Deep Sequencing MicroRNAs from Extracellular Membrane Vesicles Revealed the Association of the Vesicle Cargo with Cellular Origin
Abstract
1. Introduction
2. Results
2.1. HaCaT- and PKC-Derived EVs Exhibit Common Highly Abundant miRNAs
2.2. Specific EV miRNAs Are Correlated with Cellular Origin
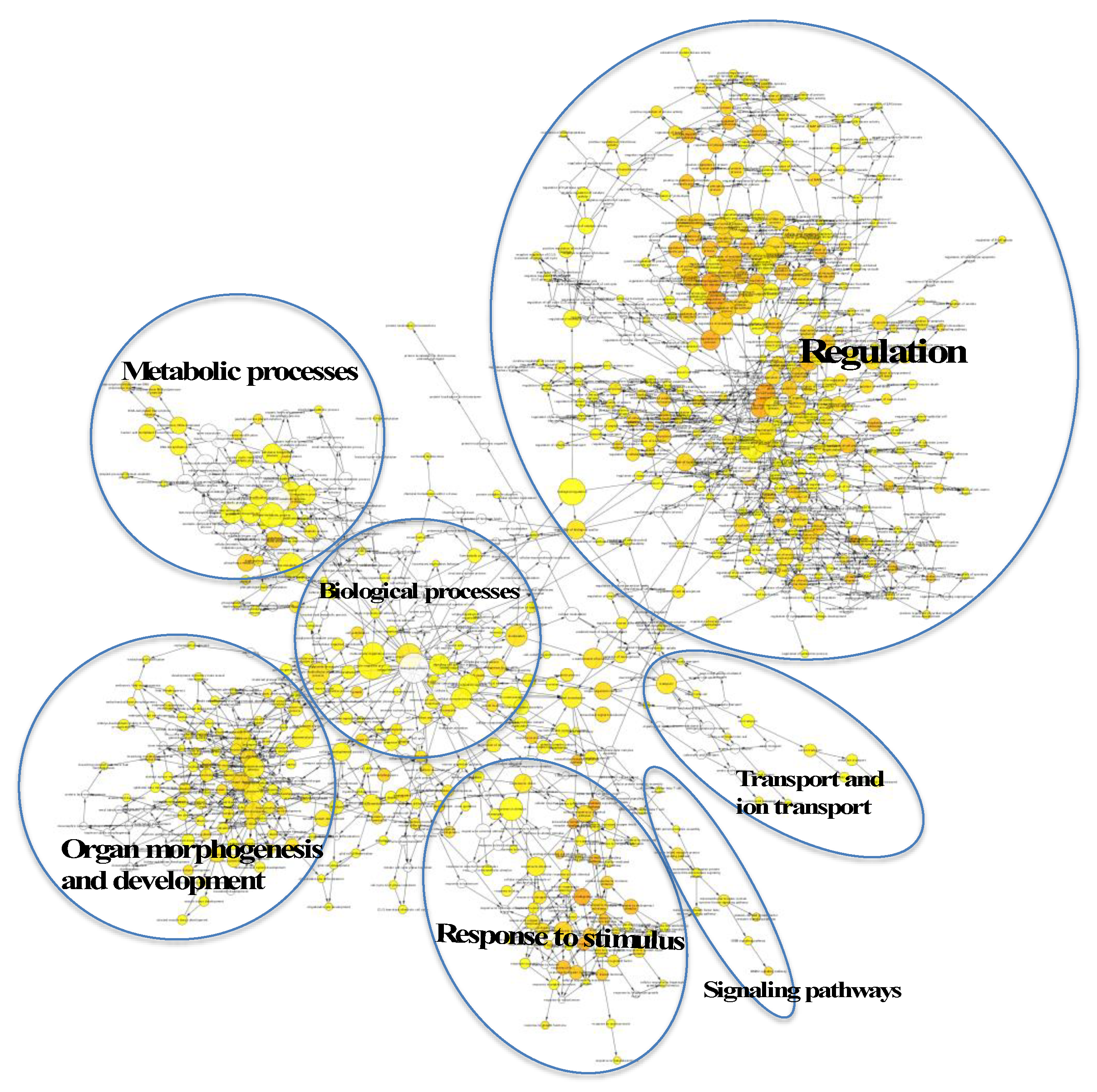
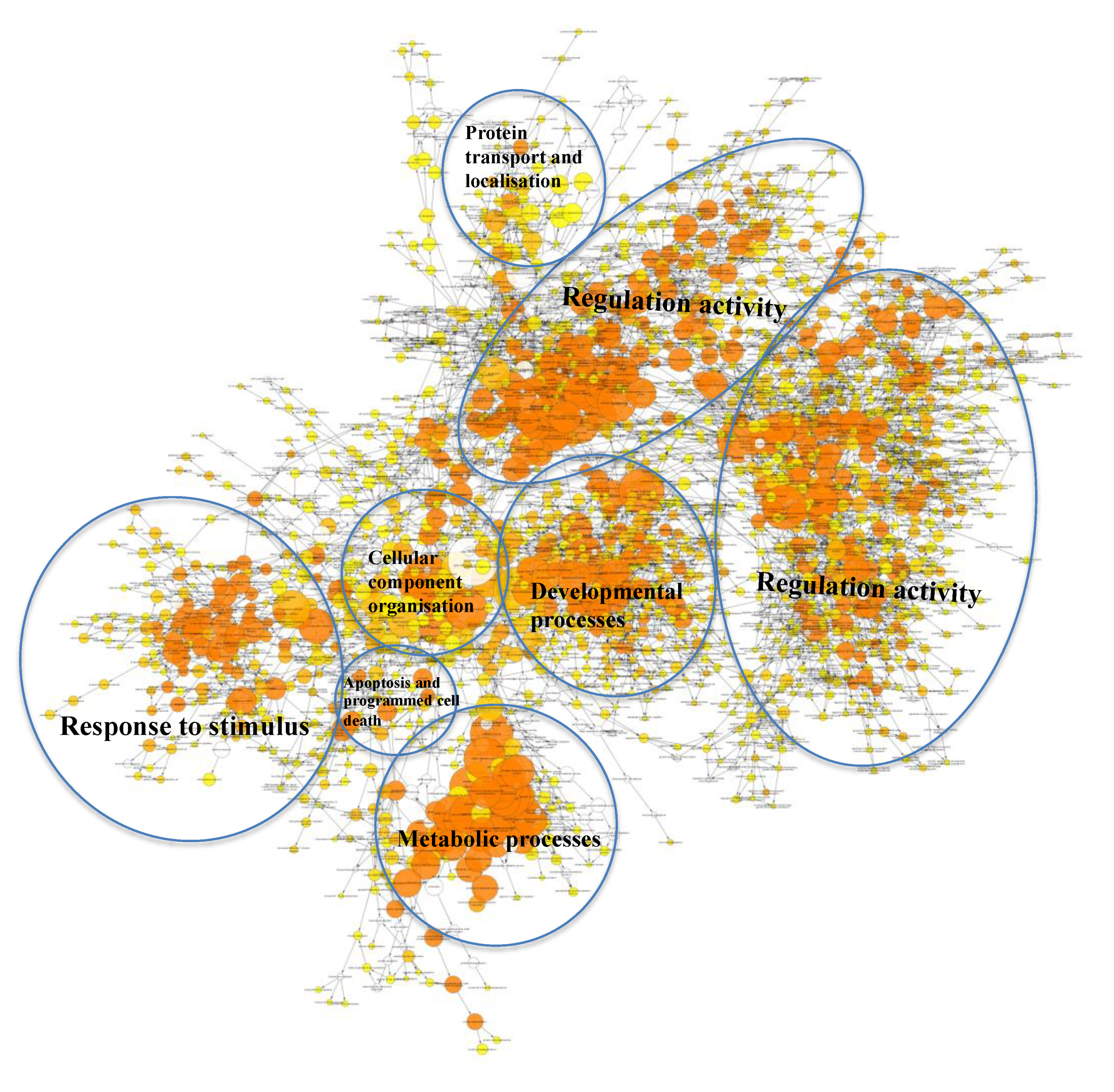
2.3. Many Target Genes Regulated by miRNAs Associated with HaCaT and PKCs
2.4. Regulatory Roles of EVs
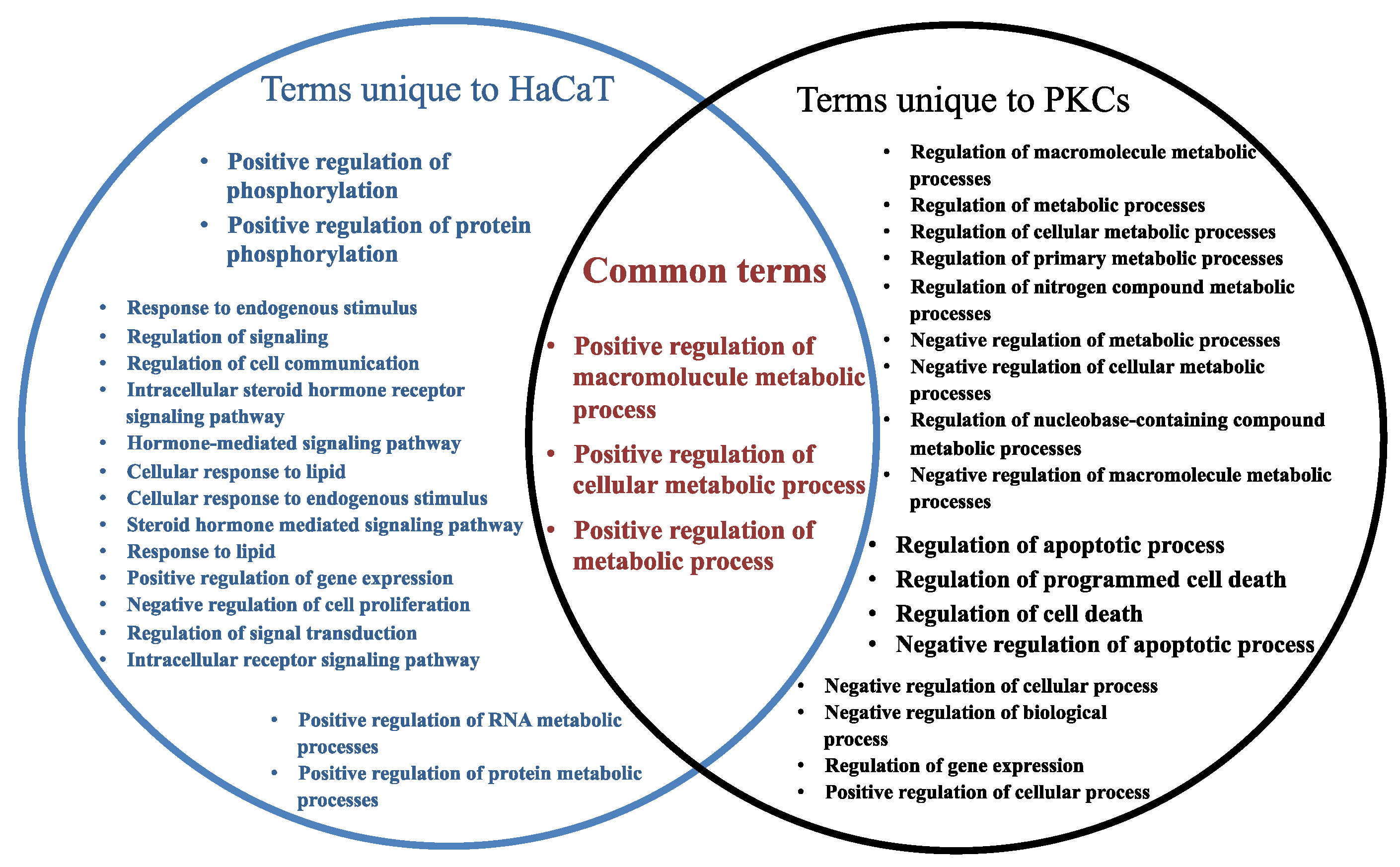
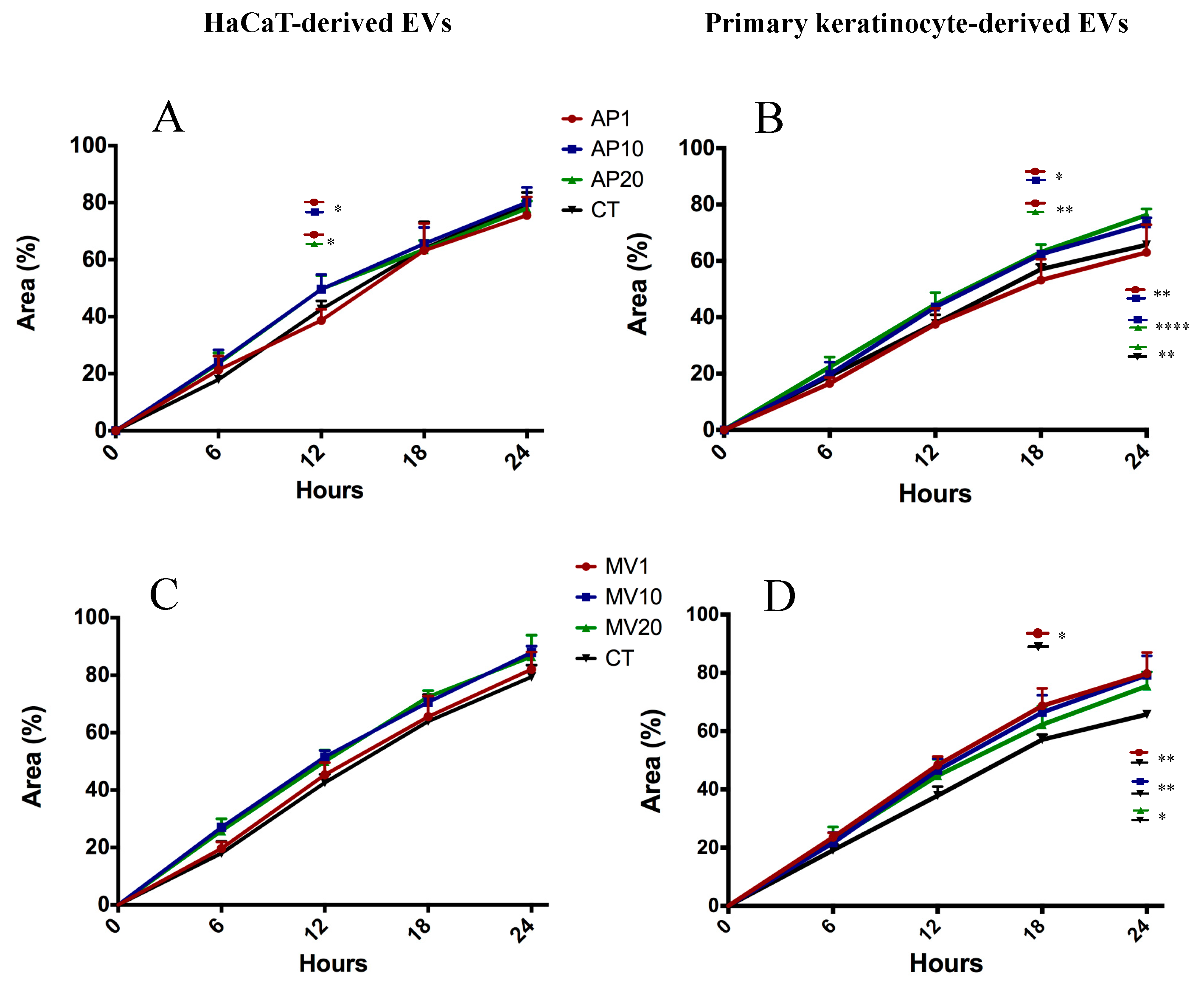
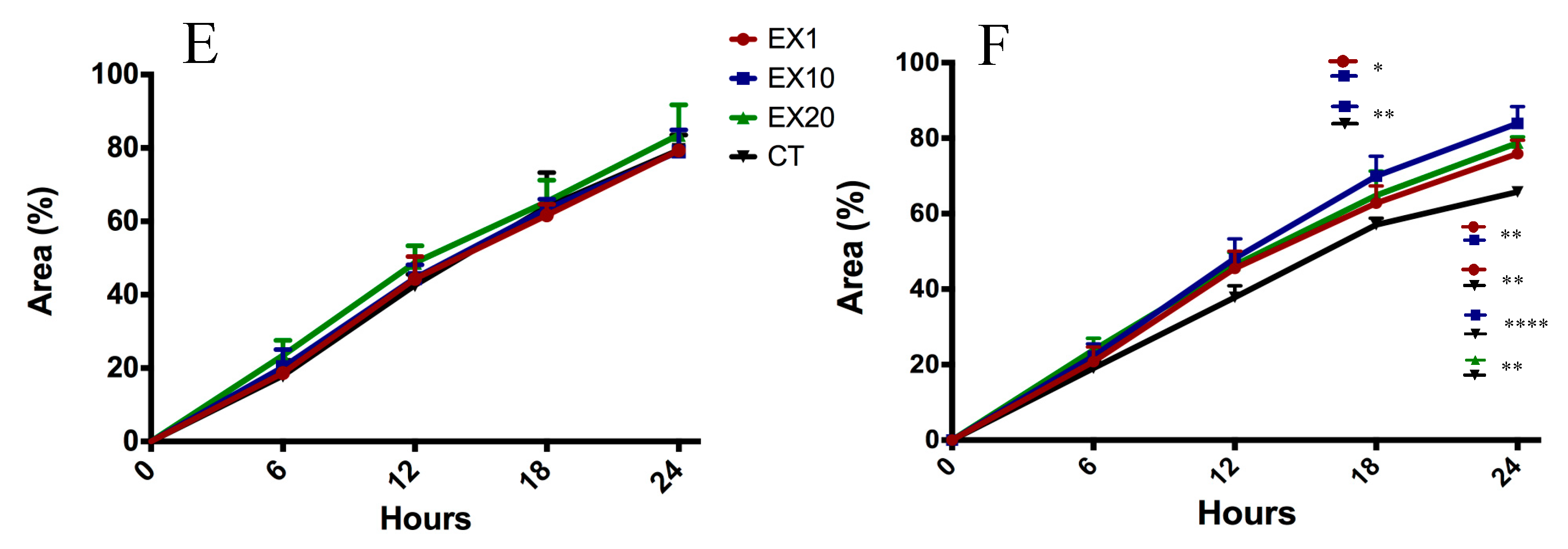
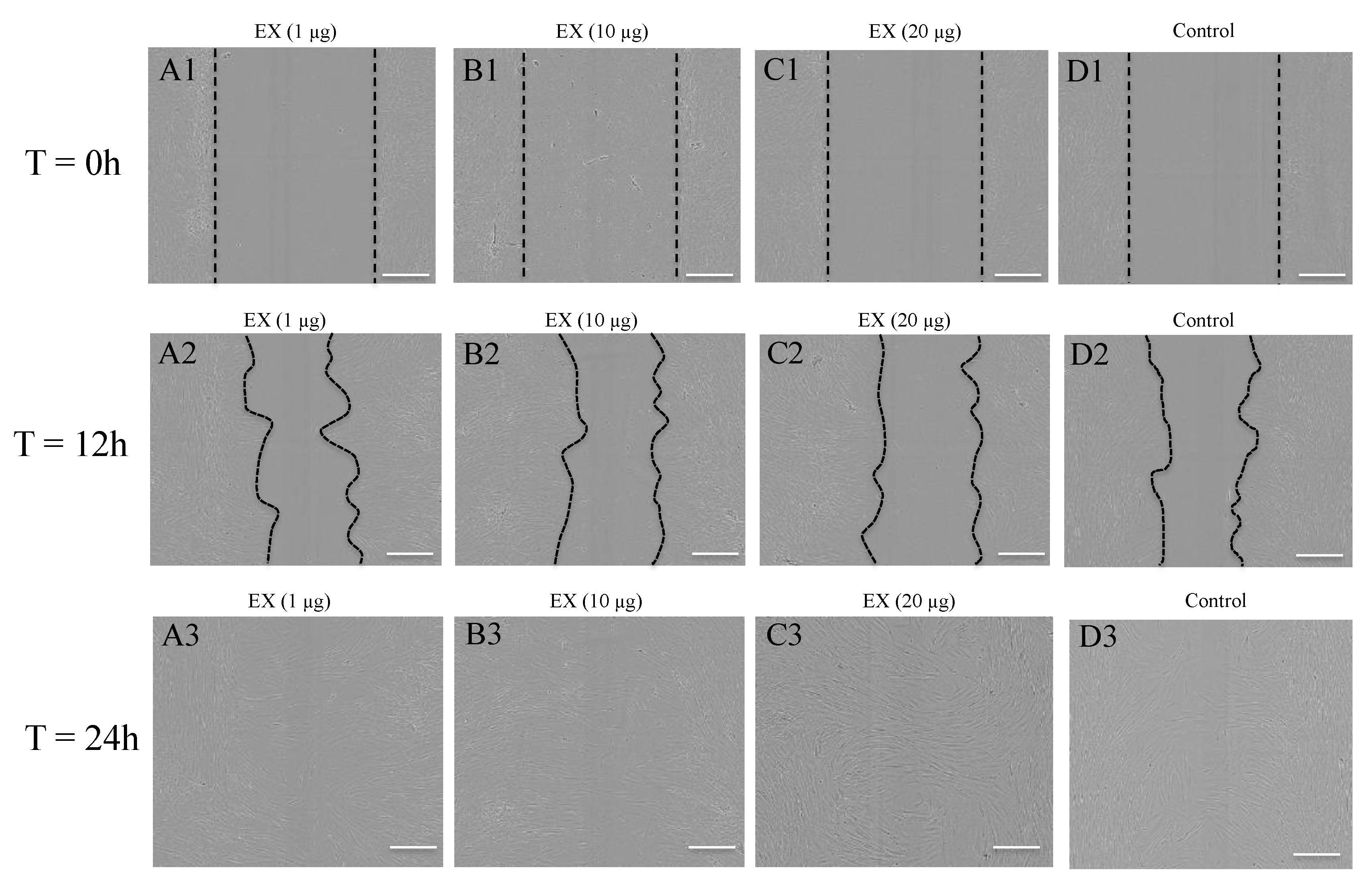
2.5. Target Genes Regulated by EV miRNAs Related to Cell Migration
3. Discussions
4. Materials and Methods
4.1. Cell Culture
4.2. EV Production and Isolation
4.3. Total RNA Extraction and miRNA Sequencing
4.4. miRNA Identification and Statistics
4.5. Analysis of miRNA Target Genes
4.6. Scratch-Wound Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AP(s) | Apoptotic Bodies |
| BP | Biological process |
| CM | Conditioned medium |
| ECM | Extracellular matrix |
| EX(s) | Exosome(s) |
| EV(s) | Extracellular membrane vesicle(s) |
| GO | Gene Ontology |
| MAPK | Mitogen-activated protein kinase |
| miRNA | microRNAs |
| MV(s) | Microvesicle(s) |
| PCA | Principal component analysis |
| PBS | Phosphate -saline |
| PKC(s) | Primary keratinocyte(s) |
| PTEN | Phosphatase and tensin homolog |
| RPM | Read per million |
| SHIP2 | SH2-domain containing inositol 5-phospatase 2 |
| TGF-beta | Transforming growth factor beta |
References
- Chávez-Muñoz, C.; Morse, J.; Kilani, R.; Ghahary, A. Primary human keratinocytes externalize stratifin protein via exosomes. J. Cell. Biochem. 2008, 104, 2165–2173. [Google Scholar] [CrossRef]
- Cicero, A.L.; Delevoye, C.; Gilles-Marsens, F.; Loew, D.; Dingli, F.; Guéré, C.; Andre, N.; Vie, K.; Van Niel, G.; Raposo, G. Exosomes released by keratinocytes modulate melanocyte pigmentation. Nat. Commun. 2015, 6, 7506. [Google Scholar] [CrossRef] [PubMed]
- Than, U.T.; Guanzon, D.; Broadbent, J.A.; Leavesley, D.I.; Salomon, C.; Parker, T.J. Differential expression of keratinocyte-derived extracellular vesicle mirRNAs discriminate exosomes from apoptotic bodies and microvesicles. Front. Endocrinol. 2018, 9, 535. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neuro-Oncol. 2013, 113, 1–11. [Google Scholar] [CrossRef] [PubMed]
- György, B.; Kittel, A.; Nagy, G.; Falus, A.; Buzás, E.I.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. CMLS 2011, 68, 2667–2688. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Than, U.T.T.; Guanzon, D.; Leavesley, D.; Parker, T. Association of extracellular membrane vesicles with cutaneous wound healing. Int. J. Mol. Sci. 2017, 18, 956. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, L.; Wang, S.; Han, Q.; Zhao, R.C. Exosomes secreted by mesenchymal stem cells promote endothelial cell angiogenesis by transferring miR-125a. J. Cell Sci. 2016, 129, 2182–2189. [Google Scholar] [CrossRef]
