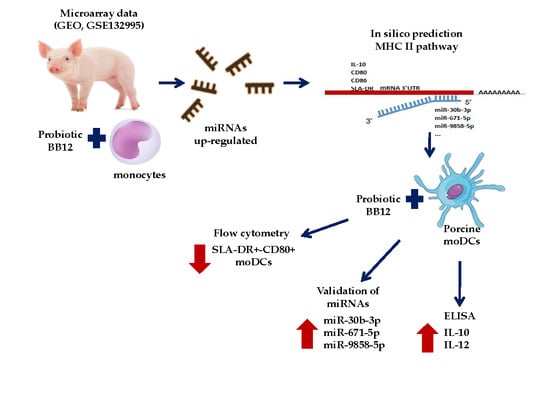
The Probiotic BB12 Induces MicroRNAs Involved in Antigen Processing and Presentation in Porcine Monocyte-Derived Dendritic Cells
Abstract
1. Introduction
2. Results
2.1. In Silico Prediction Analysis of the Hybridization Reaction between miRNAs and mRNAs
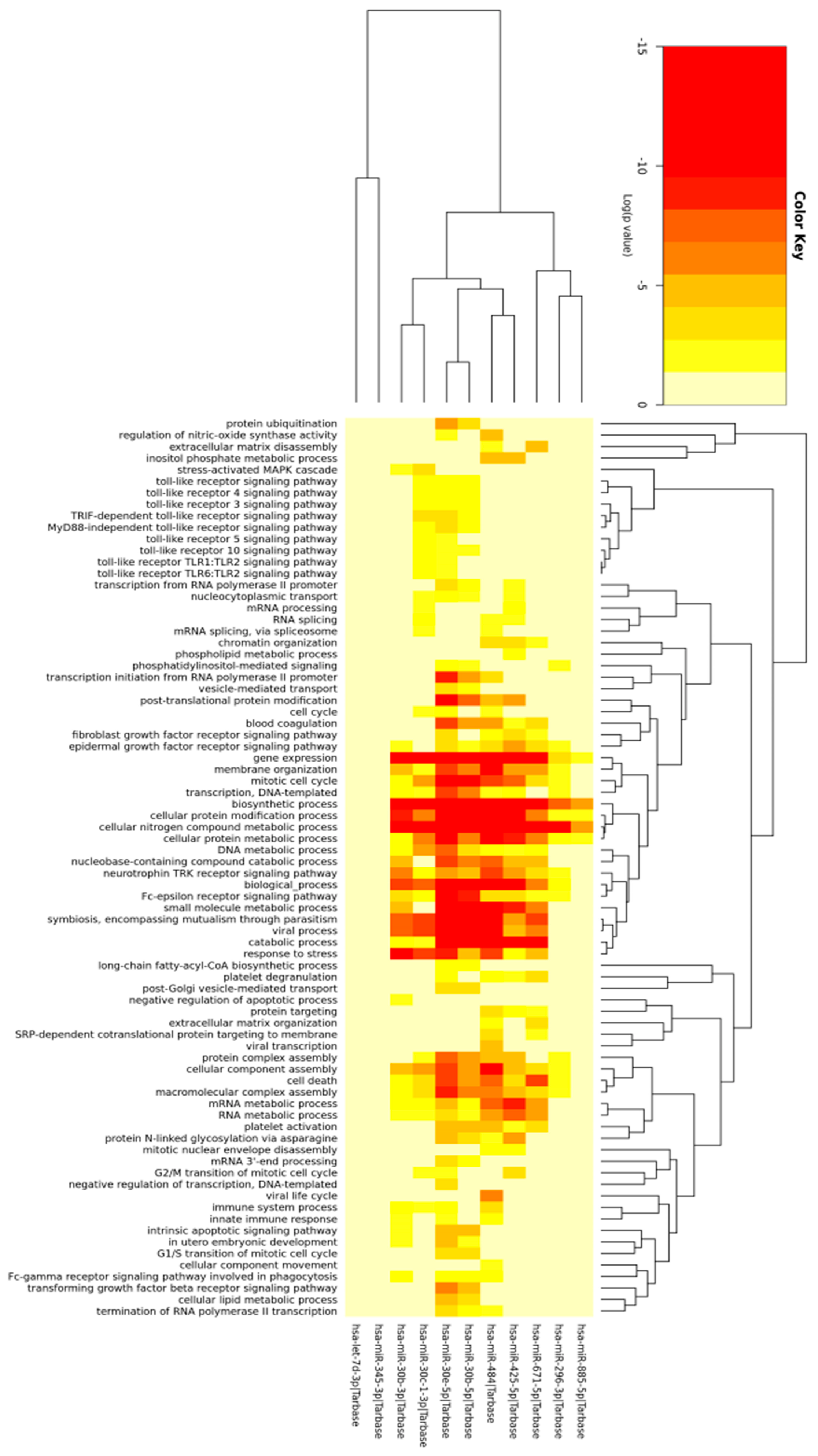
2.2. Prediction of Differentially Expressed miRNAs Target and Gene Ontology
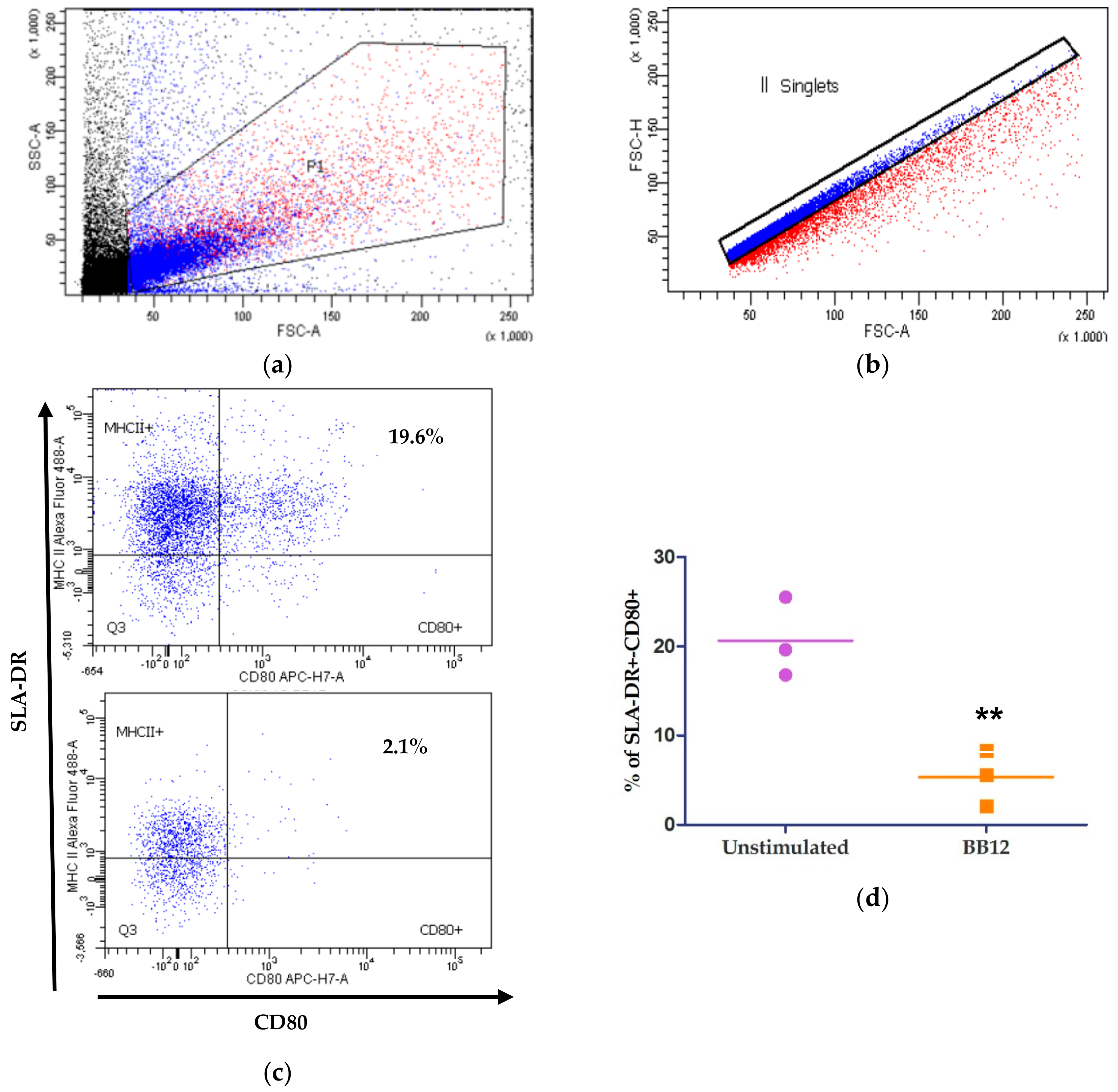
2.3. moDCs Phenotype after BB12 Stimuli
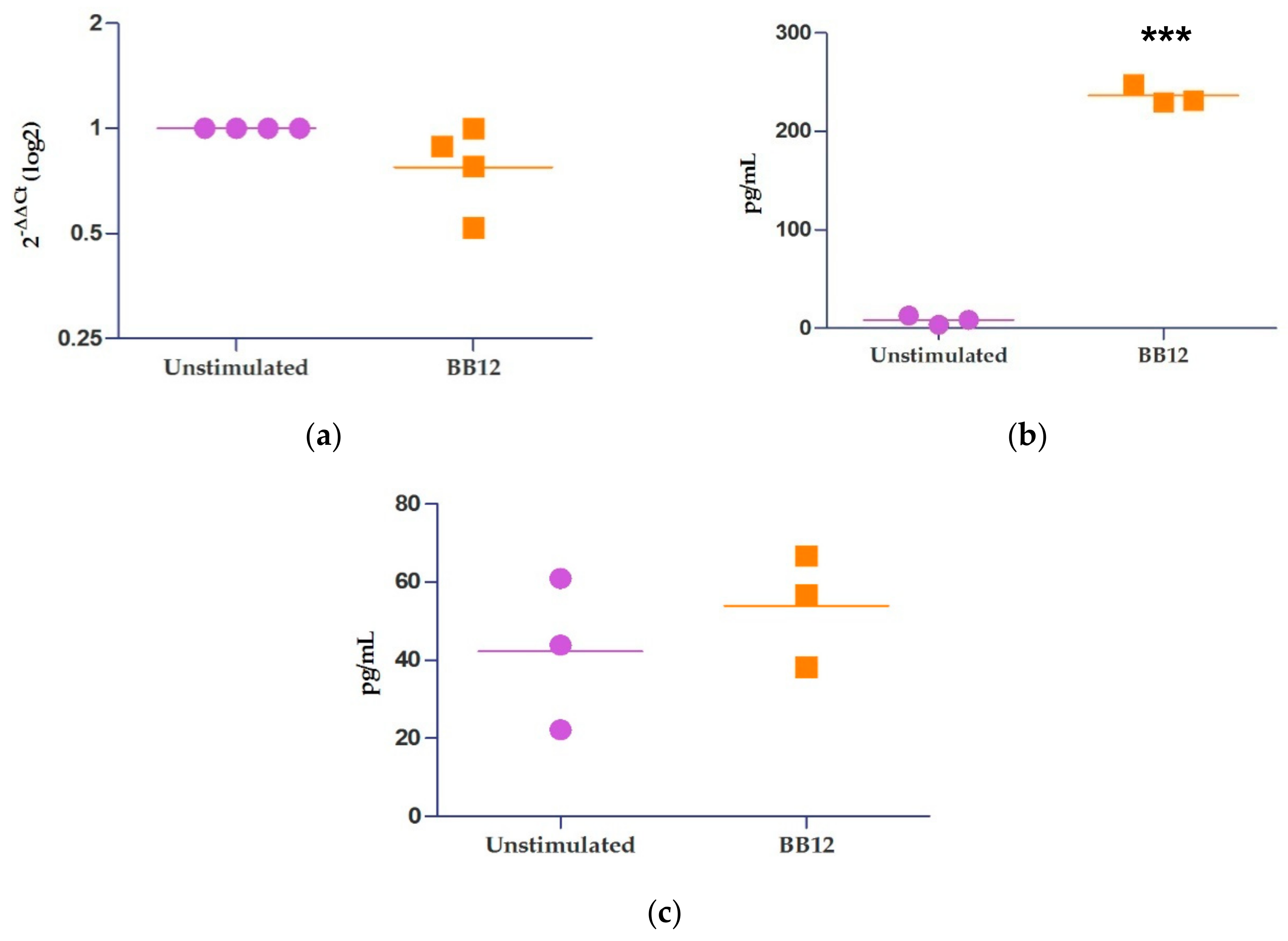
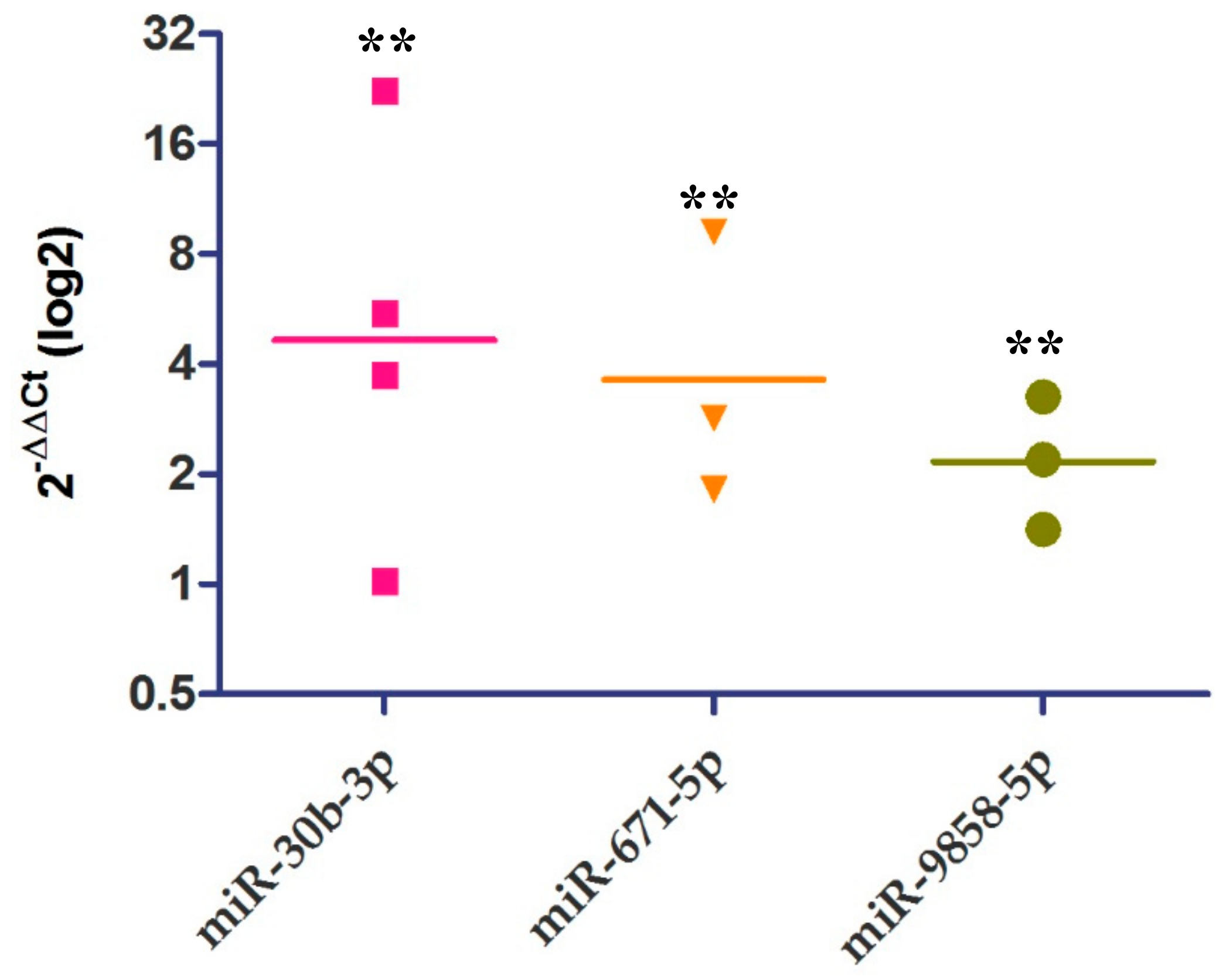
2.4. Relative Expression of IL-10 and miRNAs
3. Discussion
4. Materials and Methods
4.1. In Silico Prediction of the miRNA–mRNA 3′UTR Hybridization Reaction
4.2. Porcine Monocytes Isolation from Peripheral Blood
4.3. Monocyte Dendritic Cell (moDC) Generation
4.4. Stimulation of moDCs with BB12
4.5. Flow Cytometry Analysis
4.6. Relative Quantification of Cytokines Expression UsingRT-qPCR
4.7. Relative Quantification of miRNA Expression by RT-qPCR
4.8. MicroRNAs Primer Sequences
4.9. Quantification of Cytokines UsingELISA
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CLIP | Class II-associated invariant chain peptide |
| TLR2 | Toll-like receptor 2 |
| CD74 | SLA-DR antigens-associated invariant chain |
| RFX5 | DNA-binding protein RFX5 |
| LGMN | Legumain |
| CTSB | Cathepsin B precursor |
| SELL | L-selectin, CD62L |
| NFYA, B, C | Nuclear transcription factor Y subunit alpha, beta, and gamma |
| CREB1 | Cyclic AMP-responsive element-binding protein 1 isoform X1 |
| IFI30 | Gamma-interferon-inducible-lysosomal thiol reductase precursor |
| SLA-DRB1 | MHC class II histocompatibility antigen SLA-DRB1 precursor |
| SLA-DQB1 | SLA class II histocompatibility antigen, DQ haplotype C beta chain |
| SLA-DMA | SLA-DM alpha chain precursor |
| SLA-DMB | MHC class II, DM beta precursor |
| SLA-DMB | MHC class II, DO beta precursor |
| GO | Gene ontology |
References
