Recombination and Pol ζ Rescue Defective DNA Replication upon Impaired CMG Helicase—Pol ε Interaction
Abstract
1. Introduction
2. Results
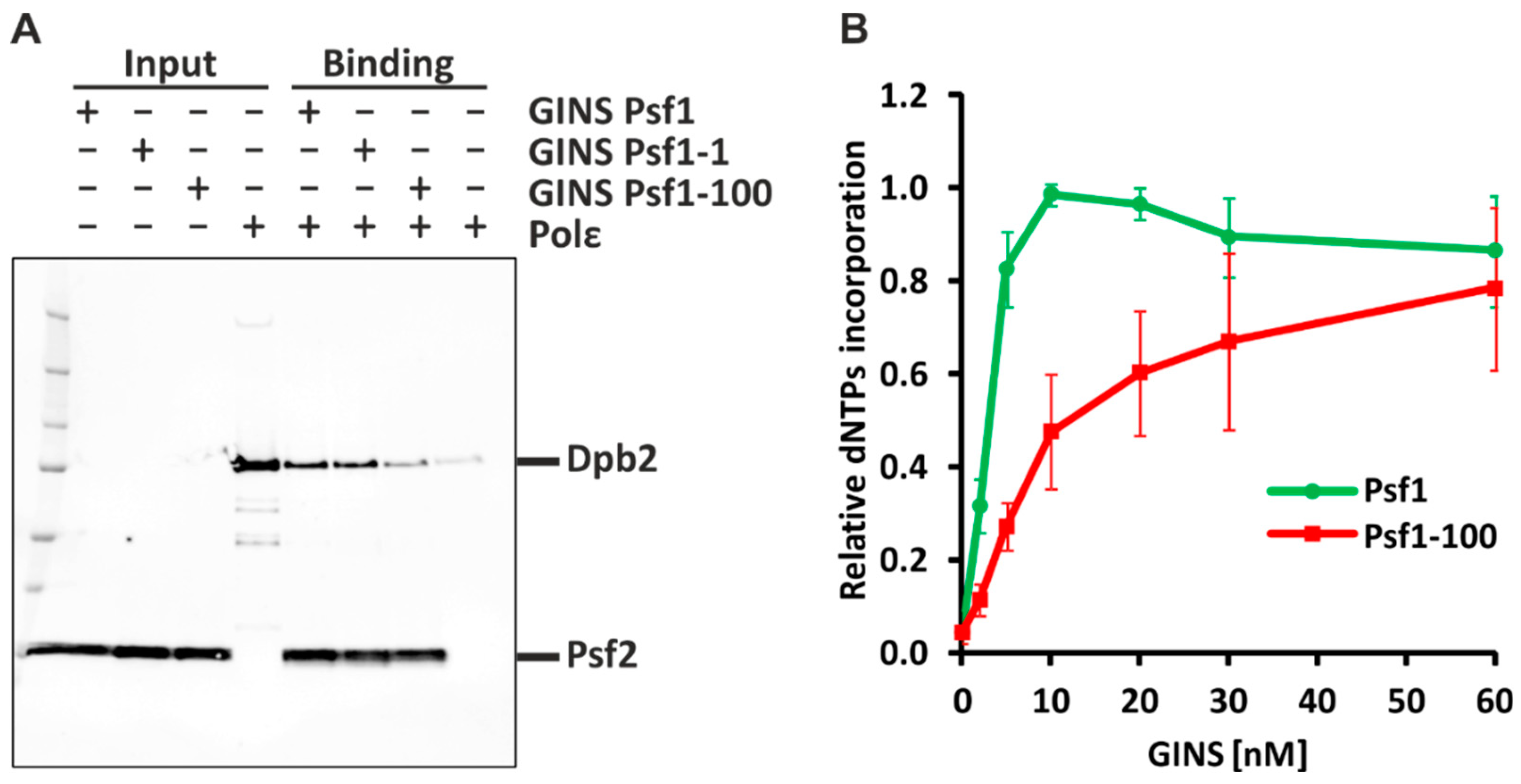
