Gene Expression Signature Predictive of Neuroendocrine Transformation in Prostate Adenocarcinoma
Abstract
1. Introduction
2. Results
2.1. Sample Selection
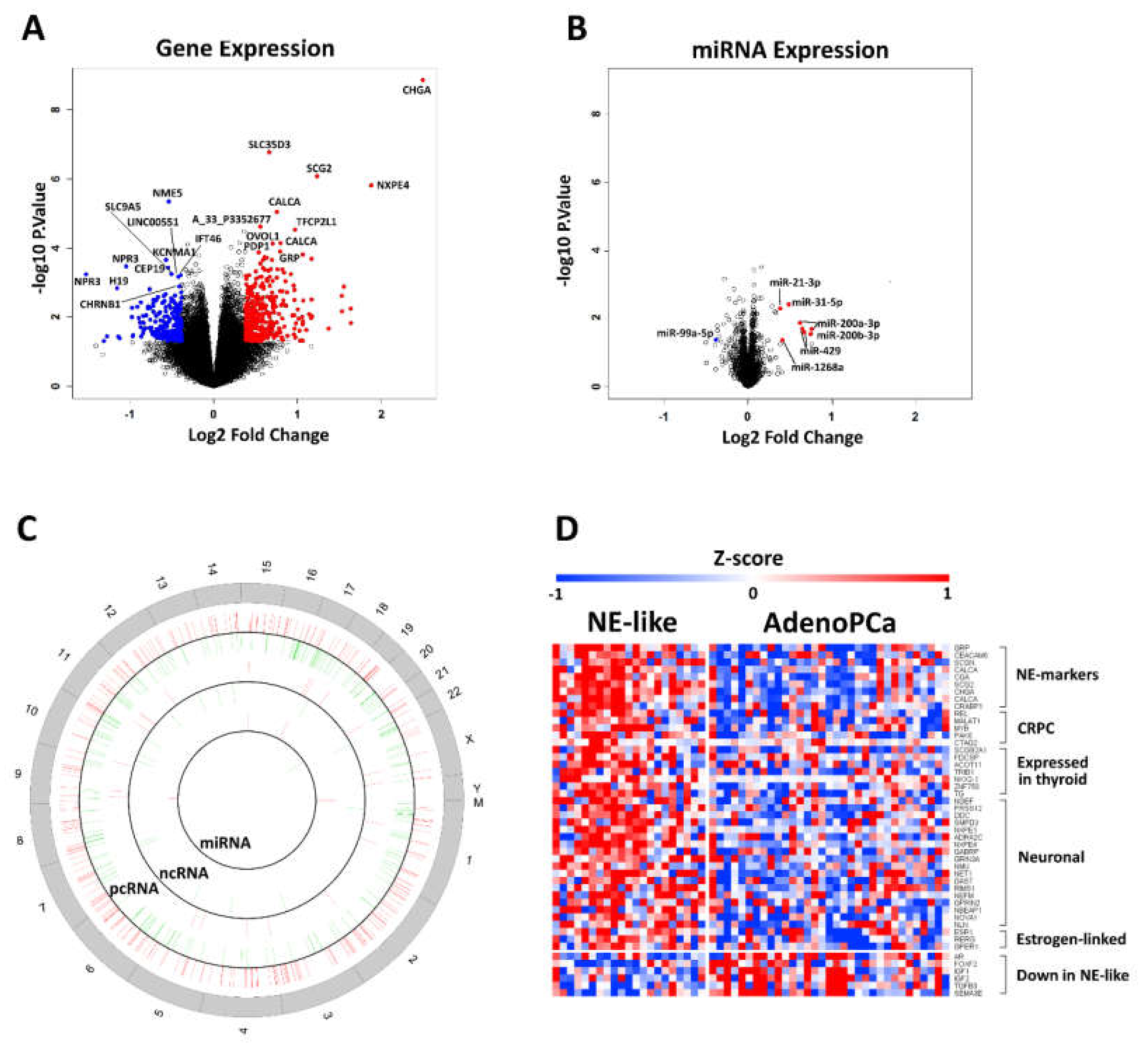
2.2. Identification of Differentially Expressed mRNAs, lncRNAs and miRs
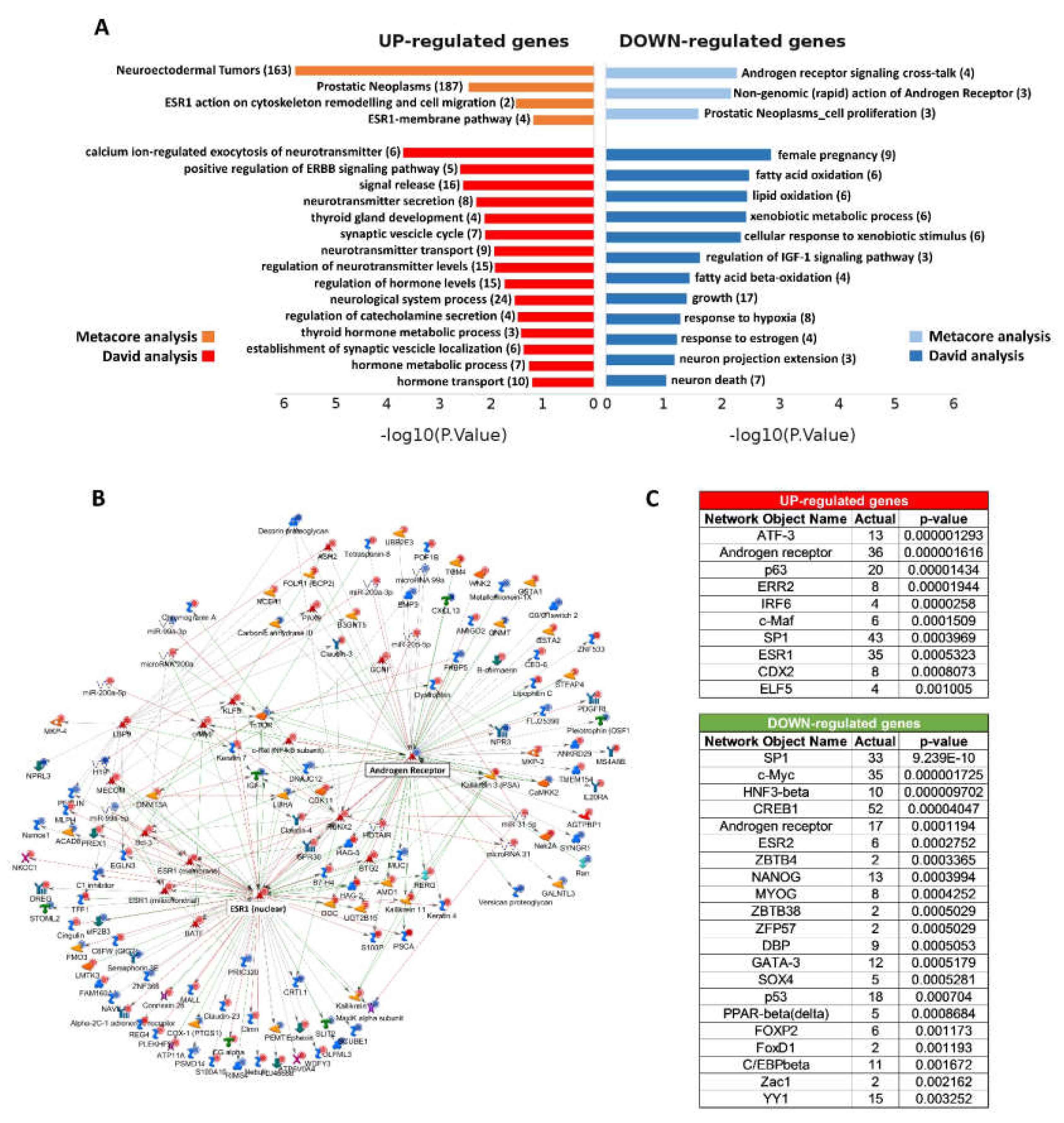
2.3. Functional Enrichment Analysis
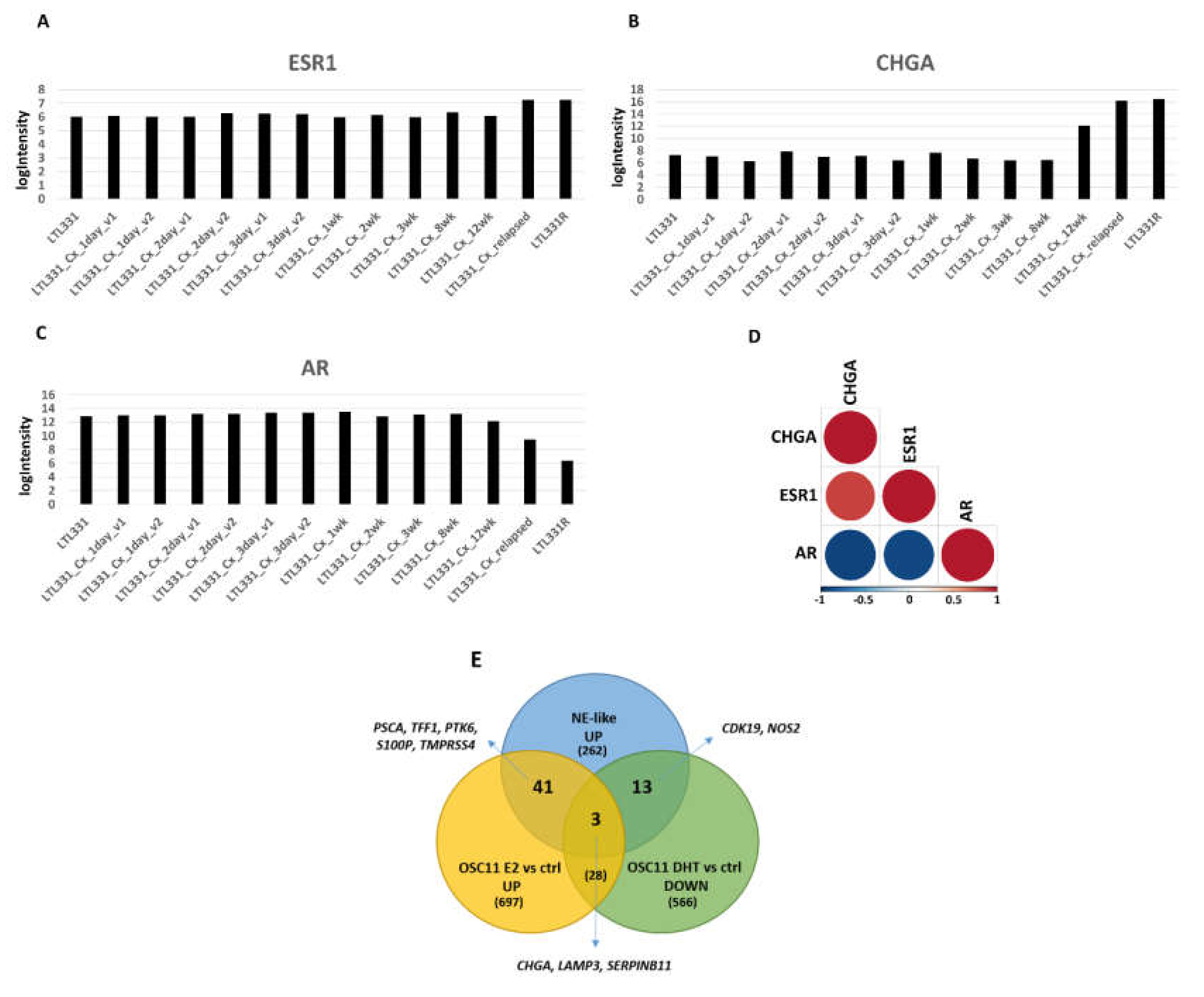
2.4. Estrogen Pathway in Neuroendocrine Transdifferentiation of Prostate Cancer
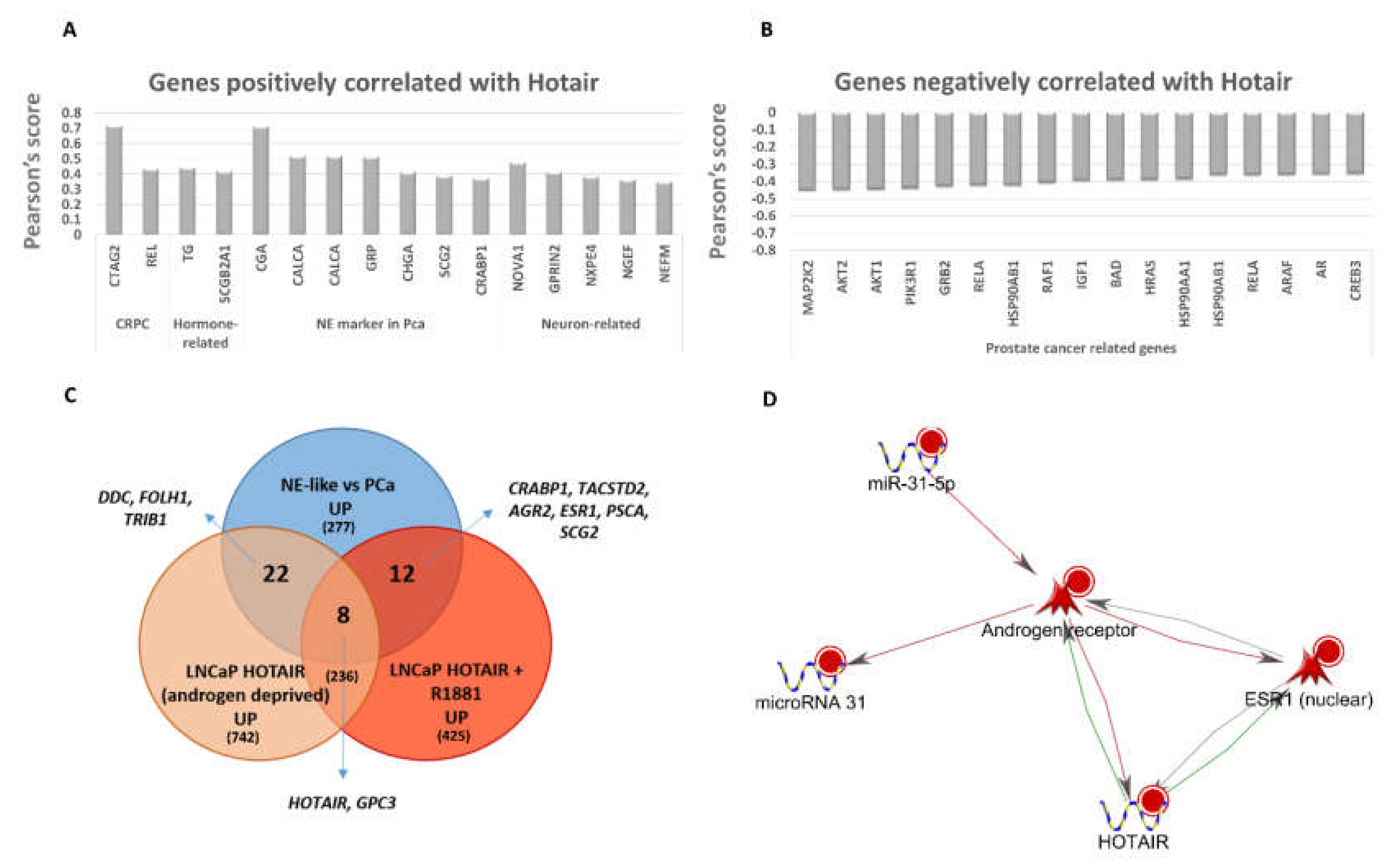
2.5. LncRNA and miRNA Expression Profiling
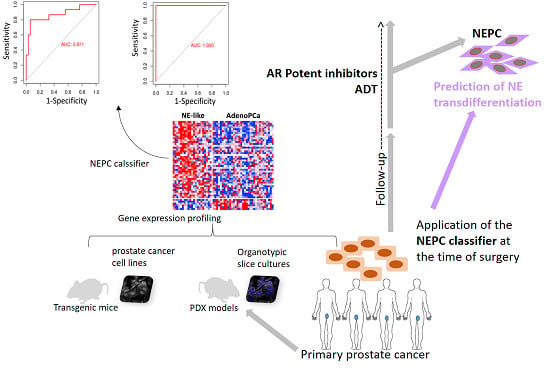
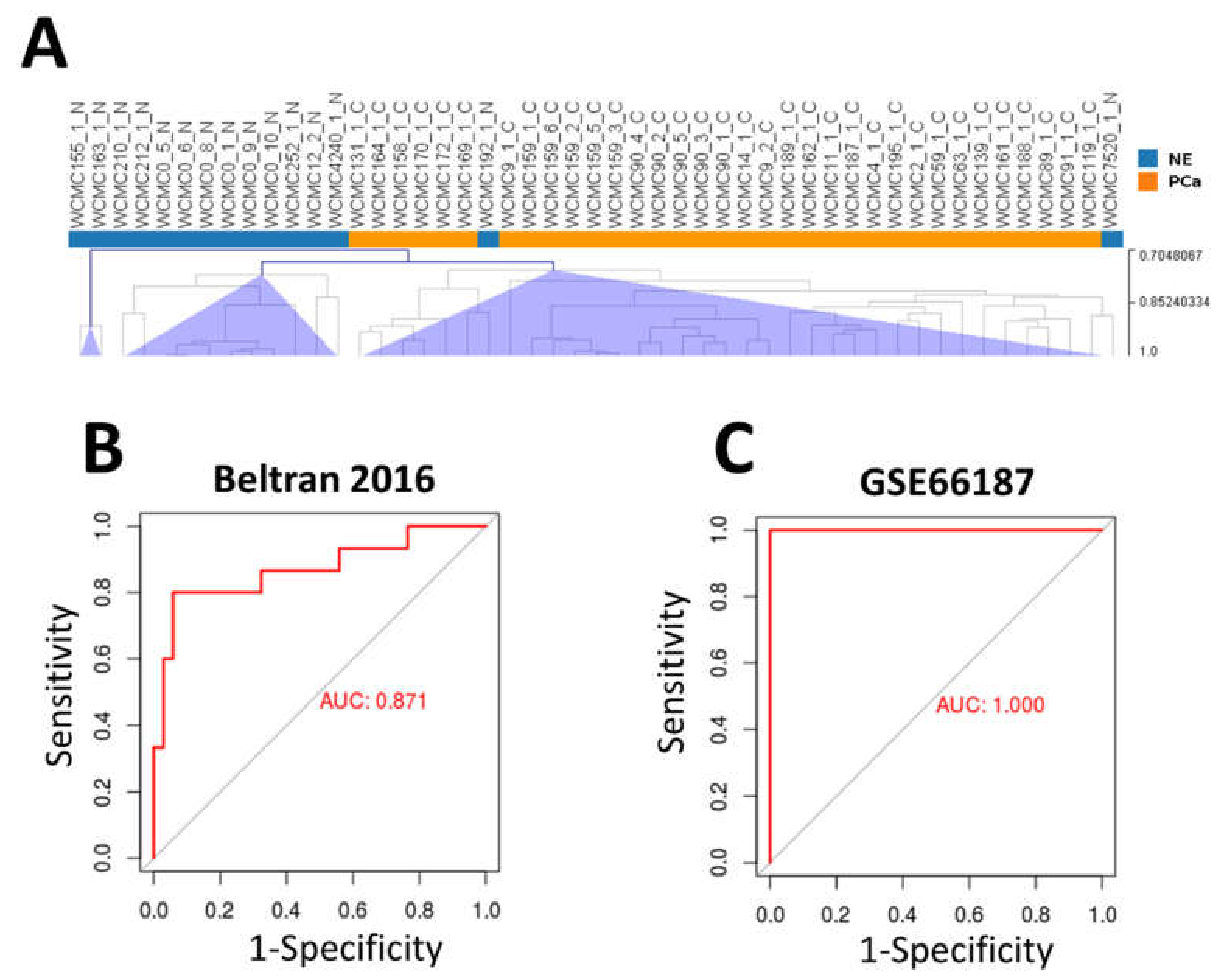
2.6. Gene Signature of Neuroendocrine Prostate Cancer
3. Discussion
4. Materials and Methods
4.1. Prostate Samples Recruitment
4.2. Organotypic Slice Culture (OSC) Preparation and Treatment
4.3. Gene Expression Analysis
4.4. Cluster and Principal Component Analysis
4.5. Functional Analysis
4.6. Gene Set Enrichment Analysis (GSEA)
4.7. External Datasets
4.8. Statistical Analysis
4.9. NEPC Classifier
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| NE | neuroendocrine |
| NEPC | neuroendocrine prostate cancer |
| PCa | prostate cancer |
| PSA | prostate specific antigen |
| pcRNA | protein-coding RNA |
| lncRNA | long non-coding RNA |
| miRNA | microRNA |
| AUC | area under the curve |
| GS | Gleason score |
| DEG | differentially expressed gene |
| PDX | patient-derived xenograft |
| OSC | organotypic slice culture |
| GO | gene ontology |
| CRPC | castration-resistant prostate cancer |
| SERMS | Selective estrogen receptor modulators |
| BCR | biochemical relapse |
| ROC | receiver operating characteristic |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Terry, S.; Beltran, H. The many faces of neuroendocrine differentiation in prostate cancer progression. Front. Oncol. 2014, 4, 60. [Google Scholar] [CrossRef]
- Teo, M.Y.; Rathkopf, D.E.; Kantoff, P. Treatment of Advanced Prostate Cancer. Annu. Rev. Med. 2019, 70, 479–499. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Prandi, D.; Mosquera, J.M.; Benelli, M.; Puca, L.; Cyrta, J.; Marotz, C.; Giannopoulou, E.; Chakravarthi, B.V.S.K.; Varambally, S.; et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 2016, 22, 298–305. [Google Scholar] [CrossRef]
- Grigore, A.D.; Ben-Jacob, E.; Farach-Carson, M.C. Prostate cancer and neuroendocrine differentiation: more neuronal, less endocrine? Front. Oncol. 2015, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, A.; Logothetis, C.J.; Maity, S.N. Understanding the lethal variant of prostate cancer: power of examining extremes. Cancer Discov. 2011, 1, 466–468. [Google Scholar] [CrossRef] [PubMed]
