Dbp5/DDX19 between Translational Readthrough and Nonsense Mediated Decay
Abstract
1. Translation Termination
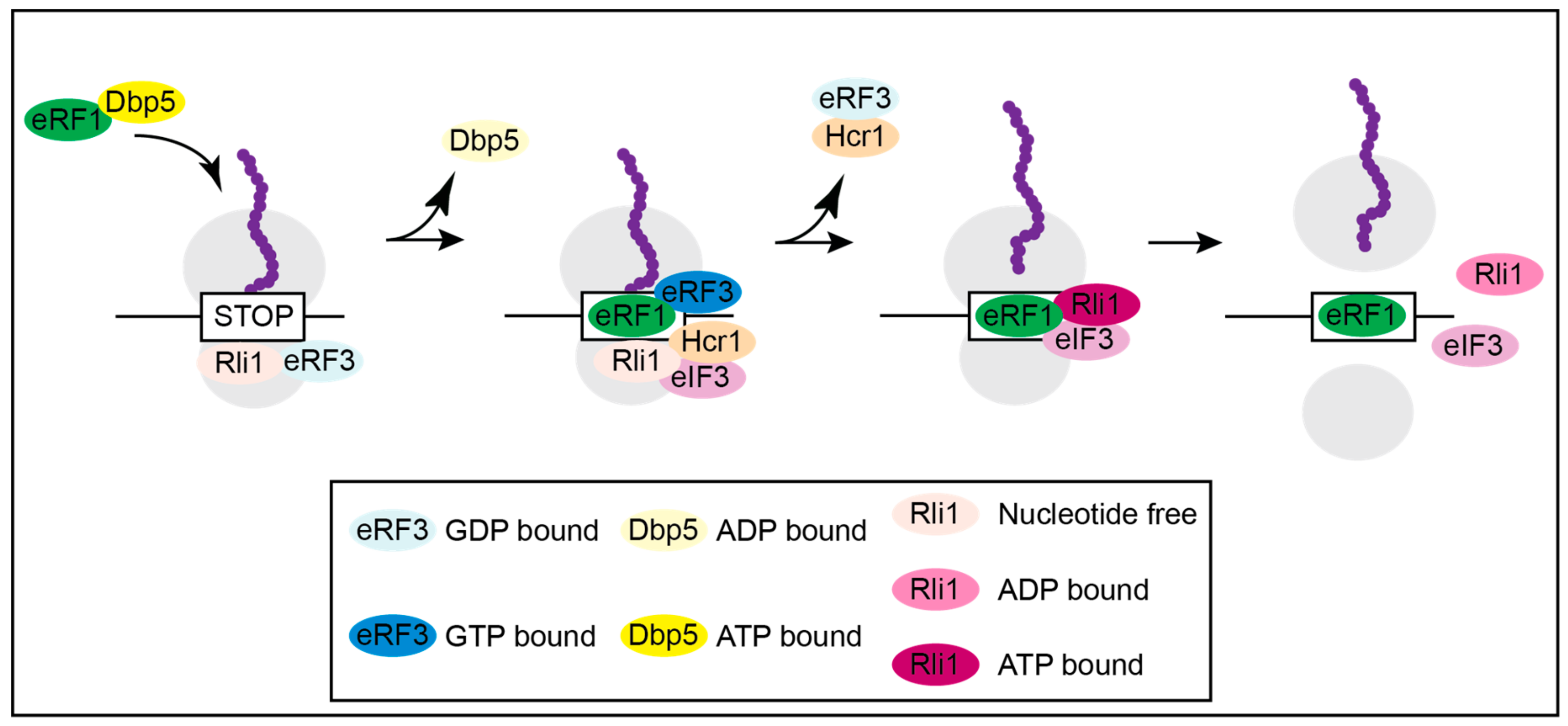
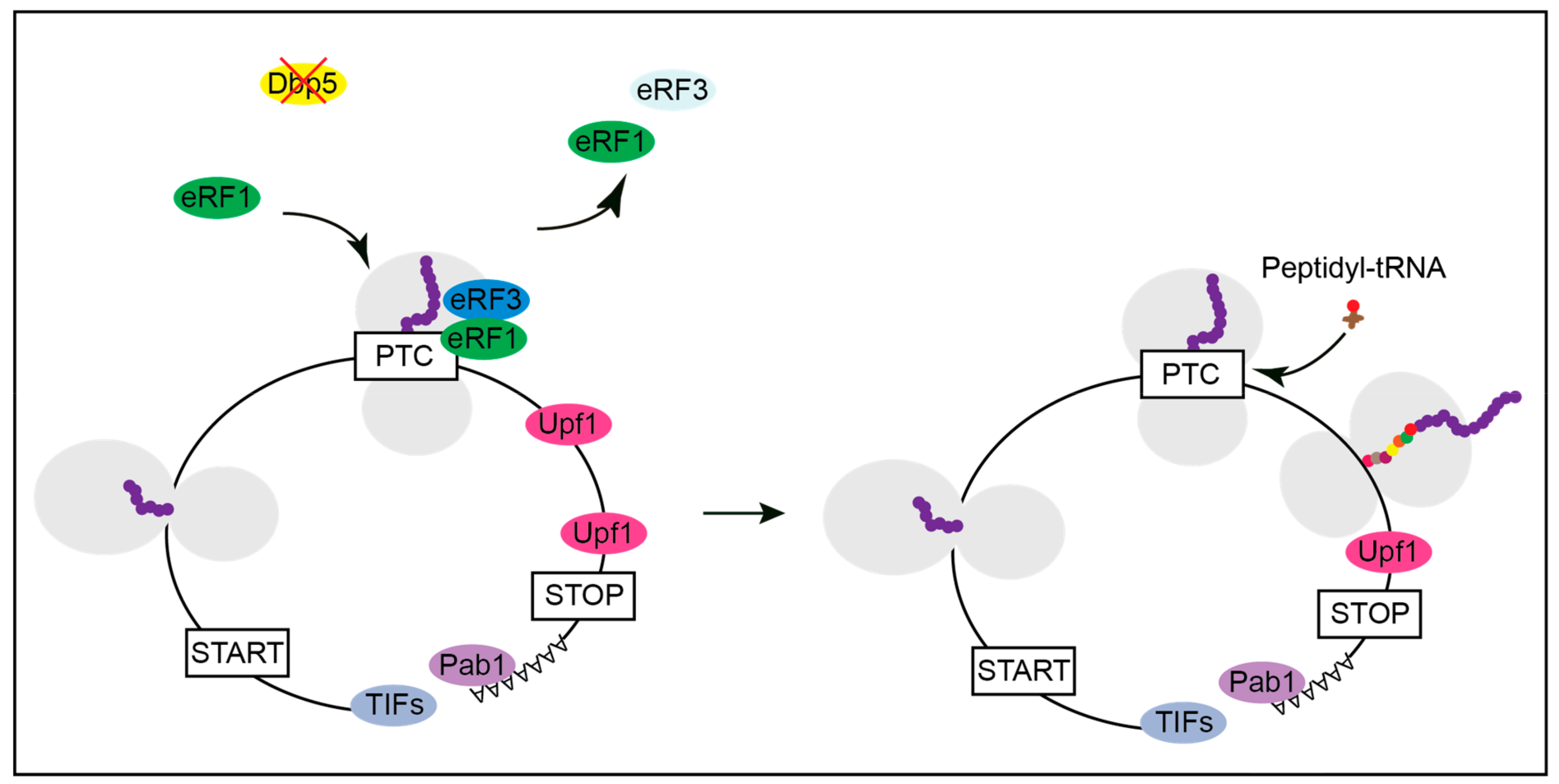
2. The Function of Dbp5 in Translation Termination
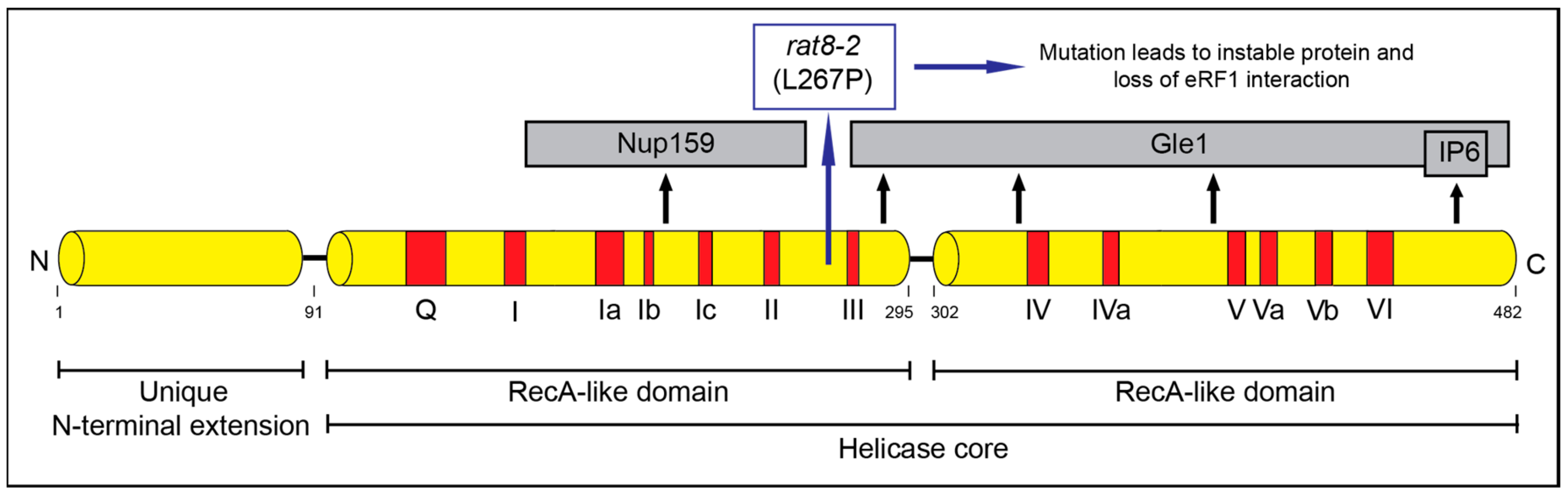
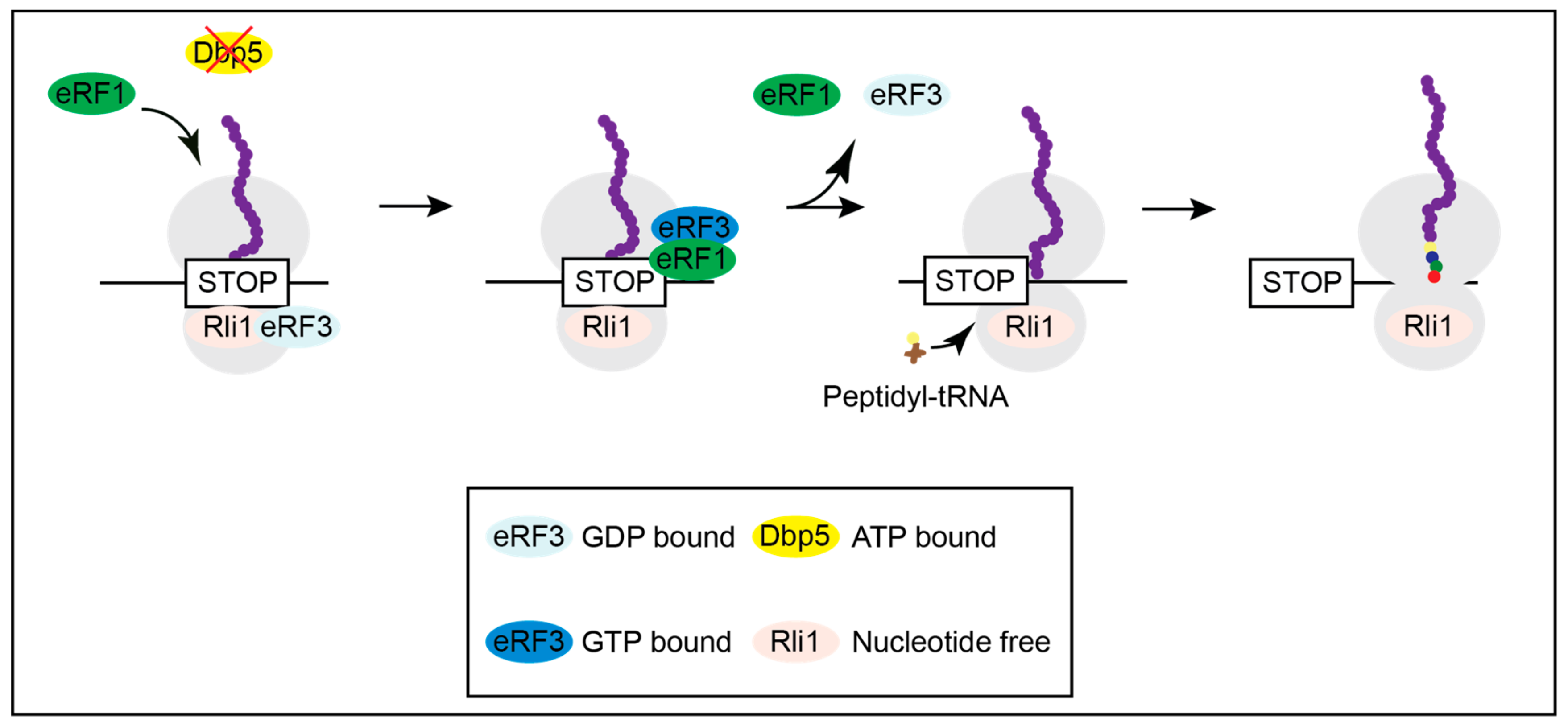
3. Mutations in DBP5 Lead to Termination Readthrough
4. Regulatory Principles in Nonsense Mediated Decay (NMD)
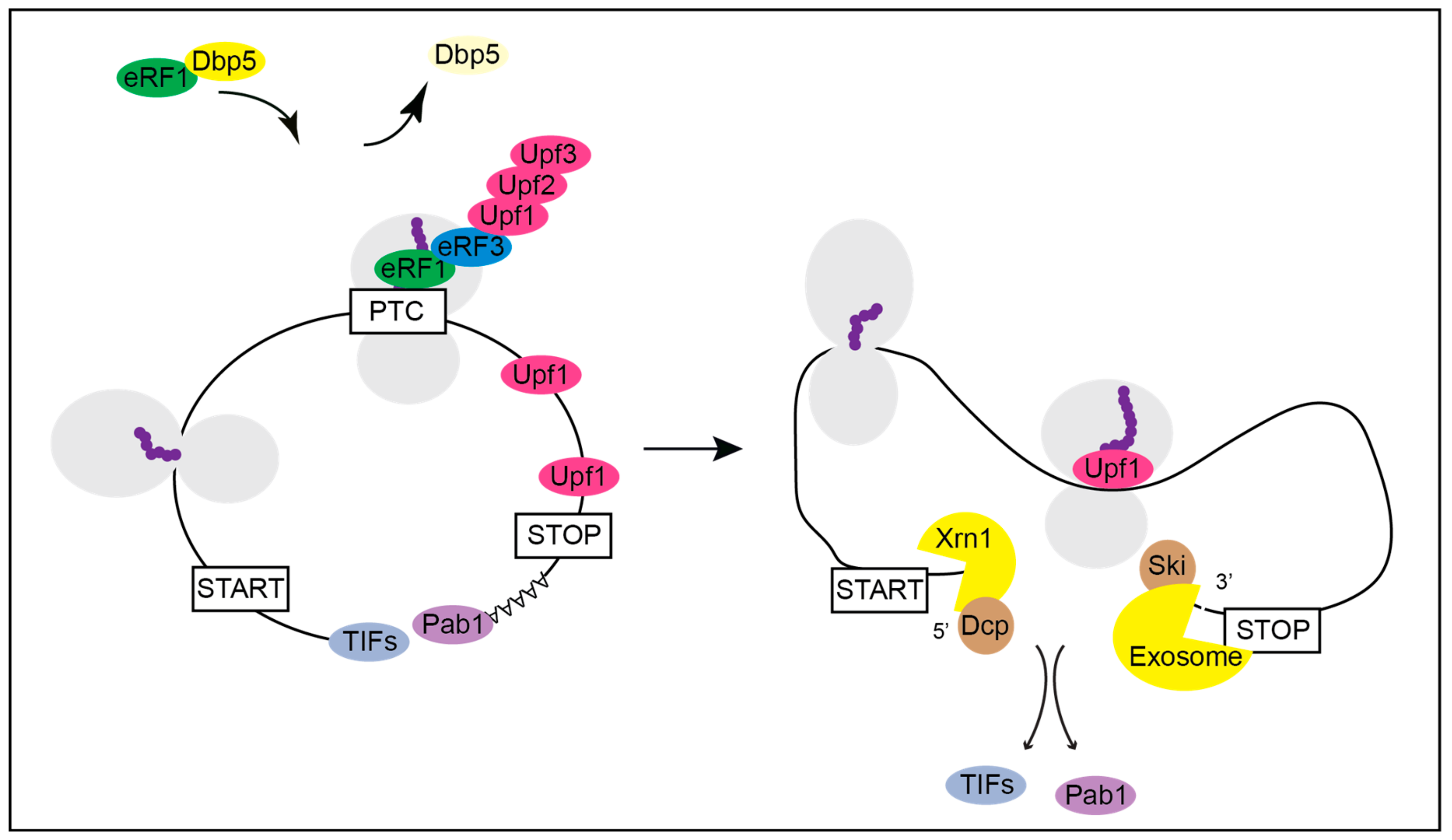
5. Termination Readthrough and Nonsense Mediated Decay (NMD)
Funding
Acknowledgments
Conflicts of Interest
References
- Doma, M.K.; Parker, R. RNA quality control in eukaryotes. Cell 2007, 131, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Soheilypour, M.; Mofrad, M.R.K. Quality control of mRNAs at the entry of the nuclear pore: Cooperation in a complex molecular system. Nucleus 2018, 9, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Zander, G.; Krebber, H. Quick or quality? How mRNA escapes nuclear quality control during stress. RNA Biol. 2017, 14, 1642–1648. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, J.; Bennett, E.J. Protecting the proteome: Eukaryotic cotranslational quality control pathways. J. Cell Biol. 2014, 204, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Mühlemann, O.; Jensen, T.H. mRNP quality control goes regulatory. Trends Genet. 2012, 28, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Dever, T.E.; Green, R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 2012, 4, a013706. [Google Scholar] [CrossRef]
- Dever, T.E.; Kinzy, T.G.; Pavitt, G.D. Mechanism and Regulation of Protein Synthesis in Saccharomyces cerevisiae. Genetics 2016, 203, 65–107. [Google Scholar] [CrossRef]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. Termination and post-termination events in eukaryotic translation. Adv. Protein Chem. Struct. Biol. 2012, 86, 45–93. [Google Scholar]
- Bertram, G.; Bell, H.A.; Ritchie, D.W.; Fullerton, G.; Stansfield, I. Terminating eukaryote translation: Domain 1 of release factor eRF1 functions in stop codon recognition. RNA 2000, 6, 1236–1247. [Google Scholar]
- Blanchet, S.; Rowe, M.; Von der Haar, T.; Fabret, C.; Demais, S.; Howard, M.J.; Namy, O. New insights into stop codon recognition by eRF1. Nucleic Acids Res. 2015, 43, 3298–3308. [Google Scholar] [CrossRef] [PubMed]
- Conard, S.E.; Buckley, J.; Dang, M.; Bedwell, G.J.; Carter, R.L.; Khass, M.; Bedwell, D.M. Identification of eRF1 residues that play critical and complementary roles in stop codon recognition. RNA 2012, 18, 1210–1221. [Google Scholar] [PubMed]
- Heurgue-Hamard, V.; Champ, S.; Mora, L.; Merkulova-Rainon, T.; Kisselev, L.L.; Buckingham, R.H. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J. Biol. Chem. 2005, 280, 2439–2445. [Google Scholar] [PubMed]
