Interplay of Heme with Macrophages in Homeostasis and Inflammation
Abstract
1. Introduction
2. The Intra- and Extracellular Pool of Labile Heme
3. Role of Extra- and Intra-Cellular Heme-Binding Proteins (HBPs) in the Regulation of Labile Heme Homeostasis
3.1. Extracellular HBPs Neutralize Labile Heme
3.2. Intracellular HBPs Regulate Bioavailability of Labile Heme
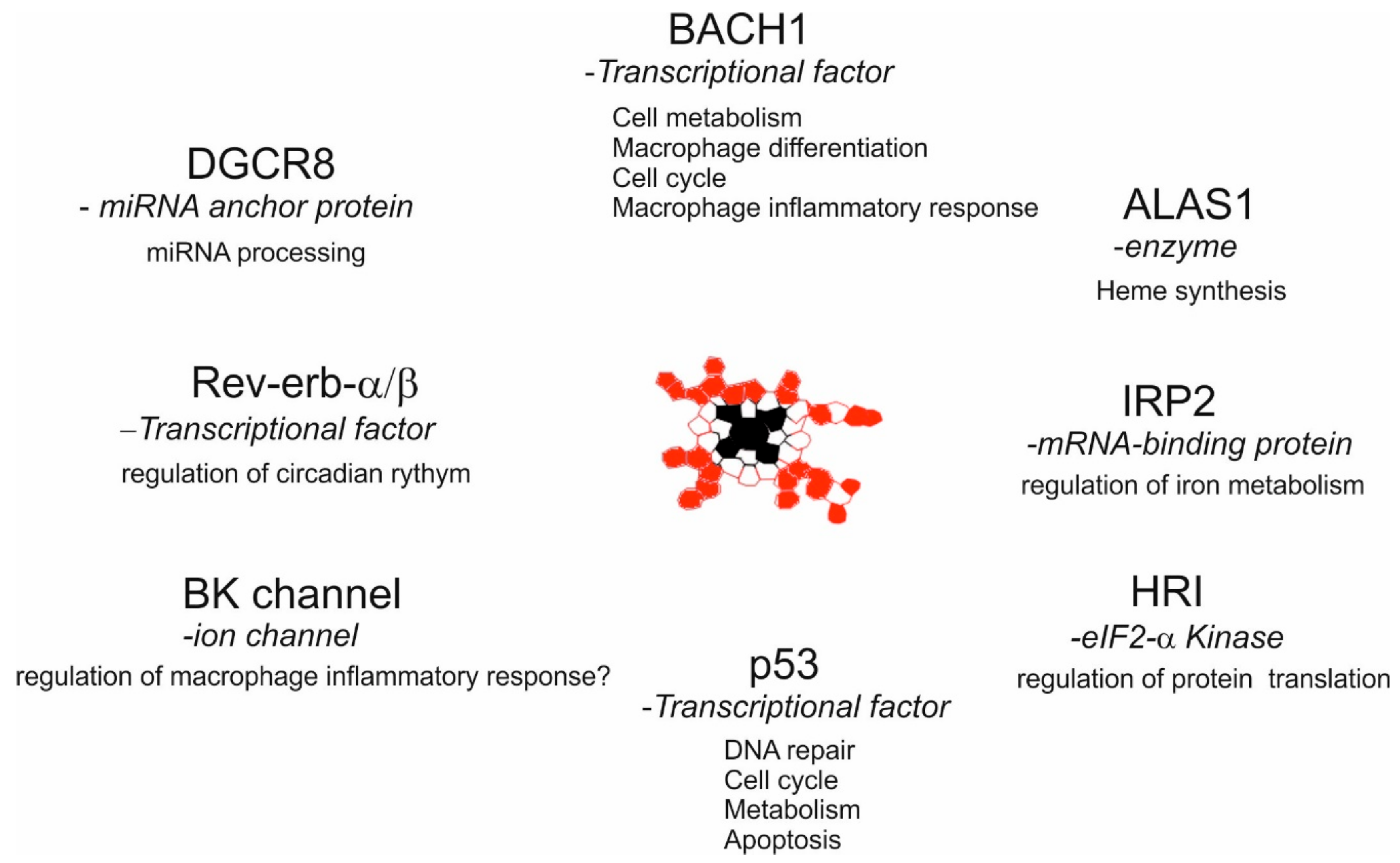
3.3. Intracellular Labile Heme Controls Cellular Functions via Heme-Sensor Proteins
4. How Heme and Macrophages Control Erythropoiesis in Steady State and during Inflammation?
Heme Sustains Erythropoiesis through Differentiation of Erythrophagocytosing Macrophages
5. Role of Heme in Inflammatory Activation of Macrophages
5.1. Heme as an Anti-Inflammatory Signal in Macrophages
5.2. Heme as a Pro-Inflammatory Signal in Macrophages
6. Conclusions and Outlook
Funding
Conflicts of Interest
References
- Gordon, S. The macrophage. BioEssays 1995, 17, 977–986. [Google Scholar] [CrossRef]
- Gordon, S.; Plüddemann, A. Tissue macrophages: Heterogeneity and functions. BMC Boil. 2017, 15, 53. [Google Scholar] [CrossRef]
- Okabe, Y.; Medzhitov, R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 2014, 157, 832–844. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, X.; Li, G.; Chong, Y.; Zhang, J.; Guo, X.; Li, B.; Bi, Z. Osteoclast regulation of osteoblasts via RANK-RANKL reverse signal transduction in vitro. Mol. Med. Rep. 2017, 16, 3994–4000. [Google Scholar] [CrossRef]
- Shibata, Y.; Berclaz, P.Y.; Chroneos, Z.C.; Yoshida, M.; Whitsett, J.A.; Trapnell, C.B. GM-CSF regulates alveolar macrophage differentiation and innate immunity in the lung through PU.1. Immunity 2001, 15, 557–567. [Google Scholar] [CrossRef]
- Haldar, M.; Kohyama, M.; So, A.Y.; Kc, W.; Wu, X.; Briseno, C.G.; Satpathy, A.T.; Kretzer, N.M.; Arase, H.; Rajasekaran, N.S.; et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell 2014, 156, 1223–1234. [Google Scholar] [CrossRef] [PubMed]
- Ponka, P. Cell biology of heme. Am. J. Med. Sci. 1999, 318, 241–256. [Google Scholar] [CrossRef]
- Igarashi, K.; Watanabe-Matsui, M. Wearing Red for Signaling: The Heme-Bach Axis in Heme Metabolism, Oxidative Stress Response and Iron Immunology. Tohoku J. Exp. Med. 2014, 232, 229–253. [Google Scholar] [CrossRef]
- Ajioka, R.S.; Phillips, J.D.; Kushner, J.P. Biosynthesis of heme in mammals. Biochim. Biophys. Acta 2006, 1763, 723–736. [Google Scholar] [CrossRef]
- Immenschuh, S. Therapeutic applications of the heme oxygenase system. Curr. Drug Targets 2010, 11, 1483–1484. [Google Scholar] [CrossRef]
- Immenschuh, S.; Schröder, H. Heme oxygenase-1 and cardiovascular disease. Histol. Histopathol. 2006, 21, 679–685. [Google Scholar]
- Immenschuh, S.; Baumgart-Vogt, E.; Tan, M.; Iwahara, S.; Ramadori, G.; Fahimi, H.D. Differential cellular and subcellular localization of heme-binding protein 23/peroxiredoxin I and heme oxygenase-1 in rat liver. J. Histochem. Cytochem. 2003, 51, 1621–1631. [Google Scholar] [CrossRef]
- Tenhunen, R.; Marver, H.S.; Schmid, R. Microsomal heme oxygenase. Characterization of the enzyme. J. Boil. Chem. 1969, 244, 6388–6395. [Google Scholar]
- Nemeth, Z.; Li, M.; Csizmadia, E.; Döme, B.; Johansson, M.; Persson, J.L.; Seth, P.; Otterbein, L.; Wegiel, B. Heme oxygenase-1 in macrophages controls prostate cancer progression. Oncotarget 2015, 6, 33675–33688. [Google Scholar] [CrossRef]