- Palmgren, J.S.; Karavadia, S.S.; Wakefield, M.R. Unusual and underappreciated: small cell carcinoma of the prostate. Semin. Oncol. 2007, 34, 22–29. [Google Scholar] [CrossRef]
- Beltran, H.; Rickman, D.S.; Park, K.; Chae, S.S.; Sboner, A.; MacDonald, T.Y.; Wang, Y.; Sheikh, K.L.; Terry, S.; Tagawa, S.T.; et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011, 1, 487–495. [Google Scholar] [CrossRef]
- Rahman, H.P.; Hofland, J.; Foster, P.A. In touch with your feminine side: how oestrogen metabolism impacts prostate cancer. Endocr. Relat. Cancer 2016, 23, R249–R266. [Google Scholar] [CrossRef]
- Hariri, W.; Sudha, T.; Bharali, D.J.; Cui, H.; Mousa, S.A. Nano-Targeted Delivery of Toremifene, an Estrogen Receptor-α Blocker in Prostate Cancer. Pharm. Res. 2015, 32, 2764–2774. [Google Scholar] [CrossRef]
- Ricke, W.A.; McPherson, S.J.; Bianco, J.J.; Cunha, G.R.; Wang, Y.; Risbridger, G.P. Prostatic hormonal carcinogenesis is mediated by in situ estrogen production and estrogen receptor alpha signaling. FASEB J. 2008, 22, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Capper, C.P.; Larios, J.M.; Sikora, M.J.; Johnson, M.D.; Rae, J.M. The CYP17A1 inhibitor abiraterone exhibits estrogen receptor agonist activity in breast cancer. Breast Cancer Res. Treat. 2016, 157, 23–30. [Google Scholar] [CrossRef]
- Chakravarty, D.; Sboner, A.; Nair, S.S.; Giannopoulou, E.; Li, R.; Hennig, S.; Mosquera, J.M.; Pauwels, J.; Park, K.; Kossai, M.; et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014, 5, 5383. [Google Scholar] [CrossRef]
- Sun, Y.; Niu, J.; Huang, J. Neuroendocrine differentiation in prostate cancer. Am. J. Transl. Res. 2009, 1, 148–162. [Google Scholar] [PubMed]
- Mauri, G.; Jachetti, E.; Comuzzi, B.; Dugo, M.; Arioli, I.; Miotti, S.; Sangaletti, S.; Di Carlo, E.; Tripodo, C.; Colombo, M.P. Genetic deletion of osteopontin in TRAMP mice skews prostate carcinogenesis from adenocarcinoma to aggressive human-like neuroendocrine cancers. Oncotarget 2016, 7, 3905–3920. [Google Scholar] [CrossRef] [PubMed]
- Ramnarine, V.R.; Alshalalfa, M.; Mo, F.; Nabavi, N.; Erho, N.; Takhar, M.; Shukin, R.; Brahmbhatt, S.; Gawronski, A.; Kobelev, M.; et al. The long noncoding RNA landscape of neuroendocrine prostate cancer and its clinical implications. Gigascience 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Zhao, J.C.; Kim, J.; Fong, K.-W.; Yang, Y.A.; Chakravarti, D.; Mo, Y.-Y.; Yu, J. LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep. 2015, 13, 209–221. [Google Scholar] [CrossRef]