- Eyler, D.E.; Green, R. Distinct response of yeast ribosomes to a miscoding event during translation. RNA 2011, 17, 925–932. [Google Scholar]
- Salas-Marco, J.; Bedwell, D.M. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 2004, 24, 7769–7778. [Google Scholar] [CrossRef]
- Beissel, C.; Neumann, B.; Uhse, S.; Hampe, I.; Karki, P.; Krebber, H. Translation termination depends on the sequential ribosomal entry of eRF1 and eRF3. Nucleic Acids Res. 2019, 47, 4798–4813. [Google Scholar] [CrossRef]
- Brown, A.; Shao, S.; Murray, J.; Hegde, R.S.; Ramakrishnan, V. Structural basis for stop codon recognition in eukaryotes. Nature 2015, 524, 493–496. [Google Scholar] [CrossRef]
- Khoshnevis, S.; Gross, T.; Rotte, C.; Baierlein, C.; Ficner, R.; Krebber, H. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010, 11, 214–219. [Google Scholar]
- Preis, A.; Heuer, A.; Barrio-Garcia, C.; Hauser, A.; Eyler, D.E.; Berninghausen, O.; Green, R.; Becker, T.; Beckmann, R. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 2014, 8, 59–65. [Google Scholar] [CrossRef]
- Pisarev, A.V.; Skabkin, M.A.; Pisareva, V.P.; Skabkina, O.V.; Rakotondrafara, A.M.; Hentze, M.W.; Hellen, C.U.; Pestova, T.V. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell 2010, 37, 196–210. [Google Scholar]
- Schuller, A.P.; Wu, C.C.; Dever, T.E.; Buskirk, A.R.; Green, R. eIF5A Functions Globally in Translation Elongation and Termination. Mol. Cell 2017, 66, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Urakov, V.N.; Mitkevich, O.V.; Safenkova, I.V.; Ter-Avanesyan, M.D. Ribosome-bound Pub1 modulates stop codon decoding during translation termination in yeast. FEBS J. 2017, 284, 1914–1930. [Google Scholar] [CrossRef] [PubMed]
- Beznoskova, P.; Cuchalova, L.; Wagner, S.; Shoemaker, C.J.; Gunisova, S.; von der Haar, T.; Valasek, L.S. Translation initiation factors eIF3 and HCR1 control translation termination and stop codon read-through in yeast cells. PLoS Genet. 2013, 9, e1003962. [Google Scholar] [CrossRef] [PubMed]
- Beznoskova, P.; Wagner, S.; Jansen, M.E.; von der Haar, T.; Valasek, L.S. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res. 2015, 43, 5099–5111. [Google Scholar] [CrossRef]
- Gross, T.; Siepmann, A.; Sturm, D.; Windgassen, M.; Scarcelli, J.J.; Seedorf, M.; Cole, C.N.; Krebber, H. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science 2007, 315, 646–649. [Google Scholar] [CrossRef]
- Folkmann, A.W.; Noble, K.N.; Cole, C.N.; Wente, S.R. Dbp5, Gle1-IP6 and Nup159: A working model for mRNP export. Nucleus 2011, 2, 540–548. [Google Scholar] [CrossRef]
- Snay-Hodge, C.A.; Colot, H.V.; Goldstein, A.L.; Cole, C.N. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998, 17, 2663–2676. [Google Scholar] [CrossRef]
- Stewart, M. Nuclear export of mRNA. Trends Biochem. Sci. 2010, 35, 609–617. [Google Scholar] [CrossRef]
