Human Pluripotent Stem Cells in Reproductive Science—A Comparison of Protocols Used to Generate and Define Male Germ Cells from Pluripotent Stem Cells
Abstract
1. Introduction
2. Frequently Used Cell Lines in the Course of In Vitro Differentiation Approaches
3. Variations in Culture Methods
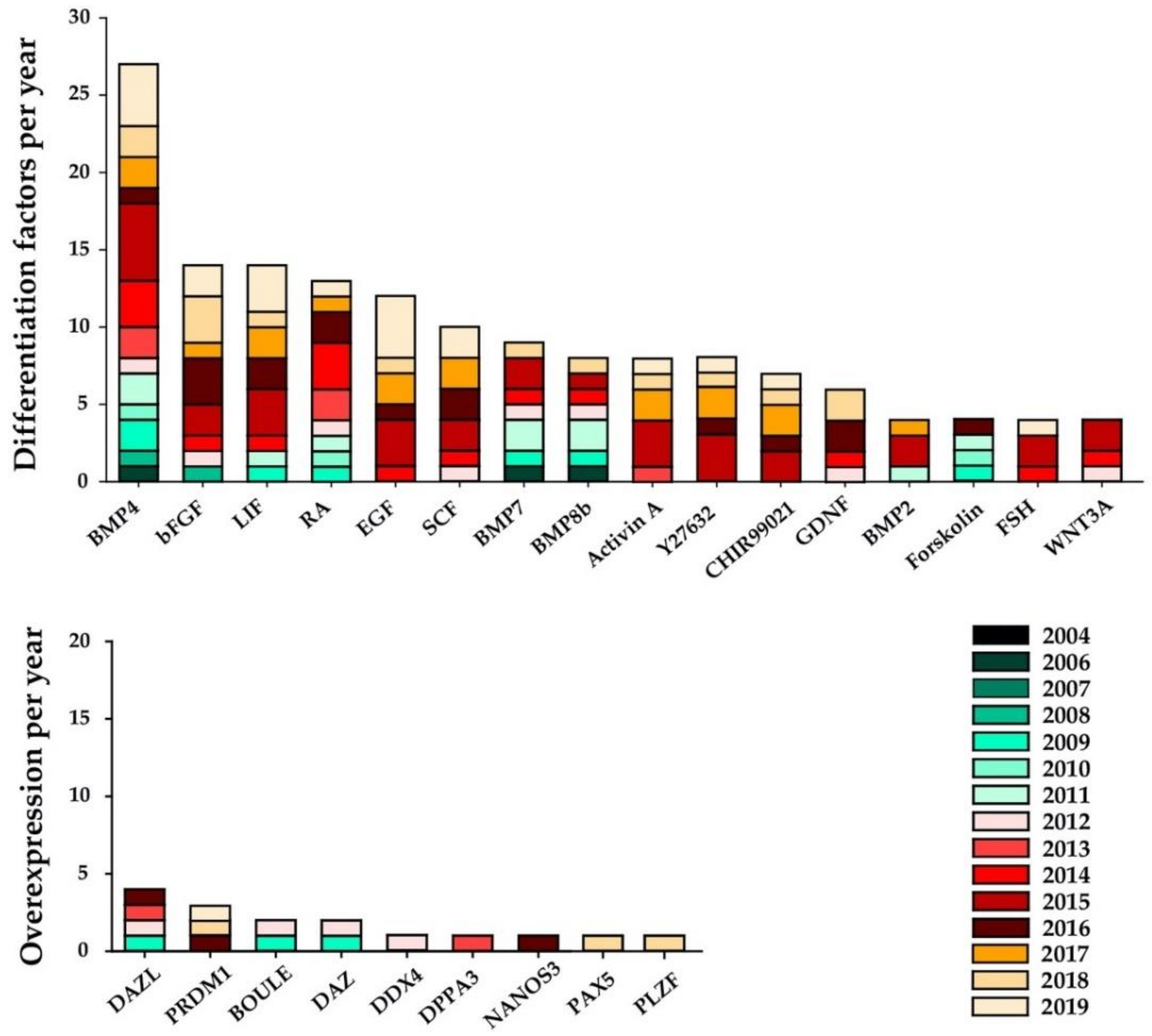
4. Variation in Growth Factors Supporting Differentiation
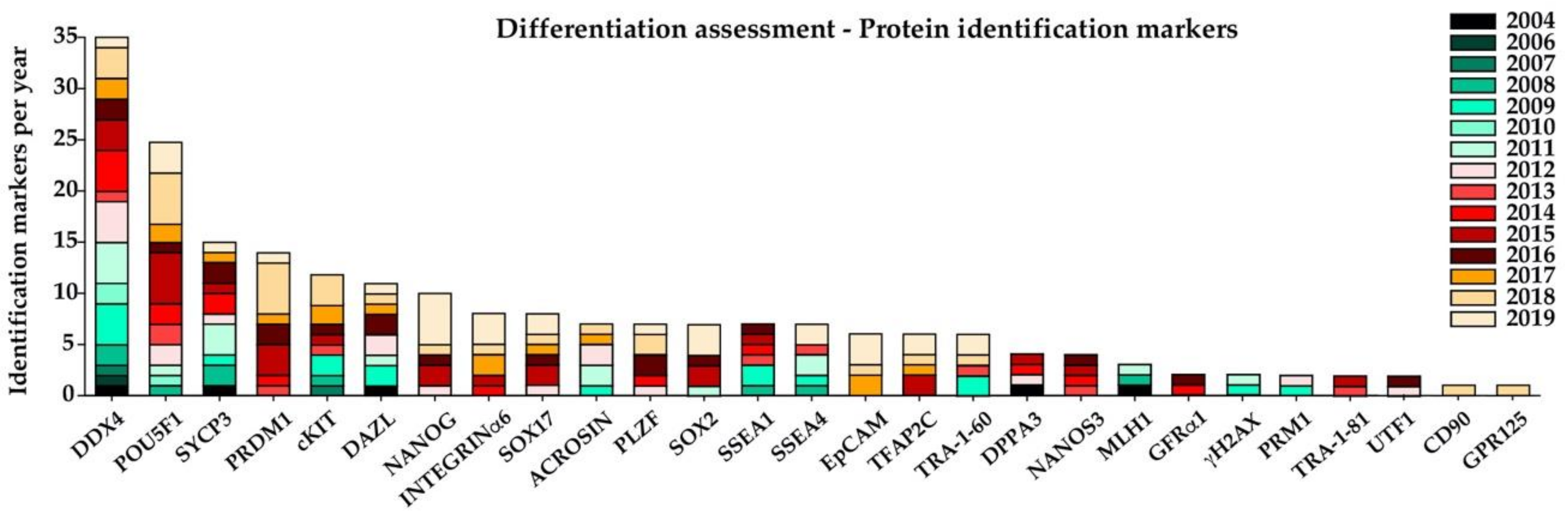
5. Assessment of In Vitro Germ-Cell Differentiation
6. Understanding of In Vitro Spermatogenesis through Single Cell RNA Sequencing
7. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, S.V.; Jones, M.J. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Shiohara, M.; Tai, T.; Nagao, K.; Nakajima, K.; Kobayashi, H. Derivation of integration-free iPSCs from a Klinefelter syndrome patient. Reprod. Med. Biol. 2016, 15, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Li, C.; Gu, J.; Tang, F.; Li, C.; Li, P.; Ping, P.; Yang, S.; Li, Z.; Jin, Y. Aberrant gene expression profiles in pluripotent stem cells induced from fibroblasts of a Klinefelter syndrome patient. J. Biol. Chem. 2012, 287, 38970–38979. [Google Scholar] [CrossRef] [PubMed]
- Panula, S.; Kurek, M.; Kumar, P.; Albalushi, H.; Padrell Sanchez, S.; Damdimopoulou, P.; Olofsson, J.I.; Hovatta, O.; Lanner, F.; Stukenborg, J.B. Human induced pluripotent stem cells from two azoospermic patients with Klinefelter syndrome show similar X chromosome inactivation behavior to female pluripotent stem cells. Hum. Reprod. 2019, 34, 2297–2310. [Google Scholar] [CrossRef]
- Schuster, J.; Sobol, M.; Fatima, A.; Khalfallah, A.; Laan, L.; Anderlid, B.M.; Nordgren, A.; Dahl, N. Mowat-Wilson syndrome: Generation of two human iPS cell lines (UUIGPi004A and UUIGPi005A) from siblings with a truncating ZEB2 gene variant. Stem Cell Res. 2019, 39, 101518. [Google Scholar] [CrossRef]
- Schuster, J.; Fatima, A.; Sobol, M.; Norradin, F.H.; Laan, L.; Dahl, N. Generation of three human induced pluripotent stem cell (iPSC) lines from three patients with Dravet syndrome carrying distinct SCN1A gene mutations. Stem Cell Res. 2019, 39, 101523. [Google Scholar] [CrossRef]
- Zhao, Y.; Ye, S.; Liang, D.; Wang, P.; Fu, J.; Ma, Q.; Kong, R.; Shi, L.; Gong, X.; Chen, W.; et al. In Vitro Modeling of Human Germ Cell Development Using Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 509–523. [Google Scholar] [CrossRef]
- Vermeulen, M.; Giudice, M.G.; Del Vento, F.; Wyns, C. Role of stem cells in fertility preservation: Current insights. Stem Cells Cloning 2019, 12, 27–48. [Google Scholar] [CrossRef]
- Rombaut, C.; Mertes, H.; Heindryckx, B.; Goossens, E. Human in vitro spermatogenesis from pluripotent stem cells: In need of a stepwise differentiation protocol? Mol. Hum. Reprod. 2018, 24, 47–54. [Google Scholar] [CrossRef]