- Lin, P.-C.; Chiu, Y.-L.; Banerjee, S.; Park, K.; Mosquera, J.M.; Giannopoulou, E.; Alves, P.; Tewari, A.K.; Gerstein, M.B.; Beltran, H.; et al. Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res. 2013, 73, 1232–1244. [Google Scholar] [CrossRef]
- Xue, X.; Yang, Y.A.; Zhang, A.; Fong, K.-W.; Kim, J.; Song, B.; Li, S.; Zhao, J.C.; Yu, J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 2016, 35, 2746–2755. [Google Scholar] [CrossRef]
- Bhan, A.; Hussain, I.; Ansari, K.I.; Kasiri, S.; Bashyal, A.; Mandal, S.S. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J. Mol. Biol. 2013, 425, 3707–3722. [Google Scholar] [CrossRef]
- Elliot, S.J.; Berho, M.; Korach, K.; Doublier, S.; Lupia, E.; Striker, G.E.; Karl, M. Gender-specific effects of endogenous testosterone: female alpha-estrogen receptor-deficient C57Bl/6J mice develop glomerulosclerosis. Kidney Int. 2007, 72, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hamilton, K.J.; Wang, T.; Coons, L.A.; Jefferson, W.N.; Li, R.; Wang, Y.; Grimm, S.A.; Ramsey, J.T.; Liu, L.; et al. DNA methylation and transcriptome aberrations mediated by ERα in mouse seminal vesicles following developmental DES exposure. Proc. Natl. Acad. Sci. USA 2018, 115, E4189–E4198. [Google Scholar] [CrossRef] [PubMed]
- Simanainen, U.; McNamara, K.; Gao, Y.R.; McPherson, S.; Desai, R.; Jimenez, M.; Handelsman, D.J. Anterior prostate epithelial AR inactivation modifies estrogen receptor expression and increases estrogen sensitivity. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E727–E735. [Google Scholar] [CrossRef] [PubMed]
- Lanzino, M.; De Amicis, F.; McPhaul, M.J.; Marsico, S.; Panno, M.L.; Andò, S. Endogenous coactivator ARA70 interacts with estrogen receptor alpha (ERalpha) and modulates the functional ERalpha/androgen receptor interplay in MCF-7 cells. J. Biol. Chem. 2005, 280, 20421–20430. [Google Scholar] [CrossRef]
- Zhang, X.; Coleman, I.M.; Brown, L.G.; True, L.D.; Kollath, L.; Lucas, J.M.; Lam, H.-M.; Dumpit, R.; Corey, E.; Chéry, L.; et al. SRRM4 Expression and the Loss of REST Activity May Promote the Emergence of the Neuroendocrine Phenotype in Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2015, 21, 4698–4708. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, D.; Zhou, T.; Song, H.; Hulsurkar, M.; Su, N.; Liu, Y.; Wang, Z.; Shao, L.; Ittmann, M.; et al. Androgen deprivation promotes neuroendocrine differentiation and angiogenesis through CREB-EZH2-TSP1 pathway in prostate cancers. Nat. Commun. 2018, 9, 4080. [Google Scholar] [CrossRef]