- Yunoki, K.; Inoue, T.; Sugioka, K.; Nakagawa, M.; Inaba, M.; Wada, S.; Ohsawa, M.; Komatsu, R.; Itoh, A.; Haze, K.; et al. Association between hemoglobin scavenger receptor and heme oxygenase–1–related anti-inflammatory mediators in human coronary stable and unstable plaques. Hum. Pathol. 2013, 44, 2256–2265. [Google Scholar] [CrossRef]
- Schaer, D.J.; Schaer, C.A.; Schoedon, G.; Imhof, A.; Kurrer, M.O. Hemophagocytic macrophages constitute a major compartment of heme oxygenase expression in sepsis. Eur. J. Haematol. 2006, 77, 432–436. [Google Scholar] [CrossRef]
- Alam, Z.; Devalaraja, S.; Haldar, M. The Heme Connection: Linking Erythrocytes and Macrophage Biology. Front. Immunol. 2017, 8, 445. [Google Scholar] [CrossRef]
- Vijayan, V.; Wagener, F.A.; Immenschuh, S. The macrophage heme-heme oxygenase-1 system and its role in inflammation. Biochem. Pharmacol. 2018, 153, 159–167. [Google Scholar] [CrossRef]
- Wagener, F.A.; Volk, H.D.; Willis, D.; Abraham, N.G.; Soares, M.P.; Adema, G.J.; Figdor, C.G. Different faces of the heme-heme oxygenase system in inflammation. Pharmacol. Rev. 2003, 55, 551–571. [Google Scholar] [CrossRef]
- Atamna, H.; Brahmbhatt, M.; Atamna, W.; Shanower, G.A.; Dhahbi, J.M. ApoHRP-based assay to measure intracellular regulatory heme. Metallomics 2015, 7, 309–321. [Google Scholar] [CrossRef]
- Hanna, D.A.; Hu, R.; Kim, H.; Martinez-Guzman, O.; Torres, M.P.; Reddi, A.R. Heme bioavailability and signaling in response to stress in yeast cells. J. Biol. Chem. 2018, 293, 12378–12393. [Google Scholar] [CrossRef]
- Sweeny, E.A.; Singh, A.B.; Chakravarti, R.; Martinez-Guzman, O.; Saini, A.; Haque, M.M.; Garee, G.; Dans, P.D.; Hannibal, L.; Reddi, A.R.; et al. Glyceraldehyde-3-phosphate dehydrogenase is a chaperone that allocates labile heme in cells. J. Biol. Chem. 2018, 293, 14557–14568. [Google Scholar] [CrossRef]
- Yuan, X.; Rietzschel, N.; Kwon, H.; Nuno, A.B.W.; Hanna, D.A.; Phillips, J.D.; Raven, E.L.; Reddi, A.R.; Hamza, I. Regulation of intracellular heme trafficking revealed by subcellular reporters. Proc. Natl. Acad. Sci. USA 2016, 113, E5144–E5152. [Google Scholar] [CrossRef]
- Donegan, R.K.; Moore, C.M.; Hanna, D.A.; Reddi, A.R. Handling heme: The mechanisms underlying the movement of heme within and between cells. Free Radic. Biol. Med. 2019, 133, 88–100. [Google Scholar] [CrossRef]
- Hanna, D.A.; Harvey, R.M.; Martinez-Guzman, O.; Yuan, X.; Chandrasekharan, B.; Raju, G.; Outten, F.W.; Hamza, I.; Reddi, A.R. Heme dynamics and trafficking factors revealed by genetically encoded fluorescent heme sensors. Proc. Natl. Acad. Sci. USA 2016, 113, 7539–7544. [Google Scholar] [CrossRef]
- Sudan, K.; Vijayan, V.; Madyaningrana, K.; Gueler, F.; Igarashi, K.; Foresti, R.; Motterlini, R.; Immenschuh, S. TLR4 activation alters labile heme levels to regulate BACH1 and heme oxygenase-1 expression in macrophages. Free Radic. Boil. Med. 2019, 137, 131–142. [Google Scholar] [CrossRef]
- Ponka, P.; Sheftel, A.D.; English, A.M.; Bohle, D.S.; Garcia-Santos, D. Do Mammalian Cells Really Need to Export and Import Heme? Trends Biochem. Sci. 2017, 42, 395–406. [Google Scholar] [CrossRef]
- Sassa, S. Why heme needs to be degraded to iron, biliverdin IXalpha, and carbon monoxide? Antioxid. Redox Signal. 2004, 6, 819–824. [Google Scholar]