- Chávez-Muñoz, C.; Kilani, R.T.; Ghahary, A. Profile of exosomes related proteins released by differentiated and undifferentiated human keratinocytes. J. Cell. Physiol. 2009, 221, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xue, L.; Gao, H.; Chang, L.; Yu, X.; Zhu, Z.; He, X.; Geng, J.; Dong, Y.; Li, H.; et al. Exosomal miRNA derived from keratinocytes regulates pigmentation in melanocytes. J. Dermatol. Sci. 2019, 93, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Mullokandov, G.; Baccarini, A.; Ruzo, A.; Jayaprakash, A.D.; Tung, N.; Israelow, B.; Evans, M.J.; Sachidanandam, R.; Brown, B.D. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods 2012, 9, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Chen, M.; Greening, D.W.; He, W.; Rai, A.; Zhang, W.; Simpson, R.J. Deep Sequencing of RNA from Three Different Extracellular Vesicle (EV) Subtypes Released from the Human LIM1863 Colon Cancer Cell Line Uncovers Distinct Mirna-Enrichment Signatures. PLoS ONE 2014, 9, e110314. [Google Scholar] [CrossRef]
- Lunavat, T.R.; Cheng, L.; Kim, D.-K.; Bhadury, J.; Jang, S.C.; Lässer, C.; Sharples, R.A.; López, M.D.; Nilsson, J.; Gho, Y.S.; et al. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells--Evidence of unique microRNA cargos. RNA Biol. 2015, 12, 810–823. [Google Scholar] [CrossRef]
- Than, U.T.; Guanzon, D.; Wager, L.; Manton, K.J.; Hollier, B.G.; Leavesley, D.I. An Analysis of Exosomes From Keratinocytes And Fibroblasts. In IFMBE_5th International Conference on Biomedical Engineering in Vietnam; Springer International Publishing: Basel, Switzerland, 2015; Volume 46. [Google Scholar]
- Brown, B.D.; Gentner, B.; Cantore, A.; Colleoni, S.; Amendola, M.; Zingale, A.; Baccarini, A.; Lazzari, G.; Galli, C.; Naldini, L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007, 25, 1457–1467. [Google Scholar] [CrossRef]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M.; Jungkamp, A.-C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef]
- Lena, A.M.; Shalom-Feuerstein, R.; Cervo, P.R.d.; Aberdam, D.; Knight, R.A.; Melino, G.; Candi, E. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008, 15, 1187. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Guo, S.-L.; Fan, K.-J.; Li, J.; Wang, Y.-L.; Teng, Y.; Yang, X. miR-21 Promotes keratinocyte migration and re-epithelialization during wound healing. Int. J. Boil. Sci. 2011, 7, 685–690. [Google Scholar] [CrossRef]
- Yu, J.; Peng, H.; Ruan, Q.; Fatima, A.; Getsios, S.; Lavker, R.M. MicroRNA-205 promotes keratinocyte migration via the lipid phosphatase SHIP2. FASEB J. 2010, 24, 3950–3959. [Google Scholar] [CrossRef]
- Wang, T.; Feng, Y.; Sun, H.; Zhang, L.; Hao, L.; Shi, C.; Wang, J.; Li, R.; Ran, X.; Su, Y.; et al. miR-21 Regulates Skin Wound Healing by Targeting Multiple Aspects of the Healing Process. Am. J. Pathol. 2012, 181, 1911–1920. [Google Scholar] [CrossRef] [PubMed]