- Volta, A.D.; Cosentini, D.; Antonelli, A.; Pedersini, R.; Simeone, C.; Volante, M.; Berruti, A. Transformation of Prostate Adenocarcinoma into Small-Cell Neuroendocrine Cancer Under Androgen Deprivation Therapy: Much Is Achieved but More Information Is Needed. J. Clin. Oncol. 2019, 37, 350–351. [Google Scholar] [CrossRef]
- Wang, H.T.; Yao, Y.H.; Li, B.G.; Tang, Y.; Chang, J.W.; Zhang, J. Neuroendocrine Prostate Cancer (NEPC) progressing from conventional prostatic adenocarcinoma: factors associated with time to development of NEPC and survival from NEPC diagnosis-a systematic review and pooled analysis. J. Clin. Oncol. 2014, 32, 3383–3390. [Google Scholar] [CrossRef]
- Chang, Y.-T.; Lin, T.-P.; Tang, J.-T.; Campbell, M.; Luo, Y.-L.; Lu, S.-Y.; Yang, C.-P.; Cheng, T.-Y.; Chang, C.-H.; Liu, T.-T.; et al. HOTAIR is a REST-regulated lncRNA that promotes neuroendocrine differentiation in castration resistant prostate cancer. Cancer Lett. 2018, 433, 43–52. [Google Scholar] [CrossRef]
- Mather, R.L.; Wang, Y.; Crea, F. Is HOTAIR really involved in neuroendocrine prostate cancer differentiation? Epigenomics 2018, 10, 1259–1261. [Google Scholar] [CrossRef]
- El Sheikh, S.S.; Domin, J.; Abel, P.; Stamp, G.; Lalani, E.-N. Androgen-independent prostate cancer: potential role of androgen and ErbB receptor signal transduction crosstalk. Neoplasia 2003, 5, 99–109. [Google Scholar] [CrossRef][Green Version]
- Kelly, K.; Balk, S.P. Reprogramming to resist. Science (New York, N.Y.) 2017, 355, 29–30. [Google Scholar] [CrossRef]
- Hung, C.-L.; Wang, L.-Y.; Yu, Y.-L.; Chen, H.-W.; Srivastava, S.; Petrovics, G.; Kung, H.-J. A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl. Acad. Sci. USA 2014, 111, 18697–18702. [Google Scholar] [CrossRef] [PubMed]
- Myung, J.-K.; Wang, G.; Chiu, H.H.L.; Wang, J.; Mawji, N.R.; Sadar, M.D. Inhibition of androgen receptor by decoy molecules delays progression to castration-recurrent prostate cancer. PLoS ONE 2017, 12, e0174134. [Google Scholar] [CrossRef]
- Hu, D.G.; Selth, L.A.; Tarulli, G.A.; Meech, R.; Wijayakumara, D.; Chanawong, A.; Russell, R.; Caldas, C.; Robinson, J.L.L.; Carroll, J.S.; et al. Androgen and Estrogen Receptors in Breast Cancer Coregulate Human UDP-Glucuronosyltransferases 2B15 and 2B17. Cancer Res. 2016, 76, 5881–5893. [Google Scholar] [CrossRef]
