Abstract
It is essential to develop and discover alternative eco-friendly antibacterial agents due to the emergence of multi-drug-resistant microorganisms. In this study, we isolated and characterized a novel bacterium named Terrabacter humi MAHUQ-38T, utilized for the eco-friendly synthesis of silver nanoparticles (AgNPs) and the synthesized AgNPs were used to control multi-drug-resistant microorganisms. The novel strain was Gram stain positive, strictly aerobic, milky white colored, rod shaped and non-motile. The optimal growth temperature, pH and NaCl concentration were 30 °C, 6.5 and 0%, respectively. Based on 16S rRNA gene sequence, strain MAHUQ-38T belongs to the genus Terrabacter and is most closely related to several Terrabacter type strains (98.2%–98.8%). Terrabacter humi MAHUQ-38T had a genome of 5,156,829 bp long (19 contigs) with 4555 protein-coding genes, 48 tRNA and 5 rRNA genes. The culture supernatant of strain MAHUQ-38T was used for the eco-friendly and facile synthesis of AgNPs. The transmission electron microscopy (TEM) image showed the spherical shape of AgNPs with a size of 6 to 24 nm, and the Fourier transform infrared (FTIR) analysis revealed the functional groups responsible for the synthesis of AgNPs. The synthesized AgNPs exhibited strong anti-bacterial activity against multi-drug-resistant pathogens, Escherichia coli and Pseudomonas aeruginosa. Minimal inhibitory/bactericidal concentrations against E. coli and P. aeruginosa were 6.25/50 and 12.5/50 μg/mL, respectively. The AgNPs altered the cell morphology and damaged the cell membrane of pathogens. This study encourages the use of Terrabacter humi for the ecofriendly synthesis of AgNPs to control multi-drug-resistant microorganisms.
1. Introduction
The genus Terrabacter was first proposed by Collins et al. [1] through the transfer of Pimelobacter tumescens to Terrabacter tumescens. The Terrabacter genus comprises 10 species with valid published names at the time of writing (https://lpsn.dsmz.de/genus/terrabacter). Species of the genus are characterized as rod or coccus shape, Gram-positive, catalase-positive, obligately aerobic bacteria and have C14:0 iso and C15:0 iso as the major fatty acids and MK- 8(H4) as the predominant respiratory quinone. The members of this genus have DNA G + C content from 70 to 73 mol% [2,3]. The members of this genus have been isolated from different sources including soil, air, stone, flowerbeds, etc. [1,2,3,4,5,6]. In the present study, we isolated a novel species of the genus Terrabacter, Terrabacter humi sp. nov., designated as MAHUQ-38T, isolated from the soil sample of a grape garden.
Nanotechnology is broadly classified into different sectors, including chemical, physical and biogenic [7]. Among these three methods, researchers are more interested in biogenic methods because of their various advantages over the other processes [8]. Both physical and chemical methods have many drawbacks, including the use of toxic chemicals, synthesis of harmful byproducts, they require enormous energy and pressure, etc. [9,10]. The biogenic method of nanoparticle (NP) synthesis is not only ecofriendly but also the biosynthesized NPs have many good properties, such as optical, chemical and photoelectrical properties that make them diverse for industrial applications [11]. The ecofriendly synthesized NPs have different biomedical applications, such as antibacterial agents, magnetic resonance imaging, carriers for targeted drug delivery, gene therapy, biosensors, cancer treatment, etc. [12].
For the ecofriendly synthesis of NPs, a number of microorganisms such as bacteria, algae, fungi, etc., have been isolated from various sources, including foods, soil, plants, etc. [13,14,15,16]. Among all of these microorganisms, bacteria-mediated NP synthesis is the favored one, because of their easy genetic manipulation and handling, such as, Paenibacillus anseongense, Bacillus subtilis, Bacillus methylotrophicus, Sporosarcina koreensis, and Microvirga rosea [10,12,13,14,17]. Ecofriendly synthesis of various metal NPs has gained considerable interest in different sectors of research and development. Among various metal NPs, silver nanoparticles (AgNPs) have achieved considerable attention, due to their strong antimicrobial, anticancer, catalytic, optical and electrical properties [14,18]. AgNPs are being used as an antimicrobial agent to control pathogens, an anticancer agent to treat different types of cancers, probes for bioimaging, an anti-inflammatory agent and targeted drug delivery system [14,16,18]. AgNPs are also being considered for the utilization in numerous industrial products including hand gel, wound dressings, medical catheter coverings, footwear, etc., due to their strong antimicrobial properties [18,19].
The emergence of multi-drug-resistant microorganisms is a serious concern for human health. So, it is essential to find out the alternative antimicrobial agent to control the multi-drug-resistant microorganisms. Ecofriendly synthesized AgNPs could be a potential candidate in this regard. The present study is an attempt of isolation and characterization of novel bacterial species T. humi MAHUQ-38T and utilization of this novel species for ecofriendly, facile synthesis of AgNPs and further subjected to characterization and evaluation of antibacterial efficacy against multi-drug-resistant human pathogens.
2. Results and Discussion
2.1. Phenotypic and Biochemical Characteristics
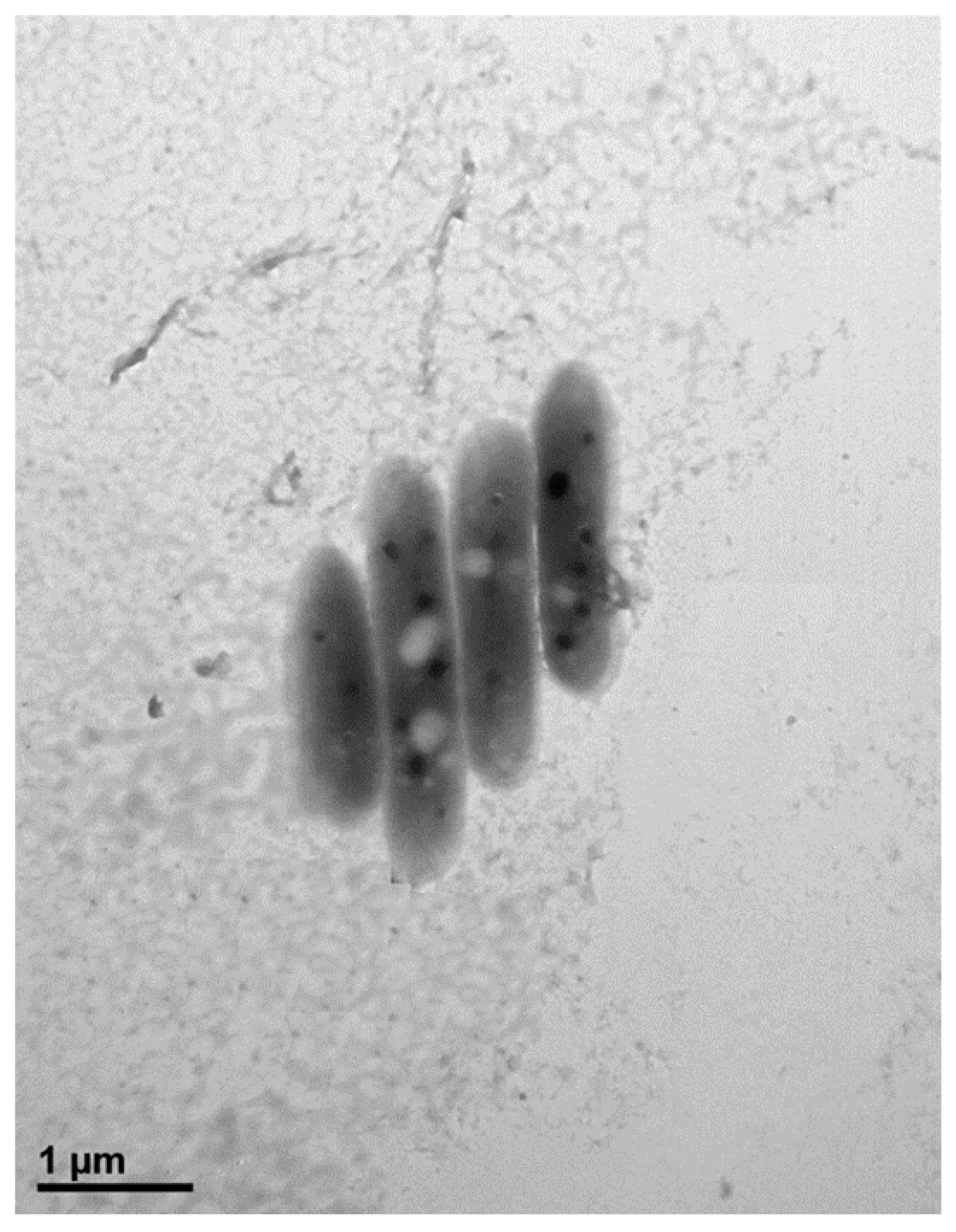
Cells of strain MAHUQ-38T were Gram positive, strictly aerobic, non-motile, and rod shaped with 0.3–0.7 μm wide and 1.6–2.9 μm long (Figure 1). Colonies of MAHUQ-38T grown on Reasoner’s 2A (R2A) agar medium were spherical, milky white colored and 0.1–0.5 mm in diameter after 48 h of incubation at 30 °C. The isolate can grow on R2A agar, Luria–Bertani (LB) agar, nutrient agar (NA) and tryptic soy agar (TSA), but did not grow on MacConkey agar. The strain was positive for catalase, but negative for oxidase. They are able to hydrolysis of casein, DNA, l-tyrosine, esculin, gelatin, Tween 80 and Tween 20, but negative for starch and urea. Phenotypic examination showed that strain MAHUQ-38T shared several traits in common with its close relatives. However, there are some morphological, physiological and biochemical differences between strain MAHUQ-38T and phylogenetically related Terrabacter species that differentiate this novel strain from related type strains (Table 1). Strain MAHUQ-38T could be clearly differentiated from the phylogenetically most closely relative, T. tumescens DSM 20308T by the ability to hydrolyse a-glucosidase; the presence of alkaline phosphatase, valine arylamidase, cystine arylamidase, a-glucosidase and a-galactosidase activities. Physiological characteristics of strain MAHUQ-38T are summarized in the species description and in Table 1.
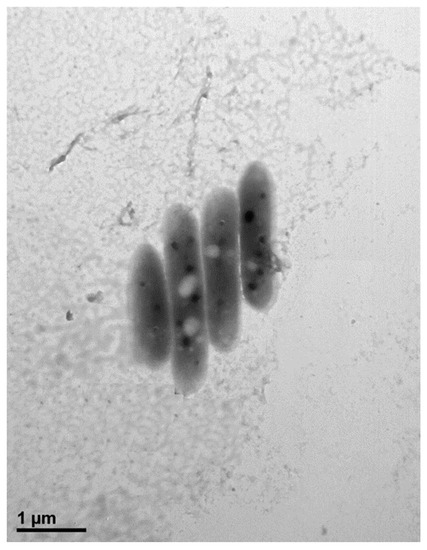
Figure 1.
Transmission electron micrograph of cells of Terrabacter humi MAHUQ-38T after negative staining with uranyl acetate, Bar, 1.0 μm.

Table 1.
Differential characteristics of Terrabacter humi MAHUQ-38T and phylogenetically closely related Terrabacter species. Strains: 1, T. humi MAHUQ-38T; 2, T. tumescens KACC 20535T; 3, T. terrae KACC 11642T; 4, T. terrigena KCTC 19602T; 5, T. lapilli LR-26T; 6, T. ginsengihumi KACC 19444T and 7, T. aerolatus KACC 20556T.