- Fang, F.; Li, Z.; Zhao, Q.; Li, H.; Xiong, C. Human induced pluripotent stem cells and male infertility: An overview of current progress and perspectives. Hum. Reprod. 2018, 33, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.T.; Bodnar, M.S.; Fox, M.; Rodriquez, R.T.; Abeyta, M.J.; Firpo, M.T.; Pera, R.A. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum. Mol. Genet. 2004, 13, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Easley, C.A.; Phillips, B.T.; McGuire, M.M.; Barringer, J.M.; Valli, H.; Hermann, B.P.; Simerly, C.R.; Rajkovic, A.; Miki, T.; Orwig, K.E.; et al. Direct differentiation of human pluripotent stem cells into haploid spermatogenic cells. Cell Rep. 2012, 2, 440–446. [Google Scholar] [CrossRef]
- Handel, M.A.; Eppig, J.J.; Schimenti, J.C. Applying “gold standards” to in-vitro-derived germ cells. Cell 2014, 157, 1257–1261. [Google Scholar] [CrossRef] [PubMed]
- De Felici, M. Origin, Migration, and Proliferation of Human Primordial Germ Cells. In Oogenesis; Springer: London, UK, 2013; pp. 19–37. [Google Scholar]
- Messerschmidt, D.M.; Knowles, B.B.; Solter, D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014, 28, 812–828. [Google Scholar] [CrossRef] [PubMed]
- Hiller, M.; Liu, C.; Blumenthal, P.D.; Gearhart, J.D.; Kerr, C.L. Bone Morphogenetic Protein 4 Mediates Human Embryonic Germ Cell Derivation. Stem Cells Dev. 2011, 20, 351–361. [Google Scholar] [CrossRef]
- Li, Y.; Hong, W.X.; Lan, B.; Xu, X.; Liu, Y.; Kong, L.; Li, Y.; Zhou, S.; Liu, Y.; Feng, R.; et al. PDGF mediates derivation of human embryonic germ cells. Differentiation 2013, 86, 141–148. [Google Scholar] [CrossRef]
- Sasaki, K.; Yokobayashi, S.; Nakamura, T.; Okamoto, I.; Yabuta, Y.; Kurimoto, K.; Ohta, H.; Moritoki, Y.; Iwatani, C.; Tsuchiya, H.; et al. Robust In Vitro Induction of Human Germ Cell Fate from Pluripotent Stem Cells. Cell Stem Cell 2015, 17, 178–194. [Google Scholar] [CrossRef]
- Chen, D.; Sun, N.; Hou, L.; Kim, R.; Faith, J.; Aslanyan, M.; Tao, Y.; Zheng, Y.; Fu, J.; Liu, W.; et al. Human Primordial Germ Cells Are Specified from Lineage-Primed Progenitors. Cell Rep. 2019, 29, 4568–4582. [Google Scholar] [CrossRef]
- Irie, N.; Weinberger, L.; Tang, W.W.; Kobayashi, T.; Viukov, S.; Manor, Y.S.; Dietmann, S.; Hanna, J.H.; Surani, M.A. SOX17 is a critical specifier of human primordial germ cell fate. Cell 2015, 160, 253–268. [Google Scholar] [CrossRef]
- Taketo, T. The role of sex chromosomes in mammalian germ cell differentiation: Can the germ cells carrying X and Y chromosomes differentiate into fertile oocytes? Asian J. Androl. 2015, 17, 360–366. [Google Scholar] [CrossRef] [PubMed]
- She, Z.Y.; Yang, W.X. Sry and SoxE genes: How they participate in mammalian sex determination and gonadal development? Semin. Cell Dev. Biol. 2017, 63, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, D.; Yang, J.X.; Thomas, P. Mammalian sex determination and gonad development. Curr. Top. Dev. Biol. 2013, 106, 89–121. [Google Scholar]