- Tieg, B.; Krebber, H. Dbp5—From nuclear export to translation. Biochim. Biophys. Acta 2013, 1829, 791–798. [Google Scholar]
- Tseng, S.S.; Weaver, P.L.; Liu, Y.; Hitomi, M.; Tartakoff, A.M.; Chang, T.H. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 1998, 17, 2651–2662. [Google Scholar]
- Fairman-Williams, M.E.; Guenther, U.P.; Jankowsky, E. SF1 and SF2 helicases: Family matters. Curr. Opin. Struct. Biol. 2010, 20, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Jin, S.B.; Bjorkroth, B.; Wieslander, L.; Daneholt, B. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 2002, 21, 1177–1187. [Google Scholar] [CrossRef]
- Lari, A.; Arul Nambi Rajan, A.; Sandhu, R.; Reiter, T.; Montpetit, R.; Young, B.P.; Loewen, C.J.; Montpetit, B. A nuclear role for the DEAD-box protein Dbp5 in tRNA export. Elife 2019, 8, e48410. [Google Scholar] [CrossRef] [PubMed]
- Linder, P.; Jankowsky, E. From unwinding to clamping—The DEAD box RNA helicase family. Nat. Rev. 2011, 12, 505–516. [Google Scholar]
- Collins, R.; Karlberg, T.; Lehtio, L.; Schutz, P.; van den Berg, S.; Dahlgren, L.G.; Hammarstrom, M.; Weigelt, J.; Schuler, H. The DEXD/H-box RNA helicase DDX19 is regulated by an {alpha}-helical switch. J. Biol. Chem. 2009, 284, 10296–10300. [Google Scholar] [PubMed]
- Lund, M.K.; Guthrie, C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol. Cell 2005, 20, 645–651. [Google Scholar] [PubMed]
- Schmitt, C.; von Kobbe, C.; Bachi, A.; Pante, N.; Rodrigues, J.P.; Boscheron, C.; Rigaut, G.; Wilm, M.; Seraphin, B.; Carmo-Fonseca, M.; et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999, 18, 4332–4347. [Google Scholar] [CrossRef]
- Weirich, C.S.; Erzberger, J.P.; Flick, J.S.; Berger, J.M.; Thorner, J.; Weis, K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 2006, 8, 668–676. [Google Scholar]
- Alcazar-Roman, A.R.; Tran, E.J.; Guo, S.; Wente, S.R. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 2006, 8, 711–716. [Google Scholar]
- Alcazar-Roman, A.R.; Bolger, T.A.; Wente, S.R. Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1. J. Biol. Chem. 2010, 285, 16683–16692. [Google Scholar]
- Montpetit, B.; Thomsen, N.D.; Helmke, K.J.; Seeliger, M.A.; Berger, J.M.; Weis, K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 2011, 472, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Tran, E.J.; Zhou, Y.; Corbett, A.H.; Wente, S.R. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell 2007, 28, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Bolger, T.A.; Wente, S.R. Gle1 is a multifunctional DEAD-box protein regulator that modulates Ded1 in translation initiation. J. Biol. Chem. 2011, 286, 39750–39759. [Google Scholar] [CrossRef] [PubMed]