2.2. 16S rRNA Gene Sequence and Phylogenetic Analysis
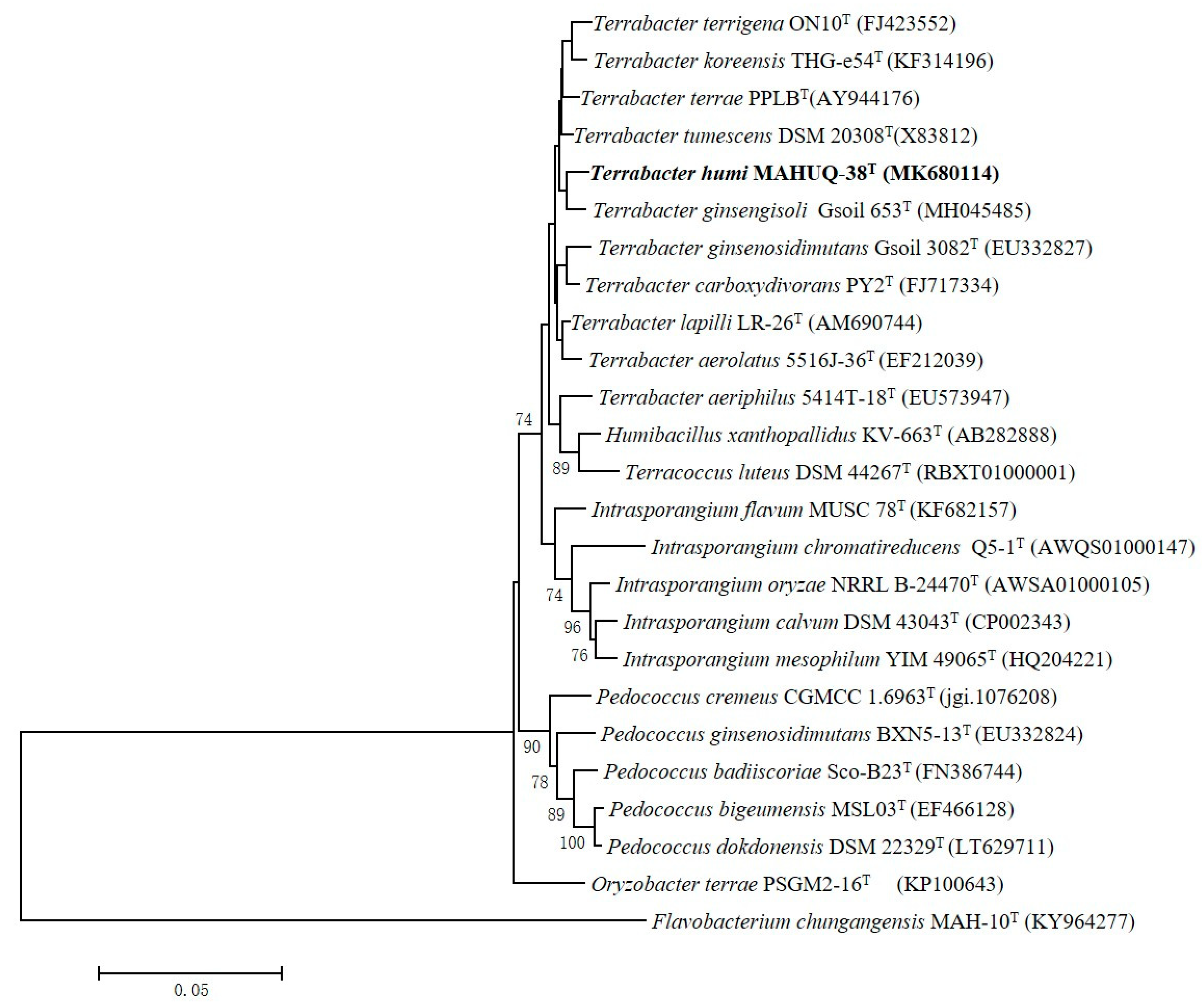
A 1456 nt long 16S rRNA gene sequence of strain MAHUQ-38T was identified and, subsequently, this sequence was used for comparative analysis (GenBank accession number, MK680114). 16S rRNA gene sequence similarity calculations indicated that strain MAHUQ-38T belongs to the genus Terrabacter and was closely related to T. tumescens DSM 20308T (98.8%), T. terrae PPLBT (98.6%), T. terrigena ON10T (98.5%), T. lapilli LR-26T (98.4%), T. ginsengihumi Gsoil 653T (98.4%) and T. aerolatus 5516J-36T (98.3%). Phylogenetic analysis using the neighbor-joining method on the basis of 16S rRNA gene sequences also showed that the strain MAHUQ-38T was clustered within the genus Terrabacter (Figure 2).
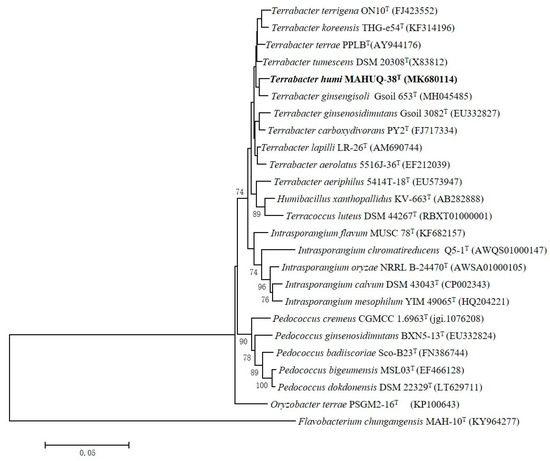
Figure 2.
Neighbor-joining (NJ) tree based on 16S rRNA gene sequence analysis showing phylogenetic relationships of strain MAHUQ-38T and members of genus Terrabacter. Bootstrap values more than 70% based on 1000 replications are shown at branching points. Scale bar, 0.05 substitutions per nucleotide position.
2.3. Draft Genome and DNA G + C Content Analysis
The draft genome of strain MAHUQ-38T contains 19 contigs with an N50 size of 910,522 bp. The total genome size was 5,156,829 bp, with an average G + C content of 70.8 mol%. Gene prediction and annotation by PGAP resulted in the identification of 4555 protein-encoding genes, 5 rRNA genes and 48 tRNA genes. The NCBI accession number for the genome sequence of strain MAHUQ-38T is JACVCU000000000. Genome sequence features of strain MAHUQ-38T are shown in Table 2. On the other hand, the closest type strain, T. tumescens JCM 1365T, contains a 4,707,860 bp long genome with an N50 size of 611,474 bp, 71.0 mol% GC, 4295 protein-encoding genes, three rRNA genes and 46 tRNA genes (http://gctype.wdcm.org/sequencing/GCM10009721). The genomic ANI (average nucleotide identity) values between strain MAHUQ-38T and the most close type strains T. tumescens JCM 1365T and T. aerolatus 5516J-36T were 83.7% and 83.4%, respectively (Table S1), which were well below (≥95–96%) to suggest a novel species. The digital DNA-DNA hybridization (dDDH) value based on the draft genomes between strain MAHUQ-38T and close relatives T. tumescens JCM 1365T and T. aerolatus 5516J-36T were 27.2% and 26.7%, respectively (Table S1), which were far below the threshold value (70%) for species delineation.

Table 2.
Genome sequence features of Terrabacter humi MAHUQ-38T.
2.4. DNA-DNA Hybridization
MAHUQ-38T showed DNA-DNA relatedness values of 38.9 ± 1.3% with T. tumescens KACC 20535T, 36.6 ± 1.0% with T. terrae KACC 11642T and 34.36 ± 1.2% with T. terrigena KCTC 19602T. Since the DNA-DNA relatedness values were much lower, strain MAHUQ-38T was determined as a novel species of the genus Terrabacter [20].
2.5. Cellular Fatty Acid, Respiratory Quinones and Polar Lipid Analysis
The major fatty acids of MAHUQ-38T were C14:0 iso (24.6%), C15:0 iso (24.4%), C16:0 iso (16.8%) and C15:0 anteiso (12.3%). Some quantitative and qualitative differences in the fatty acid profiles of close type strains that distinguished strain MAHUQ-38T from close type strains of genus Terrabacter are shown in Table 3. The major respiratory quinone of strain MAHUQ-38T was menaquinone MK-8 (H4) which is one of the common characteristics of genus Terrabacter [1,3]. Polar lipids of strain MAHUQ-38T comprised diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol and some unknown lipids (Figure S1). The similar polar lipid profiles were detected in the close relatives of the Terrabacter genus [2,3,4].

Table 3.
Cellular fatty acid composition of Terrabacter humi MAHUQ-38T and phylogenetically closely related Terrabacter species. Strains: 1, T. humi MAHUQ-38T; 2, T. tumescens KACC 20535T; 3, T. terrae KACC 11642T; 4, T. terrigena KCTC 19602T; 5, T. lapilli LR-26T; 6, T. ginsengihumi KACC 19444T and 7, T. aerolatus KACC 20556T.
2.6. Taxonomic Conclusion
The phenotypic, chemotaxonomic and biochemical features and the phylogenetic inference support the proposition that strain MAHUQ-38T represents a novel species of the genus Terrabacter, for which the name Terrabacter humi sp. nov. is proposed.
2.7. Description of Terrabacter humi sp. nov.
Terrabacter humi (hu’mi. L. gen. n. humi of/from soil)
Cells are strictly aerobic, Gram stain positive, non-motile and rod shaped and are 0.3–0.7 µm wide and 0.9–2.0 µm long. Cells grow well on R2A agar, LB agar, NA and TSA, but did not grow on MacConkey agar. Colonies on R2A agar are milky white and spherical in shape. The colony size is 0.1–5.0 mm after 48 h of growth on R2A agar. Growth occurs at 10–40 °C (optimum, 30 °C), at pH 4.5–10.0 (optimum, pH 6.5) and with 0–5% NaCl (optimum, 0%). The cells were positive for the catalase test and hydrolysis of casein, DNA, l-tyrosine, Tween 80 and Tween 20; they were negative for the oxidase test and hydrolysis of starch and urea. Flexirubin-type pigments are absent. In API 20NE tests, the cells were positive for the hydrolysis of aesculin and gelatin, nitrate reduction and β-galactosidase (PNPG) activities, assimilation of mannose, arabinose, glucose, maltose, N-acetyl-glucosamine, gluconate, and malate, but they were negative for indole production, d-glucose acidification, urease and arginine dihydrolase activities, and assimilation of mannitol, caprate, adipate, citrate and phenyl-acetate. In the API ZYM tests, valine arylamidase, acid phosphatase, esterase lipase (C8), alkaline phosphatase, C4 esterase, leucine arylamidase, trypsin, cystine arylamidase, naphthol-AS-BI-phosphohydrolase, β-glucosidase, β-galactosidase, α-glucosidase, α-galactosidase and lipase (C14) activities are present, but α-chymotrypsin, α-mannosidase, β-glucuronidase, α-fucosidase and N-acetyl-β-glucosaminidase activities are absent. The major fatty acids were C14:0 iso, C15:0 iso, C16:0 iso and C15:0 anteiso. The predominant respiratory quinone was menaquinone MK-8(H4). The main polar lipids were diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol and phosphatidylinositol. The DNA G + C content of the type strain MAHUQ-38T is 70.8 mol%.
The type strain is MAHUQ-38T (=KACC 21245T = CGMCC 1.17350T), isolated from the soil sample of a grape garden, Anseong, South Korea.