- Manku, G.; Culty, M. Mammalian gonocyte and spermatogonia differentiation: Recent advances and remaining challenges. Reproduction 2015, 149, R139–R157. [Google Scholar] [CrossRef] [PubMed]
- Fox, N.; Damjanov, I.; Martinez-Hernandez, A.; Knowles, B.B.; Solter, D. Lmmunohistochemical Localization of the Early Embryonic Antigen (SSEA-1) in Postimplantation Mouse Embryos and Fetal and Adult Tissues. Dev. Biol. 1981, 83, 391–398. [Google Scholar] [CrossRef]
- Koopman, P.; Muensterberg, M.; Capel, B.; Vivian, N.; Lovell-Badge, R. Expression of candidate sex-determining gene during mouse testis differentiation. Nature 1990, 348, 450–452. [Google Scholar] [CrossRef]
- Anderson, R.; Copeland, T.K.; Schöler, H.; Heasman, J.; Wylie, C. The onset of germ cell migration in the mouse embryo. Mech. Dev. 2000, 91, 61–68. [Google Scholar] [CrossRef]
- Pesce, M.; Schoeler, H.R. Oct-4: Control of Totipotency and Germline Determination. Mol. Reprod. Dev. 2000, 55, 452–457. [Google Scholar] [CrossRef]
- Toyooka, Y.; Tsunekawa, N.; Takahashi, Y.; Matsui, Y.; Satoh, M.; Noce, T. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech. Dev. 2000, 93, 139–149. [Google Scholar] [CrossRef]
- Sakai, Y.; Nakamura, T.; Okamoto, I.; Gyobu-Motani, S.; Ohta, H.; Yabuta, Y.; Tsukiyama, T.; Iwatani, C.; Tsuchiya, H.; Ema, M.; et al. Induction of the Germ-Cell Fate from Pluripotent Stem Cells in Cynomolgus Monkeys. Biol. Reprod. 2019. [Google Scholar] [CrossRef]
- McKinnell, C.; Mitchell, R.T.; Morris, K.; Anderson, R.A.; Kelnar, C.J.; Wallace, W.H.; Sharpe, R.M. Perinatal germ cell development and differentiation in the male marmoset (Callithrix jacchus): Similarities with the human and differences from the rat. Hum. Reprod. 2013, 28, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Mouka, A.; Tachdjian, G.; Dupont, J.; Drevillon, L.; Tosca, L. In Vitro Gamete Differentiation from Pluripotent Stem Cells as a Promising Therapy for Infertility. Stem Cells Dev. 2016, 25, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Geens, M.; Sermon, K.D.; Van de Velde, H.; Tournaye, H. Sertoli cell-conditioned medium induces germ cell differentiation in human embryonic stem cells. J. Assist. Reprod. Genet. 2011, 28, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Aflatoonian, B.; Ruban, L.; Jones, M.; Aflatoonian, R.; Fazeli, A.; Moore, H.D. In vitro post-meiotic germ cell development from human embryonic stem cells. Hum. Reprod. 2009, 24, 3150–3159. [Google Scholar] [CrossRef]
- Panula, S.; Medrano, J.V.; Kee, K.; Bergstrom, R.; Ha, N.N.; Byers, B.; Wilson, K.D.; Wu, J.C.; Simon, C.; Hovatta, O.; et al. Human germ cell differentiation from fetal- and adult-derived induced pluripotent stem cells. Hum. Mol. Genet. 2011, 20, 752–762. [Google Scholar] [CrossRef]