- Viticchiè, G.; Lena, A.M.; Cianfarani, F.; Odorisio, T.; Annicchiarico-Petruzzelli, M.; Melino, G.; Candi, E. MicroRNA-203 contributes to skin re-epithelialization. Cell Death Dis. 2012, 3, e435. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Friggeri, A.; Yang, Y.; Milosevic, J.; Ding, Q.; Thannickal, V.J.; Kaminski, N.; Abraham, E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 2010, 207, 1589–1597. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Kumar, M.; Choudhury, S.N.; Buscaglia, L.E.B.; Barker, J.R.; Kanakamedala, K.; Liu, M.-F.; Li, Y. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc. Natl. Acad. Sci. 2011, 108, 10144–10149. [Google Scholar] [CrossRef]
- Parapuram, S.K.; Shi-Wen, X.; Elliott, C.; Welch, I.D.; Jones, H.; Baron, M.; Denton, C.P.; Abraham, D.J.; Leask, A. Loss of PTEN expression by dermal fibroblasts causes skin fibrosis. J. Investig. Dermatol. 2011, 131, 1996–2003. [Google Scholar] [CrossRef]
- Bruegger, C.; Kempf, W.; Spoerri, I.; Arnold, A.W.; Itin, P.H.; Burger, B. MicroRNA expression differs in cutaneous squamous cell carcinomas and healthy skin of immunocompetent individuals. Exp. Dermatol. 2013, 22, 426–428. [Google Scholar] [CrossRef]
- Harris, S.L.; Levine, A.J. The p53 pathway: Positive and negative feedback loops. Oncogene 2005, 24, 2899–2908. [Google Scholar] [CrossRef]
- De Caestecker, M.P.; Piek, E.; Roberts, A.B. Role of transforming growth factor-beta signaling in cancer. J. Natl. Cancer Inst. 2000, 92, 1388–1402. [Google Scholar] [CrossRef]
- Seo, M.-D.; Kang, T.-J.; Lee, C.-H.; Lee, A.-Y.; Noh, M.-S. HaCaT keratinocytes and primary epidermal keratinocytes have different transcriptional profiles of cornified envelope-associated genes to T helper cell cytokines. Biomol. Ther. 2012, 20, 171–176. [Google Scholar] [CrossRef]
- Sprenger, A.; Weber, S.; Zarai, M.; Engelke, R.; Nascimento, J.M.; Gretzmeier, C.; Hilpert, M.; Boerries, M.; Has, C.; Busch, H.; et al. Consistency of the proteome in primary human keratinocytes with respect to gender, age, and skin localization. Mol. Cell. Proteom. 2013, 12, 2509–2521. [Google Scholar] [CrossRef]
- Shahjin, F.; Guda, R.S.; Schaal, V.L.; Odegaard, K.; Clark, A.; Gowen, A.; Xiao, P.; Lisco, S.J.; Pendyala, G.; Yelamanchili, S.V. Brain-derived extracellular vesicle microRNA signatures associated with in utero and postnatal oxycodone exposure. Cells 2019, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Shahjin, F.; Chand, S.; Yelamanchili, S.V. Extracellular vesicles as drug delivery vehicles to the central nervous system. J. Neuroimmune Pharmacol. 2019, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Rheinwald, J.G.; Green, H. Serial cultivation of strains of human epidermal keratinocytes: The formation of keratinizing colonies from single cells. Cell 1975, 6, 331–343. [Google Scholar] [CrossRef]
- Boukamp, P.; Petrussevska, R.T.; Breitkreutz, D.; Hornung, J.; Markham, A.; Fusenig, N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Boil. 1988, 106, 761–771. [Google Scholar] [CrossRef]
- Chiba, M.; Kimura, M.; Asari, S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol. Rep. 2012, 28, 1551–1558. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Huang, D.; Sherman, B.; Lempicki, R. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Shelke, G.V.; Lässer, C.; Gho, Y.S.; Lötvall, J. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J. Extracell. Vesicles 2014, 3, 24783. [Google Scholar] [CrossRef]