- Jungersen, M.; Wind, A.; Johansen, E.; Christensen, J.E.; Stuer-Lauridsen, B.; Eskesen, D. The Science behind the Probiotic Strain Bifidobacterium animalis subsp. Lactis BB-12((R)). Microorganisms 2014, 2, 92–110. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Polk, D.B. Probiotics and immune health. Curr. Opin. Gastroenterol. 2011, 27, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Mandal, V.; Mandal, N.C. New Health Potentials of Orally Consumed Probiotic Microorganisms. In Probiotics; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2011; Volume 21. [Google Scholar]
- Neish, A.S. The gut microflora and intestinal epithelial cells: A continuing dialogue. Microbes Infect. 2002, 4, 309–317. [Google Scholar] [CrossRef]
- Smits, H.H.; Engering, A.; van der Kleij, D.; de Jong, E.C.; Schipper, K.; van Capel, T.M.; Zaat, B.A.; Yazdanbakhsh, M.; Wierenga, E.A.; van Kooyk, Y.; et al. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbingn on integrin. J. Allergy Clin. Immunol. 2005, 115, 1260–1267. [Google Scholar] [CrossRef] [PubMed]
- Christensen, H.R.; Frokiaer, H.; Pestka, J.J. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J. Immunol. 2002, 168, 171–178. [Google Scholar] [CrossRef]
- Hart, A.L.; Lammers, K.; Brigidi, P.; Vitali, B.; Rizzello, F.; Gionchetti, P.; Campieri, M.; Kamm, M.A.; Knight, S.C.; Stagg, A.J. Modulation of human dendritic cell phenotype and function by probiotic bacteria. Gut 2004, 53, 1602–1609. [Google Scholar] [CrossRef]
- Arenas-Padilla, M.; Duarte-Gutierrez, J.L.; Mata-Haro, V. Bifidobacterium animalis ssp. Lactis Bb12 induces IL-10 through cell membrane-associated components via TLR2 in swine. J. Appl. Microbiol. 2018. [Google Scholar] [CrossRef]
- Demont, A.; Hacini-Rachinel, F.; Doucet-Ladeveze, R.; Ngom-Bru, C.; Mercenier, A.; Prioult, G.; Blanchard, C. Live and heat-treated probiotics differently modulate IL10 mRNA stabilization and microRNAexpression. J. Allergy Clin. Immunol. 2016, 137, 1264–1267. [Google Scholar] [CrossRef][Green Version]
- Yang, F.; Hou, C.; Zeng, X.; Qiao, S. The use of lactic Acid bacteria as a probiotic in Swine diets. Pathogens 2015, 4, 34–45. [Google Scholar] [CrossRef]