- Medrano, J.V.; Ramathal, C.; Nguyen, H.N.; Simon, C.; Pera, R.A.R. Divergent RNA-Binding Proteins, DAZL and VASA, Induce Meiotic Progression in Human Germ Cells Derived In Vitro. Stem Cells 2012, 30, 441–451. [Google Scholar] [CrossRef]
- Medrano, J.V.; Martinez-Arroyo, A.M.; Miguez, J.M.; Moreno, I.; Martinez, S.; Quinonero, A.; Diaz-Gimeno, P.; Marques-Mari, A.I.; Pellicer, A.; Remohi, J.; et al. Human somatic cells subjected to genetic induction with six germ line-related factors display meiotic germ cell-like features. Sci. Rep. 2016, 6, 24956. [Google Scholar] [CrossRef]
- Merkle, F.T.; Ghosh, S.; Kamitaki, N.; Mitchell, J.; Avior, Y.; Mello, C.; Kashin, S.; Mekhoubad, S.; Ilic, D.; Charlton, M.; et al. Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature 2017, 545, 229–233. [Google Scholar] [CrossRef]
- Deuse, T.; Hu, X.; Agbor-Enoh, S.; Koch, M.; Spitzer, M.H.; Gravina, A.; Alawi, M.; Marishta, A.; Peters, B.; Kosaloglu-Yalcin, Z.; et al. De novo mutations in mitochondrial DNA of iPSCs produce immunogenic neoepitopes in mice and humans. Nat. Biotechnol. 2019, 37, 1137–1144. [Google Scholar] [CrossRef]
- Richards, M.; Fong, C.Y.; Bongso, A. Comparative evaluation of different in vitro systems that stimulate germ cell differentiation in human embryonic stem cells. Fertil. Steril. 2010, 93, 986–994. [Google Scholar] [CrossRef]
- Gkountela, S.; Li, Z.W.; Vincent, J.J.; Zhang, K.X.; Chen, A.; Pellegrini, M.; Clark, A.T. The ontogeny of cKIT(+) human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat. Cell Biol. 2013, 15, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Weng, S.P.; Surrey, M.W.; Danzer, H.C.; Hill, D.L.; Chen, P.C.; Wu, T.C. Chromosome abnormalities in embryos derived from microsurgical epididymal sperm aspiration and testicular sperm extraction. Taiwan J. Obstet. Gynecol. 2014, 53, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.Y.; Lin, K.I.; Hsiao, M.; Stone, L.; Chen, H.F.; Huang, Y.H.; Lin, S.P.; Ho, H.N.; Kuo, H.C. Meiotic Competent Human Germ Cell-like Cells Derived from Human Embryonic Stem Cells Induced by BMP4/WNT3A Signaling and OCT4/EpCAM (Epithelial Cell Adhesion Molecule) Selection. J. Biol. Chem. 2012, 287, 14389–14401. [Google Scholar] [CrossRef] [PubMed]
- Lin, I.Y.; Chiu, F.L.; Yeang, C.H.; Chen, H.F.; Chuang, C.Y.; Yang, S.Y.; Hou, P.S.; Sintupisut, N.; Ho, H.N.; Kuo, H.C.; et al. Suppression of the SOX2 Neural Effector Gene by PRDM1 Promotes Human Germ Cell Fate in Embryonic Stem Cells. Stem Cell Rep. 2014, 2, 189–204. [Google Scholar] [CrossRef]
- Leng, L.; Tan, Y.; Gong, F.; Hu, L.; Ouyang, Q.; Zhao, Y.; Lu, G.; Lin, G. Differentiation of primordial germ cells from induced pluripotent stem cells of primary ovarian insufficiency. Hum. Reprod. 2015, 30, 737–748. [Google Scholar] [CrossRef]
