Root Proteomic Analysis of Two Grapevine Rootstock Genotypes Showing Different Susceptibility to Salt Stress
Abstract
1. Introduction
2. Results and Discussion
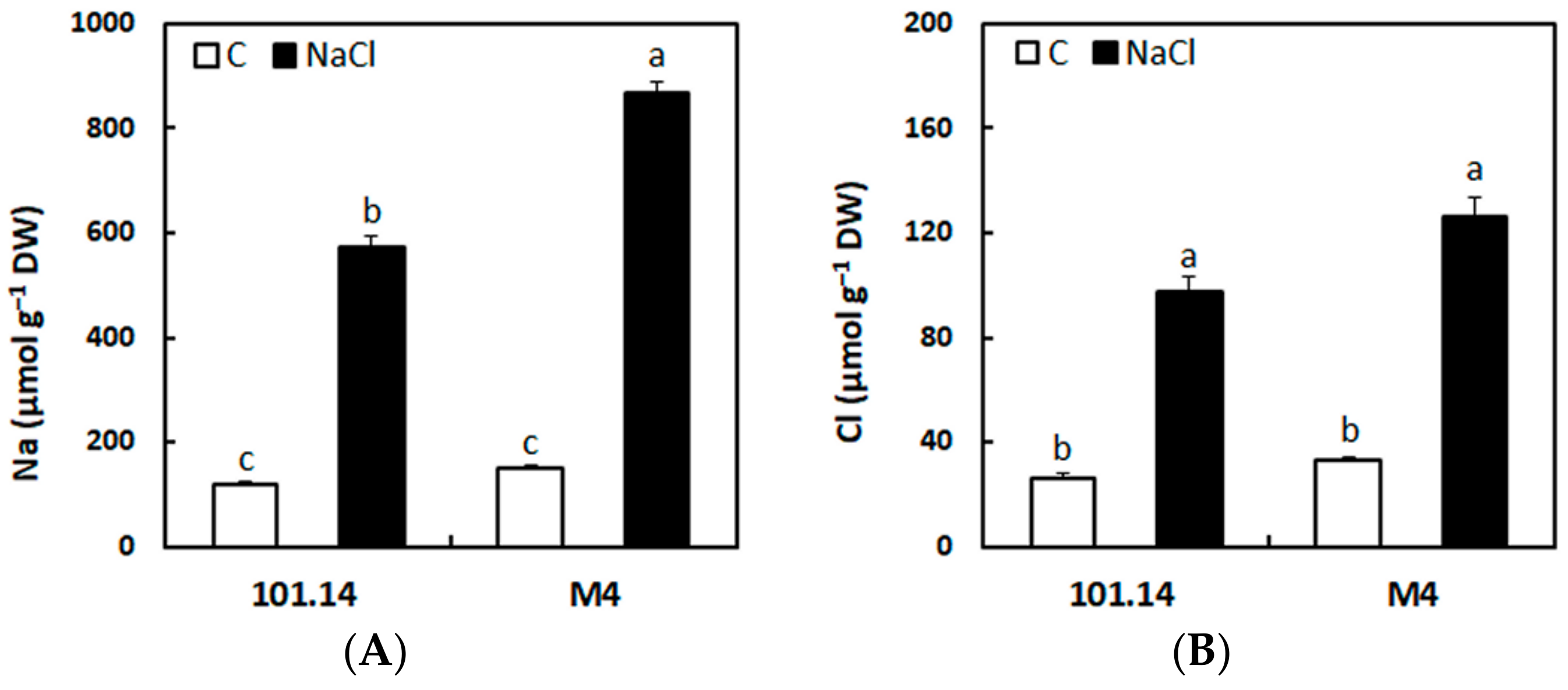
2.1. Accumulation of Na+ and Cl− in the Roots
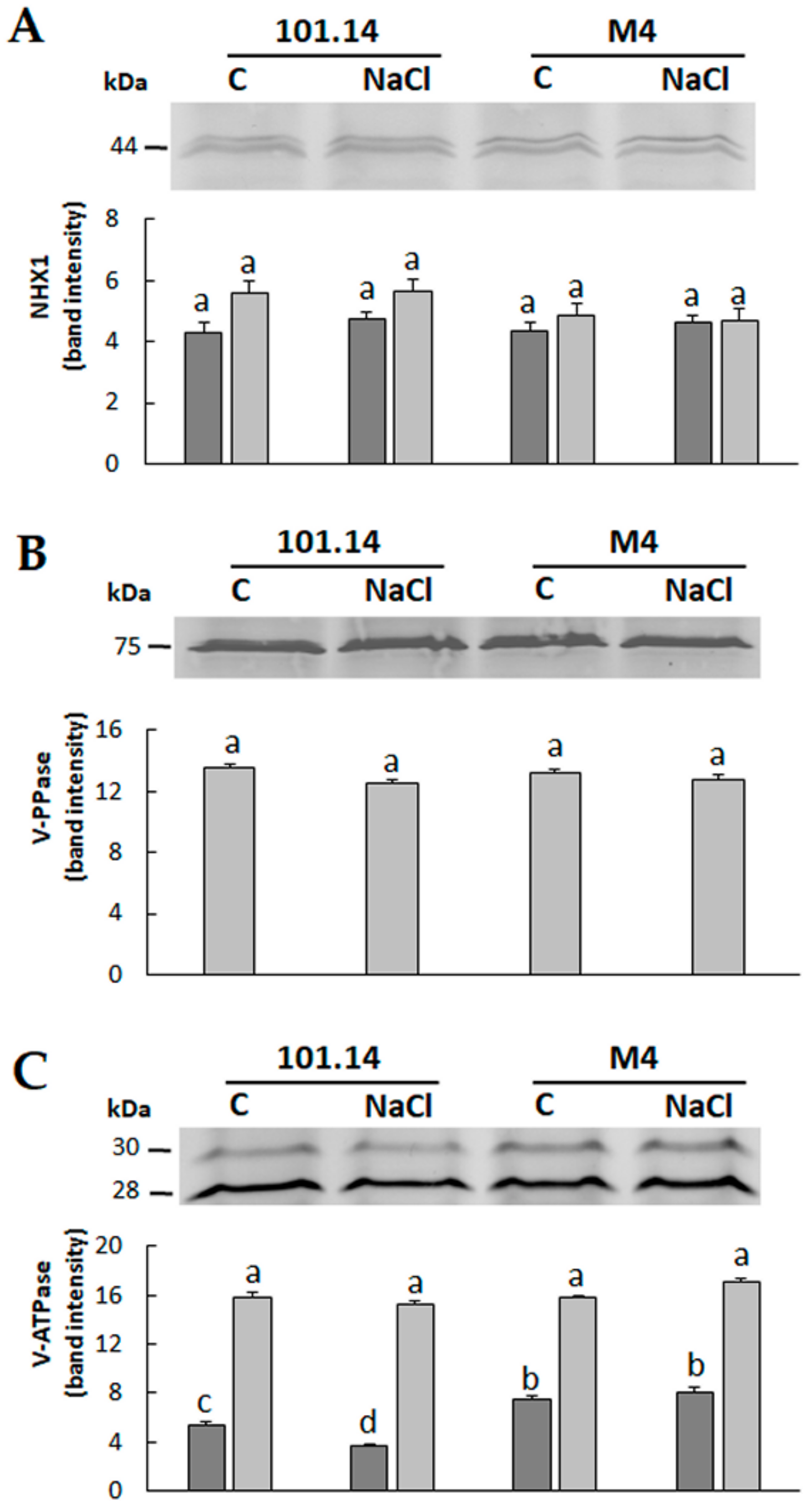
2.2. Proteomic Analyses
2.2.1. Functional Distribution of the Identified Proteins
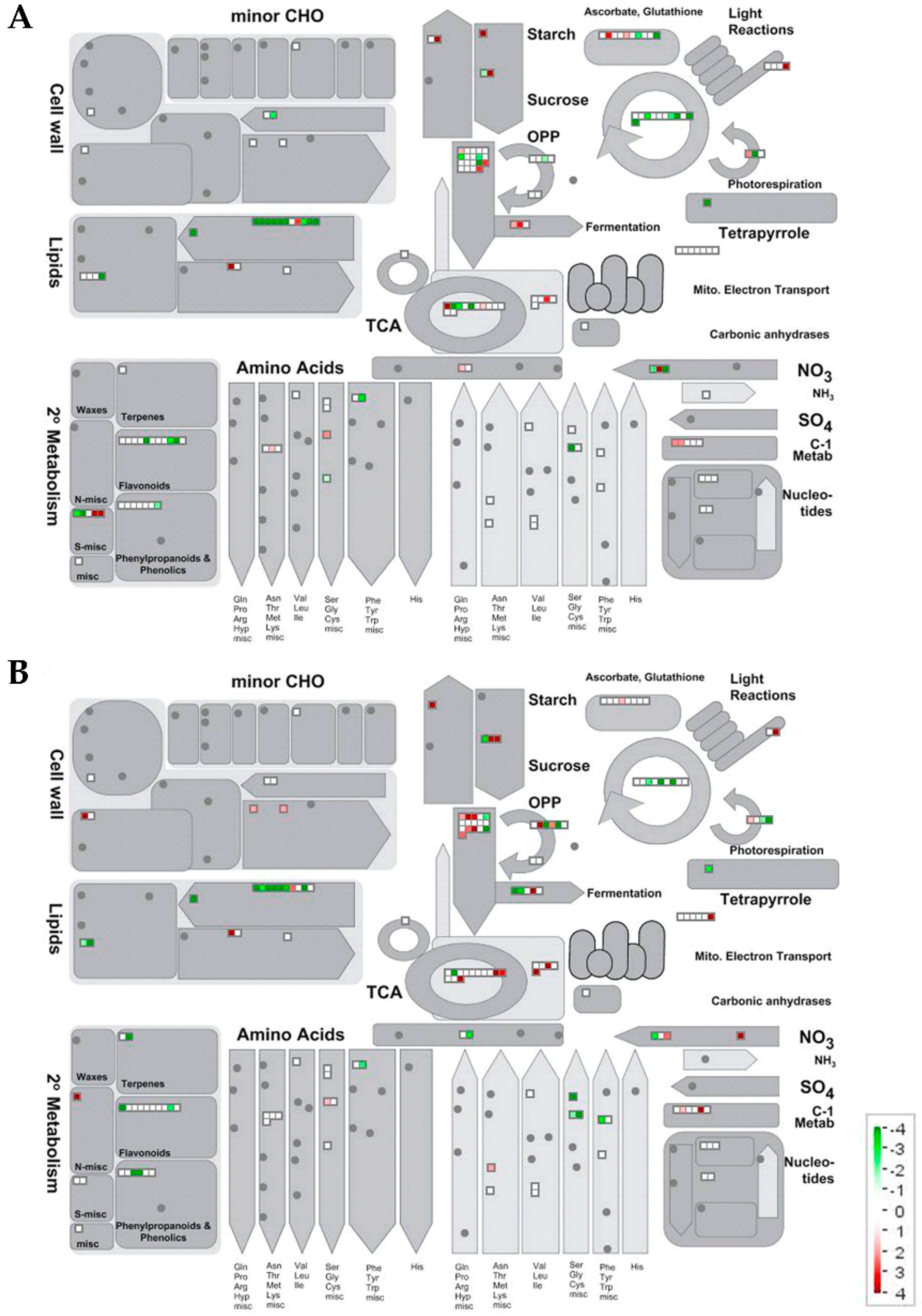
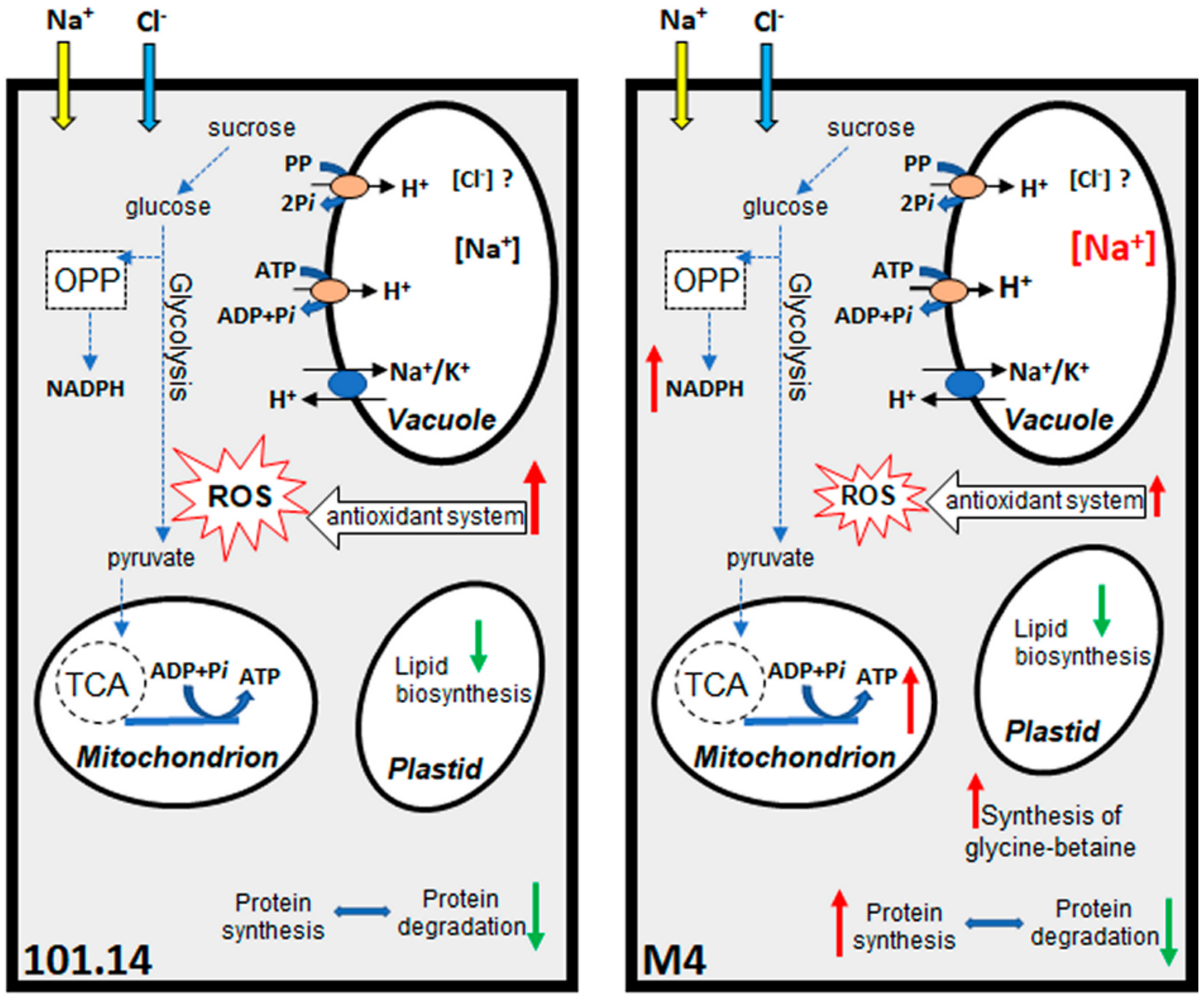
2.2.2. Metabolic Pathways Affected by Salt Stress
Proteomic Changes Involved in Carbon and Energy Metabolism
Proteomic Changes Involved in Lipid Metabolism
Proteomic Changes Involved in N, Amino Acid and Protein Metabolism
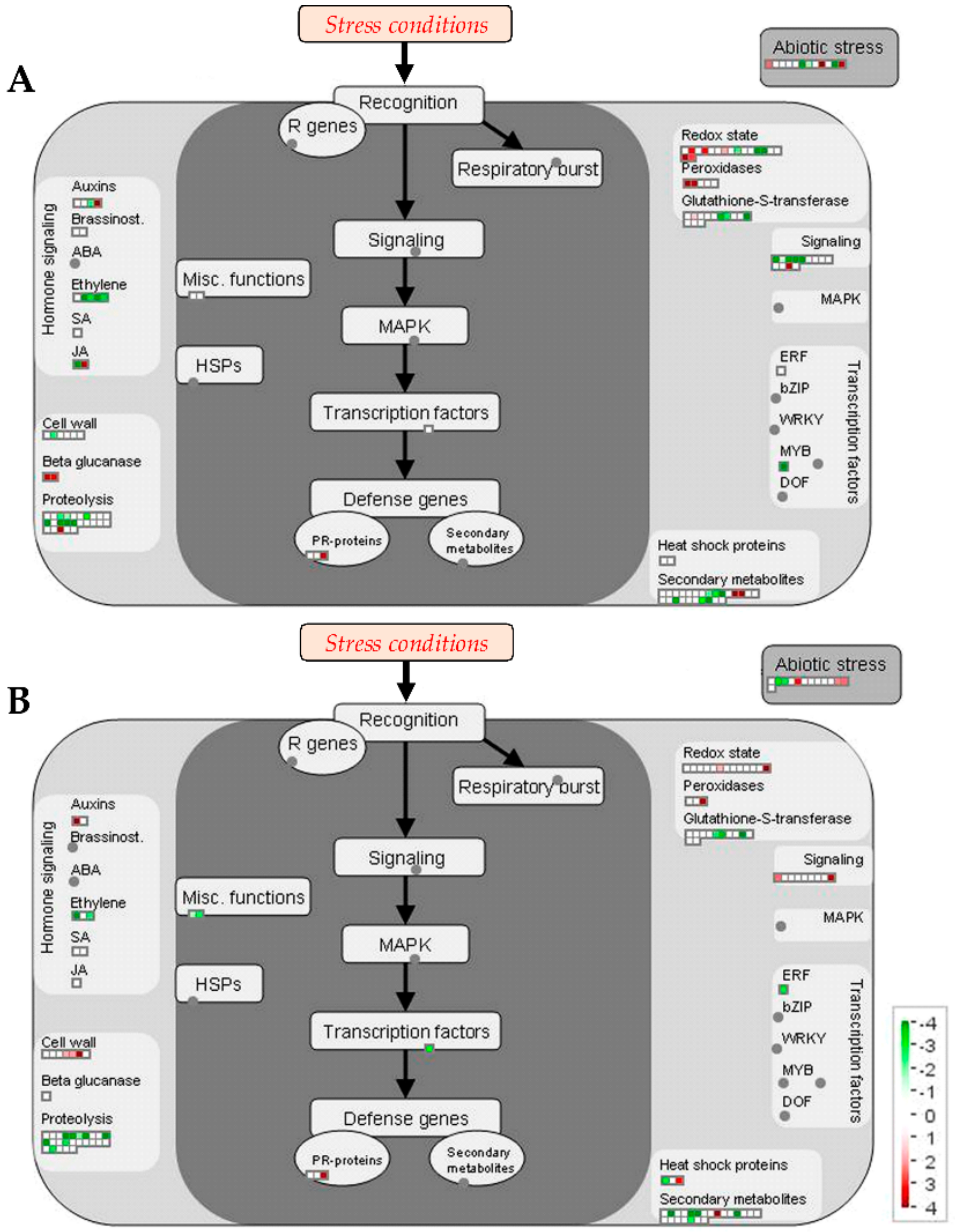
Proteomic Changes Involved in Secondary Metabolism, Stress and Redox Metabolisms
2.2.3. Final Considerations
3. Materials and Methods
3.1. Sample Material and Growth Conditions
3.2. Chloride and Sodium Quantification
3.3. Protein Extraction
3.4. Western Blot Analyses
3.5. Gel Electrophoresis, In-Gel Digestion and Mass Spectrometry Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Munns, R. Chapter 1—Plant Adaptations to Salt and Water Stress: Differences and Commonalities. In Advances in Botanical Research; Turkan, I., Ed.; Plant Responses to Drought and Salinity Stress; Academic Press: Waltham, MA, USA, 2011; Volume 57, pp. 1–32. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.-K.; Bohnert, H.J. Plant Cellular and Molecular Responses to High Salinity. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [PubMed]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant Salt-Tolerance Mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef]
- Walker, R.R.; Blackmore, D.H.; Clingeleffer, P.R.; Iacono, F. Effect of Salinity and Ramsey Rootstock on Ion Concentrations and Carbon Dioxide Assimilation in Leaves of Drip-Irrigated, Field-Grown Grapevines (Vitis vinifera L. Cv. Sultana). Aust. J. Grape Wine Res. 1997, 3, 66–74. [Google Scholar] [CrossRef]
- Walker, R.R.; Blackmore, D.H.; Clingeleffer, P.R. Impact of Rootstock on Yield and Ion Concentrations in Petioles, Juice and Wine of Shiraz and Chardonnay in Different Viticultural Environments with Different Irrigation Water Salinity. Aust. J. Grape Wine Res. 2010, 16, 243–257. [Google Scholar] [CrossRef]
- Tregeagle, J.M.; Tisdall, J.M.; Tester, M.; Walker, R.R. Cl—Uptake, Transport and Accumulation in Grapevine Rootstocks of Differing Capacity for Cl—Exclusion. Funct. Plant Biol. 2010, 37, 665–673. [Google Scholar] [CrossRef]
- Gong, H.; Blackmore, D.H.; Walker, R.R. Organic and Inorganic Anions in Shiraz and Chardonnay Grape Berries and Wine as Affected by Rootstock under Saline Conditions. Aust. J. Grape Wine Res. 2010, 16, 227–236. [Google Scholar] [CrossRef]
- Henderson, S.W.; Baumann, U.; Blackmore, D.H.; Walker, A.R.; Walker, R.R.; Gilliham, M. Shoot Chloride Exclusion and Salt Tolerance in Grapevine Is Associated with Differential Ion Transporter Expression in Roots. BMC Plant Biol. 2014, 14, 273. [Google Scholar] [CrossRef]
- Walker, R.R.; Blackmore, D.H.; Clingeleffer, P.R.; Emanuelli, D. Rootstock Type Determines Tolerance of Chardonnay and Shiraz to Long-Term Saline Irrigation. Aust. J. Grape Wine Res. 2014, 20, 496–506. [Google Scholar] [CrossRef]
- Ollat, N.; Peccoux, A.; Papura, D.; Esmenjaud, D.; Marguerit, E.; Tandonnet, J.-P.; Bordenave, L.; Cookson, S.J.; Barrieu, F.; Rossdeutsch, L.; et al. Rootstocks as a Component of Adaptation to Environment. In Grapevine in a Changing Environment; John Wiley & Sons, Ltd.: West Sussex, UK, 2016; pp. 68–108. [Google Scholar] [CrossRef]
- Gong, H.; Blackmore, D.; Clingeleffer, P.; Sykes, S.; Jha, D.; Tester, M.; Walker, R. Contrast in Chloride Exclusion between Two Grapevine Genotypes and Its Variation in Their Hybrid Progeny. J. Exp. Bot. 2011, 62, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Conde, A.; Breia, R.; Moutinho-Pereira, J.; Grimplet, J.; Gerós, H. Metabolic Rearrangements in Grapevine Response to Salt Stress. In Grapevine in a Changing Environment; John Wiley & Sons, Ltd.: West Sussex, UK, 2016; pp. 279–298. [Google Scholar] [CrossRef]