- Vestergaard, E.M.; Nexø, E.; Tørring, N.; Borre, M.; Ørntoft, T.F.; Sørensen, K.D. Promoter hypomethylation and upregulation of trefoil factors in prostate cancer. Int. J. Cancer 2010, 127, 1857–1865. [Google Scholar] [CrossRef]
- Dobbs, R.W.; Malhotra, N.R.; Greenwald, D.T.; Wang, A.Y.; Prins, G.S.; Abern, M.R. Estrogens and prostate cancer. Prostate Cancer Prostatic Dis. 2019, 22, 185–194. [Google Scholar] [CrossRef]
- Kim, I.Y.; Kim, B.-C.; Seong, D.H.; Lee, D.K.; Seo, J.-M.; Hong, Y.J.; Kim, H.-T.; Morton, R.A.; Kim, S.-J. Raloxifene, a mixed estrogen agonist/antagonist, induces apoptosis in androgen-independent human prostate cancer cell lines. Cancer Res. 2002, 62, 5365–5369. [Google Scholar]
- Shazer, R.L.; Jain, A.; Galkin, A.V.; Cinman, N.; Nguyen, K.N.; Natale, R.B.; Gross, M.; Green, L.; Bender, L.I.; Holden, S.; et al. Raloxifene, an oestrogen-receptor-beta-targeted therapy, inhibits androgen-independent prostate cancer growth: results from preclinical studies and a pilot phase II clinical trial. BJU Int. 2006, 97, 691–697. [Google Scholar] [CrossRef]
- Fujimura, T.; Takahashi, S.; Kume, H.; Urano, T.; Takayama, K.; Yamada, Y.; Suzuki, M.; Fukuhara, H.; Nakagawa, T.; Inoue, S.; et al. Toremifene, a selective estrogen receptor modulator, significantly improved biochemical recurrence in bone metastatic prostate cancer: a randomized controlled phase II a trial. BMC Cancer 2015, 15, 836. [Google Scholar] [CrossRef]
- Chadha, M.K.; Ashraf, U.; Lawrence, D.; Tian, L.; Levine, E.; Silliman, C.; Escott, P.; Payne, V.; Trump, D.L. Phase II study of fulvestrant (Faslodex) in castration resistant prostate cancer. Prostate 2008, 68, 1461–1466. [Google Scholar] [CrossRef]
- Saraon, P.; Cretu, D.; Musrap, N.; Karagiannis, G.S.; Batruch, I.; Drabovich, A.P.; van der Kwast, T.; Mizokami, A.; Morrissey, C.; Jarvi, K.; et al. Quantitative proteomics reveals that enzymes of the ketogenic pathway are associated with prostate cancer progression. Mol. Cell Proteomics 2013, 12, 1589–1601. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Gato, D.; Thysell, E.; Tyanova, S.; Crnalic, S.; Santos, A.; Lima, T.S.; Geiger, T.; Cox, J.; Widmark, A.; Bergh, A.; et al. The Proteome of Prostate Cancer Bone Metastasis Reveals Heterogeneity with Prognostic Implications. Clin. Cancer Res. 2018, 24, 5433–5444. [Google Scholar] [CrossRef] [PubMed]