- Gouveia, Z.; Carlos, A.R.; Yuan, X.; Aires-da-Silva, F.; Stocker, R.; Maghzal, G.J.; Leal, S.S.; Gomes, C.M.; Todorovic, S.; Iranzo, O.; et al. Characterization of plasma labile heme in hemolytic conditions. FEBS J. 2017, 284, 3278–3301. [Google Scholar] [CrossRef]
- Gladwin, M.T.; Ofori-Acquah, S.F. Erythroid DAMPs drive inflammation in SCD. Blood 2014, 123, 3689–3690. [Google Scholar] [CrossRef]
- Soares, M.P.; Bozza, M.T. Red alert: Labile heme is an alarmin. Curr. Opin. Immunol. 2016, 38, 94–100. [Google Scholar] [CrossRef]
- Soares, M.P.; Hamza, I. Macrophages and Iron Metabolism. Immunity 2016, 44, 492–504. [Google Scholar] [CrossRef]
- Immenschuh, S.; Vijayan, V.; Janciauskiene, S.; Gueler, F. Heme as a Target for Therapeutic Interventions. Front. Pharmacol. 2017, 146, 8. [Google Scholar] [CrossRef]
- Schaer, D.J.; Buehler, P.W.; Alayash, A.I.; Belcher, J.D.; Vercellotti, G.M. Hemolysis and free hemoglobin revisited: Exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood 2012, 121, 1276–1284. [Google Scholar] [CrossRef]
- Schaer, D.J.; Vinchi, F.; Ingoglia, G.; Tolosano, E.; Buehler, P.W. Haptoglobin, hemopexin, and related defense pathways-basic science, clinical perspectives, and drug development. Front. Physiol. 2014, 415, 5. [Google Scholar]
- Vincent, S.H. Oxidative effects of heme and porphyrins on proteins and lipids. Semin. Hematol. 1989, 26, 105–113. [Google Scholar]
- Vincent, S.H.; Grady, R.W.; Shaklai, N.; Snider, J.M.; Eberhar, M. The influence of heme-binding proteins in heme-catalyzed oxidations. Arch. Biochem. Biophys. 1988, 265, 539–550. [Google Scholar] [CrossRef]
- Eberhard, U.M. Hemopexin. N. Engl. J. Med. 1970, 283, 1090–1094. [Google Scholar] [CrossRef]
- Little, H.N.; Neilands, J.B. Binding of haematin by human serum albumin. Nature 1960, 188, 913–915. [Google Scholar] [CrossRef]
- Tolosano, E.; Altruda, F. Hemopexin: Structure, function, and regulation. DNA Cell Biol. 2002, 21, 297–306. [Google Scholar] [CrossRef]
- Allhorn, M.; Berggard, T.; Nordberg, J.; Olsson, M.L.; Akerstrom, B. Processing of the lipocalin alpha1-microglobulin by hemoglobin induces heme-binding and heme-degradation properties. Blood 2002, 99, 1894–1901. [Google Scholar] [CrossRef]
- Karnaukhova, E.; Krupnikova, S.S.; Rajabi, M.; Alayash, A.I. Heme binding to human alpha-1 proteinase inhibitor. Biochim. Biophys. Acta 2012, 1820, 2020–2029. [Google Scholar] [CrossRef]
- Muller-Eberhard, U.; Cleve, H. Immunoelectrophoretic studies of the beta1-haem-binding globulin (haemopexin) in hereditary haemolytic disorders. Nature 1963, 197, 602–603. [Google Scholar] [CrossRef]
- Adams, P.; Berman, M.C. Kinetics and mechanism of the interaction between human serum albumin and monomeric haemin. Biochem. J. 1980, 191, 95–102. [Google Scholar] [CrossRef]
- Hamza, I.; Dailey, H.A. One ring to rule them all: Trafficking of heme and heme synthesis intermediates in the metazoans. Biochim. et Biophys. Acta (BBA) Bioenerg. 2012, 1823, 1617–1632. [Google Scholar] [CrossRef]
- White, C.; Yuan, X.; Schmidt, P.J.; Bresciani, E.; Samuel, T.K.; Campagna, D.; Hall, C.; Bishop, K.; Calicchio, M.L.; Lapierre, A.; et al. HRG1 is essential for heme transport from the phagolysosome of macrophages during erythrophagocytosis. Cell Metab. 2013, 17, 261–270. [Google Scholar] [CrossRef]