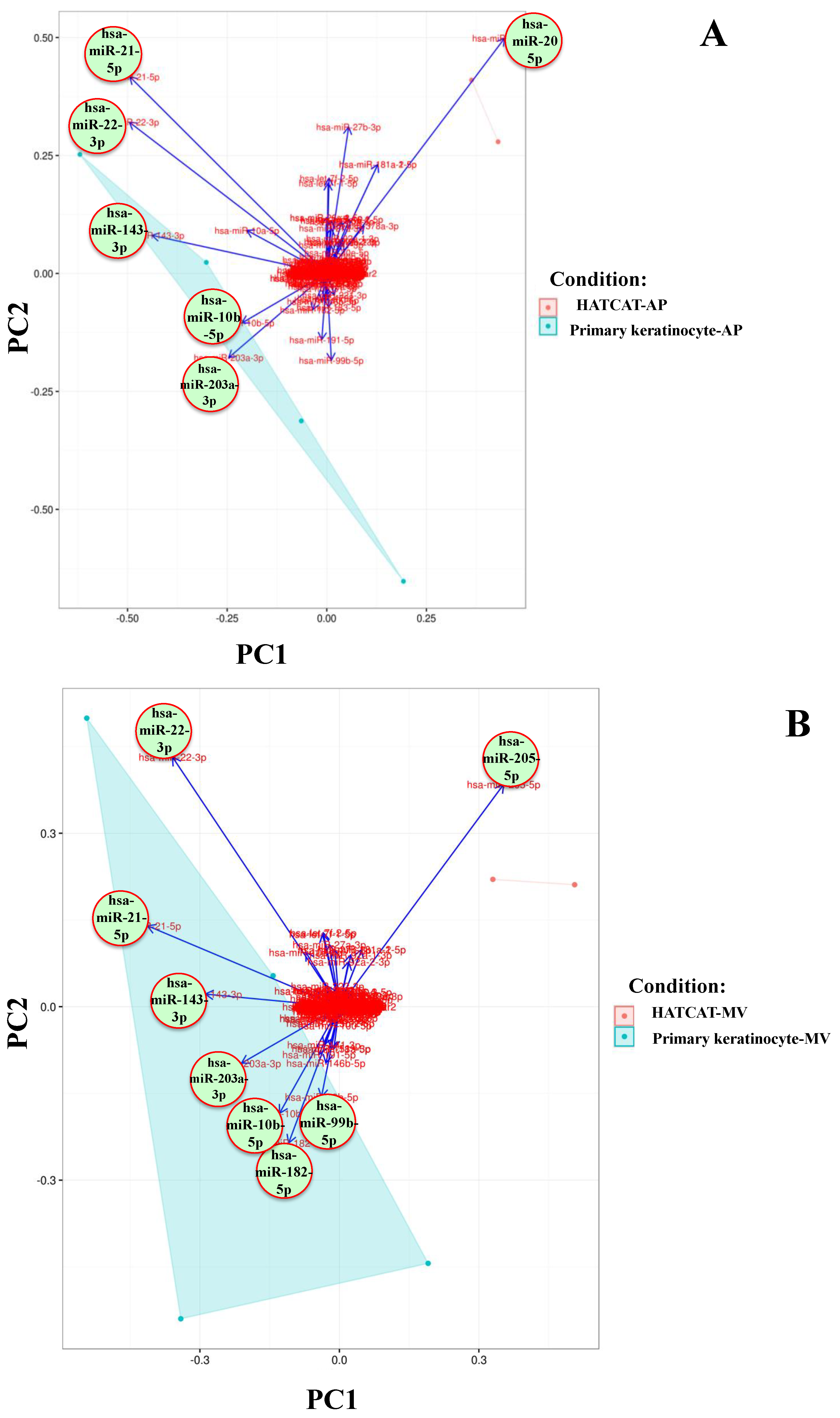
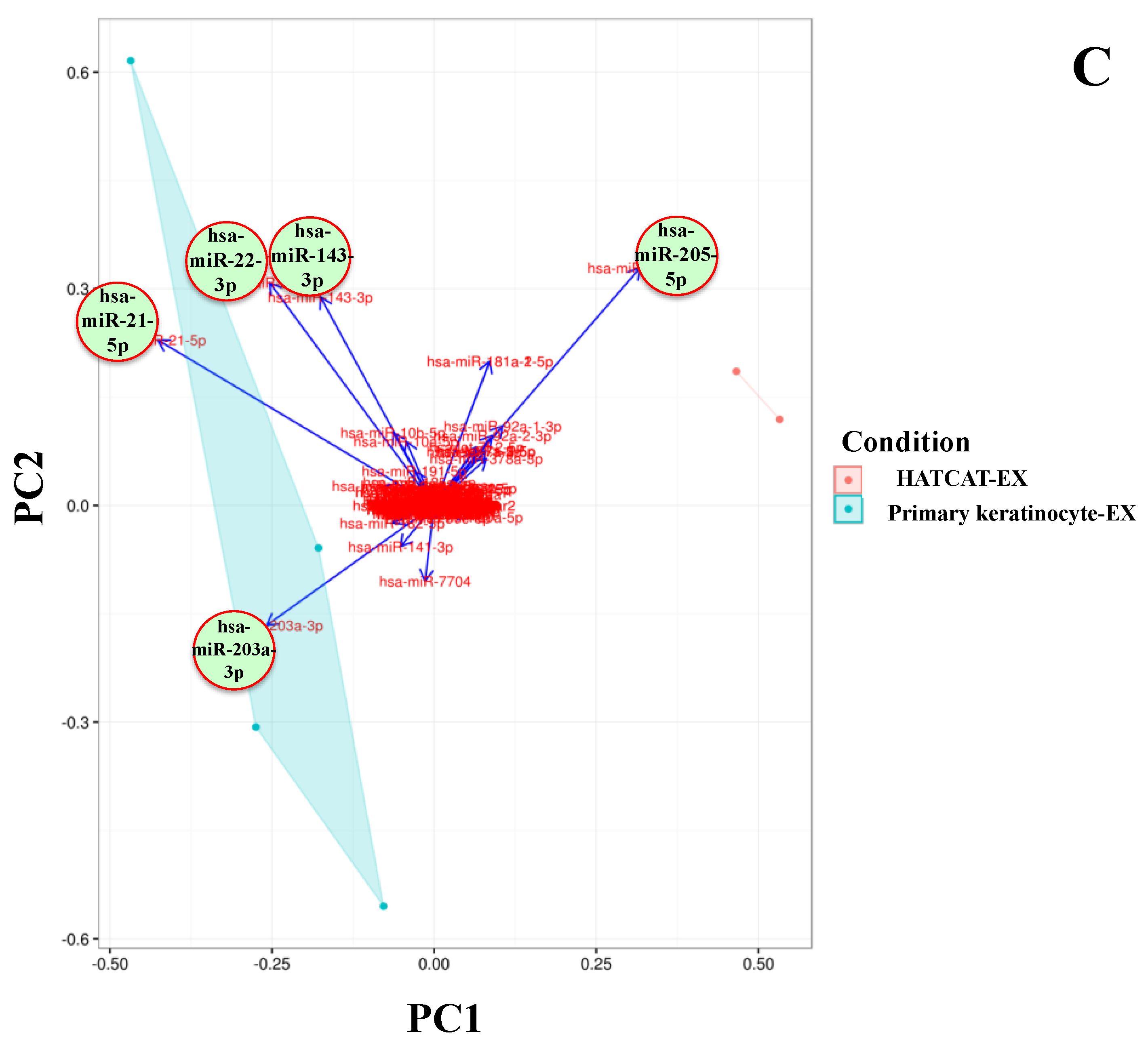

| Parental Cell Types | AP | MV | EX | |
|---|---|---|---|---|
| HaCaT | >100 RPM | 181 | 186 | 189 |
| >1000 RPM | 78 | 78 | 69 | |
| Primary keratinocytes | >100 RPM | 210 | 214 | 210 |
| >1000 RPM | 82 | 81 | 79 |
| HaCaT | |||||
| AP | MV | EX | |||
| miRNA Name | MN Counts ± SD | miRNA Name | MN Counts ± SD | miRNA Name | MN Counts ± SD |
| hsa-miR-205-5p | 106,506.6 ± 8267.9 **** | hsa-miR-205-5p | 123,329.2 ± 44,355.9 *** | hsa-miR-205-5p | 100,832.8 ± 16,630.3 * |
| hsa-miR-22-3p | 92,576.7 ± 16,680 **** | hsa-miR-27b-3p | 60,380.3 ± 11,723.7 ** | hsa-miR-22-3p | 77,505.6 ± 14,611.1 * |
| hsa-miR-27b-3p | 57,661.6 ± 7889.6 **** | hsa-miR-22-3p | 59,791.6 ± 4818.9 ** | hsa-miR-27b-3p | 51,490.9 ± 237.7 *** |
| hsa-miR-21-5p | 49,946.5 ± 6634.3 ***** | hsa-miR-21-5p | 45,496.7 ± 3357 *** | hsa-miR-21-5p | 49,943.9 ± 4831.5 *** |
| hsa-miR-181a-1-5p | 48,805.9 ± 1721.1 ***** | hsa-miR-181a-1-5p | 45,541.5 ± 3393.9 *** | hsa-miR-92a-1-3p | 37,858.4 ± 523.1 **** |
| Primary Keratinocytes | |||||
| AP | MV | EX | |||
| miRNA Name | MN Counts ± SD | miRNA Name | MN Counts ± SD | miRNA Name | MN Counts ± SD |
| hsa-miR-22-3p | 139,339.8 ± 43,597.7 *** | hsa-miR-22-3p | 93,878.6 ± 29,984.7 *** | hsa-miR-22-3p | 111,055.9 ± 9430.8 ****** |
| hsa-miR-21-5p | 81,954.5 ± 31,594.6 | hsa-miR-21-5p | 86,901.3 ± 21,422.5 | hsa-miR-21-5p | 110,112.7 ± 15,649.5 |
| hsa-miR-143-3p | 42,915.2 ± 24,838.4 ** | hsa-miR-143-3p | 38,016.7 ± 17,012.7 ** | hsa-miR-27b-3p | 52,399.1 ± 8430.5 **** |
| hsa-miR-203a-3p | 40,830.8 ± 7053.9 ** | hsa-miR-27b-3p | 45,115.2 ± 3168.5 * | hsa-miR-203a-3p | 49,640.4 ± 10,579.5 **** |
| hsa-miR-27b-3p | 40,707.7 ± 12,724.9 ** | hsa-miR-203a-3p | 34,067.1 ± 13,381.3 ** | hsa-miR-205-5p | 44,981.6 ± 10,617.4 ***** |
| Group (Enrichment Score) | Genes | Biological Function |
|---|---|---|
| Group 1 (2.72) | MAP3K9, NEK9, ICK, STK38L | Protein kinase, protein phosphorylation, binding, nucleus, cytosol, cytoplasm, membrane |
| Group 2 (2.01) | BCL6, ZEB1, ZRB2, YY1, PLAGL2 | Zinc finger, zinc, repressor, activator, transcription, binding, nucleus |
| Group 3 (1.08) | SLC38A1, TMEM123, PAPPA-AS1, F2RL2, ENPP4, MMD, LYMSMD3, SLC5A12, SHISA6, NIPA2, TM9SF2, LRRTM4, SLC39A14, TMEM201, SMIM13, TMEM239, SERINC3, PRRG4, MANMI | Plasma membrane, transmembrane, topological domain: cytoplasmic, topological domain: extracellular, blood coagulation and haemostasis |
| Group (Enrichment Score) | Genes | Biological Function |
|---|---|---|
| Group 1 (27.64) | PLEKHA2, NUDCD1, ELP5, FILIP1L, NUFIP2, SERBP1, SSFA2, DAZAP2, HYPK, MYCBP, TSC22D4 | Cytoplasm, nucleus, protein binding, phosphoprotein |