- Noble, K.N.; Tran, E.J.; Alcazar-Roman, A.R.; Hodge, C.A.; Cole, C.N.; Wente, S.R. The Dbp5 cycle at the nuclear pore complex during mRNA export II: Nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 2011, 25, 1065–1077. [Google Scholar] [CrossRef]
- Neumann, B.; Wu, H.; Hackmann, A.; Krebber, H. Nuclear Export of Pre-Ribosomal Subunits Requires Dbp5, but Not as an RNA-Helicase as for mRNA Export. PLoS ONE 2016, 11, e0149571. [Google Scholar] [CrossRef] [PubMed]
- Bolger, T.A.; Folkmann, A.W.; Tran, E.J.; Wente, S.R. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell 2008, 134, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Mikhailova, T.; Shuvalova, E.; Ivanov, A.; Susorov, D.; Shuvalov, A.; Kolosov, P.M.; Alkalaeva, E. RNA helicase DDX19 stabilizes ribosomal elongation and termination complexes. Nucleic Acids Res. 2017, 45, 1307–1318. [Google Scholar] [CrossRef]
- Takemura, R.; Inoue, Y.; Izawa, S. Stress response in yeast mRNA export factor: Reversible changes in Rat8p localization are caused by ethanol stress but not heat shock. J. Cell Sci. 2004, 117, 4189–4197. [Google Scholar] [CrossRef]
- Nasif, S.; Contu, L.; Muhlemann, O. Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin. Cell Dev. Biol. 2018, 75, 78–87. [Google Scholar] [CrossRef]
- Nickless, A.; Bailis, J.M.; You, Z. Control of gene expression through the nonsense-mediated RNA decay pathway. Cell Biosci. 2017, 7, 26. [Google Scholar] [CrossRef]
- Amrani, N.; Ganesan, R.; Kervestin, S.; Mangus, D.A.; Ghosh, S.; Jacobson, A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 2004, 432, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Keeling, K.M.; Lanier, J.; Du, M.; Salas-Marco, J.; Gao, L.; Kaenjak-Angeletti, A.; Bedwell, D.M. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 2004, 10, 691–703. [Google Scholar] [PubMed]
- Czaplinski, K.; Ruiz-Echevarria, M.J.; Paushkin, S.V.; Han, X.; Weng, Y.; Perlick, H.A.; Dietz, H.C.; Ter-Avanesyan, M.D.; Peltz, S.W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998, 12, 1665–1677. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.V.; Gehring, N.H.; Kunz, J.B.; Hentze, M.W.; Kulozik, A.E. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008, 27, 736–747. [Google Scholar] [CrossRef]
- Dehecq, M.; Decourty, L.; Namane, A.; Proux, C.; Kanaan, J.; Le Hir, H.; Jacquier, A.; Saveanu, C. Nonsense-mediated mRNA decay involves two distinct Upf1-bound complexes. EMBO J. 2018, 37, e99278. [Google Scholar] [CrossRef]
- Maquat, L.E.; Serin, G. Nonsense-mediated mRNA decay: Insights into mechanism from the cellular abundance of human Upf1, Upf2, Upf3, and Upf3X proteins. Cold Spring Symp. Quant. Biol. 2001, 66, 313–320. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Jayachandran, U.; Bonneau, F.; Fiorini, F.; Basquin, C.; Domcke, S.; Le Hir, H.; Conti, E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell 2011, 41, 693–703. [Google Scholar] [CrossRef]