- Tilgner, K.; Atkinson, S.P.; Golebiewska, A.; Stojkovic, M.; Lako, M.; Armstrong, L. Isolation of primordial germ cells from differentiating human embryonic stem cells. Stem Cells 2008, 26, 3075–3085. [Google Scholar] [CrossRef]
- West, F.D.; Machacek, D.W.; Boyd, N.L.; Pandiyan, K.; Robbins, K.R.; Stice, S.L. Enrichment and differentiation of human germ-like cells mediated by feeder cells and basic fibroblast growth factor signaling. Stem Cells 2008, 26, 2768–2776. [Google Scholar] [CrossRef]
- Kee, K.; Angeles, V.T.; Flores, M.; Nguyen, H.N.; Pera, R.A.R. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature 2009, 462, 222–225. [Google Scholar] [CrossRef]
- Julaton, V.T.A.; Pera, R.A.R. NANOS3 function in human germ cell development. Hum. Mol. Genet. 2011, 20, 2238–2250. [Google Scholar] [CrossRef]
- Park, T.S.; Galic, Z.; Conway, A.E.; Lindgren, A.; Van Handel, B.J.; Magnusson, M.; Richter, L.; Teitell, M.A.; Mikkola, H.K.A.; Lowry, W.E.; et al. Derivation of Primordial Germ Cells from Human Embryonic and Induced Pluripotent Stem Cells Is Significantly Improved by Coculture with Human Fetal Gonadal Cells. Stem Cells 2009, 27, 783–795. [Google Scholar] [CrossRef]
- Albalushi, H.; Kurek, M.; Karlsson, L.; Landreh, L.; Kjartansdottir, K.R.; Soder, O.; Hovatta, O.; Stukenborg, J.B. Laminin 521 Stabilizes the Pluripotency Expression Pattern of Human Embryonic Stem Cells Initially Derived on Feeder Cells. Stem Cells Int. 2018, 2018, 7127042. [Google Scholar] [CrossRef] [PubMed]
- Bier, E.; De Robertis, E.M. Embryo Development. BMP gradients: A paradigm for morphogen-mediated developmental patterning. Science 2015, 348, aaa5838. [Google Scholar] [CrossRef] [PubMed]
- Warmflash, A.; Sorre, B.; Etoc, F.; Siggia, E.D.; Brivanlou, A.H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods 2014, 11, 847–854. [Google Scholar] [CrossRef] [PubMed]
- West, F.D.; Roche-Rios, M.I.; Abraham, S.; Rao, R.R.; Natrajan, M.S.; Bacanamwo, M.; Stice, S.L. KIT ligand and bone morphogenetic protein signaling enhances human embryonic stem cell to germ-like cell differentiation. Hum. Reprod. 2010, 25, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Kee, K.; Gonsalves, J.M.; Clark, A.T.; Pera, R.A. Bone morphogenetic proteins induce germ cell differentiation from human embryonic stem cells. Stem Cells Dev. 2006, 15, 831–837. [Google Scholar] [CrossRef]
- Chen, D.; Liu, W.; Lukianchikov, A.; Hancock, G.V.; Zimmerman, J.; Lowe, M.G.; Kim, R.; Galic, Z.; Irie, N.; Surani, M.A.; et al. Germline competency of human embryonic stem cells depends on eomesodermin. Biol. Reprod. 2017, 97, 850–861. [Google Scholar] [CrossRef]
- Duggal, G.; Heindryckx, B.; Warrier, S.; Taelman, J.; Van der Jeught, M.; Deforce, D.; Lopes, S.C.D.; De Sutter, P. Exogenous supplementation of Activin A enhances germ cell differentiation of human embryonic stem cells. Mol. Hum. Reprod. 2015, 21, 410–423. [Google Scholar] [CrossRef][Green Version]