- Sciascia, Q.; Daş, G.; Metges, C.C. REVIEW: The pig as a model for humans: Effects of nutritional factors on intestinal function and health. J. Anim. Sci. 2016, 94, 441–452. [Google Scholar] [CrossRef]
- Hvistendahl, M. Pigs as stand-ins for microbiome studies. Science 2012, 336 (Suppl.3), 1250. [Google Scholar] [CrossRef]
- Shin, D.; Chang, S.Y.; Bogere, P.; Won, K.; Choi, J.-Y.; Choi, Y.-J.; Lee, H.K.; Hur, J.; Park, B.-Y.; Kim, Y.; et al. Beneficial roles of probiotics on the modulation of gut microbiota and immune response in pigs. PLoS ONE 2019, 14, e0220843. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Baltimore, D. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016, 16, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Masotti, A. Interplays between Gut microbiota and gene expression regulation by miRNAs. Front. Cell. Infect. Microbiol. 2012, 2, 137. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.R.; Stedtfeld, R.D.; Tiedje, J.M.; Hashsham, S.A. MicroRNAs-Based Inter-Domain Communication between the Host and Members of the Gut Microbiome. Front. Microbiol. 2017, 8, 1896. [Google Scholar] [CrossRef] [PubMed]
- Bron, P.A.; van Baarlen, P.; Kleerebezem, M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat. Rev. Microbiol. 2011, 10, 66–78. [Google Scholar] [CrossRef]
- Nedumpun, T.; Ritprajak, P.; Suradhat, S. Generation of potent porcine monocyte-derived dendritic cells (MoDCs) by modified culture protocol. Vet. Immunol. Immunopathol. 2016, 182, 63–68. [Google Scholar] [CrossRef]
- Couper, K.N.; Blount, D.G.; Riley, E.M. IL-10: The master regulator of immunity to infection. J. Immunol. 2008, 180, 5771–5777. [Google Scholar] [CrossRef]
- Arenas-Padilla, M. miRNAs Expression in Swine Monocytes Stimulated with Bifidobacterium animalis ssp. lactis Bb12. Ciencia de los Alimentos. PhD Thesis, Centro de Investigación en Alimentación y Desarrollo Hermosillo, Hermosillo, México, 2018; p. 57. [Google Scholar]
- Baerlecken, N.T.; Nothdorft, S.; Stummvoll, G.H.; Sieper, J.; Rudwaleit, M.; Reuter, S.; Matthias, T.; Schmidt, R.E.; Witte, T. Autoantibodies against CD74 in spondyloarthritis. Ann. Rheum. Dis. 2014, 73, 1211–1214. [Google Scholar] [CrossRef]