2.8. Ecofriendly Synthesis of AgNPs Using Terrabacter humi sp. nov.
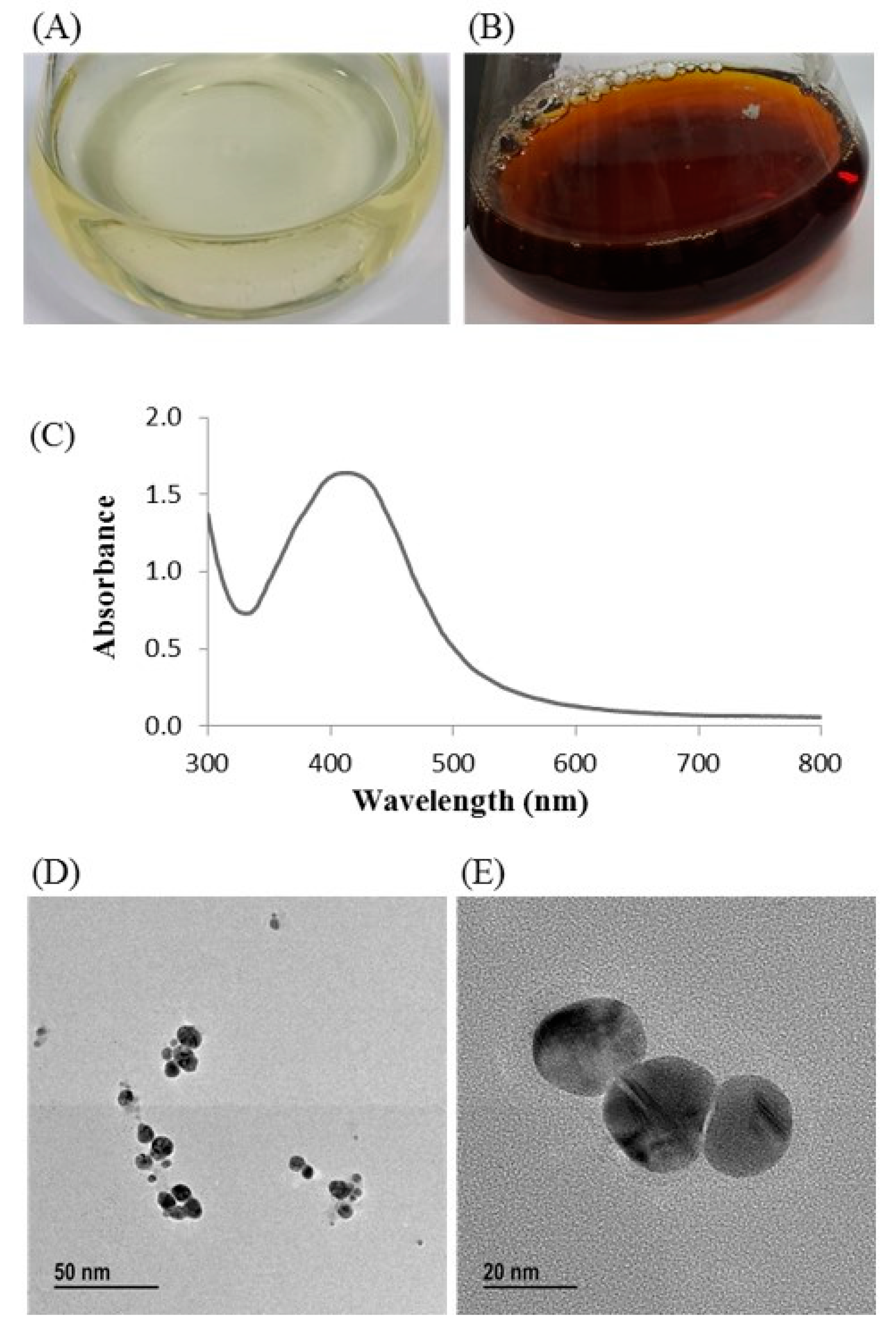
Biosynthesis of silver nanoparticles by T. humi MAHUQ-38T was confirmed by visual observation of the color of culture supernatant. The color of MAHUQ-38T culture supernatant was shifted to deep brown from watery yellow within 48 h of incubation (Figure 3B). The color change in culture supernatant indicates the synthesis of silver nanoparticles [13]. In the case of the control, there was no color change (Figure 3A). In the present study, the extracellular methodology was used for the synthesis of AgNPs using T. humi MAHUQ-38T which is simple, green, ecofriendly and cost-effective.
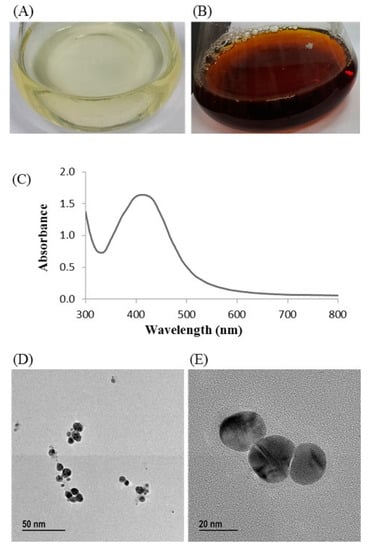
Figure 3.
Reasoner’s 2A (R2A) broth with AgNO3 as a control (A); ecofriendly synthesized silver nanoparticles (AgNPs) (B); ultraviolet–visible (UV–vis) spectra (C); and field emission-transmission electron microscopy (FE-TEM) images of ecofriendly synthesized silver nanoparticles (D,E).
2.9. Characterization of Synthesized AgNPs
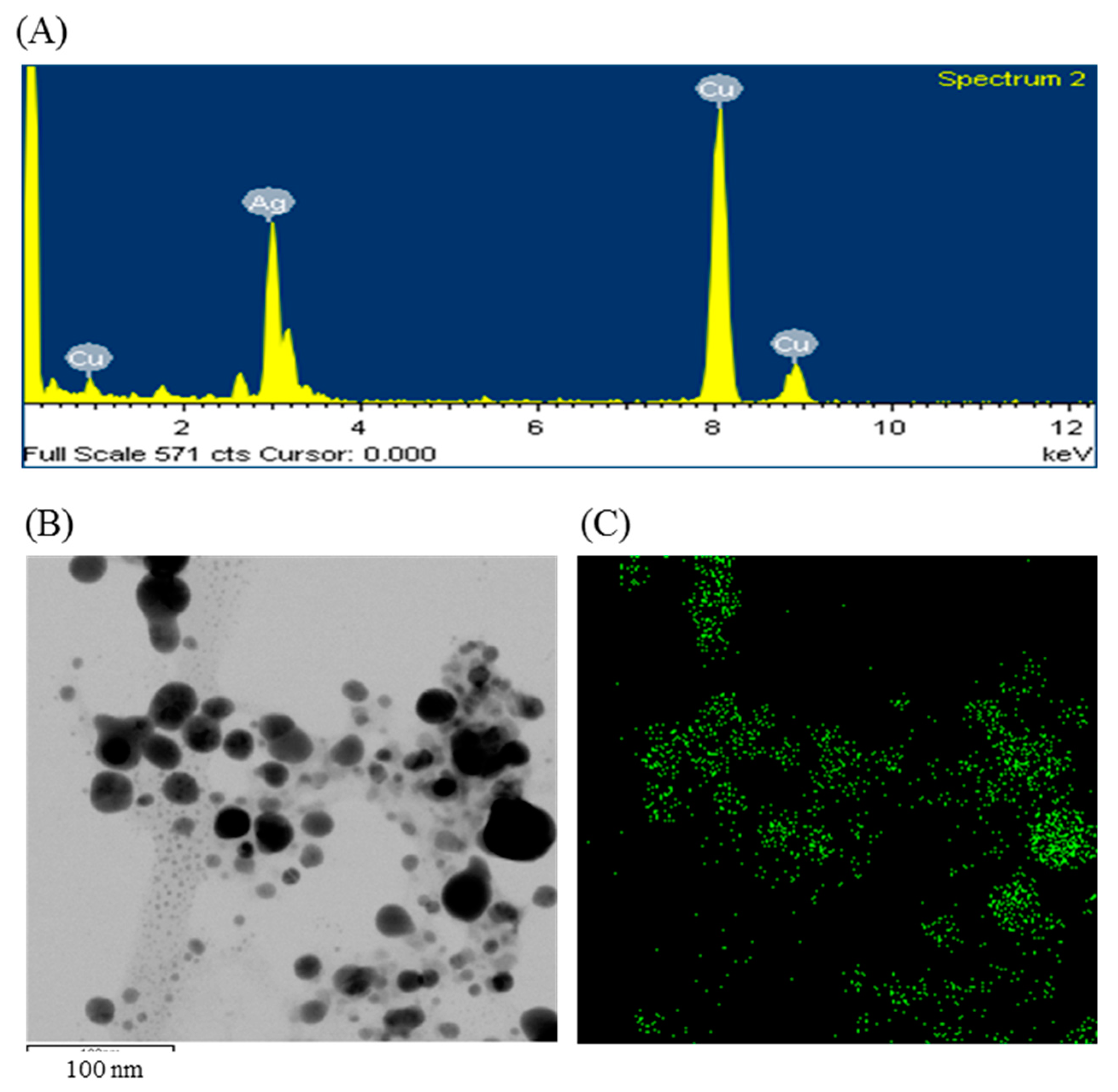
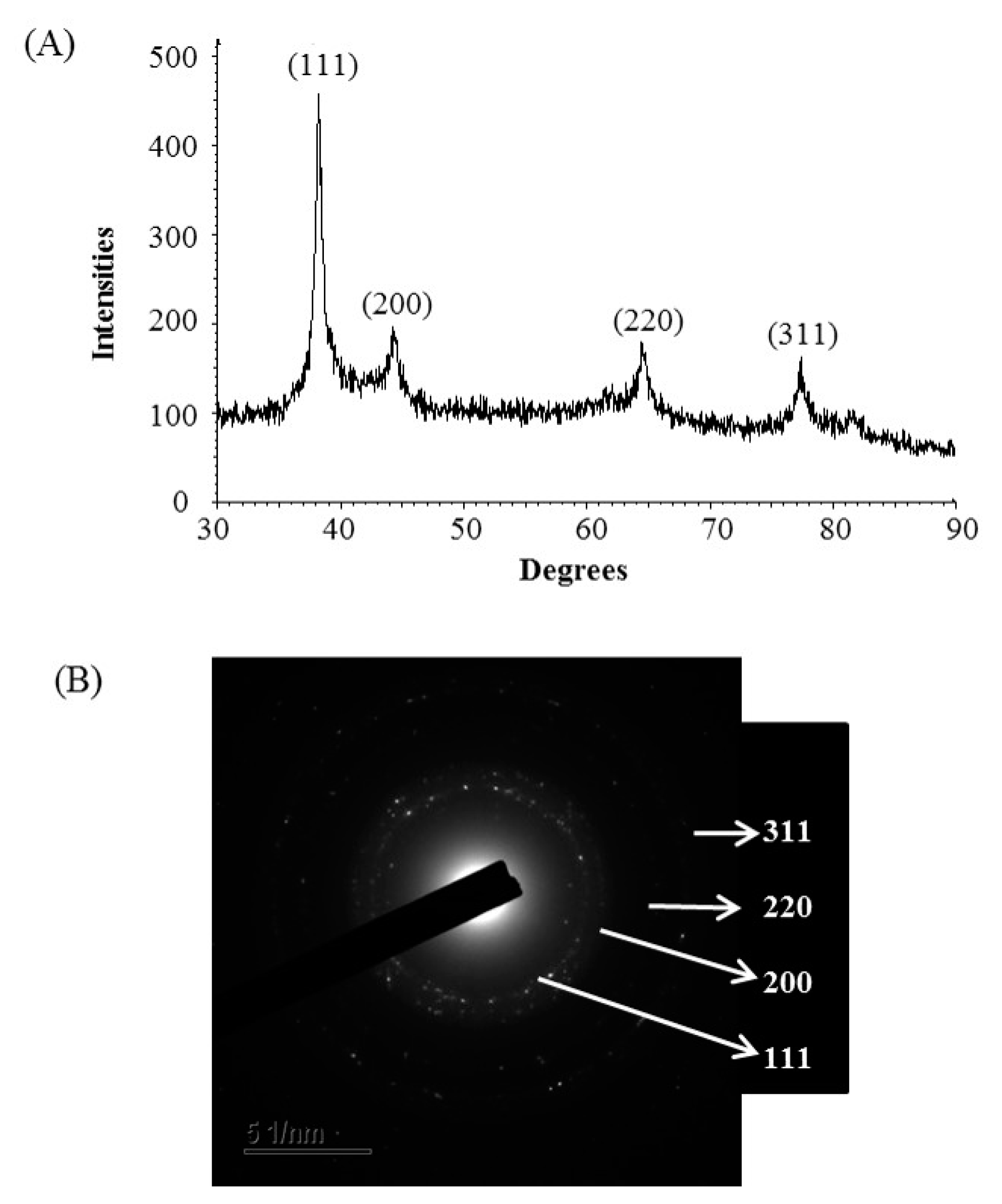
The formulation of AgNPs was assured by using an ultraviolet–visible (UV–vis) spectral scan at 300–800 nm. A strong peak at around 413 nm was observed (Figure 3C), which confirmed the synthesis of AgNPs [21,22]. FE-TEM (field emission transmission electron microscopy) analysis revealed the spherical shape of eco-friendly synthesized AgNPs and the size ranges from 6 to 24 nm (Figure 3D,E). The EDX (energy dispersive X-ray) spectrum revealed the signal for silver atoms nearly at 3 keV (Figure 4A). Through EDX spectrometer analysis, it was found that the purity of synthesized AgNPs was high (Figure 4A). Some other peaks were also found in the EDX spectrum due to the use of a copper grid in FE-TEM [10]. The elemental mapping results of synthesized silver nanoparticles exhibited the highest distribution of silver elements in the sample (Figure 4B,C). The XRD (X-ray diffraction) analysis revealed several peaks at 2θ values of 38.139°, 44.256°, 64.437° and 77.459° match the respective 111, 200, 220 and 311 lattice planes of AgNPs. These diffraction peaks may be for having the capping agents like biomolecules which are involved for stabilizing the NPs (Figure 5A). The obtained XRD results were similar with a recently reported study which showed the synthesis of silver nanoparticles using microorganisms [14]. The SAED (selected area diffraction) spectrum revealed sharp rings which indicated the crystalline structure of ecofriendly synthesized AgNPs and correlate with the following reflections at 111, 200, 220 and 311 (Figure 5B).