- Duggal, G.; Heindryckx, B.; Warrier, S.; O’Leary, T.; Van der Jeught, M.; Lierman, S.; Vossaert, L.; Deroo, T.; Deforce, D.; Lopes, S.; et al. Influence of Activin A Supplementation During Human Embryonic Stem Cell Derivation on Germ Cell Differentiation Potential. Stem Cells Dev. 2013, 22, 3141–3155. [Google Scholar] [CrossRef]
- Lim, J.J.; Shim, M.S.; Lee, J.E.; Lee, D.R. Three-Step Method for Proliferation and Differentiation of Human Embryonic Stem Cell (hESC)-Derived Male Germ Cells. PLoS ONE 2014, 9, e90454. [Google Scholar] [CrossRef]
- Kjartansdottir, K.R.; Reda, A.; Panula, S.; Day, K.; Hultenby, K.; Soder, O.; Hovatta, O.; Stukenborg, J.B. A Combination of Culture Conditions and Gene Expression Analysis Can Be Used to Investigate and Predict hES Cell Differentiation Potential towards Male Gonadal Cells. PLoS ONE 2015, 10, e0144029. [Google Scholar] [CrossRef]
- Sugawa, F.; Arauzo-Bravo, M.J.; Yoon, J.; Kim, K.P.; Aramaki, S.; Wu, G.M.; Stehling, M.; Psathaki, O.E.; Hubner, K.; Scholer, H.R. Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. EMBO J. 2015, 34, 1009–1024. [Google Scholar] [CrossRef] [PubMed]
- Von Meyenn, F.; Berrens, R.V.; Andrews, S.; Santos, F.; Collier, A.J.; Krueger, F.; Osorno, R.; Dean, W.; Rugg-Gunn, P.J.; Reik, W. Comparative Principles of DNA Methylation Reprogramming during Human and Mouse In Vitro Primordial Germ Cell Specification. Dev. Cell 2016, 39, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.F.; Jan, P.S.; Kuo, H.C.; Wu, F.C.; Lan, C.W.; Huang, M.C.; Chien, C.L.; Ho, H.N. Granulosa cells and retinoic acid co-treatment enrich potential germ cells from manually selected Oct4-EGFP expressing human embryonic stem cells. Reprod. Biomed. Online 2014, 29, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Panula, S.; Reda, A.; Stukenborg, J.B.; Ramathal, C.; Sukhwani, M.; Albalushi, H.; Edsgard, D.; Nakamura, M.; Soder, O.; Orwig, K.E.; et al. Over Expression of NANOS3 and DAZL in Human Embryonic Stem Cells. PLoS ONE 2016, 11, e0165268. [Google Scholar] [CrossRef] [PubMed]
- Eguizabal, C.; Montserrat, N.; Vassena, R.; Barragan, M.; Garreta, E.; Garcia-Quevedo, L.; Vidal, F.; Giorgetti, A.; Veiga, A.; Belmonte, J.C.I. Complete Meiosis from Human Induced Pluripotent Stem Cells. Stem Cells 2011, 29, 1186–1195. [Google Scholar] [CrossRef]
- Wongtrakoongate, P.; Jones, M.; Gokhale, P.J.; Andrews, P.W. STELLA Facilitates Differentiation of Germ Cell and Endodermal Lineages of Human Embryonic Stem Cells. PLoS ONE 2013, 8, e56893. [Google Scholar] [CrossRef]
- Liu, X.M.; Yue, J.; Xu, B.; Hu, J.; Ren, X.L.; Liu, Q.; Zhu, G.J. Retinoic acid improve germ cell differentiation from human embryonic stem cells. Iran. J. Reprod. Med. 2013, 11, 905–912. [Google Scholar]