- Nekrep, N.; Geyer, M.; Jabrane-Ferrat, N.; Peterlin, B.M. Analysis of Ankyrin Repeats Reveals How a Single Point Mutation in RFXANK Results in Bare Lymphocyte Syndrome. Mol. Cell. Biol. 2001, 21, 5566–5576. [Google Scholar] [CrossRef]
- Talpin, A.; Costantino, F.; Bonilla, N.; Leboime, A.; Letourneur, F.; Jacques, S.; Dumont, F.; Amraoui, S.; Dutertre, C.-A.; Garchon, H.-J.; et al. Monocyte-derived dendritic cells from HLA-B27+ axial spondyloarthritis (SpA) patients display altered functional capacity and deregulated gene expression. Arthritis Res. Ther. 2014, 16, 417. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, F.; Ping, Y.; Wang, L.; Chen, X.; Wang, D.; Cao, L.; Zhao, S.; Li, B.; Kalinski, P.; et al. Local production of the chemokines CCL5 and CXCL10 attracts CD8(+) T lymphocytes into esophageal squamous cell carcinoma. Oncotarget 2015, 6, 24978–24989. [Google Scholar] [CrossRef] [PubMed]
- Jabrane-Ferrat, N.; Nekrep, N.; Tosi, G.; Esserman, L.J.; Peterlin, B.M. Major histocompatibility complex class II transcriptional platform: Assembly of nuclear factor Y and regulatory factor X (RFX) on DNA requires RFX5 dimers. Mol. Cell. Biol. 2002, 22, 5616–5625. [Google Scholar] [CrossRef] [PubMed]
- Nahid, M.A.; Yao, B.; Dominguez-Gutierrez, P.R.; Kesavalu, L.; Satoh, M.; Chan, E.K. Regulation of TLR2-mediated tolerance and cross-tolerance through IRAK4 modulation by miR-132 and miR-212. J. Immunol. 2013, 190, 1250–1263. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, B.; Phan, U.T.; Geuze, H.J.; Cresswell, P. Enzymatic reduction of disulfide bonds in lysosomes: Characterization of a Gamma-interferon-inducible lysosomal thiol reductase (GILT). Proc. Natl. Acad. Sci. USA 2000, 97, 745–750. [Google Scholar] [CrossRef]
- Jakubzick, C.V.; Randolph, G.J.; Henson, P.M. Monocyte differentiation and antigen-presenting functions. Nat. Rev. Immunol. 2017, 17, 349. [Google Scholar] [CrossRef]
- Rodríguez-Gómez, I.M.; Käser, T.; Gómez-Laguna, J.; Lamp, B.; Sinn, L.; Rümenapf, T.; Carrasco, L.; Saalmüller, A.; Gerner, W. PRRSV-infected monocyte-derived dendritic cells express high levels of SLA-DR and CD80/86 but do not stimulate PRRSV-naïve regulatory T cells to proliferate. Vet. Res. 2015, 46, 54. [Google Scholar] [CrossRef]