| Group 2 (19.96) | ZBTB44, FAM208A, FOXK2, VGLL4, ZBTB8A, MAFK, MGA, LRRFIP1, MED9, NFAT5, ZNF654, ARID3B, MYCBP, HEXIM1, FOXN3, TAF1D, ATF7IP, DNTTIP2, CSRNP2, MIER3, NFYB, NFYA, ANP32A, FUBP1, ZNF646, TRAPPC2, PCGF6, ZNF367, NFIC, LCORL, TCEAL1, ELP5, HOMEZ, NFIA, BTF3, TSC22D4, SERTAD3, FOXN2, PURB, MACC1, PURA, FOXK1, HMGB3, DMTF1, COMMD2, PHTF1, SOX5, LCOR | Transcription, nucleus, protein binding, DNA binding, phosphoprotein |
| Group 3 (13.56) | BUB1B, LATS1, CSNK1A1, RPS6KA3, SGK3, LIMK1, IRAK1, PRKCE, CDK6, TESK2, DYRK3, MAP3K1, WNK3, MAP3K2, WNK1, ROCK2, MAP3K7, CDKL2, CIT, ACVR1C, CDK19, SLK, MAP2K3, SNRK, PIM3, SRPK2, SRPK1, MAP3K13, HIPK3, PDIK1L, MAPK7, MKNK2, NEK1, FRK, CSNK2A1 | Kinase activity, transferase, phosphoprotein, binding (protein, ATP, nucleotide), cytoplasm, nucleus, cytosol, protein phosphorylation, proton receptor |
| Group 4 (11.94) | TGIF1, HOMEZ, ZEB2, SATB1 | Transcription activity, nucleus, binding, acetylation, homeobox, phosphoprotein, repressor |
| Group 5 (11.74) | RAPH1, OSBPL3, PLEKHA8, SKAP2, EXOC8, OSBP, SPATA13 | Membrane, transport, cytosol, cytoplasm, protein binding, pleckstrin homology, phosphoprotein |
| Group 6 (11.44) | RLIM, RNF103, TRIM38, BMI1, RFFL, UBR3, RNF6, TRIM33, TRIM59, RNF185, PCGF6, RNF111, MARCH3, CBLL1, RNF11, DTX3L, RSPRY1, TOPORS, TRIM4, TRIM2, RNF141, SCAF11, MYCBP2 | Zinc, zinc finger, binding (metal, protein, ion), ligase activity, phosphoprotein, nucleus, Ubl conjugation pathway, RING |
| Group 7 (11.15) | NCL, CPEB4, RBM27, MYEF2, MSI2, ELAVL4, RBM39, SREK1, SRSF7, HNRNPH1, CPEB3, CELF2, PTBP3, PPIL4, HNRNPA3, SRSF11, HNRNPR | RNA/nucleotide/nucleic binding, protein binding, nucleus, cytoplasm, acetylation, mRNA processing, nucleoplasm |
| Group 8 (9.92) | ZMAT5, MBNL1, SNRNP48, SCAF11 | Spliceosome, metal/zinc/ion binding, protein binding, nucleus, cytoplasm, RNA binding, RNA splicing, mRNA processing, phosphoprotein, zinc finger |
| Group 9 (9.74) | ZNF667, RSF1, ZBTB44, ZBTB8A, PHF20L1, WHSC1L1, ZNF662, ZBTB47, REST, ZNF654, ZNF35, ZNF587, ZNF326, ZNF573, ZNF704, ZNF217, ARID2, ZNF429, ZNF24, RREB1, ZNF277, ZNF207, SNAI1, ZNF268, ZNF460, NR2C2, ZNF200, ZEB2, ZUFSP, SUZ12, ZNF292, TSHZ3, THAP1, SNAI2, ZNF646, ZNF607, HIC2, BCL11B, ZNF652, BCL6, ZNF367, PCGF6, KAT7, GLIS3, ATMIN, PHF20, ADNP, KLF5, KLF9, ZNF264, SP1, GLIS2, KAT6A, ZBTB20, ZNF148, ZNF532, TRPS1, TRIM33, BAZ1B, IKZF3, ZNF451, ZNF440, LCOR, ZBTB38, ZEB1 | Zinc finger, zinc, nucleus, transcription, binding (meta, protein, ion, DNA, nucleic acid), phosphoprotein |
| Group 10 (9.29) | USP7, USP47, USP42, USP34 | Cytosol, cytoplasm, nucleus, DNA repair, ubiquitin activity, protease, hydrolase, phosphoprotein, acetylation |
| Group 11 (9.13) | NR2C2, ESR1, PPARA, NR2F2, NR3C1, HNF4A | Regulation of transcription, acetylation, cytoplasm, nucleus, protein/metal/DNA/ion binding, receptor, lipid metabolic process, activator, phosphoroprotein, disease mutation, nucleoplasm |
| Group 12 (8.74) | SMAD2, SMAD3, SMAD4, SMAD7, SMAD9 | Signalling pathway, binding, cell cycle, transcription, nucleus, cytoplasm, cancer, disease mutation |