- Schulze, J.; Albers, J.; Baranowsky, A.; Keller, J.; Spiro, A.; Streichert, T.; Zustin, J.; Amling, M.; Schinke, T. Osteolytic prostate cancer cells induce the expression of specific cytokines in bone-forming osteoblasts through a Stat3/5-dependent mechanism. Bone 2010, 46, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Clegg, N.; Ferguson, C.; True, L.D.; Arnold, H.; Moorman, A.; Quinn, J.E.; Vessella, R.L.; Nelson, P.S. Molecular characterization of prostatic small-cell neuroendocrine carcinoma. Prostate 2003, 55, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Kainov, Y.; Favorskaya, I.; Delektorskaya, V.; Chemeris, G.; Komelkov, A.; Zhuravskaya, A.; Trukhanova, L.; Zueva, E.; Tavitian, B.; Dyakova, N.; et al. CRABP1 provides high malignancy of transformed mesenchymal cells and contributes to the pathogenesis of mesenchymal and neuroendocrine tumors. Cell Cycle 2014, 13, 1530–1539. [Google Scholar] [CrossRef]
- Zhang, Z.; Hao, C.-J.; Li, C.-G.; Zang, D.-J.; Zhao, J.; Li, X.-N.; Wei, A.-H.; Wei, Z.-B.; Yang, L.; He, X.; et al. Mutation of SLC35D3 causes metabolic syndrome by impairing dopamine signaling in striatal D1 neurons. PLoS Genet. 2014, 10, e1004124. [Google Scholar] [CrossRef]
- Farach, A.; Ding, Y.; Lee, M.; Creighton, C.; Delk, N.A.; Ittmann, M.; Miles, B.; Rowley, D.; Farach-Carson, M.C.; Ayala, G.E. Neuronal Trans-Differentiation in Prostate Cancer Cells. Prostate 2016, 76, 1312–1325. [Google Scholar] [CrossRef]
- Takahashi, S.; Shiraishi, T.; Trock, B.J.; Kulkarni, P.; Getzenberg, R.; Tominaga, T.; Homma, Y. 1048 CSAG4 and NOL4 are novel biomarkers to indicate aggressiveness of prostate cancer. Eur. Urol. Suppl. 2013, 12, e1048. [Google Scholar] [CrossRef]
- Aggarwal, R.; Huang, J.; Alumkal, J.J.; Zhang, L.; Feng, F.Y.; Thomas, G.V.; Weinstein, A.S.; Friedl, V.; Zhang, C.; Witte, O.N.; et al. Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J. Clin. Oncol. 2018, 36, 2492–2503. [Google Scholar] [CrossRef]
- Tsai, H.K.; Lehrer, J.; Alshalalfa, M.; Erho, N.; Davicioni, E.; Lotan, T.L. Gene expression signatures of neuroendocrine prostate cancer and primary small cell prostatic carcinoma. BMC Cancer 2017, 17, 759. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Yu, X. Bioinformatics analyses of publicly available NEPCa datasets. Am. J. Clin. Exp. Urol. 2019, 7, 327–340. [Google Scholar] [PubMed]
- Alshalalfa, M.; Liu, Y.; Wyatt, A.W.; Gibb, E.A.; Tsai, H.K.; Erho, N.; Lehrer, J.; Takhar, M.; Ramnarine, V.R.; Collins, C.C.; et al. Characterization of transcriptomic signature of primary prostate cancer analogous to prostatic small cell neuroendocrine carcinoma. Int. J. Cancer 2019, 145, 3453–3461. [Google Scholar] [CrossRef]