2.1. Psf1-100 Impairs the Interaction between GINS and Pol ε, Which Results in Impeded DNA Synthesis
2.2. Increased Formation of Single-Stranded DNA in psf1-100 Cells
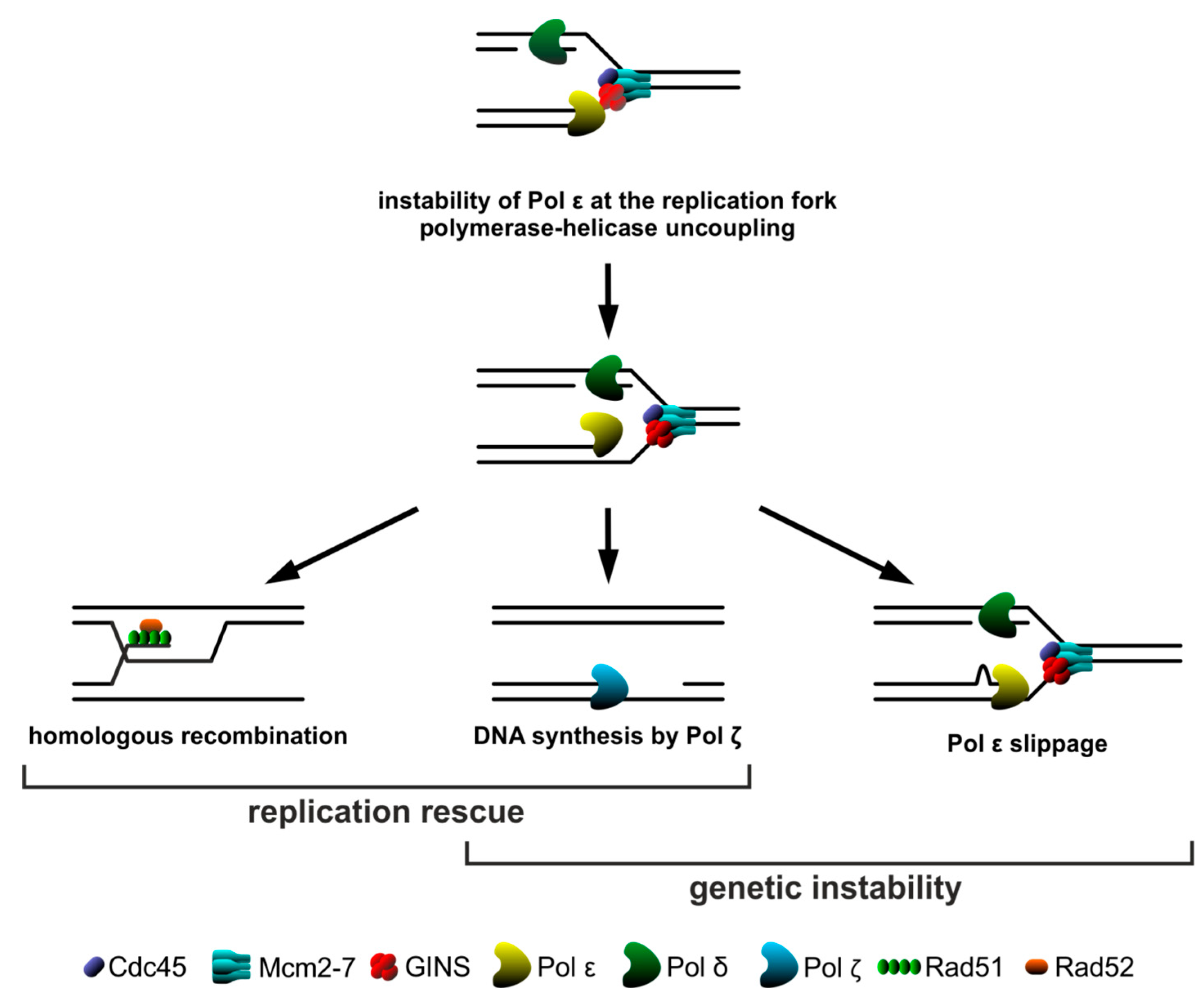