| Group 13 (8.55) | RPS4X, RPS7, RPS19, RPS27, RPS2, RPL35A, RPSA, RPL24 | Nucleus, cytoplasm, phosphoprotein, extracellular matrix, binding translation, ribonucleoprotein, ribosomal protein, membrane, cytosol, exosome |
| Group 14 (8.54) | DDX3X, EIF4A2, SMARCA4, DHX33, ATRX, DDX46, DDX55, DDX3Y, CHD9, DDX6 | Cytoplasm, nucleus, binding, phosphoprotein, helicase, hydrolase, ubl conjugation |
| Group 15 (8.26) | TTC38, TTC33, SGTB, FKBP5 | Extracellular exosome, chaperone, acetylation, phosphoprotein, tetratricopeptide, protein binding |
| Group 16 (7.89) | TGIF1, MEIS1, HOXA1, EN2, PBX1, HOXA9, PKNOX1 | Transcription, DNA binding, organism development, homeobox, nucleus |
| Group 17 (7.74) | ANKRD9, CLIP4, RAI14, SOWAHC, HECTD1, ANKRD46, ANKRD13B | Phosphoprotein, Ankyrin |
| Group 18 (7.74) | SETD2, WHSC1, WHSC1L1, TRIM33, SETD1B | Transcription regulation, zinc finger, chromosome, transferase, nucleus, phosphoprotein, isopeptide bond, Ubl conjugation, associated with SET domain |
| Group 19 (7.00) | ARHGEF28, FGD6, RALGPS2, NET1, SPATA13, ARHGEF12 | Cell membrane, cytosol, cytoplasm, signal transduction, Rho guanyl-nucleotide exchange factor, phosphoprotein |
| Group 20 (6.74) | DOCK4, RALGPS2, DOCK7, DOCK10, DOCK5 | Cytoplasm, cell membrane, acetylation, dedicator of cytokinesis, GTPase activity, phosphoprotein, DOCK-homology region, intracellular |
| Group 21 (6.48) | KIF13A, KIF5B, KIF1C, KIF2A, DYNC1LI2 | Membrane, centrosome, methylation, ATPase activity, kinesin, cytoplasm, ATP binding, cytoskeleton, coiled coil, microtube |
| Group 22 (6.13) | AKT2, PIK3CA, HRAS, AKT1, MAPK1, PIK3R1, MAPK9, MAPK8 | Signalling pathway (GnRH, MAPKinase, EGF, PDGF, IGF-1, insulin, CXCR4, Rrk, PI3, ERBB2, Trka receptor, Ras, Jak-STAT, AMPK, ErbB, cAMP, Toll-line receptor), binding, disease, cancer, cytosol, kinase activity, nucleus, cytoplasm, acetylation, infection, apoptosis, |
| Group 23 (5.99) | BMPR2, BMPR1B, BMPR1A, TGFBR2, ACVR1C, ACVR2B, DDR2 | Phosphoprotein, phosphorylation, kinase activity, signalling pathway (Hippo, BMP, regulation of stem cell, TGF-beta), binding (ATP, metal, nucleotide, ion, protein) TGF-beta receptor, phosphorylation, disease mutation, disulphide bond, transferase, receptor, membrane, transmembrane, signal, extracellular |
| Group 24 (5.41) | EGFR, PDGFRA, EPHA4, ERBB3, ERBB2, CSF1R, DDR2, IGF1R | Membrane, transmembrane, receptor, signal, binding (ATP, nucleotide, protein), kinase, signalling pathway (Ras, Rap1, calcium, cancer), wound healing, cancer, endocytosis, transferase, microRNAs in cancer, cytoplasm, phosphorylation activity, glycoprotein, extracellular, cytoplasm, disease mutation |
| Group 25 (5.19) | RASGRP3, RALGPS2, RAPGEF6, RGL2 | Cytoplasm, membrane, signalling pathway (Ras, Rap1,) binding (Ras GTPase, protein) signal transduction, GTPase activity, phosphoprotein, Ras guanine nucleotide exchange factor |
| Group 26 (4.74) | STXBP5, WDR7, WSB1, CORO2A, DCAF10, WDR77, PHIP, BRWD3, GNB4, NBEA, ELP2, DCAF8, FBXW7, TBL1XR1, TAF5 | WD (1, 2,3,4,5,6,7, 40), WD repeat, phosphoprotein |