- Mirás-Avalos, J.M.; Intrigliolo, D.S. Grape Composition under Abiotic Constrains: Water Stress and Salinity. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Henderson, S.W.; Dunlevy, J.D.; Wu, Y.; Blackmore, D.H.; Walker, R.R.; Edwards, E.J.; Gilliham, M.; Walker, A.R. Functional Differences in Transport Properties of Natural HKT1;1 Variants Influence Shoot Na+ Exclusion in Grapevine Rootstocks. New Phytol. 2018, 217, 1113–1127. [Google Scholar] [CrossRef] [PubMed]
- Meggio, F.; Prinsi, B.; Negri, A.S.; Lorenzo, G.S.D.; Lucchini, G.; Pitacco, A.; Failla, O.; Scienza, A.; Cocucci, M.; Espen, L. Biochemical and Physiological Responses of Two Grapevine Rootstock Genotypes to Drought and Salt Treatments. Aust. J. Grape Wine Res. 2014, 20, 310–323. [Google Scholar] [CrossRef]
- Shavrukov, Y. Salt Stress or Salt Shock: Which Genes Are We Studying? J. Exp. Bot. 2013, 64, 119–127. [Google Scholar] [CrossRef]
- Assaha, D.V.M.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The Role of Na+ and K+ Transporters in Salt Stress Adaptation in Glycophytes. Front. Physiol. 2017, 8. [Google Scholar] [CrossRef]
- Hamaji, K.; Nagira, M.; Yoshida, K.; Ohnishi, M.; Oda, Y.; Uemura, T.; Goh, T.; Sato, M.H.; Morita, M.T.; Tasaka, M.; et al. Dynamic Aspects of Ion Accumulation by Vesicle Traffic under Salt Stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2023–2033. [Google Scholar] [CrossRef]
- Undurraga, S.F.; Santos, M.P.; Paez-Valencia, J.; Yang, H.; Hepler, P.K.; Facanha, A.R.; Hirschi, K.D.; Gaxiola, R.A. Arabidopsis Sodium Dependent and Independent Phenotypes Triggered by H+-PPase up-Regulation Are SOS1 Dependent. Plant Sci. 2012, 183, 96–105. [Google Scholar] [CrossRef]
- Dietz, K.J.; Tavakoli, N.; Kluge, C.; Mimura, T.; Sharma, S.S.; Harris, G.C.; Chardonnens, A.N.; Golldack, D. Significance of the V-type ATPase for the Adaptation to Stressful Growth Conditions and Its Regulation on the Molecular and Biochemical Level. J. Exp. Bot. 2001, 52, 1969–1980. [Google Scholar] [CrossRef]
- Zhang, M.; Fang, Y.; Liang, Z.; Huang, L. Enhanced Expression of Vacuolar H+-ATPase Subunit E in the Roots Is Associated with the Adaptation of Broussonetia papyrifera to Salt Stress. PLoS ONE 2012, 7, e48183. [Google Scholar] [CrossRef]
- Gilmore, J.M.; Washburn, M.P. Advances in Shotgun Proteomics and the Analysis of Membrane Proteomes. J. Proteom. 2010, 73, 2078–2091. [Google Scholar] [CrossRef] [PubMed]
- Prinsi, B.; Negri, A.S.; Failla, O.; Scienza, A.; Espen, L. Root Proteomic and Metabolic Analyses Reveal Specific Responses to Drought Stress in Differently Tolerant Grapevine Rootstocks. BMC Plant Biol. 2018, 18, 126. [Google Scholar] [CrossRef] [PubMed]
- Prinsi, B.; Espen, L. Time-Course of Metabolic and Proteomic Responses to Different Nitrate/Ammonium Availabilities in Roots and Leaves of Maize. Int. J. Mol. Sci. 2018, 19, 2202. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A User-Driven Tool to Display Genomics Data Sets onto Diagrams of Metabolic Pathways and Other Biological Processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yang, B.; Harris, N.S.; Deyholos, M.K. Comparative Proteomic Analysis of NaCl Stress-Responsive Proteins in Arabidopsis Roots. J. Exp. Bot. 2007, 58, 3591–3607. [Google Scholar] [CrossRef]
- Zhang, H.; Han, B.; Wang, T.; Chen, S.; Li, H.; Zhang, Y.; Dai, S. Mechanisms of Plant Salt Response: Insights from Proteomics. J. Proteome Res. 2012, 11, 49–67. [Google Scholar] [CrossRef]
- Kruger, N.J.; Dennis, D.T. Molecular Properties of Pyrophosphate: Fructose-6-Phosphate Phosphotransferase from Potato Tuber. Arch. Biochem. Biophys. 1987, 256, 273–279. [Google Scholar] [CrossRef]
- Wong, J.H.; Kiss, F.; Wu, M.-X.; Buchanan, B.B. Pyrophosphate Fructose-6-P 1-Phosphotransferase from Tomato Fruit: Evidence for Change during Ripening. Plant Physiol. 1990, 94, 499–506. [Google Scholar] [CrossRef]