- Silva-Cardoso, S.C.; Affandi, A.J.; Spel, L.; Cossu, M.; van Roon, J.A.G.; Boes, M.; Radstake, T.R.D.J. CXCL4 Exposure Potentiates TLR-Driven Polarization of Human Monocyte-Derived Dendritic Cells and Increases Stimulation of T Cells. J. Immunol. 2017, 1602020. [Google Scholar] [CrossRef]
- Hubo, M.; Trinschek, B.; Kryczanowsky, F.; Tuettenberg, A.; Steinbrink, K.; Jonuleit, H. Costimulatory molecules on immunogenic versus tolerogenic human dendritic cells. Front. Immunol. 2013, 4, 82. [Google Scholar] [CrossRef]
- Jaeger, A.; Hadlich, F.; Kemper, N.; Lubke-Becker, A.; Murani, E.; Wimmers, K.; Ponsuksili, S. MicroRNA expression profiling of porcine mammary epithelial cells after challenge with Escherichia coli in vitro. BMC Genom. 2017, 18, 660. [Google Scholar] [CrossRef]
- Dalman, M.R.; Deeter, A.; Nimishakavi, G.; Duan, Z.-H. Foldchange and p-value cutoffs significantly alter microarray interpretations. BMC Bioinform. 2012, 13, S11. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J.; Zhu, J.; van Oudenaarden, A. MicroRNA-Mediated Feedback and Feedforward Loops Are Recurrent Network Motifs in Mammals. Mol. Cell 2007, 26, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Fu, Y.; Chen, L.; Lee, W.; Lai, Y.; Rezaei, K.; Tabbara, S.; Latham, P.; Teal, C.B.; Man, Y.G.; et al. miR-671-5p inhibits epithelial-to-mesenchymal transition by downregulating FOXM1 expression in breast cancer. Oncotarget 2016, 7, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.R.; Fordham, J.B.; Ganesh, B.; Nares, S. miR-24, miR-30b and miR-142-3p interfere with antigen processing and presentation by primary macrophages and dendritic cells. Sci. Rep. 2016, 6, 32925. [Google Scholar] [CrossRef]
- Holscher, H.D.; Czerkies, L.A.; Cekola, P.; Litov, R.; Benbow, M.; Santema, S.; Alexander, D.D.; Perez, V.; Sun, S.; Saavedra, J.M.; et al. Bifidobacterium lactis Bb12 Enhances Intestinal Antibody Response in Formula-Fed Infants: A Randomized, Double-Blind, Controlled Trial. J. Parenter. Enter. Nutr. 2012, 36, 106S–117S. [Google Scholar] [CrossRef]