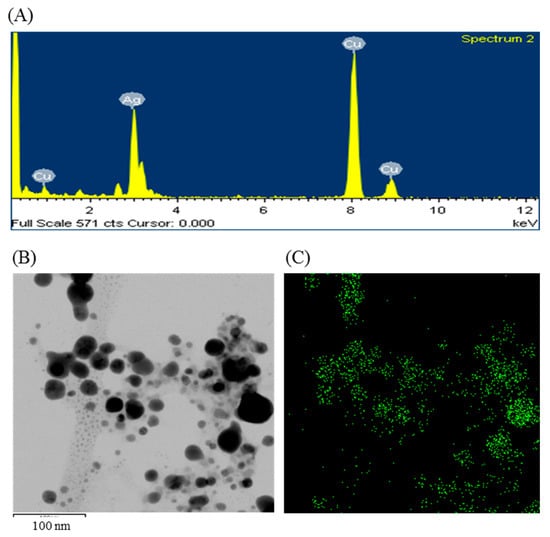
Figure 4.
Energy dispersive X-ray (EDX) spectrum of ecofriendly synthesized AgNPs (A); TEM image (electron micrograph region) used for mapping (B); and the distribution of silver in elemental mapping (C).
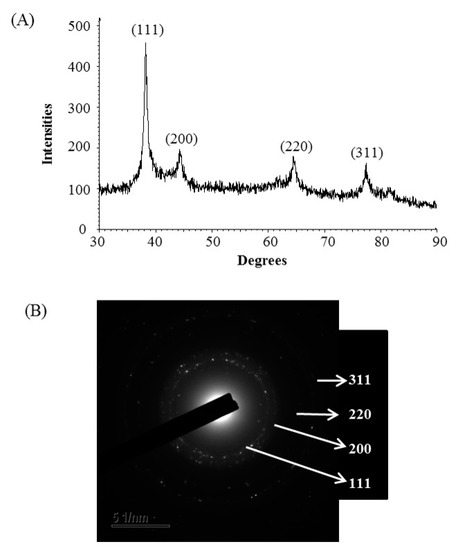
Figure 5.
X-ray diffraction pattern (A) and selected area diffraction (SAED) pattern (B) of ecofriendly synthesized AgNPs.
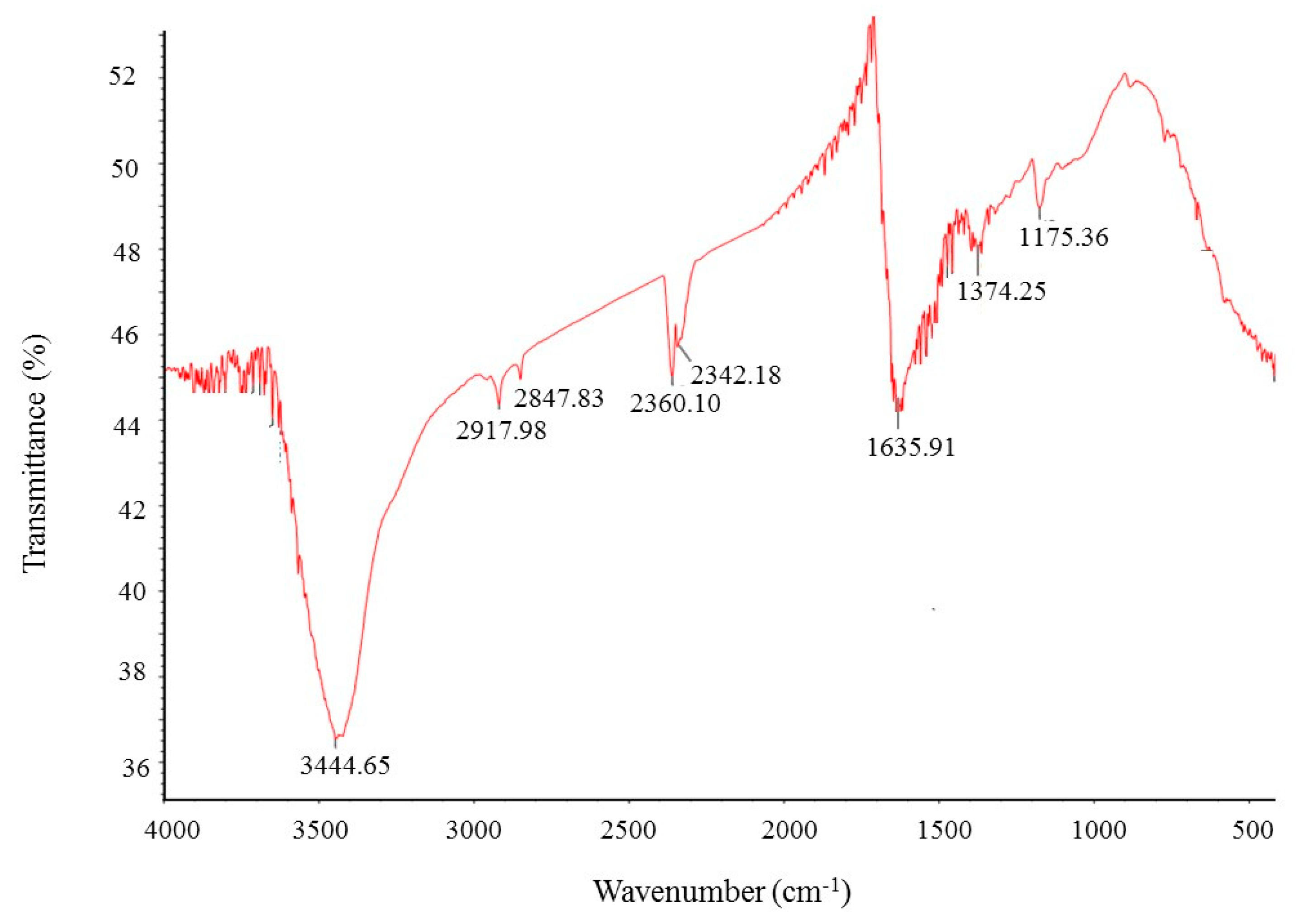
FT-IR (Fourier transform infrared) analysis was conducted to investigate the bio-active compounds, which may be responsible for the synthesis and stability of AgNPs as a capping agent. The FT-IR pattern showed different peaks throughout the entire range of analysis. FTIR analysis revealed visible bands at 3444.65, 2917.98, 2847.83, 2360.10, 2342.18, 1635.91, 1374.25 and 1175.36 cm−1 for biosynthesized silver nanoparticles (Figure 6). The band observed at 3444.65 cm−1 indicates O–H (alcohol) and/or N–H (amine) stretching. Bands observed at 2917.98 and 2847.83 cm−1 are attributed to C–H (alkane) stretching. The bands seen at 2360.10 and 2342.18 cm−1 represent O=C=O (carbonyl bond group). The band at 1635.91 cm−1 represents the N–H (amine) group. The bands observed at 1374.25 and 1175.36 cm−1 attributed to the C–H (alkane) and C–O (alcohol/ether) group, respectively. The FT-IR analysis revealed the presence of bio-active compounds, which may be responsible for the synthesis and stability of AgNPs. The present study was supported by the previous reports [23,24].
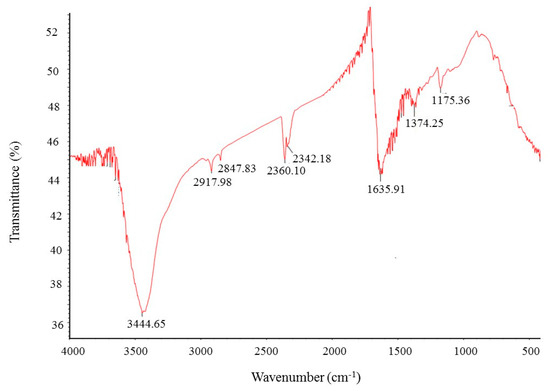
Figure 6.
Fourier transform infrared (FT-IR) spectra of ecofriendly synthesized silver nanoparticles.