- Schaer, C.A.; Vallelian, F.; Imhof, A.; Schoedon, G.; Schaer, D.J. Heme carrier protein HCP-1 spatially interacts with the CD163 hemoglobin uptake pathway and is a target of inflammatory macrophage activation. J. Leukoc. Biol. 2008, 83, 325–333. [Google Scholar] [CrossRef]
- Keel, S.B.; Doty, R.T.; Yang, Z.; Quigley, J.G.; Chen, J.; Knoblaugh, S.; Kingsley, P.D.; de Domenico, I.; Vaughn, M.B.; Kaplan, J.; et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science 2008, 319, 825–828. [Google Scholar] [CrossRef]
- Khan, A.A.; Quigley, J.G. Heme and FLVCR-related transporter families SLC48 and SLC49. Mol. Aspects Med. 2013, 343, 669–682. [Google Scholar] [CrossRef]
- Petrillo, S.; Chiabrando, D.; Genova, T.; Fiorito, V.; Ingoglia, G.; Vinchi, F.; Mussano, F.; Carossa, S.; Silengo, L.; Altruda, F.; et al. Heme accumulation in endothelial cells impairs angiogenesis by triggering paraptosis. Cell Death Differ. 2018, 25, 573–588. [Google Scholar] [CrossRef]
- Chiabrando, D.; Marro, S.; Mercurio, S.; Giorgi, C.; Petrillo, S.; Vinchi, F.; Fiorito, V.; Fagoonee, S.; Camporeale, A.; Turco, E.; et al. The mitochondrial heme exporter FLVCR1b mediates erythroid differentiation. J. Clin. Investig. 2012, 122, 4569–4579. [Google Scholar] [CrossRef]
- Duffy, S.P.; Shing, J.; Saraon, P.; Berger, L.C.; Eiden, M.V.; Wilde, A.; Tailor, C.S. The Fowler Syndrome-Associated Protein FLVCR2 Is an Importer of Heme. Mol. Cell. Boil. 2010, 30, 5318–5324. [Google Scholar] [CrossRef]
- Tudor, C.; Lerner-Marmarosh, N.; Engelborghs, Y.; Gibbs, P.E.M.; Maines, M.D. Biliverdin reductase is a transporter of haem into the nucleus and is essential for regulation of HO-1 gene expression by haematin. Biochem. J. 2008, 413, 405–416. [Google Scholar] [CrossRef]
- Vincent, S.H.; Eberhard, U.M. A protein of the Z class of liver cytosolic proteins in the rat that preferentially binds heme. J. Biol. Chem. 1985, 260, 14521–14528. [Google Scholar]
- Chakravarti, R.; Aulak, K.S.; Fox, P.L.; Stuehr, D.J. GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc. Natl. Acad. Sci. USA 2010, 107, 18004–18009. [Google Scholar] [CrossRef]
- Ketley, J.N.; Habig, W.H.; Jakoby, W.B. Binding of nonsubstrate ligands to the glutathione S-transferases. J. Biol. Chem. 1975, 250, 8670–8673. [Google Scholar]
- Taketani, S.; Adachi, Y.; Kohno, H.; Ikehara, S.; Tokunaga, R.; Ishii, T. Molecular characterization of a newly identified heme-binding protein induced during differentiation of murine erythroleukemia cells. J. Biol. Chem. 1998, 273, 31388–31394. [Google Scholar] [CrossRef]
- Iwahara, S.; Satoh, H.; Song, D.-X.; Webb, J.; Burlingame, A.L.; Nagae, Y.; Muller-Eberhard, U. Purification, characterization and cloning of a heme-binding protein 23kDa in rat liver cytosol. Biochemistry 1995, 34, 13398–13406. [Google Scholar] [CrossRef]
- Zylka, M.J.; Reppert, S.M. Discovery of a putative heme-binding protein family SOUL/HBP by two-tissue suppression subtractive hybridization and database searches. Brain Res Mol. Brain Res. 1999, 742, 175–181. [Google Scholar] [CrossRef]
- Reddi, A.R.; Hamza, I. Heme Mobilization in Animals: A Metallolipid’s Journey. Acc. Chem. Res. 2016, 49, 1104–1110. [Google Scholar] [CrossRef]
- Galmozzi, A.; Kok, B.P.; Kim, A.S.; Montenegro-Burke, J.R.; Lee, J.Y.; Spreafico, R.; Mosure, S.; Albert, V.; Cintron-Colon, R.; Godio, C.; et al. PGRMC2 is an intracellular haem chaperone critical for adipocyte function. Nature 2019, 576, 138–142. [Google Scholar] [CrossRef]