- Singleton, H.; Graham, S.P.; Bodman-Smith, K.B.; Frossard, J.-P.; Steinbach, F. Establishing Porcine Monocyte-Derived Macrophage and Dendritic Cell Systems for Studying the Interaction with PRRSV-1. Front. Microbiol. 2016, 7, 832. [Google Scholar] [CrossRef]
- Mohamadzadeh, M.; Olson, S.; Kalina, W.V.; Ruthel, G.; Demmin, G.L.; Warfield, K.L.; Bavari, S.; Klaenhammer, T.R. Lactobacilli activate human dendritic cells that skew T cells toward T helper 1 polarization. Proc. Natl. Acad. Sci. USA 2005, 102, 2880–2885. [Google Scholar] [CrossRef]
- Pérez, I.; Falco, A.; Tapia, M.; Alonso, G. Aislamiento e identificación de cepas del género Bifidobacterium presentes en productos lácteos fermentados tipo yogur. Revista de la Sociedad Venezolana de Microbiología 2012, 32, 29–36. [Google Scholar]
- Mentzel, C.M.J.; Skovgaard, K.; Cordoba, S.; Herrera, J.; Busk, P.K.; Cirera, S. Wet-lab Tested MicroRNA Assays for qPCR Studies with SYBR®Green and DNA Primers in Pig Tissues. MicroRNA 2014, 3, 174–188. [Google Scholar] [CrossRef]
- Busk, P.K. A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinform. 2014, 15, 29. [Google Scholar] [CrossRef]
| miRNA | Target | Length | Position | Mfe(kcal/mol) |
|---|---|---|---|---|
| ssc-miR-671-5p | IL10 | 735 | 435 | −41.2 |
| ssc-miR-671-5p | CD74 | 2784 | 1507 | −38.0 |
| ssc-miR-671-5p | RFXANK | 1758 | 163 | −38.0 |
| ssc-miR-345-3p | SLA-DRB1 | 341 | 111 | −37.8 |
| ssc-miR-671-5p | SLA-DQB1 | 357 | 121 | −32.0 |
| ssc-miR-671-5p | NFYA | 2590 | 1722 | −31.8 |
| ssc-miR-671-5p | LGMN | 1544 | 720 | −31.4 |
| ssc-miR-671-5p | NFYC | 743 | 590 | −31.3 |
| ssc-miR-4334-5p | RFXANK | 1758 | 828 | −31.0 |
| ssc-miR-4334-5p | CTSB | 948 | 876 | −30.7 |
| ssc-miR-7138-5p | NFYB | 1802 | 1342 | −29.9 |
| ssc-miR-9816-3p | RFXANK | 1758 | 693 | −29.5 |
| ssc-miR-671-5p | TNF | 1926 | 197 | −29.4 |
| ssc-miR-185 | IL1B2 | 583 | 170 | −29.4 |
| ssc-miR-4334-5p | CD74 | 2784 | 1507 | −29.3 |
| ssc-miR-4334-5p | SLA-DMA | 225 | 131 | −29.0 |
| ssc-miR-30b-3p | CD80 | 1492 | 920 | −28.8 |