2.10. Antimicrobial Activity of Synthesized AgNPs
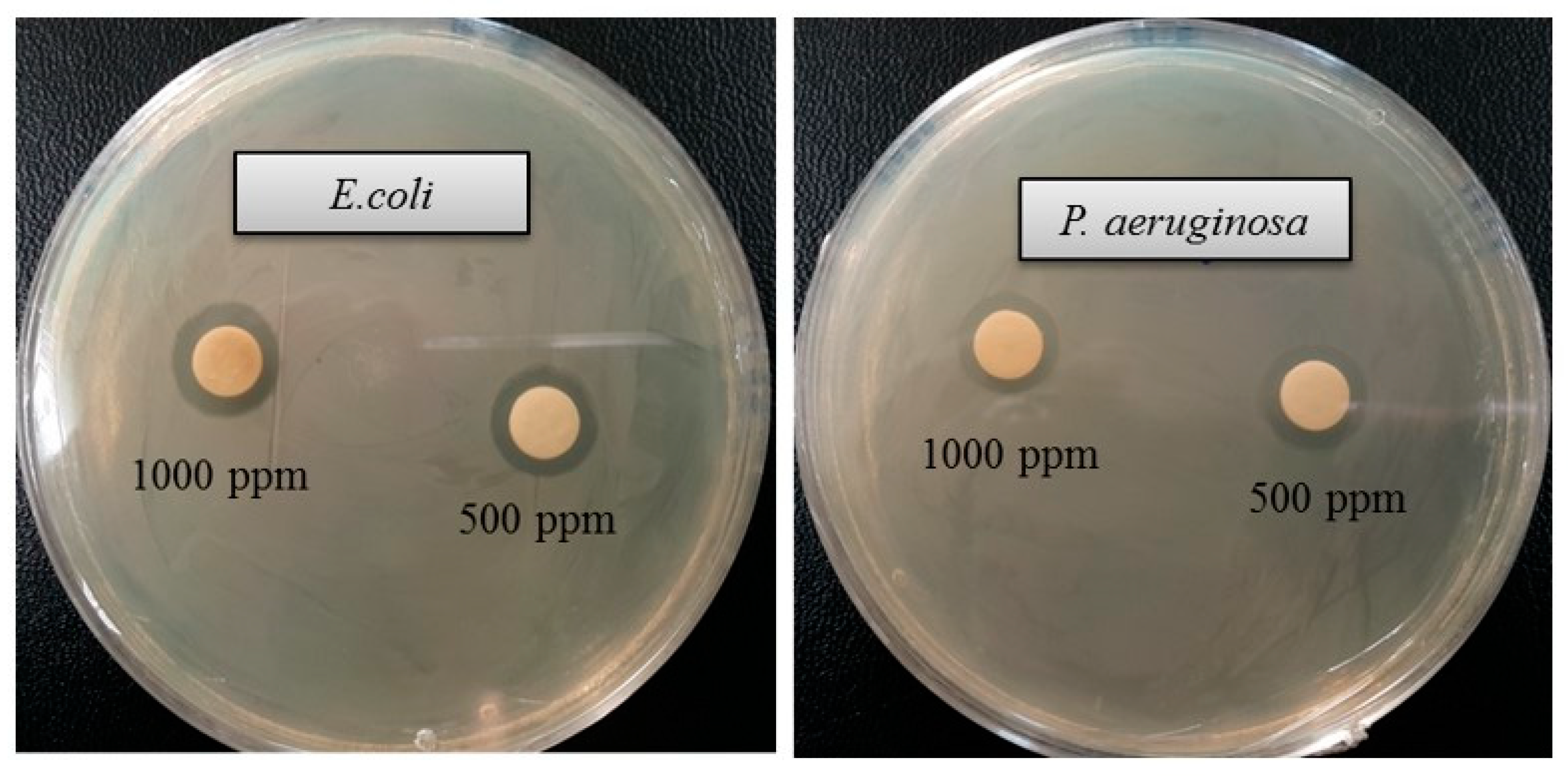
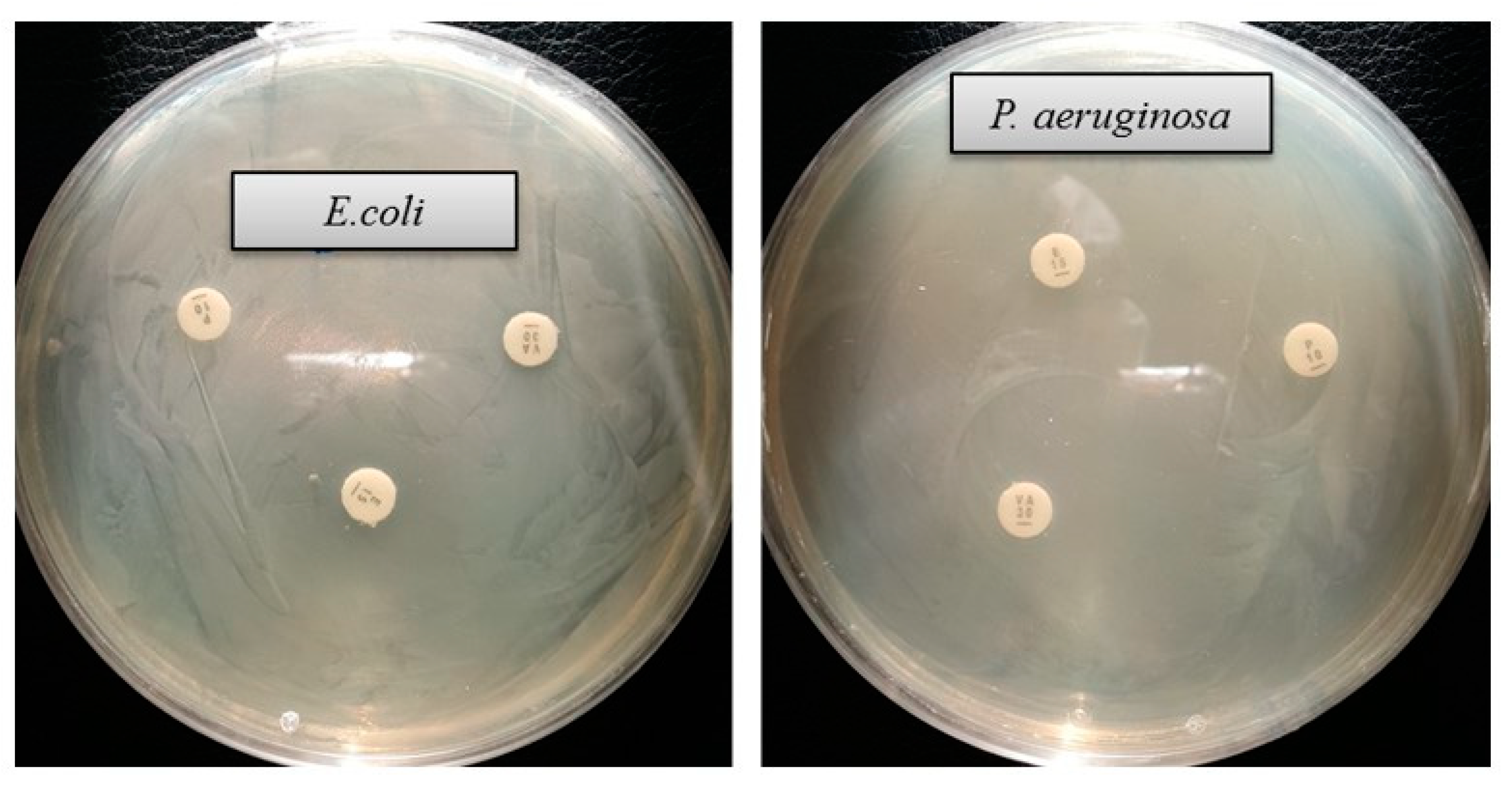
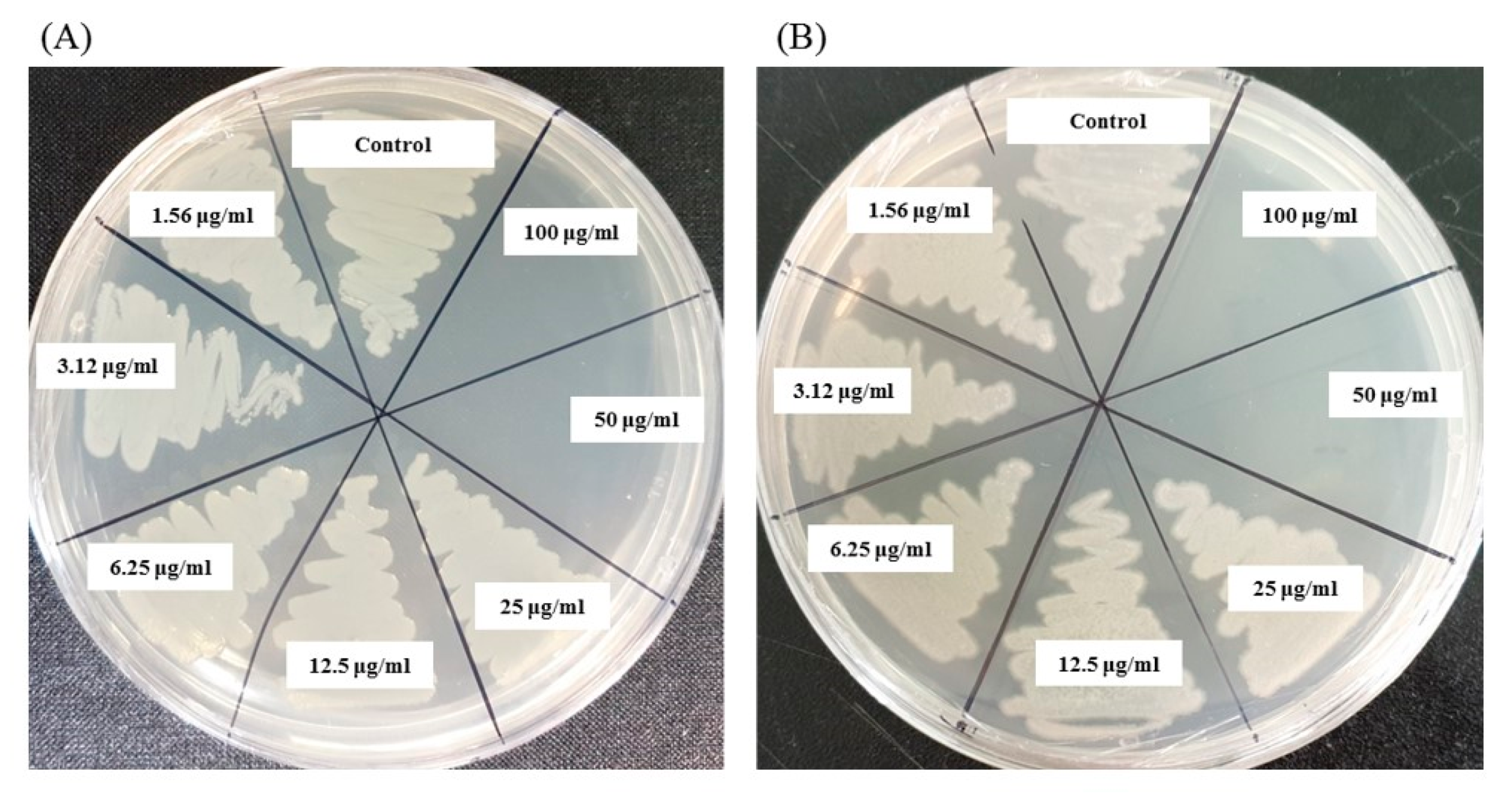
The emergence of multi-drug-resistant microorganisms is a serious concern for the medical sector, as it is difficult to control them by available antibiotics. So, it is essential to find out an alternative antimicrobial agent to control these multi-drug-resistant microorganisms. Ecofriendly synthesized AgNPs could be a potential candidate to control them. In the present study, AgNPs were synthesized using green technique and their antibacterial activity was investigated against drug-resistant pathogenic microorganisms, such as Pseudomonas aeruginosa and Escherichia coli. The results show that eco-friendly synthesized AgNPs have strong antibacterial efficacy against tested pathogens such as E. coli and P. aeruginosa (Figure 7). Figure 7 reveals the inhibition zones around the paper discs treated with AgNPs. The average results of the zone of inhibition are shown in Table 4. The antibacterial activity against pathogens was measured by calculating the diameter of the inhibition zone. The ecofriendly synthesized AgNPs showed highest antimicrobial activity against antibiotic-resistant E. coli followed by P. aeruginosa. The results suggest that Terrabacter humi MAHUQ-38T mediated AgNPs could be useful as an antimicrobial agent to control drug-resistant pathogens. The results of the present study were consistent with previously reported studies [22,25]. Both pathogens, P. aeruginosa and E. coli, were resistant to the three tested commercial antibiotics. The commercial antibiotics that were used in this study did not show any activity against tested pathogens (Figure 8, Table 5).
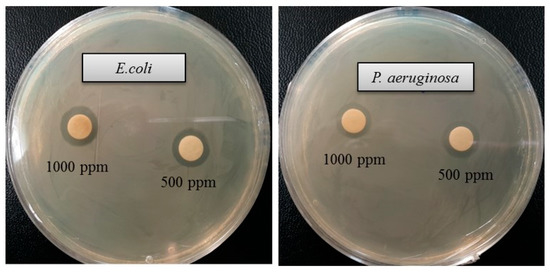
Figure 7.
Zones of inhibition of ecofriendly synthesized AgNPs (30 μL) at 500 and 1000 ppm concentrations in water against E. coli and P. aeruginosa.

Table 4.