- Kashima, I.; Yamashita, A.; Izumi, N.; Kataoka, N.; Morishita, R.; Hoshino, S.; Ohno, M.; Dreyfuss, G.; Ohno, S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006, 20, 355–367. [Google Scholar] [CrossRef]
- Yamashita, A.; Izumi, N.; Kashima, I.; Ohnishi, T.; Saari, B.; Katsuhata, Y.; Muramatsu, R.; Morita, T.; Iwamatsu, A.; Hachiya, T.; et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009, 23, 1091–1105. [Google Scholar] [CrossRef]
- Hug, N.; Caceres, J.F. The RNA helicase DHX34 activates NMD by promoting a transition from the surveillance to the decay-inducing complex. Cell Rep. 2014, 8, 1845–1856. [Google Scholar] [CrossRef]
- Yamashita, A.; Ohnishi, T.; Kashima, I.; Taya, Y.; Ohno, S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001, 15, 2215–2228. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Li, W.; Hoque, M.; Popp, M.W.; Ermolenko, D.N.; Tian, B.; Maquat, L.E. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 2014, 28, 1900–1916. [Google Scholar] [CrossRef] [PubMed]
- Lasalde, C.; Rivera, A.V.; Leon, A.J.; Gonzalez-Feliciano, J.A.; Estrella, L.A.; Rodriguez-Cruz, E.N.; Correa, M.E.; Cajigas, I.J.; Bracho, D.P.; Vega, I.E.; et al. Identification and functional analysis of novel phosphorylation sites in the RNA surveillance protein Upf1. Nucleic Acids Res. 2014, 42, 1916–1929. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cajigas, I.J.; Peltz, S.W.; Wilkinson, M.F.; Gonzalez, C.I. Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay. Mol. Cell. Biol. 2006, 26, 3390–3400. [Google Scholar] [CrossRef]
- Kurosaki, T.; Popp, M.W.; Maquat, L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. 2019, 20, 406–420. [Google Scholar] [CrossRef]
- Le Hir, H.; Izaurralde, E.; Maquat, L.E.; Moore, M.J. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000, 19, 6860–6869. [Google Scholar] [CrossRef]
- Gehring, N.H.; Kunz, J.B.; Neu-Yilik, G.; Breit, S.; Viegas, M.H.; Hentze, M.W.; Kulozik, A.E. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell 2005, 20, 65–75. [Google Scholar] [CrossRef]
- Lykke-Andersen, J. mRNA quality control: Marking the message for life or death. Curr. Biol. 2001, 11, 88–91. [Google Scholar] [CrossRef]
- Nagy, E.; Maquat, L.E. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem. Sci. 1998, 23, 198–199. [Google Scholar] [CrossRef]
- Le Hir, H.; Gatfield, D.; Izaurralde, E.; Moore, M.J. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001, 20, 4987–4997. [Google Scholar] [CrossRef]
- Kim, V.N.; Kataoka, N.; Dreyfuss, G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 2001, 293, 1832–1836. [Google Scholar] [CrossRef]
- Eberle, A.B.; Stalder, L.; Mathys, H.; Orozco, R.Z.; Muhlemann, O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008, 6, e92. [Google Scholar] [CrossRef]