- Ghosh, D.; Xu, J. Abiotic Stress Responses in Plant Roots: A Proteomics Perspective. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef]
- Munns, R.; Day, D.A.; Fricke, W.; Watt, M.; Arsova, B.; Barkla, B.J.; Bose, J.; Byrt, C.S.; Chen, Z.-H.; Foster, K.J.; et al. Energy Costs of Salt Tolerance in Crop Plants. New Phytol. 2019. [Google Scholar] [CrossRef]
- Zorrilla-Fontanesi, Y.; Rouard, M.; Cenci, A.; Kissel, E.; Do, H.; Dubois, E.; Nidelet, S.; Roux, N.; Swennen, R.; Carpentier, S.C. Differential Root Transcriptomics in a Polyploid Non-Model Crop: The Importance of Respiration during Osmotic Stress. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef]
- Stein, O.; Granot, D. An Overview of Sucrose Synthases in Plants. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B. NADPH-Generating Dehydrogenases: Their Role in the Mechanism of Protection against Nitro-Oxidative Stress Induced by Adverse Environmental Conditions. Front. Environ. Sci. 2014, 2. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, B.; Ding, H.; Zhang, J.; Li, S. Review: The Role of NADP-Malic Enzyme in Plants under Stress. Plant Sci. 2019, 281, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, Y.; Kawakami, N.; Sasakuma, T. Isolation of Novel Early Salt-Responding Genes from Wheat (Triticum aestivum L.) by Differential Display. Theor. Appl. Genet. 1999, 98, 673–678. [Google Scholar] [CrossRef]
- Nemoto, Y.; Sasakuma, T. Specific Expression of Glucose-6-Phosphate Dehydrogenase (G6PDH) Gene by Salt Stress in Wheat (Triticum aestivum L.). Plant Sci. 2000, 158, 53–60. [Google Scholar] [CrossRef]
- Cardi, M.; Castiglia, D.; Ferrara, M.; Guerriero, G.; Chiurazzi, M.; Esposito, S. The Effects of Salt Stress Cause a Diversion of Basal Metabolism in Barley Roots: Possible Different Roles for Glucose-6-Phosphate Dehydrogenase Isoforms. Plant Physiol. Biochem. 2015, 86, 44–54. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, J.; Singh, S.; Singh, V.P.; Prasad, S.M. Roles of Osmoprotectants in Improving Salinity and Drought Tolerance in Plants: A Review. Rev. Environ. Sci. Biotechnol. 2015, 14, 407–426. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, W.; Wang, X. Phospholipase D and Phosphatidic Acid Signalling in Plant Response to Drought and Salinity. Plant Cell Environ. 2010, 33, 627–635. [Google Scholar] [CrossRef]
- Hou, Q.; Ufer, G.; Bartels, D. Lipid Signalling in Plant Responses to Abiotic Stress. Plant Cell Environ. 2016, 39, 1029–1048. [Google Scholar] [CrossRef]
- Troncoso-Ponce, M.A.; Nikovics, K.; Marchive, C.; Lepiniec, L.; Baud, S. New Insights on the Organization and Regulation of the Fatty Acid Biosynthetic Network in the Model Higher Plant Arabidopsis thaliana. Biochimie 2016, 120, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Joyard, J.; Ferro, M.; Masselon, C.; Seigneurin-Berny, D.; Salvi, D.; Garin, J.; Rolland, N. Chloroplast Proteomics Highlights the Subcellular Compartmentation of Lipid Metabolism. Prog. Lipid Res. 2010, 49, 128–158. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; He, Y.; Dai, Y.; Liu, X.; Li, J. Deficiency in Fatty Acid Synthase Leads to Premature Cell Death and Dramatic Alterations in Plant Morphology. Plant Cell 2000, 12, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, T.M.; Nunes Nesi, A.; Araújo, W.L.; Braun, H.-P. Amino Acid Catabolism in Plants. Mol. Plant 2015, 8, 1563–1579. [Google Scholar] [CrossRef]
- Batista-Silva, W.; Heinemann, B.; Rugen, N.; Nunes-Nesi, A.; Araújo, W.L.; Braun, H.-P.; Hildebrandt, T.M. The Role of Amino Acid Metabolism during Abiotic Stress Release. Plant Cell Environ. 2019, 42, 1630–1644. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Hofius, D.; Hajirezaei, M.-R.; Fernie, A.R.; Börnke, F.; Sonnewald, U. Functional Analysis of the Essential Bifunctional Tobacco Enzyme 3-Dehydroquinate Dehydratase/Shikimate Dehydrogenase in Transgenic Tobacco Plants. J. Exp. Bot. 2007, 58, 2053–2067. [Google Scholar] [CrossRef]