Antimicrobial activity of eco-friendly synthesized AgNPs against E. coli and P. aeruginosa.
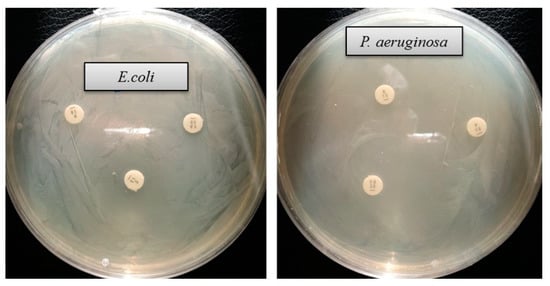
Figure 8.
Zones of inhibition of commercial antibiotics against E. coli and P. aeruginosa. Abbreviation: VA (vancomycin, 30 μg/disc), E (erythromycin, 15 μg/disc) and P (penicillin, G 10 μg/disc).

Table 5.
Antimicrobial activity of commercial antibiotics against E. coli and P. aeruginosa.
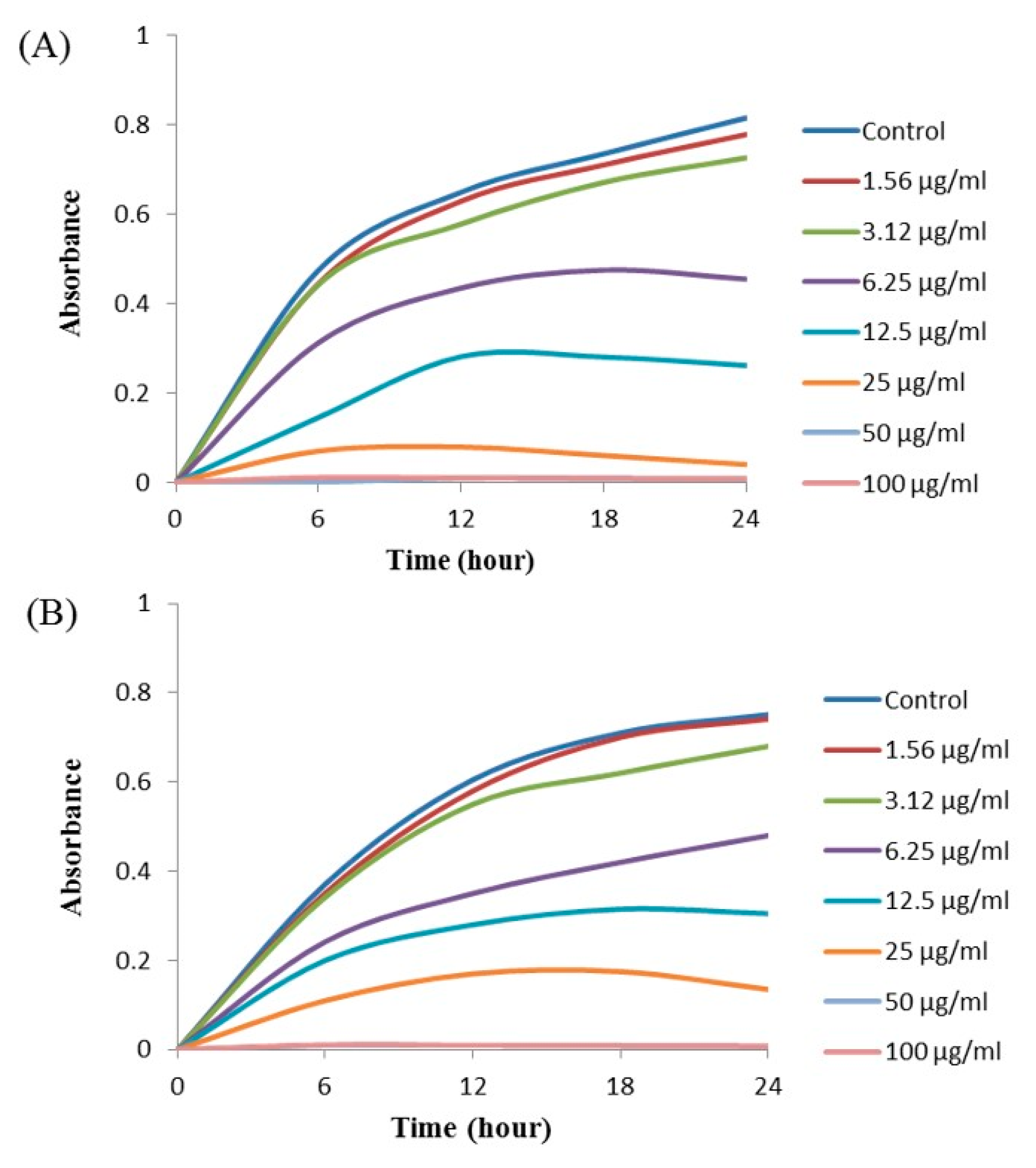
2.11. Minimal Inhibitory Concentration (MIC) and Minimal Bactericidal Concentration (MBC)
The MIC and MBC of AgNPs against E. coli and P. aeruginosa were determined by a standard broth microdilution method. From the results, it was noted that the green synthesized AgNPs showed an MIC of 6.25 μg/mL and 12.5 μg/mL for E. coli and P. aeruginosa, respectively, indicating that the synthesized AgNPs strongly suppressed the proliferation of these pathogens (Figure 9). The MIC of the AgNPs synthesized by strain MAHUQ-38T against E. coli were much lower than some other antimicrobial agents, including biosynthesized CuO-NPs and ZnO-NPs which were 0.5 mg/mL and 8 mg/mL, respectively, against E. coli [26,27]. The MICs of the AgNPs synthesized by strain MAHUQ-38T against P. aeruginosa were also significantly lower than some other antimicrobial agents, including biogenic nanoparticles, which were in the range 0.375–1.6 mg/mL against P. aeruginosa [28,29]. The MBCs of biosynthesized AgNPs for both E. coli and P. aeruginosa were 50 μg/mL (Figure 10), which was also much lower than those of some other antimicrobial agents [26,27,28].
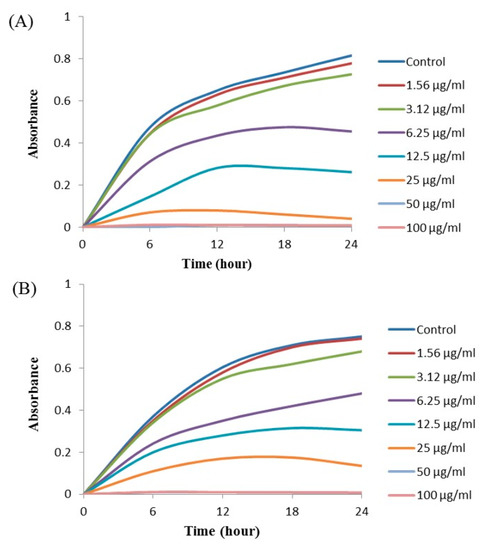
Figure 9.
Growth curves of E. coli (A) and P. aeruginosa (B) cultured in Mueller–Hinton broth (MHB) with different concentrations of the synthesized AgNPs to determine the Minimal Inhibitory Concentration (MIC).
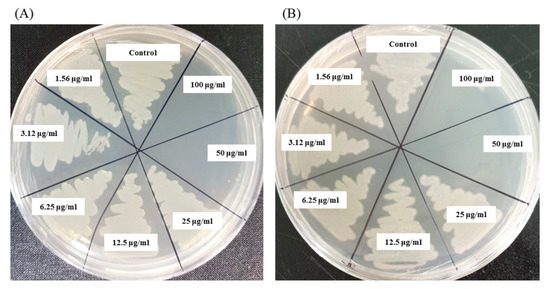
Figure 10.
MBC of synthesized AgNPs against E. coli (A) and P. aeruginosa (B).
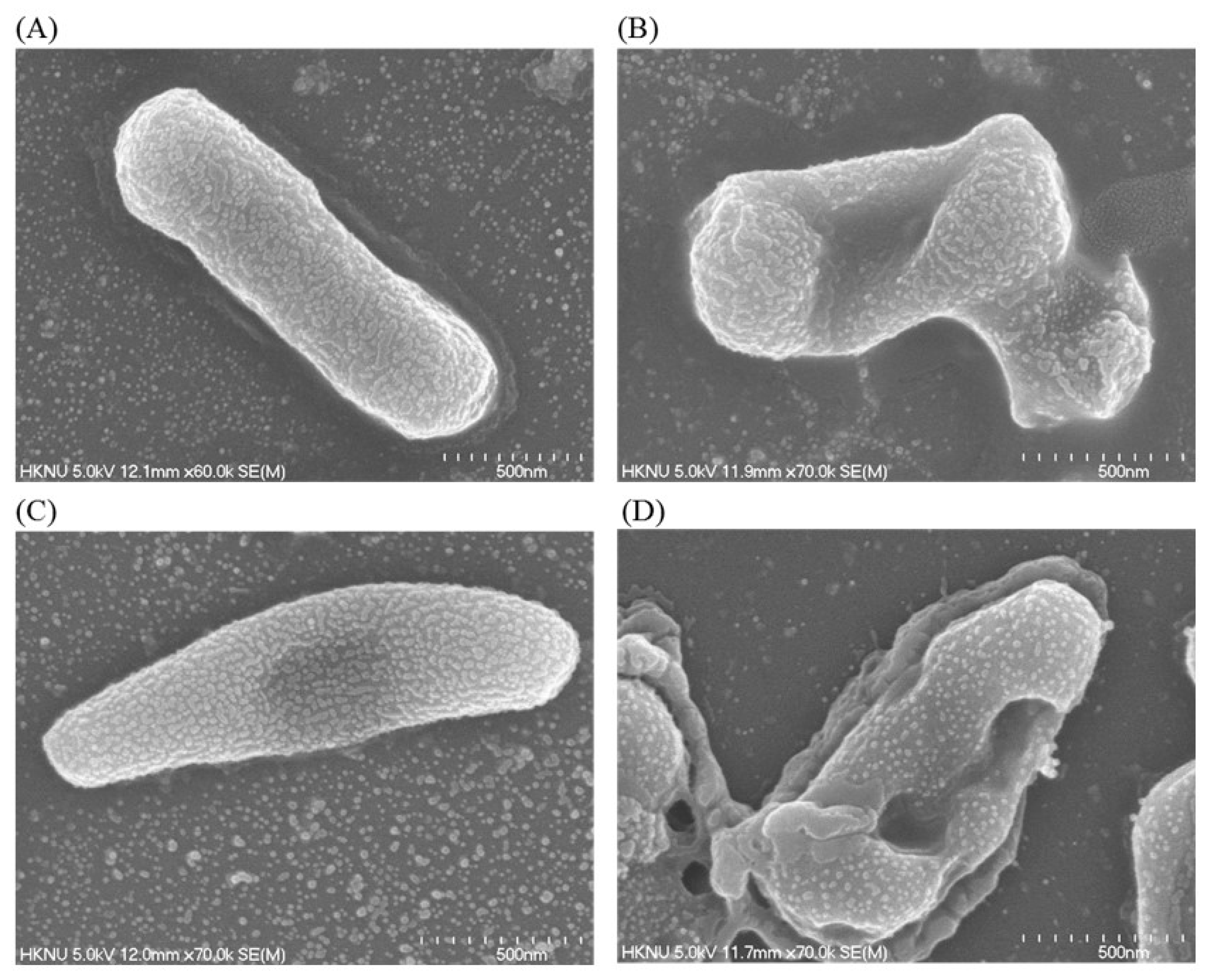
2.12. Investigation of Structural Changes by FE-SEM
Structural and morphological changes of E. coli and P. aeruginosa cells that were treated by ecofriendly synthesized AgNPs were seen by FE-SEM (field emission scanning electron microscope) (Figure 11). By examining the FE-SEM, the untreated E. coli cells were found as a normal rod-shaped and intact cell surface with no damage (Figure 11A). However, E. coli cells treated with 1 × MBC of synthesized AgNPs were displayed irregularly wrinkled, damaged, deformed and cracked outer surface, and most of the cell membranes were collapsed (Figure 11B). Similarly, P. aeruginosa cells without treatment were observed as the normal rod shape, and the cell surface was intact without any damage (Figure 11C). However, the cells treated with 1×MBC of ecofriendly synthesized AgNPs were displayed as irregularly wrinkled, damaged, deformed and with a cracked outer surface with collapsed cell membranes (Figure 11D). The treated cells depicted a distorted cell structure, indicating the loss of membrane integrity of bacterial cells. The morphological changes, structural alterations and damages of the bacterial cell wall indicated that the ecofriendly synthesized AgNPs might disturb the normal cell functions of both pathogenic strains E. coli and P. aeruginosa, which would lead to the death of cells. The results of the present study are consistent with previously reported studies [30,31].