| Group 27 (4.14) | DUSP5, PTPDC1, DUSP8, DUSP10 | Phosphatase activity, cytoplasm nucleus, nucleoplasm, hydrolase, MAPK signalling pathway, Rhodanese, dephosphorylation |
| Group 28 (3.49) | RHOB, RAB6C, RHOQ, RAB22A, RAB33B, RAB6A, RAB5B, RAP2B, RAB44 | Cytosol, membrane, transport, methylation, GTPase activity, extracellular exosome, binding (protein, nucleotide, GTP, phosphate, lipid), lipoprotein, prenylation |
| Group 29 (3.12) | SOCS5, SOCS4, SOCS3, SOCS6 | Protein binding, growth regulation, regulation of Jak-STAT, signal transduction inhibitor, signalling pathway (STAT, prolactin, cytokine-mediated, insulin), cytoplasm, SOCS box, SH2 domain, Ubl conjugation pathway, intracellular, inflammation response, suppressor of cytokine signalling, type II diabetes mellitus, protein ubiquitination |
| Group 30 (2.77) | LCLAT1, SERINC1, SERAC1, LPGAT1, TMEM147 | Membrane, transmembrane, lipid metabolism, lipid biosynthesis, phospholipid metabolism, phospholipid biosynthesis, protein binding, endoplasmic reticulum membrane |
| Group 31 (2.73) | LRRC20, FBXL2, LRRC57, TBCEL, CEP97, SKP2, FBXL5, VASN, FBXL13, FMOD, GP5, ZYG11B, FBXL3, LRRC1, CNTRL, LRRC2 | Leucine-rich repeat (1, 2, 3, 4, 5, 6, 7, 8), protein binding |
| Group 32 (2.57) | COL4A1, COL3A1, COL5A2, COL5A1, COL1A1 | Extracellular matrix, signal, disease mutation, extracellular region, fibrillar collagen, focal adhesion, collagen, secreted, binding (metal, ion, platelet-derived growth factor), PI3-Akt signalling pathway, calcium, glycoprotein, skin development, disulphide bond, glycosylation, skeletal system development |
| Group 33 (1.50) | IVNS1ABP, KLHL24, KBTBD6, KBTBD7, IPP, KLHL15, KLHL28 | Kelch (1,2,3,4,5,6), kelch repeat, kelch-like protein, protein ubiquitination, ubiquitineous activity, BTB domain, BTB/POZ |
| Group 34 (1.25) | MMP13, PAPPA, MMP10, MMP9, ADAMTS4, MMP2, MMP1 | Zinc, calcium, signal, membrane, extracellular matrix, collagen degradation, hemopexin-like domain, metallopeptidase, glycoprotein, zymogen, protease, hydrolase, disulphide bond, metal binding |
| Group 35 (0.93) | CERS4, SPTLC3, SGPL1, SPTLC2, CERS6 | Transmembrane, metabolic pathway, sphingolipid activity, pyridoxal phosphate, endoplasmic reticulum, ceramide biosynthetic process |
| Group 36 (0.01) | EDNRA, C15orf48, ARMCX3, VOPP1, ANKRD46, TMPPE, SLC2A14, SACM1L, TMEM2, ST6GAL1, FAM20B, SAMD5, SLC31A1, GLRA3, SLC12A5, TMEM178B, BOC, SLC16A10, FGFRL1, TM9SF3, IL13RA1, LIFR, B3GNT5, LMBR1, TMEM170A, IL10RB, PIGX, GALNT6, CNNM3, SLC17A5, DSE, SLC44A1, SGCB, SLC39A9, CERS6, FAXC, FAXDC2, TMEM56, CHST10, TMED4, MCTP1, SLC26A2, TMEM147, B3GALNT1, HTR2C, MRAP2, PGRMC2, BSG, SFXN1, CADM2, SFT2D2, SLC45A4, COX20, PEAR1, TOR1AIP2, CCR7, CCR6, CCR5, CCR1, NCSTN, NETO2, HEPHL1, OR7D2, BTN3A3, GJD2, CYBRD1, PIGN, LRIT3, CLPTM1L, MGAT4A, CDH7, HERPUD2, MXRA7, TMEM245, MEGF9, GPR156, SLMAP, PIGP, HSP40, GXYLT2, TMEM97, SLC5A3, EXT1, MOXD1, SERINC1, SCN2B, CD151, HS3ST3B1, GXYLT1, TNFRSF10B, TMEM120B, ORAI2, OLR1, TNFRSF10D, PTGFR, CD47, CD44, RER1 | Membrane, transmembrane, glycoprotein, cytoplasmic domain, glycosylation |
| Terms | p-adj | Terms | p-adj |
|---|---|---|---|