- Immenschuh, S.; Iwahara, S.-I.; Satoh, H.; Nell, C.; Katz, N.; Muller-Eberhard, U. Expression of the mRNA of Heme-Binding Protein 23 Is Coordinated with That of Heme Oxygenase-1 by Heme and Heavy Metals in Primary Rat Hepatocytes and Hepatoma Cells. Biochemistry 1995, 34, 13407–13411. [Google Scholar] [CrossRef]
- Harvey, J.W.; Beutler, E. Binding of heme by glutathione S-transferase: A possible role of the erythrocyte enzyme. Blood 1982, 60, 1227–1230. [Google Scholar] [CrossRef]
- Ogawa, K.; Sun, J.; Taketani, S.; Nakajima, O.; Nishitani, C.; Sassa, S.; Hayashi, N.; Yamamoto, M.; Shibahara, S.; Fujita, H.; et al. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 2001, 20, 2835–2843. [Google Scholar] [CrossRef]
- Zenke-Kawasaki, Y.; Dohi, Y.; Katoh, Y.; Ikura, T.; Ikura, M.; Asahara, T.; Tokunaga, F.; Iwai, K.; Igarashi, K. Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol. Cell. Biol. 2007, 27, 6962–6971. [Google Scholar] [CrossRef]
- Sun, J.; Hoshino, H.; Takaku, K.; Nakajima, O.; Muto, A.; Suzuki, H.; Tashiro, S.; Takahashi, S.; Shibahara, S.; Alam, J.; et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002, 21, 5216–5224. [Google Scholar] [CrossRef]
- Wiel, C.; le Gal, K.; Ibrahim, M.X.; Jahangir, C.A.; Kashif, M.; Yao, H.; Ziegler, D.V.; Xu, X.; Ghosh, T.; Mondal, T.; et al. BACH1 Stabilization by Antioxidants Stimulates Lung Cancer Metastasis. Cell 2019, 178, 330–345. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, J.; Wei, X.; Niu, C.; Jia, M.; Li, Q.; Meng, D. Bach1: Function, Regulation, and Involvement in Disease. Oxid. Med. Cell. Longev. 2018, 2018, 1347969. [Google Scholar] [CrossRef]
- Igarashi, K.; Kurosaki, T.; Roychoudhuri, R. BACH transcription factors in innate and adaptive immunity. Nat. Rev. Immunol. 2017, 17, 437–450. [Google Scholar] [CrossRef]
- Patsalos, A.; Tzerpos, P.; Halasz, L.; Nagy, G.; Pap, A.; Giannakis, N.; Lyroni, K.; Koliaraki, V.; Pintye, E.; Dezso, B.; et al. The BACH1-HMOX1 Regulatory Axis Is Indispensable for Proper Macrophage Subtype Specification and Skeletal Muscle Regeneration. J. Immunol. 2019, 203, 1532–1547. [Google Scholar] [CrossRef]
- Carter, E.L.; Gupta, N.; Ragsdale, S.W. High Affinity Heme Binding to a Heme Regulatory Motif on the Nuclear Receptor Rev-erbbeta Leads to Its Degradation and Indirectly Regulates Its Interaction with Nuclear Receptor Corepressor. J. Biol. Chem. 2016, 291, 2196–2222. [Google Scholar] [CrossRef]
- Carter, E.L.; Ramirez, Y.; Ragsdale, S.W. The heme-regulatory motif of nuclear receptor Rev-erbbeta is a key mediator of heme and redox signaling in circadian rhythm maintenance and metabolism. J. Biol. Chem. 2017, 292, 11280–11299. [Google Scholar] [CrossRef]
- Shen, J.; Sheng, X.; Chang, Z.; Wu, Q.; Wang, S.; Xuan, Z.; Li, D.; Wu, Y.; Shang, Y.; Kong, X.; et al. Iron metabolism regulates p53 signaling through direct heme-p53 interaction and modulation of p53 localization, stability, and function. Cell Rep. 2014, 7, 180–193. [Google Scholar] [CrossRef]
- Pflaum, J.; Schlosser, S.; Muller, M. p53 Family and Cellular Stress Responses in Cancer. Front. Oncol. 2014, 285, 4. [Google Scholar] [CrossRef]
- Labuschagne, C.F.; Zani, F.; Vousden, K.H. Control of metabolism by p53—Cancer and beyond. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 32–42. [Google Scholar] [CrossRef]