- Sasaki, T.; Komiya, A.; Suzuki, H.; Shimbo, M.; Ueda, T.; Akakura, K.; Ichikawa, T. Changes in chromogranin a serum levels during endocrine therapy in metastatic prostate cancer patients. Eur. Urol. 2005, 48, 224–229, discussion 229–230. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Jendrisak, A.; Landers, M.; Mosquera, J.M.; Kossai, M.; Louw, J.; Krupa, R.; Graf, R.P.; Schreiber, N.A.; Nanus, D.M.; et al. The Initial Detection and Partial Characterization of Circulating Tumor Cells in Neuroendocrine Prostate Cancer. Clin. Cancer Res. 2016, 22, 1510–1519. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- An, J.; Lai, J.; Sajjanhar, A.; Batra, J.; Wang, C.; Nelson, C.C. J-Circos: an interactive Circos plotter. Bioinformatics 2015, 31, 1463–1465. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
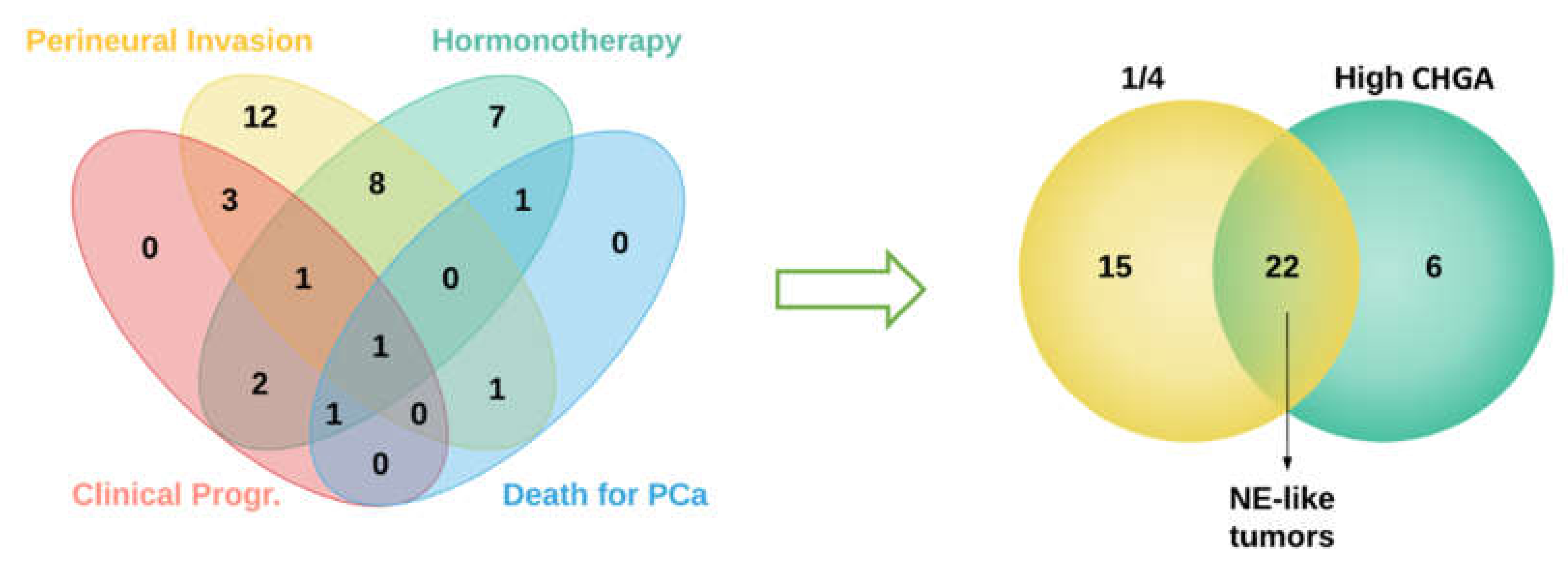
| NE-like (n = 22) | AdenoPCa (n = 32) | Fisher’s Exact Test p-Value | |
|---|---|---|---|
| Biochemical relapse | 12 (55%) | 15 (47%) | 0.78 |
| Gleason Score | 0.02 | ||
| 6 | 3 (14%) | 8 (25%) | |
| 7 | 11 (50%) | 22 (69%) | |
| 8–9 | 8 (36%) | 2 (6%) | |
| Grade | 0.067 | ||
| 2 | 12 (55%) | 26 (81%) | |
| 3 | 10 (45%) | 6 (19%) | |
| Age at surgery | 65.91 ± 5.1 | 65.86 ± 5.08 | 0.5 |
| Initial PSA | 14.2 ± 15.8 | 12.9 ± 12.98 | 0.37 |
| # pcRNAs | # lncRNAs | # miRNAs | |
|---|---|---|---|
| UP | 295 | 24 | 6 |
| DOWN | 206 | 15 | 1 |
| NAME | EDGES |
|---|---|
| ESR1 (NUCLEAR) ↑ | 88 |
| ANDROGEN RECEPTOR ↓ | 82 |
| TFCP2L1 ↑ | 46 |
| RUNX2 ↑ | 30 |
| CDX2 ↑ | 28 |
| C-MYB ↑ | 16 |
| DNMT3A ↑ | 15 |
| KLF5 ↑ | 14 |
| MTOR ↓ | 13 |
| NKX2-1 ↑ | 12 |
| BCL-3 ↑ | 9 |
| MIR-200A-3P ↑ | 9 |
| MIR-200B-3P ↑ | 9 |
| C-REL ↑ | 8 |
| TAF7L ↓ | 8 |
| ASCL2 ↑ | 7 |
| NR6A1↑ | 7 |
| MECOM ↑ | 7 |
| BATF ↑ | 6 |
| LDHA ↓ | 6 |
| MALAT1 ↑ | 6 |
| MIR-99A-5P ↓ | 6 |
| HOTAIR ↑ | 5 |
| IGF-1 ↓ | 5 |
| UP-Regulated Genes | DOWN-Regulated Genes |
|---|---|
| HCAR3 (Nicotinic acid) CXCR2 (Elubrixin, Navarixin, Reparixin, SB332235) KCNQ1 (Azimilide, Indapamide) KCND2 (Dalfampridine) REL (Apilimod) AMD1 (N(1),N(11)-Diethylnorspermine) DDC (Carbidopa) ESR1 (Afimoxifene, Clomifene, Diethylstilbestrol, Megestrol, Raloxifene) NOS2 (GW274150) ADRA2C (Apraclonidine, Azepexole, Besipirdine, Brimonidine, Deriglidole, Dipivefrine, Efaroxan, Fipamezole, Guanethidine, Idazoxan, Lusaperidone, Mianserin, Naphazoline, OPC28326, Piperoxan, Tramazoline, Yohimbine) LPL (Gemfibrozil, Ibrolipim) GPNMB (Glembatumumab vedotin) CACNA1H (Mibefradil, Sipatrigine) GPC3 (Codrituzumab) | PPIE (Ciclosporin) S1PR1 (Fingolimod) CA3 (Acetazolamide) KCNMA1 (Cromoglicic acid, Diazoxide, Hydroflumethiazide, Trichlormethiazide) AR (Androstanolone, Bicalutamide, BMS564929, Cyproterone acetate, Dehydroepiandrosterone, Diethylstilbestrol, Drospirenone, Finasteride, Flutamide, HE3235, LGD-2226, Megestrol O-acetate, Metandienone, Methyltestosterone, Osaterone, Oxandrolone, Oxendolone, RU58841, Silibinin, Testosterone, Zanoterone) MTOR (AZD8055, Everolimus, Ridaforolimus, Sirolimus, Temsirolimus) PTGS1 (Acemetacin, Aspirin, Benoxaprofen, Bromfenac, butibufen, Celecoxib, Diflunisal, Droxicam, Etodolac, Fenbufen, Fenflumizol, Fenoprofen, Ibufenac, Ibuprofen, Indoprofen, Ketorolac, Lornoxicam, Loxoprofen, Meclofenamic acid, Meloxicam, Mesalazine, Nabumetone, NCX701, Nitroaspirin, Oxaprozin, Parsalmide, Phenacetin, Piroxicam, Salsalate, Sulindac, Tarenflurbil, Tenidap, Tenoxicam, Tiaprofenic acid, Timegadine, Tolfenamic acid, Tolmetin, Zaltoprofen, Zomepirac) EPHX2 (AR9281) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ostano, P.; Mello-Grand, M.; Sesia, D.; Gregnanin, I.; Peraldo-Neia, C.; Guana, F.; Jachetti, E.; Farsetti, A.; Chiorino, G. Gene Expression Signature Predictive of Neuroendocrine Transformation in Prostate Adenocarcinoma. Int. J. Mol. Sci. 2020, 21, 1078. https://doi.org/10.3390/ijms21031078
Ostano P, Mello-Grand M, Sesia D, Gregnanin I, Peraldo-Neia C, Guana F, Jachetti E, Farsetti A, Chiorino G. Gene Expression Signature Predictive of Neuroendocrine Transformation in Prostate Adenocarcinoma. International Journal of Molecular Sciences. 2020; 21(3):1078. https://doi.org/10.3390/ijms21031078
Chicago/Turabian StyleOstano, Paola, Maurizia Mello-Grand, Debora Sesia, Ilaria Gregnanin, Caterina Peraldo-Neia, Francesca Guana, Elena Jachetti, Antonella Farsetti, and Giovanna Chiorino. 2020. "Gene Expression Signature Predictive of Neuroendocrine Transformation in Prostate Adenocarcinoma" International Journal of Molecular Sciences 21, no. 3: 1078. https://doi.org/10.3390/ijms21031078
APA StyleOstano, P., Mello-Grand, M., Sesia, D., Gregnanin, I., Peraldo-Neia, C., Guana, F., Jachetti, E., Farsetti, A., & Chiorino, G. (2020). Gene Expression Signature Predictive of Neuroendocrine Transformation in Prostate Adenocarcinoma. International Journal of Molecular Sciences, 21(3), 1078. https://doi.org/10.3390/ijms21031078