| Target Genes Associated with HaCaT Cell-Derived EV miRNAs | |||
| Regulation of cell migration | 0.0033 | Epithelium migration | 0.0382 |
| Negative regulation of cell migration | 0.0301 | Regulation of epithelial cell migration | 0.0398 |
| Epithelial cell migration | 0.0352 | Tissue migration | 0.0465 |
| Target Genes Associated with Primary Keratinocyte-Derived EV miRNAs | |||
| Regulation of cell migration | 0.0001 | Regulation of mononuclear cell migration | 0.0032 |
| Positive regulation of cell migration | 0.0001 | Regulation of vascular associated smooth muscle cell migration | 0.0063 |
| Positive regulation of epithelial cell migration | 0.0001 | Regulation of blood vessel endothelial cell migration | 0.0088 |
| Regulation of epithelial cell migration | 0.0001 | Regulation of fibroblast migration | 0.0092 |
| Positive regulation of endothelial cell migration | 0.0001 | Regulation of leukocyte migration | 0.012 |
| Regulation of endothelial cell migration | 0.0001 | Positive regulation of vascular associated smooth muscle cell migration | 0.0161 |
| Cell migration | 0.0001 | Leukocyte migration | 0.0169 |
| Positive regulation of mononuclear cell migration | 0.0002 | Thymocyte migration | 0.0192 |
| Regulation of smooth muscle cell migration | 0.0007 | Positive regulation of fibroblast migration | 0.0211 |
| Positive regulation of leukocyte migration | 0.0011 | Regulation of trophoblast cell migration | 0.0411 |
| Negative regulation of cell migration | 0.0011 | Positive regulation of cell migration involved in sprouting angiogenesis | 0.0411 |
| Positive regulation of blood vessel endothelial cell migration | 0.0017 | Dendritic cell migration | 0.0423 |
| Positive regulation of smooth muscle cell migration | 0.0032 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Than, U.T.T.; Guanzon, D.; Broadbent, J.A.; Parker, T.J.; Leavesley, D.I. Deep Sequencing MicroRNAs from Extracellular Membrane Vesicles Revealed the Association of the Vesicle Cargo with Cellular Origin. Int. J. Mol. Sci. 2020, 21, 1141. https://doi.org/10.3390/ijms21031141
Than UTT, Guanzon D, Broadbent JA, Parker TJ, Leavesley DI. Deep Sequencing MicroRNAs from Extracellular Membrane Vesicles Revealed the Association of the Vesicle Cargo with Cellular Origin. International Journal of Molecular Sciences. 2020; 21(3):1141. https://doi.org/10.3390/ijms21031141
Chicago/Turabian StyleThan, Uyen Thi Trang, Dominic Guanzon, James A Broadbent, Tony J Parker, and David I Leavesley. 2020. "Deep Sequencing MicroRNAs from Extracellular Membrane Vesicles Revealed the Association of the Vesicle Cargo with Cellular Origin" International Journal of Molecular Sciences 21, no. 3: 1141. https://doi.org/10.3390/ijms21031141
APA StyleThan, U. T. T., Guanzon, D., Broadbent, J. A., Parker, T. J., & Leavesley, D. I. (2020). Deep Sequencing MicroRNAs from Extracellular Membrane Vesicles Revealed the Association of the Vesicle Cargo with Cellular Origin. International Journal of Molecular Sciences, 21(3), 1141. https://doi.org/10.3390/ijms21031141