- Tran, N.D.; Kissner, M.; Subramanyam, D.; Parchem, R.J.; Laird, D.J.; Blelloch, R.H. A miR-372/let-7 Axis Regulates Human Germ Versus Somatic Cell Fates. Stem Cells 2016, 34, 1985–1991. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, H.J.; Wu, J.; Li, N.; Hua, J.L. Characterization of embryonic stem-like cells derived from HEK293T cells through miR302/367 expression and their potentiality to differentiate into germ-like cells. Cytotechnology 2014, 66, 729–740. [Google Scholar] [CrossRef][Green Version]
- Kjartansdottir, K.R.; Gabrielsen, A.; Reda, A.; Soder, O.; Bergstrom-Tengzelius, R.; Andersen, C.Y.; Hovatta, O.; Stukenborg, J.B.; Fedder, J. Differentiation of stem cells upon deprivation of exogenous FGF2: A general approach to study spontaneous differentiation of hESCs in vitro. Syst. Biol. Reprod. Med. 2012, 58, 330–338. [Google Scholar] [CrossRef]
- Martyn, I.; Kanno, T.Y.; Ruzo, A.; Siggia, E.D.; Brivanlou, A.H. Self-organization of a human organizer by combined Wnt and Nodal signalling. Nature 2018, 558, 132–135. [Google Scholar] [CrossRef]
- Yoney, A.; Etoc, F.; Ruzo, A.; Carroll, T.; Metzger, J.J.; Martyn, I.; Li, S.; Kirst, C.; Siggia, E.D.; Brivanlou, A.H. WNT signaling memory is required for ACTIVIN to function as a morphogen in human gastruloids. Elife 2018, 7, e38279. [Google Scholar] [CrossRef]
- Yan, L.; Della Coletta, L.; Powell, K.L.; Shen, J.; Thames, H.; Aldaz, C.M.; MacLeod, M.C. Activation of the canonical Wnt/beta-catenin pathway in ATF3-induced mammary tumors. PLoS ONE 2011, 6, e16515. [Google Scholar]
- Singh, D.; Paduch, D.A.; Schlegel, P.N.; Orwig, K.E.; Mielnik, A.; Bolyakov, A.; Wright, W.W. The production of glial cell line-derived neurotrophic factor by human sertoli cells is substantially reduced in sertoli cell-only testes. Hum. Reprod. 2017, 32, 1108–1117. [Google Scholar] [CrossRef]
- Spinnler, K.; Kohn, F.M.; Schwarzer, U.; Mayerhofer, A. Glial cell line-derived neurotrophic factor is constitutively produced by human testicular peritubular cells and may contribute to the spermatogonial stem cell niche in man. Hum. Reprod. 2010, 25, 2181–2187. [Google Scholar] [CrossRef]
- Mansour, A.A.; Gafni, O.; Weinberger, L.; Zviran, A.; Ayyash, M.; Rais, Y.; Krupalnik, V.; Zerbib, M.; Amann-Zalcenstein, D.; Maza, I.; et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature 2012, 488, 409–413. [Google Scholar] [CrossRef]
- Stukenborg, J.B.; Kjartansdottir, K.R.; Reda, A.; Colon, E.; Albersmeier, J.P.; Soder, O. Male germ cell development in humans. Horm. Res. Paediatr. 2014, 81, 2–12. [Google Scholar] [CrossRef]
- Muciaccia, B.; Boitani, C.; Berloco, B.P.; Nudo, F.; Spadetta, G.; Stefanini, M.; De Rooij, D.G.; Vicini, E. Novel stage classification of human spermatogenesis based on acrosome development. Biol. Reprod. 2013, 89, 60. [Google Scholar] [CrossRef]