- Ivanov, A.; Mikhailova, T.; Eliseev, B.; Yeramala, L.; Sokolova, E.; Susorov, D.; Shuvalov, A.; Schaffitzel, C.; Alkalaeva, E. PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res. 2016, 44, 7766–7776. [Google Scholar] [CrossRef] [PubMed]
- Hurt, J.A.; Robertson, A.D.; Burge, C.B. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013, 23, 1636–1650. [Google Scholar] [CrossRef]
- Kurosaki, T.; Maquat, L.E. Rules that govern UPF1 binding to mRNA 3′ UTRs. Proc. Natl. Acad. Sci. USA 2013, 110, 3357–3362. [Google Scholar] [CrossRef] [PubMed]
- Hoek, T.A.; Khuperkar, D.; Lindeboom, R.G.H.; Sonneveld, S.; Verhagen, B.M.P.; Boersma, S.; Vermeulen, M.; Tanenbaum, M.E. Single-Molecule Imaging Uncovers Rules Governing Nonsense-Mediated mRNA Decay. Mol. Cell 2019, 75, 324–339. [Google Scholar] [CrossRef] [PubMed]
- Peltz, S.W.; Brown, A.H.; Jacobson, A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993, 7, 1737–1754. [Google Scholar] [CrossRef]
- Ruiz-Echevarria, M.J.; Gonzalez, C.I.; Peltz, S.W. Identifying the right stop: Determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998, 17, 575–589. [Google Scholar] [CrossRef]
- Toma, K.G.; Rebbapragada, I.; Durand, S.; Lykke-Andersen, J. Identification of elements in human long 3′ UTRs that inhibit nonsense-mediated decay. RNA 2015, 21, 887–897. [Google Scholar]
- Hosoda, N.; Kim, Y.K.; Lejeune, F.; Maquat, L.E. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2005, 12, 893–901. [Google Scholar] [CrossRef]
- Hwang, J.; Sato, H.; Tang, Y.; Matsuda, D.; Maquat, L.E. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol. Cell 2010, 39, 396–409. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Das, B.; Sherman, F.; Maquat, L.E. Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast. Proc. Natl. Acad. Sci. USA 2005, 102, 4258–4263. [Google Scholar] [CrossRef] [PubMed]
- Rufener, S.C.; Muhlemann, O. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2013, 20, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Atkin, A.L.; Schenkman, L.R.; Eastham, M.; Dahlseid, J.N.; Lelivelt, M.J.; Culbertson, M.R. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem. 1997, 272, 22163–22172. [Google Scholar] [CrossRef] [PubMed]
- Bordeira-Carrico, R.; Pego, A.P.; Santos, M.; Oliveira, C. Cancer syndromes and therapy by stop-codon readthrough. Trends Mol. Med. 2012, 18, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Holbrook, J.A.; Neu-Yilik, G.; Hentze, M.W.; Kulozik, A.E. Nonsense-mediated decay approaches the clinic. Nat. Genet. 2004, 36, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Mort, M.; Ivanov, D.; Cooper, D.N.; Chuzhanova, N.A. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 2008, 29, 1037–1047. [Google Scholar]
- Frischmeyer, P.A.; Dietz, H.C. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999, 8, 1893–1900. [Google Scholar]