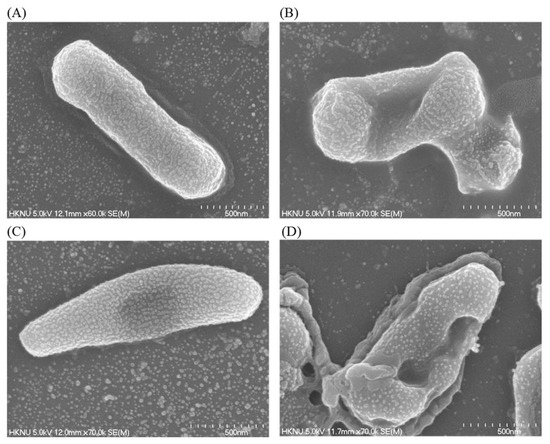
Figure 11.
SEM images of normal E. coli cells (A), 1 × MBC AgNPs treated E. coli cells (B), normal P. aeruginosa cells cells (C), and 1 × MBC AgNPs treated P. aeruginosa cells cells (D).
3. Materials and Methods
3.1. Materials
Standard antibiotic discs such as penicillin G (P10) 10 μg/disc, erythromycin (E15) 15 μg/disc and vancomycin (VA30) 30 μg/disc were obtained from Oxoid Ltd., England. Bacterial growth media and Analytical-grade AgNO3 (silver nitrate) were bought from Difco, MB Cell (Seoul, Korea) and Sigma-Aldrich Chemicals (USA), respectively. The pathogenic strains Escherichia coli (ATCC 10798) and Pseudomonas aeruginosa (ATCC 10145) were collected from American Type Culture Collection (ATCC).
3.2. Isolation of Bacteria
Strain MAHUQ-38T was isolated from the soil sample of a grape garden, located in Anseong, Republic of Korea (37°00′74″ N 127°20′10″ E). The samples were collected in 15 mL conical tubes and suspended in sterile NaCl solution (0.85%, w/v). Strain MAHUQ-38T was isolated using R2A agar medium through the serial dilution technique after incubation at 30 °C according to the method described by Huq [17]. Then, the isolated strain was sub-cultured on R2A agar and stored at −80 °C in R2A broth with 30% (v/v) glycerol. Strain MAHUQ-38T has been deposited to Korean Agriculture Culture Collection (KACC) and China General Microbiological Culture Collection Center (CGMCC).
3.3. Phenotypic and Biochemical Characteristics
The Gram reaction was examined according to the previous description [17]. Cell motility was tested using Sulfide-indole-motility (SIM; Difco) medium. Cell morphology was determined by transmission electron microscope, using cells grown on R2A agar (MB-R1129, MB-cell) medium for 2 days at 30 °C. Growth on different media, such as nutrient agar (213000, BD-Difco), R2A agar, Luria–Bertani agar (22700-025, Invitrogen, Carlsbad, CA, USA), MacConkey agar (211387, BD-Difco) and tryptic soy agar (C7121, Criterion), was investigated after 7 days of incubation at 30 °C. Growth at various temperatures (from 5 to 40 °C) and different pH values (from 4 to 10) was checked on R2A agar and R2A broth medium, respectively, after 5 days of incubation at 30 °C. NaCl tolerance test and anaerobic growth ability of strain MAHUQ-38T were conducted according to the previous description [17]. Availability of flexirubin type of pigments were examined according to Sheu et al. [32]. Activities of oxidase, catalase, urease DNase, and hydrolysis of gelatin, casein, starch, Tweens 80 and 20 were investigated according to previous description [17]. Additional biochemical tests were conducted using API 20NE and API ZYM kits, according to the manufacturer’s instructions (bioMérieux). The close type strains, T. tumescens KACC 20535T, T. terrae KACC 11642T, T. terrigena KCTC 19602T, T. lapilli LR-26T, T. ginsengihumi KACC 19444T and T. aerolatus KACC 20556T were used as reference strains and were grown under the same experimental conditions of novel strain MAHUQ-38T.
3.4. 16S rRNA Gene Sequencing and Phylogenetic Analysis
The genomic DNA of strain MAHUQ-38T was extracted and purified using a commercial genomic DNA extraction kit (PDC09-0100, BiO–Helix, Keelung, Taiwan) according to the manufacturer’s instructions. The 16S rRNA gene of strain MAHUQ-38T was amplified from Genomic DNA using bacterial universal primers 27F and 1492R [33], and then the PCR product was purified and sequenced using same primers 27F and 1492R [33] by Solgent Co. Ltd. (Daejeon, Korea). 16S rRNA gene sequences of close type strains were collected from EzBiocloud site (http://www.ezbiocloud.net/eztaxon) [34]. The CLUSTAL_X program was used for multiple sequence alignments [35]. Gaps were edited in the BioEdit program [36]. A Kimura two-parameter model was used to calculate the evolutionary distances [37]. A phylogenic tree was constructed using the neighbor-joining [38] algorithm and MEGA6 program [39] with bootstrap values of 1000 replications [40].
3.5. Genome Sequence Analysis
The draft-genome sequence of novel bacterium T. humi MAHUQ-38T was analyzed by Illumina HiSeq X Ten and the assembly was completed using de novo assembler (SOAPdenovo v. 3.10.1). The genome annotation was carried out using NCBI prokaryotic genome annotation pipeline (PGAP). The genomic DNA G + C content of strain MAHUQ-38T was directly calculated from its genome sequence. To determine the degree of pairwise relatedness between MAHUQ-38T and close relatives, BLAST-based average nucleotide identity (ANI) was measured according to Yoon et al. [34]. While the digital DNA-DNA hybridization (dDDH) value was evaluated using the genome-to-genome distance calculator (http://ggdc.dsmz.de/ggdc.php) as stated by Meier-Kolthoff et al. [41]. DNA-DNA hybridization was also carried out fluorometrically by the method of Ezaki et al. [42], according to the previous description [43].
3.6. Cellular Fatty Acid, Respiratory Quinones and Polar Lipid Analysis
Cellular fatty acid profiles of strain MAHUQ-38T and the reference strains were identified after 48 h of growth on R2A agar medium at 30 °C. The cellular fatty acids were extracted, purified and analyzed according to the previous description [17,44]. The isoprenoid quinones of strain MAHUQ-38T were extracted and purified using the method of Collins and then, analyzed by HPLC [45]. The polar lipids of strain MAHUQ-38T were extracted and analyzed using two-dimensional thin layer chromatography according to the previous description [46].
3.7. Ecofriendly Synthesis of AgNPs Using Terrabacter humi sp. nov.
For the synthesis of AgNPs, strain MAHUQ-38T was cultured in 100 mL of R2A broth media for two days at 30 °C using a shaking incubator with 180 rpm. Then, the culture supernatant was separated through centrifuge for 10 min at 10,000 rpm. A total of 100 μL AgNO3 solution (1 M/mL) was mixed with 100 mL of culture supernatant, and then the mixture was incubated at 30 °C in a shaking incubator with dark conditions for 48 h. The biosynthesis of AgNPs was confirmed through visual observation by shifting the color of the culture medium. The biosynthesized AgNPs were collected by centrifugation at 13,500 rpm for 25 min and purified twice through washing by distilled water [13]. Finally, the pellets of AgNPs were air dried and collected for characterization.
3.8. Characterization of Synthesized AgNPs
UV–vis spectrophotometer from 300 to 800 nm was used to observe and record the reduction of Ag+ ions. The morphology, size, elemental compositions and SAED (selected area diffraction) pattern of ecofriendly synthesized AgNPs were recorded using FE-TEM (field emission-transmission electron microscopy). A drop of AgNPs solution was placed on the carbon-coated TEM grid, dried at room temperature and finally observed by microscope. Air-dried samples were used for XRD (X-ray diffraction) analysis using an X-ray diffractometer over the 2θ range of 30–90° (D8 Advance, Bruker, Karlsruhe, Germany). The FT-IR (Fourier transform-infrared) spectrum shows the biomolecules which are involved for both the synthesis and stabilization of nanoparticles. The FT-IR spectrum of synthesized AgNPs was recorded by using FTIR spectroscopy in a range of 400–4000 cm−1.
3.9. Antimicrobial Activity of Synthesized AgNPs
The antimicrobial activity of ecofriendly synthesized AgNPs against pathogenic microbes was investigated by the Disc diffusion method. The tested pathogenic microbes (Pseudomonas aeruginosa and Escherichia coli) were grown overnight in MHB (Mueller–Hinton broth). Then, both pathogenic strains (100 μL) were spread completely on an MHA (Mueller–Hinton agar) plate. The sterile paper discs (8 mm) were placed on the surface of the MHA plate, and then 30 μL (both 500 and 1000 ppm) of eco-friendly synthesized AgNPs was given on the paper discs. Finally, the plates were incubated for 24 h at 37 °C. Similarly, the antimicrobial activity of some commercial antibiotics, including vancomycin (30 mg/disc), penicillin G (10 mg/disc) and erythromycin (15 mg/disc), has been evaluated against P. aeruginosa and E. coli. Inhibition zones were measured after 24 h of incubation. This test was conducted three times.
3.10. Determination of Minimal Inhibitory Concentration and Minimal Bactericidal Concentration
The MIC of ecofriendly synthesized AgNPs was also determined by following the broth micro dilution technique. The pathogenic strains P. aeruginosa and E. coli were grown in MH (Muller–Hinton) broth (70192, Sigma-Aldrich, St. Louis, MO, USA) at 37 °C overnight, and then the optical density was fixed around 1 × 106 CFUs/mL. A total of 100 uL of test bacterial (1 × 106 CFUs/mL) suspension was added into a 96-well ELISA plate followed by an equal volume of AgNPs solution with various concentrations (1.56 to 100 μg/mL), and then the plates were incubated at 37 °C for 24 h. The incubated ELISA plates were measured for absorbance (at 600 nm) using an ELISA plate reader (LabTech 4000). The MBC was determined by streaking of 10 μL of each set on MHA plate and further incubated at 37 °C for 24 h. The culture plates were watched by the naked eye to determine the lowest concentration (MBC) that blocked bacterial growth [47].
3.11. Investigation of Structural Changes by FE-SEM
The morphological and structural changes of pathogenic strains E. coli and P. aeruginosa were examined by FE-SEM. Cells of logarithmic growth phase (1 × 107 CFU/mL) were treated with the biosynthesized AgNPs at the concentration of 1 × MBC. In the control, bacterial cultures were treated with 0.85% NaCl solution. PBS buffer was used to wash the overnight treated cells. At first, the cells were fixed using 2.5% glutaraldehyde and then washed several times using PBS. Once more, the cells were fixed by osmium tetroxide (1%) and subsequently washed with PBS. Fixed cells were dehydrated at room temperature by various concentrations of ethanol (from 30% to 100%) for 10 min. The resulted samples were dried using a desiccator. Finally, the cells were installed on metallic stubs and coated with gold. The structural and morphological changes of the cells were seen by FE-SEM (JSM-7100F, JEOL, Akishima, Tokyo, Japan) [47].