- Tolbert, N.E.; Yamazaki, R.K.; Oeser, A. Localization and Properties of Hydroxypyruvate and Glyoxylate Reductases in Spinach Leaf Particles. J. Biol. Chem. 1970, 245, 5129–5136. [Google Scholar]
- Bernard, S.M.; Habash, D.Z. The Importance of Cytosolic Glutamine Synthetase in Nitrogen Assimilation and Recycling. New Phytol. 2009, 182, 608–620. [Google Scholar] [CrossRef]
- Prinsi, B.; Espen, L. Mineral Nitrogen Sources Differently Affect Root Glutamine Synthetase Isoforms and Amino Acid Balance among Organs in Maize. BMC Plant Biol. 2015, 15, 96. [Google Scholar] [CrossRef]
- Moreno, J.I.; Martín, R.; Castresana, C. Arabidopsis SHMT1, a Serine Hydroxymethyltransferase That Functions in the Photorespiratory Pathway Influences Resistance to Biotic and Abiotic Stress. Plant J. 2005, 41, 451–463. [Google Scholar] [CrossRef]
- Terol, J.; Bargues, M.; Carrasco, P.; Pérez-Alonso, M.; Paricio, N. Molecular Characterization and Evolution of the Protein Phosphatase 2A B′ Regulatory Subunit Family in Plants. Plant Physiol. 2002, 129, 808–822. [Google Scholar] [CrossRef] [PubMed]
- Máthé, C.; Garda, T.; Freytag, C. The Role of Serine-Threonine Protein Phosphatase PP2A in Plant Oxidative Stress Signaling—Facts and Hypotheses. Int. J. Mol. Sci. 2019, 20, 3028. [Google Scholar] [CrossRef] [PubMed]
- Machingura, M.; Salomon, E.; Jez, J.M.; Ebbs, S.D. The β-Cyanoalanine Synthase Pathway: Beyond Cyanide Detoxification. Plant Cell Environ. 2016, 39, 2329–2341. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.-S. Drought Stress Increases Both Cyanogenesis and β-Cyanoalanine Synthase Activity in Tobacco. Plant Sci. 2003, 165, 1109–1115. [Google Scholar] [CrossRef]
- Machingura, M.; Sidibe, A.; Wood, A.J.; Ebbs, S.D. The β-Cyanoalanine Pathway Is Involved in the Response to Water Deficit in Arabidopsis thaliana. Plant Physiol. Biochem. 2013, 63, 159–169. [Google Scholar] [CrossRef]
- Prinsi, B.; Negri, A.S.; Fedeli, C.; Morgutti, S.; Negrini, N.; Cocucci, M.; Espen, L. Peach Fruit Ripening: A Proteomic Comparative Analysis of the Mesocarp of Two Cultivars with Different Flesh Firmness at Two Ripening Stages. Phytochemistry 2011, 72, 1251–1262. [Google Scholar] [CrossRef]
- Lorenz, W.W.; Sun, F.; Liang, C.; Kolychev, D.; Wang, H.; Zhao, X.; Cordonnier-Pratt, M.-M.; Pratt, L.H.; Dean, J.F.D. Water Stress-Responsive Genes in Loblolly Pine (Pinus taeda) Roots Identified by Analyses of Expressed Sequence Tag Libraries. Tree Physiol. 2006, 26, 1–16. [Google Scholar] [CrossRef]
- Gregorová, Z.; Kováčik, J.; Klejdus, B.; Maglovski, M.; Kuna, R.; Hauptvogel, P.; Matušíková, I. Drought-Induced Responses of Physiology, Metabolites, and PR Proteins in Triticum aestivum. J. Agric. Food Chem. 2015, 63, 8125–8133. [Google Scholar] [CrossRef]
- Davidson, R.M.; Reeves, P.A.; Manosalva, P.M.; Leach, J.E. Germins: A Diverse Protein Family Important for Crop Improvement. Plant Sci. 2009, 177, 499–510. [Google Scholar] [CrossRef]
- Lu, M.; Han, Y.-P.; Gao, J.-G.; Wang, X.-J.; Li, W.-B. Identification and Analysis of the Germin-like Gene Family in Soybean. BMC Genom. 2010, 11, 620. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Dai, X.-F. Cloning and Characterization of the Gossypium hirsutum Major Latex Protein Gene and Functional Analysis in Arabidopsis thaliana. Planta 2010, 231, 861–873. [Google Scholar] [CrossRef]
- Demidchik, V. Mechanisms of Oxidative Stress in Plants: From Classical Chemistry to Cell Biology. Environ. Exp. Bot. 2015, 109, 212–228. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive Oxygen Species, Abiotic Stress and Stress Combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive Oxygen Gene Network of Plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Csiszár, J.; Horváth, E.; Váry, Z.; Gallé, Á.; Bela, K.; Brunner, S.; Tari, I. Glutathione Transferase Supergene Family in Tomato: Salt Stress-Regulated Expression of Representative Genes from Distinct GST Classes in Plants Primed with Salicylic Acid. Plant Physiol. Biochem. 2014, 78, 15–26. [Google Scholar] [CrossRef]