| ssc-miR-31 | CXCL10 | 3643 | 1848 | −28.8 |
| ssc-miR-9802-3p | RFX5 | 1521 | 139 | −28.7 |
| ssc-miR-9858-5p | TNF | 1926 | 916 | −28.7 |
| ssc-miR-30c-1-3p | CD74 | 2784 | 1084 | −28.5 |
| ssc-miR-29b | IFI30 | 228 | 164 | −28.5 |
| ssc-miR-9858-5p | CD86 | 3967 | 252 | −28.5 |
| ssc-miR-30b-3p | SELL | 2974 | 509 | −28.1 |
| ssc-miR-30c-1-3p | SELL | 2974 | 2652 | −28.0 |
| ssc-miR-9857-5p | NFYC | 743 | 16 | −28.0 |
| ssc-miR-31 | CD74 | 2784 | 27 | −27.6 |
| ssc-miR-31 | RFX5 | 1521 | 117 | −27.6 |
| ssc-miR-185 | SELL | 2974 | 1301 | −27.5 |
| ssc-miR-185 | RFXANK | 1758 | 1591 | −27.4 |
| ssc-miR-7138-5p | CD74 | 2784 | 2265 | −27.4 |
| ssc-miR-31 | SLA-DQB1 | 357 | 40 | −27.3 |
| ssc-miR-185 | IL10 | 735 | 120 | −27.1 |
| ssc-miR-9816-3p | TNF | 1926 | 1745 | −27.1 |
| ssc-miR-30b-3p | CD74 | 2784 | 1084 | −27.0 |
| ssc-miR-9858-5p | CD80 | 1492 | 928 | −27.0 |
| ssc-miR-362 | CD74 | 2784 | 124 | −26.8 |
| ssc-miR-7138-5p | TNF | 1926 | 203 | −26.6 |
| ssc-miR-4334-5p | CXCL2 | 712 | 400 | −26.5 |
| ssc-miR-185 | SLA-DMA | 225 | 64 | −26.4 |
| ssc-miR-9846-3p | CXCL2 | 712 | 401 | −26.4 |
| ssc-miR-31 | NFYC | 743 | 479 | −26.4 |
| ssc-miR-9857-5p | SLA-DOB | 1030 | 254 | −26.4 |
| ssc-miR-362 | TNF | 1926 | 1624 | −26.3 |
| ssc-miR-9813-5p | RFXANK | 1758 | 688 | −26.1 |
| ssc-miR-9858-5p | SLA-DOB | 1030 | 532 | −26.0 |
| ssc-miR-9816-3p | CXCL10 | 3643 | 876 | −26.0 |
| ssc-miR-9858-5p | CXCL10 | 3643 | 2094 | −25.9 |
| ssc-miR-185 | CTSB | 948 | 829 | −25.9 |
| ssc-miR-185 | CD74 | 2784 | 1086 | −25.9 |
| ssc-miR-9846-3p | CD86 | 3967 | 2423 | −25.9 |
| ssc-miR-181b | CREB1 | 3509 | 3382 | −25.8 |
| ssc-miR-362 | CTSB | 948 | 718 | −25.8 |
| ssc-miR-185 | LGMN | 1544 | 122 | −25.7 |
| ssc-miR-185 | SLA-DRB1 | 341 | 89 | −25.7 |
| ssc-miR-9813-5p | CD74 | 2784 | 1416 | −25.6 |
| ssc-miR-7 | IL1B2 | 583 | 412 | −25.6 |
| ssc-miR-671-5p | SELL | 2974 | 309 | −25.5 |
| ssc-miR-9813-5p | TNF | 1926 | 1378 | −25.5 |
| ssc-miR-362 | LGMN | 1544 | 1280 | −25.4 |
| ssc-miR-9846-3p | CD80 | 1492 | 829 | −25.4 |
| ssc-miR-9846-3p | RFXANK | 1758 | 1362 | −25.3 |