- Lee, H.L.; Dougherty, J.P. Pharmaceutical therapies to recode nonsense mutations in inherited diseases. Pharmacol. Ther. 2012, 136, 227–266. [Google Scholar]
- Linde, L.; Kerem, B. Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet. 2008, 24, 552–563. [Google Scholar]
- Keeling, K.M.; Xue, X.; Gunn, G.; Bedwell, D.M. Therapeutics based on stop codon readthrough. Annu. Rev. Genom. Hum. Genet. 2014, 15, 371–394. [Google Scholar] [CrossRef] [PubMed]
- Jackson, K.L.; Dayton, R.D.; Orchard, E.A.; Ju, S.; Ringe, D.; Petsko, G.A.; Maquat, L.E.; Klein, R.L. Preservation of forelimb function by UPF1 gene therapy in a rat model of TDP-43-induced motor paralysis. Gene Ther. 2015, 22, 20–28. [Google Scholar] [PubMed]
- Miller, J.N.; Pearce, D.A. Nonsense-mediated decay in genetic disease: Friend or foe? Mutat. Res. Rev. Mutat. Res. 2014, 762, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Fearon, K.; McClendon, V.; Bonetti, B.; Bedwell, D.M. Premature translation termination mutations are efficiently suppressed in a highly conserved region of yeast Ste6p, a member of the ATP-binding cassette (ABC) transporter family. J. Biol. Chem. 1994, 269, 17802–17808. [Google Scholar]
- Keeling, K.M.; Bedwell, D.M. Suppression of nonsense mutations as a therapeutic approach to treat genetic diseases. Wiley Interdiscip. Rev. RNA 2011, 2, 837–852. [Google Scholar] [CrossRef]
- Anczukow, O.; Ware, M.D.; Buisson, M.; Zetoune, A.B.; Stoppa-Lyonnet, D.; Sinilnikova, O.M.; Mazoyer, S. Does the nonsense-mediated mRNA decay mechanism prevent the synthesis of truncated BRCA1, CHK2, and p53 proteins? Hum. Mutat. 2008, 29, 65–73. [Google Scholar]
- Pinyol, M.; Bea, S.; Pla, L.; Ribrag, V.; Bosq, J.; Rosenwald, A.; Campo, E.; Jares, P. Inactivation of RB1 in mantle-cell lymphoma detected by nonsense-mediated mRNA decay pathway inhibition and microarray analysis. Blood 2007, 109, 5422–5429. [Google Scholar] [CrossRef]
- Ware, M.D.; DeSilva, D.; Sinilnikova, O.M.; Stoppa-Lyonnet, D.; Tavtigian, S.V.; Mazoyer, S. Does nonsense-mediated mRNA decay explain the ovarian cancer cluster region of the BRCA2 gene? Oncogene 2006, 25, 323–328. [Google Scholar] [CrossRef]
- Chernikov, V.G.; Terekhov, S.M.; Krokhina, T.B.; Shishkin, S.S.; Smirnova, T.D.; Kalashnikova, E.A.; Adnoral, N.V.; Rebrov, L.B.; Denisov-Nikol’skii, Y.I.; Bykov, V.A. Comparison of cytotoxicity of aminoglycoside antibiotics using a panel cellular biotest system. Bull. Exp. Biol. Med. 2003, 135, 103–105. [Google Scholar]
- Du, L.; Damoiseaux, R.; Nahas, S.; Gao, K.; Hu, H.; Pollard, J.M.; Goldstine, J.; Jung, M.E.; Henning, S.M.; Bertoni, C.; et al. Nonaminoglycoside compounds induce readthrough of nonsense mutations. J. Exp. Med. 2009, 206, 2285–2297. [Google Scholar] [CrossRef]