4. Conclusions
The present study reported the characterization of a novel bacterial species Terrabacter humi MAHUQ-38T and also the biologically facile and green synthesis of AgNPs using this novel species and also demonstrated the significant antimicrobial activity of biosynthesized AgNPs against multi-drug-resistant pathogenic microbes. The methodology is ecofriendly, safe, facile, effective and economical. The strain MAHUQ-38T is capable of producing valuable AgNPs within 48 h of incubation. These biosynthesized AgNPs were confirmed by UV–vis spectroscopy, FE-TEM, XRD and FT-IR analysis. Extracellular methodology was used to synthesis AgNPs using Terrabacter humi MAHUQ-38T which is simple, green, ecofriendly and cost-effective for large-scale production. Moreover, the biosynthesized AgNPs show significant antimicrobial activity against antibiotic-resistant human pathogenic strain E. coli and P. aeruginosa. Minimal inhibitory/bactericidal concentration against E. coli and P. aeruginosa were 6.25/50 and 12.5/50 μg/mL, respectively. The AgNPs altered the cell morphology and damaged the cell membrane of pathogens. Thus, the novel species Terrabacter humi MAHUQ-38T could be useful for utilization in the ecofriendly synthesis of AgNPs for various applications in both medical and non-medical sectors, especially as an antimicrobial agent to control drug-resistant pathogens.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/24/9746/s1.
Author Contributions
M.A.H. conceived the study, designed and supervised the experiments, analyzed the results and wrote the article. S.A. performed most of the experiments. S.-Y.L. checked the manuscript. M.Z.S., S.R.B.; M.A. and E.J.R. performed some experiments. All authors thoroughly revised the manuscript and approved its submission. All authors have read and agreed to the published version of the manuscript.
Funding
This study was performed with the support of the National Research Foundation (NRF) of Korea grant (Project no. NRF-2018R1C1B5041386, Grant Recipient: M.A.H.) funded by the Korean government, Korea.
Acknowledgments
I would like to give special thanks to the CGM 10K project for analyzing the draft genome sequence of strain MAHUQ-38T (GCM60012305).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Collins, M.D.; Dorsch, M.; Stackebrandt, E. Transfer of Pimelobacter tumescens to Terrabacter gen. nov. as Terrabacter tumescens comb. nov. and of Pimelobacter jensenii to Nocardioides as Nocardioides jensenii comb. nov. Int. J. Syst. Bacteriol. 1989, 39, 1–6. [Google Scholar] [CrossRef]
- Weon, H.Y.; Schumann, P.; Reiner, M.; Kroppenstedt, R.M.; Kim, B.Y.; Song, J.; Kwon, S.W.; Go, S.J.; Stackebrandt, E. Terrabacter aerolatus sp. nov., isolated from an air sample. Int. J. Syst. Evol. Microbiol. 2007, 55, 2491–2495. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.F.; Quan, X.T.; Siddiqi, M.Z.; Liu, Q.Z.; Yu, H.S.; Im, W.T. Terrabacter ginsengihumi sp. nov., isolated from ginseng cultivating soil. J. Microbiol. 2018, 56, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Montero-Barrientos, M.; Rivas, R.; Velazquez, E.; Monte, E.; Roig, M.G. Terrabacter terrae sp. nov., a novel actinomycete isolated from soil in Spain. Int. J. Syst. Evol. Microbiol. 2005, 55, 2491–2495. [Google Scholar] [CrossRef]
- Lee, J.E.; Seo, J.P.; Lee, D.W.; Ko, Y.H.; Lee, S.D. Terrabacter lapilli sp. nov., an actinomycete isolated from stone. Int. J. Syst. Evol. Microbiol. 2008, 58, 1084–1088. [Google Scholar] [CrossRef]
- Yoon, J.H.; Park, S.; Kang, S.J.; Jung, Y.T.; Kim, W. Terrabacter terrigena sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2009, 59, 2798–2802. [Google Scholar] [CrossRef]
- Al-Dhabi, N.A.; Arasu, M.V. Environmentally-friendly green approach for the production of zinc oxide nanoparticles and their anti-fungal, ovicidal, and larvicidal properties. Nanomaterials 2018, 8, 500. [Google Scholar] [CrossRef]
- Ojo, S.A.; Lateef, A.; Azeez, M.A. Biomedical and catalytic applications of gold and silver-gold alloy nanoparticles biosynthesized using cell-free extract of Bacillus safensis LAU 13: Antifungal, dye degradation, anti-coagulant and thrombolytic activities. IEEE. Transon. Nanobiosci. 2016, 15, 433–442. [Google Scholar] [CrossRef]
- Taner, M.; Sayar, N.; Yulug, I.G. Synthesis, characterization and antibacterial investigation of silver–copper nanoalloys. J. Mater. Chem. 2011, 21, 13150–13154. [Google Scholar] [CrossRef]
- Wang, C.; Kim, Y.J.; Singh, P. Green synthesis of silver nanoparticles by Bacillus methylotrophicus, and their antimicrobial activity. Artific. Cells. Nanomed. Biotechnol. 2016, 44, 1127–1132. [Google Scholar]
- Dhandapani, P.; Maruthamuthu, S.; Rajagopal, G. Bio-mediated synthesis of TiO2 nanoparticles and its photocatalytic effect on aquatic biofilm. J. Photochem. Photobiol. B Biol. 2012, 110, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.A.; Asif, M.A.; Mashrai, A.M. Green synthesis of ZnO nanoparticles using Bacillus subtilis and their catalytic performance in the one-pot synthesis of steroidal thiophenes. Eur. Chem. Bull. 2014, 3, 939–945. [Google Scholar]
- Huq, M.A. Paenibacillus anseongense sp. nov. a Silver Nanoparticle Producing Bacterium Isolated from Rhizospheric Soil. Curr. Microbiol. 2020, 77, 2023–2030. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Singh, H.; Kim, Y.J. Extracellular synthesis of silver and gold nanoparticles by Sporosarcina koreensis DC4 and their biological applications. Enzym. Microb. Technol. 2016, 86, 75–83. [Google Scholar] [CrossRef]
- Vigneshwaran, N.; Kathe, A.A.; Varadarajan, P.V. Silve-protein (core-shell) nanoparticle production using spent mushroom substrate. Langmuir 2007, 23, 7113–7117. [Google Scholar] [CrossRef]
- Castro-Aceituno, V.; Ahn, S.; Simu, S.Y.; Singh, P.; Mathiyalagan, R.; Lee, H.A.; Yang, D.C. Anticancer activity of silver nanoparticles from Panax ginseng fresh leaves in human cancer cells. Biomed. Pharmacother. 2016, 84, 158–165. [Google Scholar] [CrossRef]
- Huq, M.A. Microvirga rosea sp. nov.: A nanoparticle producing bacterium isolated from soil of rose garden. Arch. Microbiol. 2018, 200, 1439–1445. [Google Scholar] [CrossRef]
- Akter, S.; Huq, M.A. Biologically rapid synthesis of silver nanoparticles by Sphingobium sp. MAH-11 T and their antibacterial activity and mechanisms investigation against drug-resistant pathogenic microbes. Artif. Cells Nanomed. Biotechnol. 2020, 48, 672–682. [Google Scholar] [CrossRef]
- Chaloupka, K.; Malam, Y.; Seifalian, A.M. Nanosilver as a new generation of nanoproduct in biomedical applications. Trends Biotechnol. 2010, 28, 580–588. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Singh, N.; Khanna, P.K. In situ synthesis of silver nano-particles in polymethylmethacrylate. Mater. Chem. Phys. 2007, 104, 367–372. [Google Scholar] [CrossRef]
- Du, J.; Sing, H.; Yi, T.H. Biosynthesis of silver nanoparticles by Novosphingobium sp. THG-C3 and their antimicrobial potential. Artif. Cells Nanomed. Biotechnol. 2017, 45, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Sing, H.; Yi, T.H. Antibacterial, anti-biofilm and anticancer potentials of green synthesized silver nanoparticles using benzoin gum (Styrax benzoin) extract. Bioprocess. Biosyst. Eng. 2016, 39, 1923–1931. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Kim, Y.J.; Singh, H. Biosynthesis, characterization, and antimicrobial applications of silver nanoparticles. Int. J. Nanomed. 2015, 10, 2567–2577. [Google Scholar]
- Kalishwaralal, K.; BarathManiKanth, S.; Pandian, S.R. Silver nanoparticles impede the biofilm formation by Pseudomonas aeruginosa and Staphylococcus epidermidis. Colloids Surf. B Biointerfaces. 2010, 79, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Cherian, T.; Ali, K.; Saquib, Q.; Faisal, M.; Wahab, R.; Musarrat, J. Cymbopogon Citratus Functionalized Green Synthesis of CuO-Nanoparticles: Novel Prospects as Antibacterial and Antibiofilm Agents. Biomolecules 2020, 10, 169. [Google Scholar] [CrossRef]
- Rehman, S.; Jermy, B.R.; Akhtar, S.; Borgio, J.F. Isolation and characterization of a novel thermophile; Bacillus haynesii, applied for the green synthesis of ZnO nanoparticles. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2072–2082. [Google Scholar] [CrossRef]
- Bayroodi, E.; Jala, R. Modulation of antibiotic resistance in Pseudomonas aeruginosa by ZnO nanoparticles. Iran J. Microbiol. 2016, 8, 85–92. [Google Scholar]
- Ali, S.G.; Ansari, M.A.; Alzohairy, M.A.; Alomary, M.N.; Jalal, M.; AlYahya, S.; Asiri, S.M.M.; Khan, H.M. Effect of Biosynthesized ZnO Nanoparticles on Multi-Drug Resistant Pseudomonas Aeruginosa. Antibiotics 2020, 9, 260. [Google Scholar] [CrossRef]
- Shankar, S.; Rhim, J.W. Amino acid mediated synthesis of silver nanoparticles and preparation of antimicrobial agar/silver nanoparticles composite films. Carbohydr. Polym. 2015, 130, 353–363. [Google Scholar] [CrossRef]
- Kim, J.S.; Kuk, E.; Yu, K.N. Antimicrobial effects of silver nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Sheu, S.Y.; Su, C.L.; Kwon, S.W.; Chen, W.M. Flavobacterium amniphilum sp. nov., isolated from a stream. Int. J. Syst. Evol. Microbiol. 2017, 67, 5179–5186. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The Clustal_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 24, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kimura, M. The Neutral Theory of Molecular Evolution; Cambridge University Press: New York, NY, USA, 1983. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Bio. Evol. 1987, 4, 406–425. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Ezaki, T.; Hashimoto, Y.; Yabuuchi, E. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Evol. Microbiol. 1989, 39, 224–229. [Google Scholar] [CrossRef]
- Huq, M.A.; Akter, S.; Lee, S.Y. Mucilaginibacter formosus sp. nov., a bacterium isolated from road-side soil. Antonie van Leeuwenhoek 2019, 112, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; MIDI Technical Note 101; MIDI Inc.: Newark, DE, USA, 1990. [Google Scholar]
- Collins, M.D. Isoprenoid quinones. In Chemical Methods in Prokaryotic Systematics; Goodfellow, M., O’Donnell, A.G., Eds.; Wilye: Chichester, UK, 1994; pp. 265–309. [Google Scholar]
- Akter, S.; Huq, M.A. Sphingomonas chungangi sp. nov., a bacterium isolated from soil sample of a garden. Int. J. Syst. Evol. Microbiol. 2020, 70, 4151–4157. [Google Scholar] [CrossRef] [PubMed]
- Ansari, M.A.; Baykal, A.; Asiri, S. Synthesis and characterization of antibacterial activity of spinel chromium-substituted copper ferrite nanoparticles for biomedical application. J. Inorg. Organomet. Polym. Mater. 2018, 28, 2316–2327. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).