- Cruz-Rus, E.; Botella, M.A.; Valpuesta, V.; Gomez-Jimenez, M.C. Analysis of Genes Involved in L-Ascorbic Acid Biosynthesis during Growth and Ripening of Grape Berries. J. Plant Physiol. 2010, 167, 739–748. [Google Scholar] [CrossRef]
- Tao, J.; Wu, H.; Li, Z.; Huang, C.; Xu, X. Molecular Evolution of GDP-D-Mannose Epimerase (GME), a Key Gene in Plant Ascorbic Acid Biosynthesis. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Xu, J.; Xing, X.-J.; Tian, Y.-S.; Peng, R.-H.; Xue, Y.; Zhao, W.; Yao, Q.-H. Transgenic Arabidopsis Plants Expressing Tomato Glutathione S-Transferase Showed Enhanced Resistance to Salt and Drought Stress. PLoS ONE 2015, 10, e0136960. [Google Scholar] [CrossRef]
- Peng, X.; Yu, D.; Yan, J.; Zhang, N.; Lin, J.; Wang, J. Physiological and Proteomic Analyses Reveal Adaptive Mechanisms of Ryegrass (Annual vs. Perennial) Seedlings to Salt Stress. Agronomy 2019, 9, 843. [Google Scholar] [CrossRef]
- Prinsi, B.; Negri, A.S.; Quattrocchio, F.M.; Koes, R.E.; Espen, L. Proteomics of Red and White Corolla Limbs in Petunia Reveals a Novel Function of the Anthocyanin Regulator ANTHOCYANIN1 in Determining Flower Longevity. J. Proteom. 2016, 131, 38–47. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Prinsi, B.; Negri, A.S.; Pesaresi, P.; Cocucci, M.; Espen, L. Evaluation of Protein Pattern Changes in Roots and Leaves of Zea Mays plants in Response to Nitrate Availability by Two-Dimensional Gel Electrophoresis Analysis. BMC Plant Biol. 2009, 9, 113. [Google Scholar] [CrossRef] [PubMed]
| Parameter | 101.14 | M4 |
|---|---|---|
| n. of peptides per genotype | 15,105 | 15,131 |
| Average of unique peptide per protein (±SE) | 5.0 ± 0.2 | 5.2 ± 0.2 |
| Average protein intensity | 1.43 ± 0.039 (×106) | 1.72 ± 0.049 (×106) |
| Dynamic range of protein intensity | 4.2 × 103–2.3 × 107 | 7.7 × 103–2.5 × 107 |
| Average protein score (Spectrum Mill) | 78.4 ± 3.7 | 79.7 ± 3.9 |
| Average amino acid coverage % (±SE) | 17.4 ± 0.7 | 17.4 ± 0.7 |
| n. of identified proteins (i.e. protein groups) | 280 | 271 |
| n. of differentially accumulated proteins (%) | 87 (31%) | 90 (34%) |
| # | Accession | Name (f.c.) | ΔSS/C |
|---|---|---|---|
| Carbon and energy metabolism (1-9, 25) | |||
| 184 | A5C6H7 | Sucrose synthase (2) | 150.46 |
| 185 | F6I1P0 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial (8) | 3.49 |
| 146 | F6H710 | Galactokinase, putative (3) | 2.17 |
| 64 | F6I0F6 | Alcohol dehydrogenase (5, 26) | 1.78 |
| 7 | A0A1Z2THL4 | NADP-dependent malic enzyme (8) | 1.75 |
| 58 | C5DB68 | Pyruvate kinase, cytosolic isozyme (4, 11) | 1.70 |
| 175 | F6GX20 | 4-hydroxy-4-methyl-2-oxoglutarate aldolase (25) | 1.52 |
| 86 | D7TJI9 | Pyruvate decarboxylase 1 (5) | 1.42 |
| 101 | A5BEM8 | Putative oxidoreductase GLYR1 (7) | −1.43 |
| 24 | F6HFL6 | Fructose-bisphosphate aldolase (4) | −1.59 |
| 120 | F6HFN8 | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex (8, 11) | −1.74 |
| 61 | D7T0U8 | Glyceraldehyde-3-phosphate dehydrogenase (1) | −1.77 |
| 144 | F6HI27 | Pyruvate dehydrogenase E1 component subunit beta-2, chloroplastic (8, 1, 11) | −4.93 |
| 60 | F6GTG3 | Enolase 1, chloroplastic-like (4) | −6.75 |
| 168 | D7SY46 | Dihydrolipoyl dehydrogenase 2, chloroplastic (8, 11, 21) | −26.09 |
| Lipid metabolism (11) | |||
| 100 | F6HXC8 | Phospholipase D (11, 1, 27) | 3.90 |
| 41 | F6H9P9 | Biotin carboxylase 1, chloroplastic (11) | −2.57 |
| 70 | F6HLJ7 | Enoyl-[acyl-carrier-protein] reductase [NADH] 1, chloroplastic (11) | −2.13 |
| 182 | D7STF0 | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ (11) | −3.22 |
| 211 | D7T4I1 | Biotin carboxyl carrier protein of acetyl-CoA carboxylase 2, chloroplastic (11) | −4.35 |
| 105 | D7TAP7 | 3-oxoacyl-[acyl-carrier-protein] reductase 2, chloroplastic (11, 26) | −12.24 |
| 173 | A5C5V3 | Dihydroceramide fatty acyl 2-hydroxylase FAH1 (11) | −32.25 |
| 188 | D7TVI4 | 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic (11) | d. |