- Kovtunovych, G.; Eckhaus, M.A.; Ghosh, M.C.; Ollivierre-Wilson, H.; Rouault, T.A. Dysfunction of the heme recycling system in heme oxygenase 1-deficient mice: Effects on macrophage viability and tissue iron distribution. Blood 2010, 116, 6054–6062. [Google Scholar] [CrossRef]
- Kovtunovych, G.; Ghosh, M.C.; Ollivierre, W.; Weitzel, R.P.; Eckhaus, M.A.; Tisdale, J.F.; Yachie, A.; Rouault, T.A. Wild-type macrophages reverse disease in heme oxygenase 1-deficient mice. Blood 2014, 124, 1522–1530. [Google Scholar] [CrossRef]
- Li, W.; Wang, Y.; Zhao, H.; Zhang, H.; Xu, Y.; Wang, S.; Guo, X.; Huang, Y.; Zhang, S.; Han, Y.; et al. Identification and transcriptome analysis of erythroblastic island macrophages. Blood 2019, 134, 480–491. [Google Scholar] [CrossRef]
- Ganz, T.; Nemeth, E. Iron sequestration and anemia of inflammation. Semin. Hematol. 2009, 46, 387–393. [Google Scholar] [CrossRef]
- Recalcati, S.; Locati, M.; Marini, A.; Santambrogio, P.; Zaninotto, F.; De Pizzol, M.; Zammataro, L.; Girelli, D.; Cairo, G. Differential regulation of iron homeostasis during human macrophage polarized activation. Eur. J. Immunol. 2010, 40, 824–835. [Google Scholar] [CrossRef]
- Bennett, L.F.; Liao, C.; Quickel, M.D.; Yeoh, B.S.; Vijay-Kumar, M.; Hankey-Giblin, P.; Prabhu, K.S.; Paulson, R.F. Inflammation induces stress erythropoiesis through heme-dependent activation of SPI-C. Sci. Signal. 2019, 12, eaap7336. [Google Scholar] [CrossRef]
- Kato, H.; Itoh-Nakadai, A.; Matsumoto, M.; Ishii, Y.; Watanabe-Matsui, M.; Ikeda, M.; Ebina-Shibuya, R.; Sato, Y.; Kobayashi, M.; Nishizawa, H.; et al. Infection perturbs Bach2- and Bach1-dependent erythroid lineage ‘choice’ to cause anemia. Nat. Immunol. 2018, 19, 1059–1070. [Google Scholar] [CrossRef]
- Schultze, J.L.; Mass, E.; Schlitzer, A. Emerging Principles in Myelopoiesis at Homeostasis and during Infection and Inflammation. Immunity 2019, 50, 288–301. [Google Scholar] [CrossRef]
- Kato, H.; Igarashi, K. To be red or white: Lineage commitment and maintenance of the hematopoietic system by the “inner myeloid”. Haematologica 2019, 104, 1919–1927. [Google Scholar] [CrossRef]
- Schaer, C.A.; Schoedon, G.; Imhof, A.; Kurrer, M.O.; Schaer, D.J. Constitutive endocytosis of CD163 mediates hemoglobin-heme uptake and determines the noninflammatory and protective transcriptional response of macrophages to hemoglobin. Circ. Res. 2006, 99, 943–950. [Google Scholar] [CrossRef]
- Philippidis, P.; Mason, J.C.; Evans, B.J.; Nadra, I.; Taylor, K.M.; Haskard, D.O.; Landis, R.C. Hemoglobin scavenger receptor CD163 mediates interleukin-10 release and heme oxygenase-1 synthesis: Antiinflammatory monocyte-macrophage responses in vitro, in resolving skin blisters in vivo, and after cardiopulmonary bypass surgery. Circ. Res. 2004, 94, 119–926. [Google Scholar] [CrossRef]
- Wan, S.; Cheng, Y.; Jin, H.; Guo, D.; Hua, Y.; Keep, R.F.; Xi, G. Microglia Activation and Polarization After Intracerebral Hemorrhage in Mice: The Role of Protease-Activated Receptor-1. Transl. Stroke Res. 2016, 7, 478–487. [Google Scholar] [CrossRef]
- WLiu, Q.; Meng, H.-M.; Xie, W.-J.; Yu, H.-Q.; Zhang, Y. CD163 promotes hematoma absorption and improves neurological functions in patients with intracerebral hemorrhage. Neural Regen. Res. 2016, 11, 1122–1127. [Google Scholar]
- Ryter, S.W.; Alam, J.; Choi, A.M. Heme oxygenase-1/carbon monoxide: From basic science to therapeutic applications. Physiol. Rev. 2006, 86, 583–650. [Google Scholar] [CrossRef]
