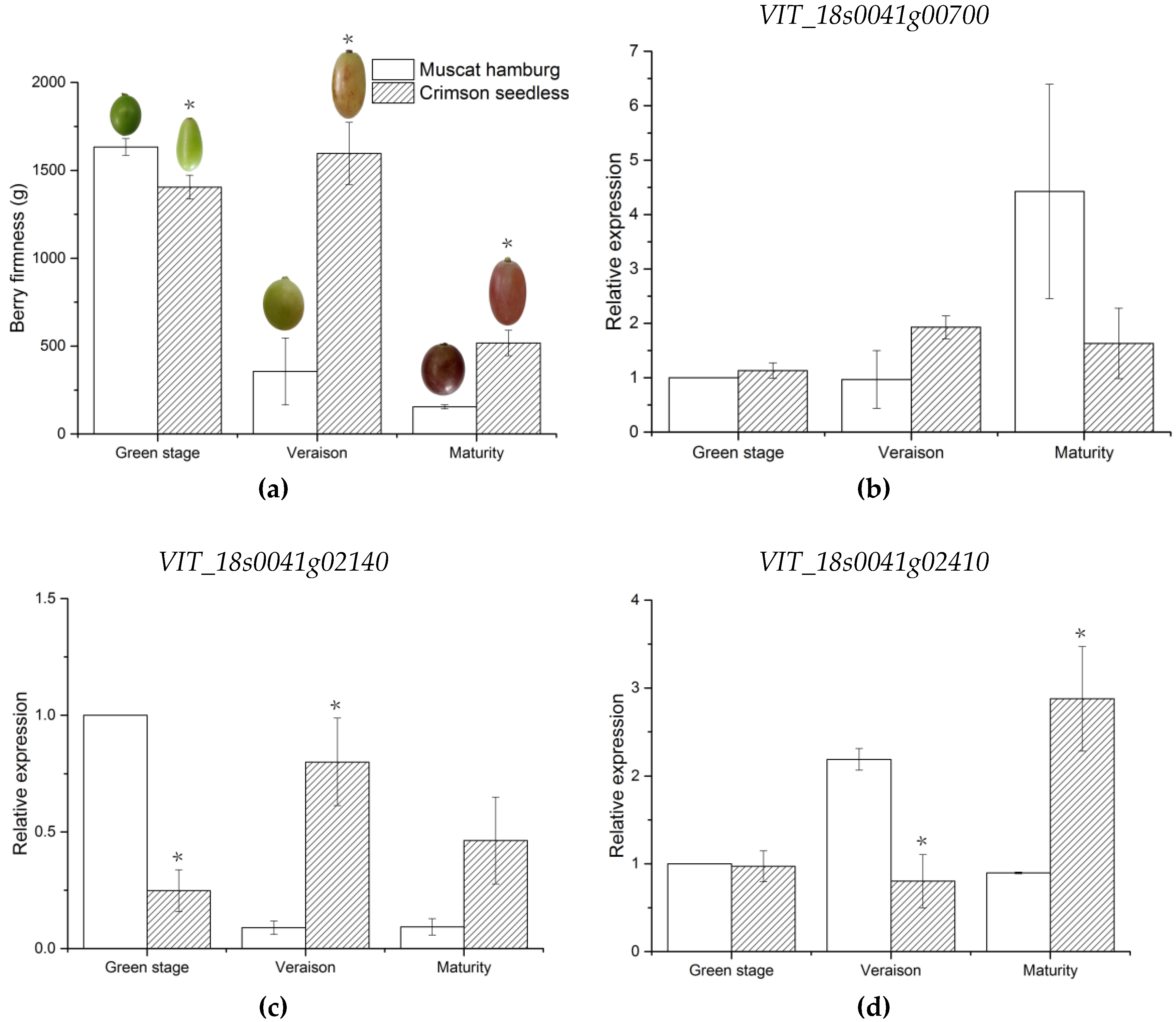
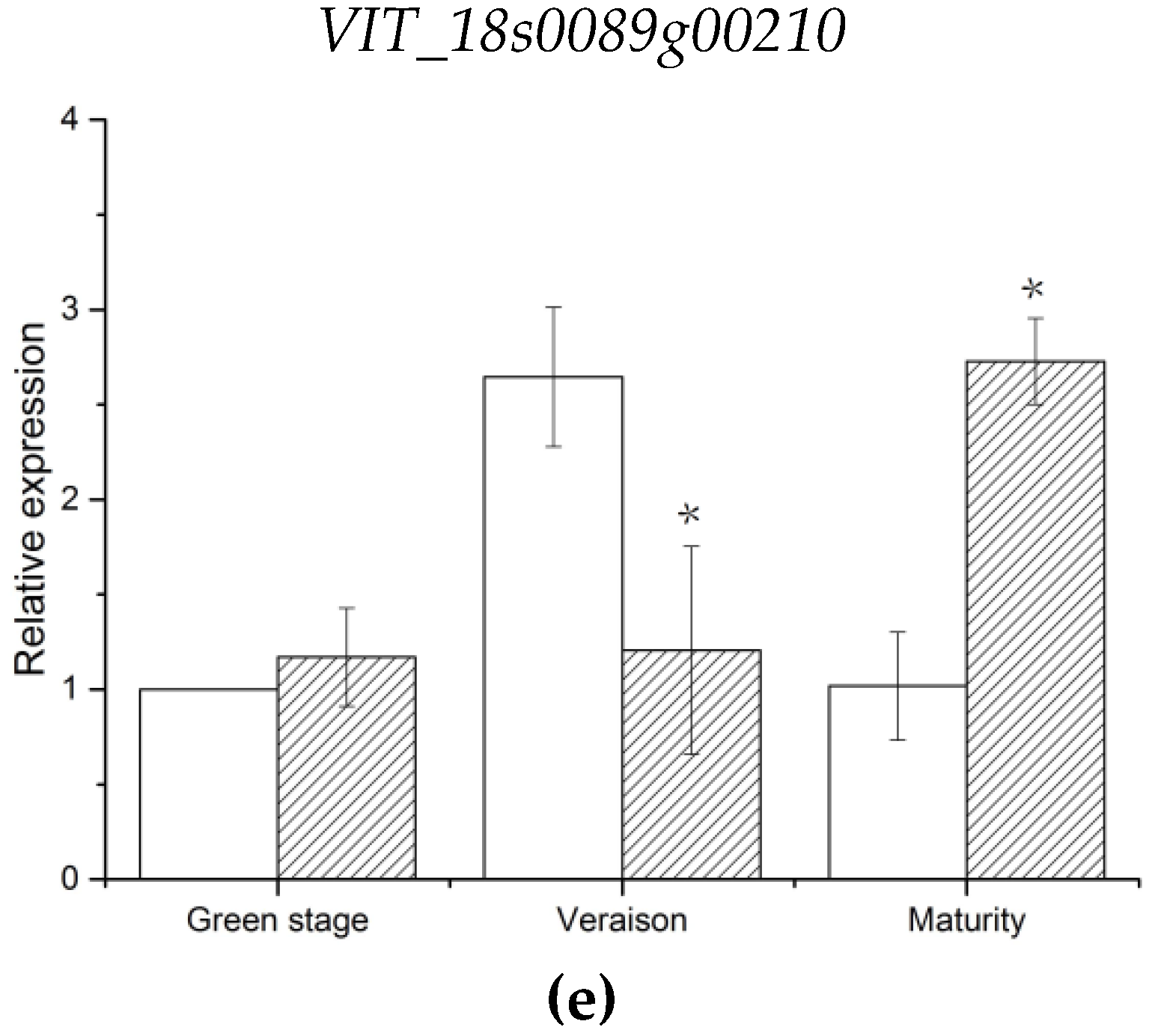
VIT_18s0089g00210 and
VIT_18s0041g02410, annotated to endoglucanase 3 and abscisic-aldehyde oxidase-like genes, were involved in cell wall assembly during cell elongation and ABA biosynthesis, respectively [
100,
101]. The expression levels of these two genes were consistent with that of parent berry firmness. Thus,
VIT_18s0089g00210 and
VIT_18s0041g02410 are likely the candidate genes associated with berry firmness of grapes. Notably, the genes were located in stable candidate regions with high LOD values in this study. Given that there was no direct evidence that these genes control these characteristics, further experiments will be required to verify the function of these candidate genes. An increase in population size can increase the number of recombination events of offspring, which is conducive to improve QTL precision [
102,
103], The F1 population in this study was relatively small (105 offspring) and the population size can be increased to delimit QTL interval in future research. Our results provide valuable tools for future candidate gene identification, map-based gene cloning, and MAS for grape berry firmness.