| N and amino acid metabolism (12, 13) | |||
| 27 | F6GWF3 | Serine hydroxymethyltransferase (13, 1) | 1.49 |
| 36 | P51119 | Glutamine synthetase cytosolic isozyme 2 (12) | −1.54 |
| 191 | A5ATW2 | Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase (13) | −1.79 |
| 116 | A5CAL1 | Glyoxylate/hydroxypyruvate reductase A HPR2 (13, 1, 26) | −2.59 |
| Secondary metabolism (16) | |||
| 256 | F6HHQ2 | Nitrile-specifier protein 5 (16) | New |
| 261 | F6I080 | Bifunctional nitrilase/nitrile hydratase NIT4B-like (16, 26) | New |
| 161 | D7TRU0 | Cinnamyl alcohol dehydrogenase 8 (16) | −1.50 |
| 215 | D7U0Q6 | Probable plastid-lipid-associated protein 1, chloroplastic (16, 17, 26) | −1.78 |
| 93 | A0A0M5I8D0 | Flavanone 3-hydroxylase (16) | −1.78 |
| 155 | O22519 | Chalcone synthase (16) | −14.75 |
| 250 | S5FNE7 | Protein SRG1 (16, 17) | d. |
| Hormone metabolism (17) | |||
| 85 | D7TUK1 | Perakine reductase isoform X1 (17, 26) | 5.71 |
| 207 | A3QRC1 | Allene oxide cyclase 2, chloroplastic (17, 20) | −2.06 |
| 224 | F6HX49 | Gibberellin 20 oxidase 1 (17, 16, 26) | −2.39 |
| Stress (20) | |||
| 201 | Q0MYQ7 | Germin-like protein 2 (20, 15, 31) | 44.61 |
| 51 | A3QRB7 | Chitinase class I basic (20) | 2.47 |
| 214 | F6HZ19 | Germin-like protein subfamily 1 member 17 (20, 12, 27, 34) | 2.46 |
| 96 | A5ASS2 | Thaumatin (20) | 1.60 |
| 128 | A5BAY1 | Germin-like protein 9-2 (20, 15) | −1.42 |
| 165 | D7TKM8 | Putative germin-like protein 2-1 (20, 12, 27, 30, 34) | −2.42 |
| Redox (21) | |||
| 222 | F6HTY5 | Superoxide dismutase [Cu-Zn] (21) | 2.11 |
| 59 | A5C8L8 | Monodehydroascorbate reductase 5, mitochondrial isoform X1 (21) | 1.84 |
| 29 | E0CR49 | Protein disulfide-isomerase (21) | 1.81 |
| 12 | Q8S568 | Catalase (21) | 1.69 |
| 44 | A5JPK7 | Monodehydroascorbate reductase (21) | 1.43 |
| 45 | F6HDW4 | GDP-mannose 3,5-epimerase (10, 21) | −1.61 |
| Miscellaneous enzyme families (26) | |||
| 153 | A7NY33 | Peroxidase 4 (26, 16) | New |
| 156 | Q69D51 | Beta-1,3-glucanase (26) | New |
| 94 | F6GWS4 | Peroxidase (26) | 4.88 |
| 22 | Q9M563 | Beta-1,3-glucanase (26) | 1.90 |
| 90 | A5AKD8 | Peptidyl-prolyl cis-trans isomerase (26) | 1.44 |
| 30 | F6HR72 | Glutathione S-transferase (26) | −1.66 |
| 139 | F6GT84 | Glutathione S-transferase U9 (26) | −1.97 |
| 104 | D7TE48 | Soluble epoxide hydrolase (26) | −2.41 |
| 134 | A5AZ36 | Glutathione S-transferase U25 (26) | −2.85 |
| 249 | F6HL77 | Tropinone reductase homolog At1g07440 (34, 2) | −29.52 |
| 190 | D7T8G2 | Purple acid phosphatase (26) | −38.33 |
| 170 | F6HZD8 | Short-chain dehydrogenase reductase 3b-like (26) | d. |
| DNA/RNA (27,28) | |||
| 121 | A5B427 | Cyclase (28) | 1,41 |
| 223 | A5AXT8 | Pentatricopeptide repeat-containing protein At5g66520-like (27, 26) | d. |
| Protein (29) | |||
| 277 | F6H2W4 | Aspartate--tRNA ligase 2, cytoplasmic (29) | New |
| 266 | D7SIX7 | Serine/threonine-prot. phosphatase 2A 65 kDa regulatory sub. A isoform (29) | 5.60 |
| 149 | E0CTI4 | 26S proteasome non-ATPase regulatory subunit 2 homolog (29) | 2.92 |
| 56 | F6H7H1 | Procardosin-A (29) | −1.40 |
| 157 | F6GZY7 | Cysteine proteinase RD21A (29) | −1.54 |
| 135 | A5BIH7 | Peptidase_S10 domain-containing protein (29) | −1.82 |
| 195 | F6H1D7 | Carboxypeptidase (29) | −2.08 |
| 186 | D7SHN2 | Heme-binding protein 2-like (29, 19) | −2.63 |
| 48 | F6GWA8 | Chaperonin 60 subunit alpha 2, chloroplastic (29, 1) | −3.30 |
| 122 | D7SLM9 | Chaperonin 60 subunit beta 2, chloroplastic (29, 1) | −3.48 |
| Cell/signaling/development (30, 31, 33) | |||
| 218 | A5ARE0 | Glutelin type-A 1-like (33, 28) | 6.08 |
| 89 | A5BXT5 | Guanosine nucleotide diphosphate dissociation inhibitor (30) | 2.05 |
| 131 | D7T2N7 | Late embryogenesis abundant protein Lea14-A (33) | 1.52 |
| 73 | A5AKB1 | Plastid-lipid-associated protein 1, chloroplastic (31) | −1.85 |
| 202 | D7T9L8 | Coatomer subunit delta (31) | −2.21 |
| Transport (34) | |||
| 246 | F6H9B5 | Glucose-6-phosphate/phosphate translocator 1, chloroplastic | 4.78 |