- Motterlini, R.; Foresti, R. Heme Oxygenase-1 As a Target for Drug Discovery. Antioxid. Redox Signal. 2014, 20, 1810–1826. [Google Scholar] [CrossRef]
- Nakamichi, I.; Habtezion, A.; Zhong, B.; Contag, C.H.; Butcher, E.C.; Omary, M.B. Hemin-activated macrophages home to the pancreas and protect from acute pancreatitis via heme oxygenase-1 induction. J. Clin. Investig. 2005, 115, 3007–3014. [Google Scholar] [CrossRef]
- Kayama, H.; Kohyama, M.; Okuzaki, D.; Motooka, D.; Barman, S.; Okumura, R.; Muneta, M.; Hoshino, K.; Sasaki, I.; Ise, W.; et al. Heme ameliorates dextran sodium sulfate-induced colitis through providing intestinal macrophages with noninflammatory profiles. Proc. Natl. Acad. Sci. USA 2018, 115, 8418–8423. [Google Scholar] [CrossRef]
- Ben-Mordechai, T.; Kain, D.; Holbova, R.; Landa, N.; Levin, L.-P.; Elron-Gross, I.; Glucksam-Galnoy, Y.; Feinberg, M.S.; Margalit, R.; Leor, J. Targeting and modulating infarct macrophages with hemin formulated in designed lipid-based particles improves cardiac remodeling and function. J. Control. Release 2017, 257, 21–31. [Google Scholar] [CrossRef]
- Dutra, F.F.; Alves, L.S.; Rodrigues, D.; Fernandez, P.L.; de Oliveira, R.B.; Golenbock, D.T.; Zamboni, D.S.; Bozza, M.T. Hemolysis-induced lethality involves inflammasome activation by heme. Proc. Natl. Acad. Sci. USA 2014, 111, E4110–E4118. [Google Scholar] [CrossRef]
- Dutra, F.F.; Bozza, M.T. Heme on innate immunity and inflammation. Front. Pharmacol. 2014, 115, 5. [Google Scholar] [CrossRef]
- Figueiredo, R.T.; Fernandez, P.L.; Mourao-Sa, D.S.; Porto, B.N.; Dutra, F.F.; Alves, L.S.; Oliveira, M.F.; Oliveira, P.L.; Graca-Souza, A.V.; Bozza, M.T. Characterization of heme as activator of Toll-like receptor 4. J. Biol. Chem. 2007, 282, 20221–20229. [Google Scholar] [CrossRef]
- Vallelian, F.; Schaer, C.A.; Deuel, J.W.; Ingoglia, G.; Humar, R.; Buehler, P.W.; Schaer, D.J. Revisiting the putative role of heme as a trigger of inflammation. Pharmacol. Res. Perspect. 2018, 6, e00392. [Google Scholar] [CrossRef]
- Nath, K.A.; Balla, J.; Croatt, A.J.; Vercellotti, G.M. Heme protein-mediated renal injury: A protective role for 21-aminosteroids in vitro and in vivo. Kidney Int. 1995, 47, 592–602. [Google Scholar] [CrossRef]
- Nath, K.A.; Croatt, A.J.; Haggard, J.J.; Grande, J.P. Renal response to repetitive exposure to heme proteins: Chronic injury induced by an acute insult. Kidney Int. 2000, 57, 2423–2433. [Google Scholar] [CrossRef][Green Version]
- Nath, K.A.; Grande, J.P.; Croatt, A.J.; Likely, S.; Hebbel, R.P.; Enright, H. Intracellular targets in heme protein-induced renal injury. Kidney Int. 1998, 53, 100–111. [Google Scholar] [CrossRef]
- Wang, L.; Vijayan, V.; Jang, M.S.; Thorenz, A.; Greite, R.; Rong, S.; Chen, R.; Shushakova, N.; Tudorache, I.; Derlin, K.; et al. Labile heme aggravates renal inflammation and complement actvation after ischemia reperfusion injury. Front. Immunol. 2019, 10, 2975. [Google Scholar] [CrossRef]
- Frimat, M.; Tabarin, F.; Dimitrov, J.D.; Poitou, C.; Halbwachs-Mecarelli, L.; Fremeaux-Bacchi, V.; Roumenina, L.T. Complement activation by heme as a secondary hit for atypical hemolytic uremic syndrome. Blood 2013, 122, 282–292. [Google Scholar] [CrossRef]