- Baccetti, B.; Renieri, T.; Rosati, F.; Selmi, M.G.; Casanova, S. Further Observations on the Morphogenesis of the Round Headed Human Spermatozoa. Andrologia 1977, 9, 255–264. [Google Scholar] [CrossRef]
- De Jong, J.; Stoop, H.; Gillis, A.J.; Van Gurp, R.J.; Van de Geijn, G.J.; Boer, M.; Hersmus, R.; Saunders, P.T.; Anderson, R.A.; Oosterhuis, J.W.; et al. Differential expression of SOX17 and SOX2 in germ cells and stem cells has biological and clinical implications. J. Pathol. 2008, 215, 21–30. [Google Scholar] [CrossRef]
- Guo, F.; Yan, L.; Guo, H.; Li, L.; Hu, B.; Zhao, Y.; Yong, J.; Hu, Y.; Wang, X.; Wei, Y.; et al. The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell 2015, 161, 1437–1452. [Google Scholar] [CrossRef]
- Sohni, A.; Tan, K.; Song, H.W.; Burow, D.; De Rooij, D.G.; Laurent, L.; Hsieh, T.C.; Rabah, R.; Hammoud, S.S.; Vicini, E.; et al. The Neonatal and Adult Human Testis Defined at the Single-Cell Level. Cell Rep. 2019, 26, 1501–1517. [Google Scholar] [CrossRef]
- Guo, J.; Grow, E.J.; Mlcochova, H.; Maher, G.J.; Lindskog, C.; Nie, X.; Guo, Y.; Takei, Y.; Yun, J.; Cai, L.; et al. The adult human testis transcriptional cell atlas. Cell Res. 2018, 28, 1141–1157. [Google Scholar] [CrossRef]
- Hermann, B.P.; Cheng, K.; Singh, A.; Roa-De La Cruz, L.; Mutoji, K.N.; Chen, I.C.; Gildersleeve, H.; Lehle, J.D.; Mayo, M.; Westernstroer, B.; et al. The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Rep. 2018, 25, 1650–1667. [Google Scholar] [CrossRef]
- Oliver, E.; Stukenborg, J.B. Rebuilding the human testis in vitro. Andrology 2019. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurek, M.; Albalushi, H.; Hovatta, O.; Stukenborg, J.-B. Human Pluripotent Stem Cells in Reproductive Science—A Comparison of Protocols Used to Generate and Define Male Germ Cells from Pluripotent Stem Cells. Int. J. Mol. Sci. 2020, 21, 1028. https://doi.org/10.3390/ijms21031028
Kurek M, Albalushi H, Hovatta O, Stukenborg J-B. Human Pluripotent Stem Cells in Reproductive Science—A Comparison of Protocols Used to Generate and Define Male Germ Cells from Pluripotent Stem Cells. International Journal of Molecular Sciences. 2020; 21(3):1028. https://doi.org/10.3390/ijms21031028
Chicago/Turabian StyleKurek, Magdalena, Halima Albalushi, Outi Hovatta, and Jan-Bernd Stukenborg. 2020. "Human Pluripotent Stem Cells in Reproductive Science—A Comparison of Protocols Used to Generate and Define Male Germ Cells from Pluripotent Stem Cells" International Journal of Molecular Sciences 21, no. 3: 1028. https://doi.org/10.3390/ijms21031028
APA StyleKurek, M., Albalushi, H., Hovatta, O., & Stukenborg, J.-B. (2020). Human Pluripotent Stem Cells in Reproductive Science—A Comparison of Protocols Used to Generate and Define Male Germ Cells from Pluripotent Stem Cells. International Journal of Molecular Sciences, 21(3), 1028. https://doi.org/10.3390/ijms21031028