| 133 | F6HXK4 | Plasma membrane ATPase (34) | 2.32 |
| 210 | F6HS56 | Potassium channel beta, putative (34, 17) | −1.59 |
| Others (15, 18) | |||
| 227 | F6H2P8 | Protein DJ-1 homolog B (18) | 6.81 |
| Not assigned (35) | |||
| 87 | F6HHU9 | Uncharacterized protein (35) | 16.51 |
| 37 | F6HUS6 | Uncharacterized protein (35) | 1.78 |
| 136 | F6HJB9 | Uncharacterized protein (35) | −2.30 |
| 238 | F6H0J2 | DPP6 N-terminal domain-like protein (35) | −2.53 |
| 219 | A5B729 | Uncharacterized protein | d. |
| # | Accession | Name (f.c.) | ΔSS/C |
|---|---|---|---|
| Carbon and energy metabolism (1-9, 25) | |||
| 89 | A5C6H7 | Sucrose synthase (2) | 10.91 |
| 235 | D7SHY3 | Betaine aldehyde dehydrogenase 1, chloroplastic (5, 16) | 8.62 |
| 64 | F6HFF7 | Phosphoglucomutase, cytoplasmic 1 (4) | 3.46 |
| 227 | A5BF93 | Succinate–CoA ligase [ADP-forming] subunit beta, mitochondrial (8) | 3.44 |
| 152 | F6HGZ9 | Sucrose synthase (2) | 3.11 |
| 133 | D7T300 | ATP synthase subunit O, mitochondrial (9) | 3.06 |
| 259 | F6HHP3 | Glucose-6-phosphate 1-dehydrogenase (7, 30) | 3.01 |
| 130 | F6H9T6 | Succinate-semialdehyde dehydrogenase, mitochondrial (8) | 2.45 |
| 18 | A0A1Z2THL4 | NADP-dependent malic enzyme (8) | 2.24 |
| 187 | F6I5I7 | Methylenetetrahydrofolate reductase (25) | 2.24 |
| 91 | F6I6W5 | Pyrophosphate–fructose 6-phosphate 1-phosphotransferase subunit alpha (4) | 1.97 |
| 172 | D7SPF1 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial (8) | 1.85 |
| 97 | C5DB68 | Pyruvate kinase (4, 11) | 1.60 |
| 31 | F6HGH4 | 6-phosphogluconate dehydrogenase, decarboxylating (7) | 1.53 |
| 23 | F6I0H8 | UTP–glucose-1-phosphate uridylyl transferase (4) | 1.44 |
| 42 | D7T0U8 | Glyceraldehyde-3-phosphate dehydrogenase (1) | −1.56 |
| 45 | F6HFL6 | Fructose-bisphosphate aldolase (4) | −1.72 |
| 13 | A5B8T3 | Fructokinase (2) | −1.86 |
| 72 | D7TJI9 | Pyruvate decarboxylase 1 (5) | −1.90 |
| 132 | A5BEM8 | Oxidoreductase GLYR1 (7) | −2.60 |
| 100 | D7TR81 | Pyrophosphate–fructose 6-phosphate 1-phosphotransferase subunit beta (4) | −2.96 |
| 46 | F6I134 | Triosephosphate isomerase, chloroplastic (1, 21) | −3.06 |
| 207 | C0KY93 | Leucoanthocyanidin dioxygenase (1, 7, 13, 16, 17, 26) | −3.29 |
| 192 | F6HI27 | Pyruvate dehydrogenase E1 component subunit beta-2, chloroplastic (1, 8, 11) | −3.75 |
| 265 | F6GY71 | Pyruvate decarboxylase 1 (5) | −18.60 |
| Cell Wall (10) | |||
| 233 | F6I390 | Pectinesterase (10) | 5.87 |
| 57 | F6I6R4 | Beta-xylosidase/alpha-L-arabinofuranosidase 2 (10) | 1.44 |
| Lipid metabolism (11) | |||
| 111 | F6HXC8 | Phospholipase D (11,1) | 3.51 |
| 73 | A5AS18 | Putative quinone reductase (11) | −1.42 |
| 60 | F6HLJ7 | Enoyl-[acyl-carrier-protein] reductase [NADH] 1, chloroplastic isoform X1 (11) | −1.68 |
| 65 | F6H9P9 | Biotin carboxylase 1, chloroplastic () | −1.76 |
| 168 | D7STF0 | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ (11) | −1.94 |
| 203 | D7T4I1 | Biotin carboxyl carrier protein of acetyl-CoA carboxylase 2, chloroplastic (11) | −2.98 |
| 88 | D7TAP7 | 3-oxoacyl-[acyl-carrier-protein] reductase 2, chloroplastic (11, 26) | −3.19 |
| 125 | A5C5V3 | Dihydroceramide fatty acyl 2-hydroxylase FAH1 (11) | −7.02 |
| 141 | D7TVI4 | 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic (11) | d. |
| N and amino acid metabolism (12, 13) | |||
| 231 | F6HQA7 | Nitrite reductase 1 (12) | 4.11 |
| 25 | A5C5K3 | Adenosyl homocysteinase (13) | 1.43 |
| 139 | A5CAL1 | Glyoxylate/hydroxypyruvate reductase A HPR2 (13, 1, 26) | −1.45 |
| 155 | A5ATW2 | Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase (13) | −1.61 |
| 26 | P51119 | Glutamine synthetase cytosolic isozyme 2 (12) | −1.72 |
| 160 | D7SW04 | Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase isoform X2 (13) | −1.80 |
| Secondary metabolism (16) | |||
| 101 | A0A0M5I8D0 | Flavanone 3-hydroxylase (16, 17, 29) | −1.63 |
| 50 | F6H775 | Class I-like SAM-binding methyltransferase superfamily (16, 26) | −4.95 |
| 257 | F6GX19 | Isopentenyl-diphosphate Delta-isomerase I (16) | −6.85 |