- Merle, N.S.; Grunenwald, A.; Rajaratnam, H.; Gnemmi, V.; Frimat, M.; Figueres, M.L.; Knockaert, S.; Bouzekri, S.; Charue, D.; Noe, R.; et al. Intravascular hemolysis activates complement via cell-free heme and heme-loaded microvesicles. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- Thomas, A.M.; Gerogianni, A.; McAdam, M.B.; Fløisand, Y.; Lau, C.; Espevik, T.; Nilsson, P.H.; Mollnes, T.E.; Barratt-Due, A. Complement Component C5 and TLR Molecule CD14 Mediate Heme-Induced Thromboinflammation in Human Blood. J. Immunol. 2019, 203, 1571–1578. [Google Scholar] [CrossRef]
- Larsen, R.; Gozzelino, R.; Jeney, V.; Tokaji, L.; Bozza, F.A.; Japiassu, A.M.; Bonaparte, D.; Cavalcante, M.M.; Chora, A.; Ferreira, A.; et al. A central role for free heme in the pathogenesis of severe sepsis. Sci. Transl. Med. 2010, 2, 51ra71. [Google Scholar] [CrossRef]
- Leite, G.G.F.; Scicluna, B.P.; Van Der Poll, T.; Salomão, R. Genetic signature related to heme-hemoglobin metabolism pathway in sepsis secondary to pneumonia. NPJ Syst. Boil. Appl. 2019, 5, 26–29. [Google Scholar] [CrossRef]
| Heme Binding Protein | Heme Affinity (Kd) (M) | Serum Concentration | Ref. |
|---|---|---|---|
| Hemopexin | 1 × 10−14 | 0.6–1.2 g/L | [38,43] |
| Albumin | 1.2 × 10−8 | 35–53 g/L | [39,44] |
| α1-Microglobulin | 1 × 10−6 | 0.03 g/L | [41] |
| α1-Antitrypsin | 2 × 10−8 | 1.3–2.5 g/L | [42] |
| Heme binding protein | Function | Ref |
|---|---|---|
| Putative heme transporters | ||
| Feline leukemia virus subgroup C receptor 1a (FLVCR1a) | export of heme to extracellular space | [48,50] |
| Feline leukemia virus subgroup C receptor 1a (FLVCR1b) | export of heme from the mitochondria to cytosol | [51] |
| Feline leukemia virus subgroup C receptor 2 (FLVCR2) | import of heme from extracellular space | [52] |
| Heme responsive gene-1 (HRG-1) | export of heme from phagolysosome to cytosol | [46] |
| Heme carrier protein-1 (HCP-1) | export of heme from lysosome to cytosol (?) | [47] |
| Putative heme chaperones | ||
| Biliverdin reductase (BVR) | heme trafficking to nucleus (?) | [53] |
| Fatty acid binding protein (FABP) | heme trafficking in cytosol (?) | [54] |
| Glceraldehyde phosphate dehydrogenase (GAPDH) | heme trafficking in cytosol | [22], [55] |
| GSH-S-transferase | heme trafficking in cytosol (?) | [56] |
| Heme binding protein 22 (HBP22) | heme trafficking in cytosol (?) | [57] |
| Heme binding protein 23 (HBP23) | heme trafficking in cytosol (?) | [58] |
| SOUL | heme trafficking in cytosol (?) | [59] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pradhan, P.; Vijayan, V.; Gueler, F.; Immenschuh, S. Interplay of Heme with Macrophages in Homeostasis and Inflammation. Int. J. Mol. Sci. 2020, 21, 740. https://doi.org/10.3390/ijms21030740
Pradhan P, Vijayan V, Gueler F, Immenschuh S. Interplay of Heme with Macrophages in Homeostasis and Inflammation. International Journal of Molecular Sciences. 2020; 21(3):740. https://doi.org/10.3390/ijms21030740
Chicago/Turabian StylePradhan, Pooja, Vijith Vijayan, Faikah Gueler, and Stephan Immenschuh. 2020. "Interplay of Heme with Macrophages in Homeostasis and Inflammation" International Journal of Molecular Sciences 21, no. 3: 740. https://doi.org/10.3390/ijms21030740
APA StylePradhan, P., Vijayan, V., Gueler, F., & Immenschuh, S. (2020). Interplay of Heme with Macrophages in Homeostasis and Inflammation. International Journal of Molecular Sciences, 21(3), 740. https://doi.org/10.3390/ijms21030740