| ssc-miR-31 | CD86 | 3967 | 1545 | −25.3 |
| ssc-miR-9816-3p | CD74 | 2784 | 757 | −25.3 |
| ssc-miR-9802-3p | SLA-DMB | 355 | 52 | −25.2 |
| ssc-miR-99b | LGMN | 1544 | 909 | −25.2 |
| ssc-miR-99b | CD74 | 2784 | 294 | −25.2 |
| ssc-miR-9802-3p | CTSB | 948 | 39 | −25.0 |
| ssc-miR-181b | RFXANK | 1758 | 308 | −25.0 |
| miR-9858-5p: forward (5′–3′) GCAGTTCCTGAGTCGGA | reverse (3′–5′) GGTCCAGTTTTTTTTTTTTTTTAGC |
| miR-671-5p: forward (5′–3′) CCCTGGAGGGGCTG | reverse (3′–5′) GGTCCAGTTTTTTTTTTTTTTTCCT |
| miR-30b-3p: forward (5′–3′) GCTGGGAGGTGGATGT | reverse (3′–5′) CAGGTCCAGTTTTTTTTTTTTTTTAAG |
| miR-181a: forward (5′–3′) CATTCAACGCTGTCGGT | reverse (3′–5′) GTCCAGTTTTTTTTTTTTTTTAACTCA |
| miR-30b-5p: forward (5′–3′) GCAGTGTAAACATCCTCGAC | reverse (3′–5′) TCCAGTTTTTTTTTTTTTTTCTTCCA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bravo-Parra, M.; Arenas-Padilla, M.; Bárcenas-Preciado, V.; Hernández, J.; Mata-Haro, V. The Probiotic BB12 Induces MicroRNAs Involved in Antigen Processing and Presentation in Porcine Monocyte-Derived Dendritic Cells. Int. J. Mol. Sci. 2020, 21, 687. https://doi.org/10.3390/ijms21030687
Bravo-Parra M, Arenas-Padilla M, Bárcenas-Preciado V, Hernández J, Mata-Haro V. The Probiotic BB12 Induces MicroRNAs Involved in Antigen Processing and Presentation in Porcine Monocyte-Derived Dendritic Cells. International Journal of Molecular Sciences. 2020; 21(3):687. https://doi.org/10.3390/ijms21030687
Chicago/Turabian StyleBravo-Parra, Marlene, Marina Arenas-Padilla, Valeria Bárcenas-Preciado, Jesús Hernández, and Verónica Mata-Haro. 2020. "The Probiotic BB12 Induces MicroRNAs Involved in Antigen Processing and Presentation in Porcine Monocyte-Derived Dendritic Cells" International Journal of Molecular Sciences 21, no. 3: 687. https://doi.org/10.3390/ijms21030687
APA StyleBravo-Parra, M., Arenas-Padilla, M., Bárcenas-Preciado, V., Hernández, J., & Mata-Haro, V. (2020). The Probiotic BB12 Induces MicroRNAs Involved in Antigen Processing and Presentation in Porcine Monocyte-Derived Dendritic Cells. International Journal of Molecular Sciences, 21(3), 687. https://doi.org/10.3390/ijms21030687