| 213 | A5BVM7 | O-methyltransferase YrrM (16) | −7.23 |
| Hormone metabolism (17) | |||
| 241 | A5B174 | Perakine reductase (17) | 10.76 |
| Stress (20) | |||
| 54 | A3QRB7 | Chitinase class I basic (20) | 2.16 |
| 43 | D7TS57 | Chaperonin CPN60-2, mitochondrial (20, 29) | 1.82 |
| 163 | D7TKM8 | Germin-like protein 2-1 (12, 20, 27, 30, 34) | 1.59 |
| 110 | D7T8R2 | MLP-like protein 34 (20) | 1.54 |
| 264 | D7TNE5 | Hypersensitive-induced response protein 1-like isoform X1 (20) | −1.64 |
| 252 | A5AKX5 | SOUL heme-binding protein (19, 29) | −1.71 |
| 71 | F6GTP0 | Heat shock protein, putative (20, 27) | −1.73 |
| 251 | D7UE33 | PLAT domain-containing protein 3-like (20) | −1.81 |
| Redox (21) | |||
| 16 | Q8S568 | Catalase (21) | 2.61 |
| Miscellaneous enzyme families (26) | |||
| 263 | F6GUF3 | Peroxidase 53 (26) | New |
| 218 | F6HIC8 | Dienelactone hydrolase (26) | 1.42 |
| 48 | F6I4V3 | ADP-ribosylation factor 1-like 2 (26, 33) | −1.51 |
| 266 | D7TUE8 | Glycosyltransferase (26) | −1.56 |
| 86 | F6GT84 | Glutathione S-transferase U9 (26) | −1.66 |
| 36 | F6HR72 | Glutathione S-transferase (26) | −1.93 |
| 67 | A0A024FS61 | Polyphenol_oxidase (26) | −1.90 |
| 107 | A5AZ36 | Glutathione S-transferase U25 (26) | −3.16 |
| 234 | D7TE48 | Soluble epoxide hydrolase (26) | −4.66 |
| 167 | F6HZD8 | Short-chain dehydrogenase reductase 3b-like (26) | −10.74 |
| 188 | D7T8G2 | Purple acid phosphatase (26) | −54.69 |
| DNA/RNA (27,28) | |||
| 225 | A5ARE0 | Glutelin type-A 1-like (28, 33) | 7.40 |
| 28 | D7TCM7 | UPI00053F79C7 (RNA helicase) (27) | 1.48 |
| 118 | A5B427 | Cyclase (28) | 1.41 |
| Protein (29) | |||
| 260 | D7SIX7 | Serine/threonine-prot. phosphatase 2A 65 kDa regulatory sub. A isoform (29) | New |
| 105 | F6I455 | Probable elongation factor 1-gamma 2 (29) | 4.34 |
| 30 | F6H4T7 | Elongation factor 2 (29) | 2.16 |
| 80 | A5BUU4 | 40S ribosomal protein SA (29) | 1.95 |
| 134 | E0CV68 | Importin subunit beta-1 (29) | 1.52 |
| 121 | F6GZY7 | Cysteine proteinase RD21A (29, 34) | −1.47 |
| 165 | E0CR38 | Proteasome subunit beta type (29) | −1.71 |
| 63 | F6H7H1 | Procardosin-A (29) | −2.12 |
| 185 | F6H1D7 | Carboxypeptidase (29) | −5.53 |
| 221 | A5BIH7 | Peptidase_S10 domain-containing protein (29) | −7.81 |
| 197 | A5AKL4 | Cysteine protease, putative (29, 34) | d. |
| 201 | D7TW90 | Cucumsin (29) | d. |
| Cell/signaling/development (30, 31, 33) | |||
| 224 | D7SJV3 | Clathrin heavy chain (31) | New |
| 12 | A5BTZ8 | Annexin (31) | 1.43 |
| 8 | F6I0I5 | Actin-8 (31) | −1.42 |
| 190 | D7T9L8 | Coatomer subunit delta (31) | −2.20 |
| Transport (34) | |||
| 39 | F6HBF2 | ADP, ATP carrier protein, mitochondrial (34, 2) | 3.37 |
| Not assigned (35) | |||
| 228 | D7T9K4 | Uncharacterized protein (35) | 3.04 |
| 226 | D7SJF5 | Uncharacterized protein (35) | 1.96 |
| 174 | A5B729 | Uncharacterized protein (35) | −21.21 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prinsi, B.; Failla, O.; Scienza, A.; Espen, L. Root Proteomic Analysis of Two Grapevine Rootstock Genotypes Showing Different Susceptibility to Salt Stress. Int. J. Mol. Sci. 2020, 21, 1076. https://doi.org/10.3390/ijms21031076
Prinsi B, Failla O, Scienza A, Espen L. Root Proteomic Analysis of Two Grapevine Rootstock Genotypes Showing Different Susceptibility to Salt Stress. International Journal of Molecular Sciences. 2020; 21(3):1076. https://doi.org/10.3390/ijms21031076
Chicago/Turabian StylePrinsi, Bhakti, Osvaldo Failla, Attilio Scienza, and Luca Espen. 2020. "Root Proteomic Analysis of Two Grapevine Rootstock Genotypes Showing Different Susceptibility to Salt Stress" International Journal of Molecular Sciences 21, no. 3: 1076. https://doi.org/10.3390/ijms21031076
APA StylePrinsi, B., Failla, O., Scienza, A., & Espen, L. (2020). Root Proteomic Analysis of Two Grapevine Rootstock Genotypes Showing Different Susceptibility to Salt Stress. International Journal of Molecular Sciences, 21(3), 1076. https://doi.org/10.3390/ijms21031076