- Lykke-Andersen, S.; Jensen, T.H. Nonsense-mediated mRNA decay: An intricate machinery that shapes transcriptomes. Nat. Rev. 2015, 16, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Liu, X.; Welch, E.M.; Hirawat, S.; Peltz, S.W.; Bedwell, D.M. PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR-G542X nonsense allele in a CF mouse model. Proc. Natl. Acad. Sci. USA 2008, 105, 2064–2069. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Hilarion, S.; Beghyn, T.; Jia, J.; Debreuck, N.; Berte, G.; Mamchaoui, K.; Mouly, V.; Gruenert, D.C.; Deprez, B.; Lejeune, F. Rescue of nonsense mutations by amlexanox in human cells. Orphanet J. Rare Dis. 2012, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Du, L.; Tunuguntla, R.; Fike, F.; Cavalieri, S.; Morio, T.; Mizutani, S.; Brusco, A.; Gatti, R.A. Functional characterization and targeted correction of ATM mutations identified in Japanese patients with ataxia-telangiectasia. Hum. Mutat. 2012, 33, 198–208. [Google Scholar] [CrossRef]
- Shalev, M.; Baasov, T. When Proteins Start to Make Sense: Fine-tuning Aminoglycosides for PTC Suppression Therapy. Medchemcomm 2014, 5, 1092–1105. [Google Scholar] [CrossRef]
- Welch, E.M.; Barton, E.R.; Zhuo, J.; Tomizawa, Y.; Friesen, W.J.; Trifillis, P.; Paushkin, S.; Patel, M.; Trotta, C.R.; Hwang, S.; et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature 2007, 447, 87–91. [Google Scholar] [CrossRef]
- Mehta, J.; Tuteja, R. Inhibition of unwinding and ATPase activities of Plasmodium falciparum Dbp5/DDX19 homolog. Commun. Integr. Biol. 2011, 4, 299–303. [Google Scholar]
- Aslam, A.A.; Higgins, C.; Sinha, I.P.; Southern, K.W. Ataluren and similar compounds (specific therapies for premature termination codon class I mutations) for cystic fibrosis. Cochrane Database Syst. Rev. 2017. [Google Scholar] [CrossRef]
- DeFrancesco, L. Drug pipeline: 1Q17. Nat. Biotechnol. 2017, 35, 400. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beißel, C.; Grosse, S.; Krebber, H. Dbp5/DDX19 between Translational Readthrough and Nonsense Mediated Decay. Int. J. Mol. Sci. 2020, 21, 1085. https://doi.org/10.3390/ijms21031085
Beißel C, Grosse S, Krebber H. Dbp5/DDX19 between Translational Readthrough and Nonsense Mediated Decay. International Journal of Molecular Sciences. 2020; 21(3):1085. https://doi.org/10.3390/ijms21031085
Chicago/Turabian StyleBeißel, Christian, Sebastian Grosse, and Heike Krebber. 2020. "Dbp5/DDX19 between Translational Readthrough and Nonsense Mediated Decay" International Journal of Molecular Sciences 21, no. 3: 1085. https://doi.org/10.3390/ijms21031085
APA StyleBeißel, C., Grosse, S., & Krebber, H. (2020). Dbp5/DDX19 between Translational Readthrough and Nonsense Mediated Decay. International Journal of Molecular Sciences, 21(3), 1085. https://doi.org/10.3390/ijms21031085