2.3. Homologous Recombination Rescues psf1-100 Cells
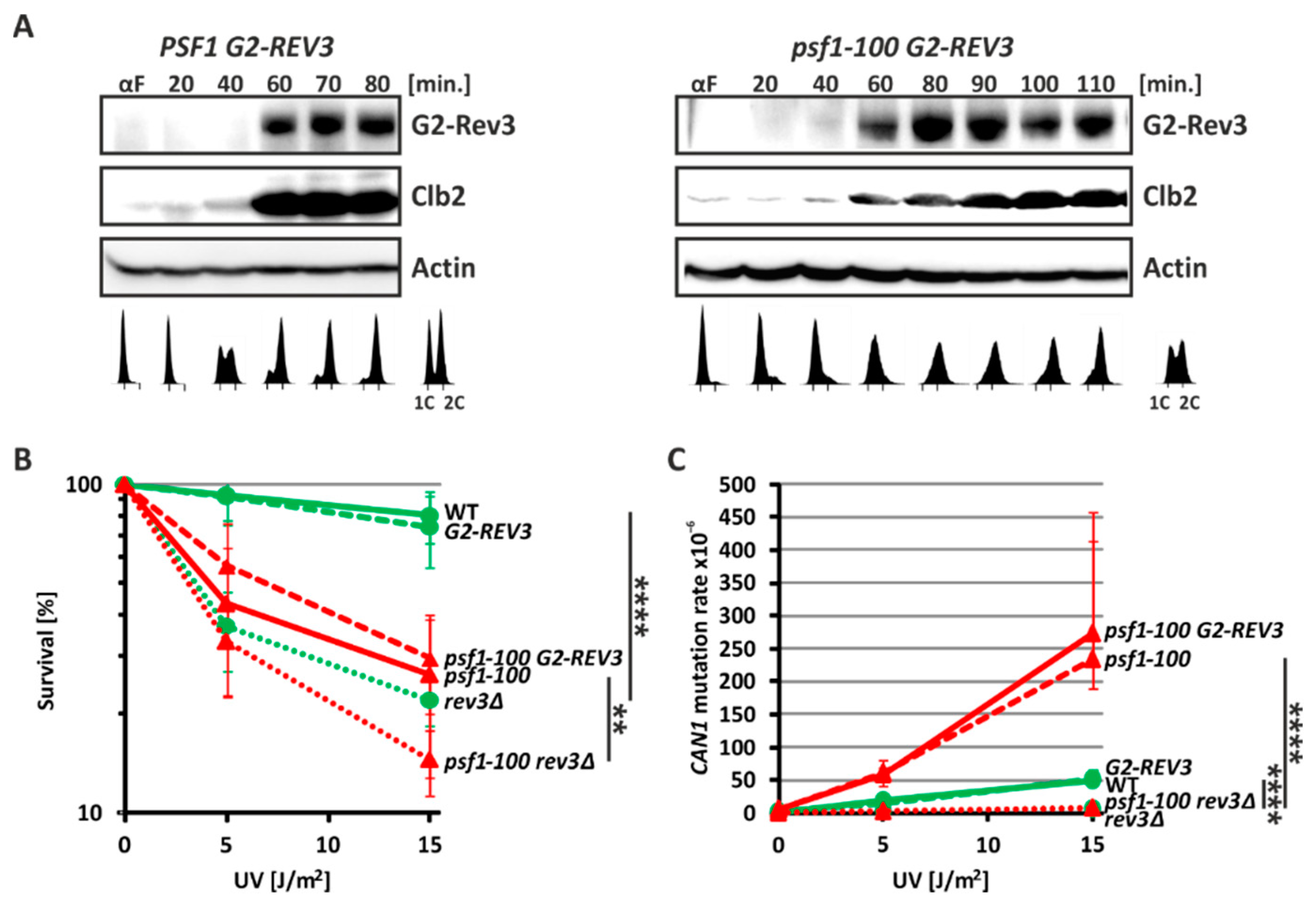
2.4. Pol ζ Exerts Its Mutagenic Activity in psf1-100 Cells Mainly in G2 Phase
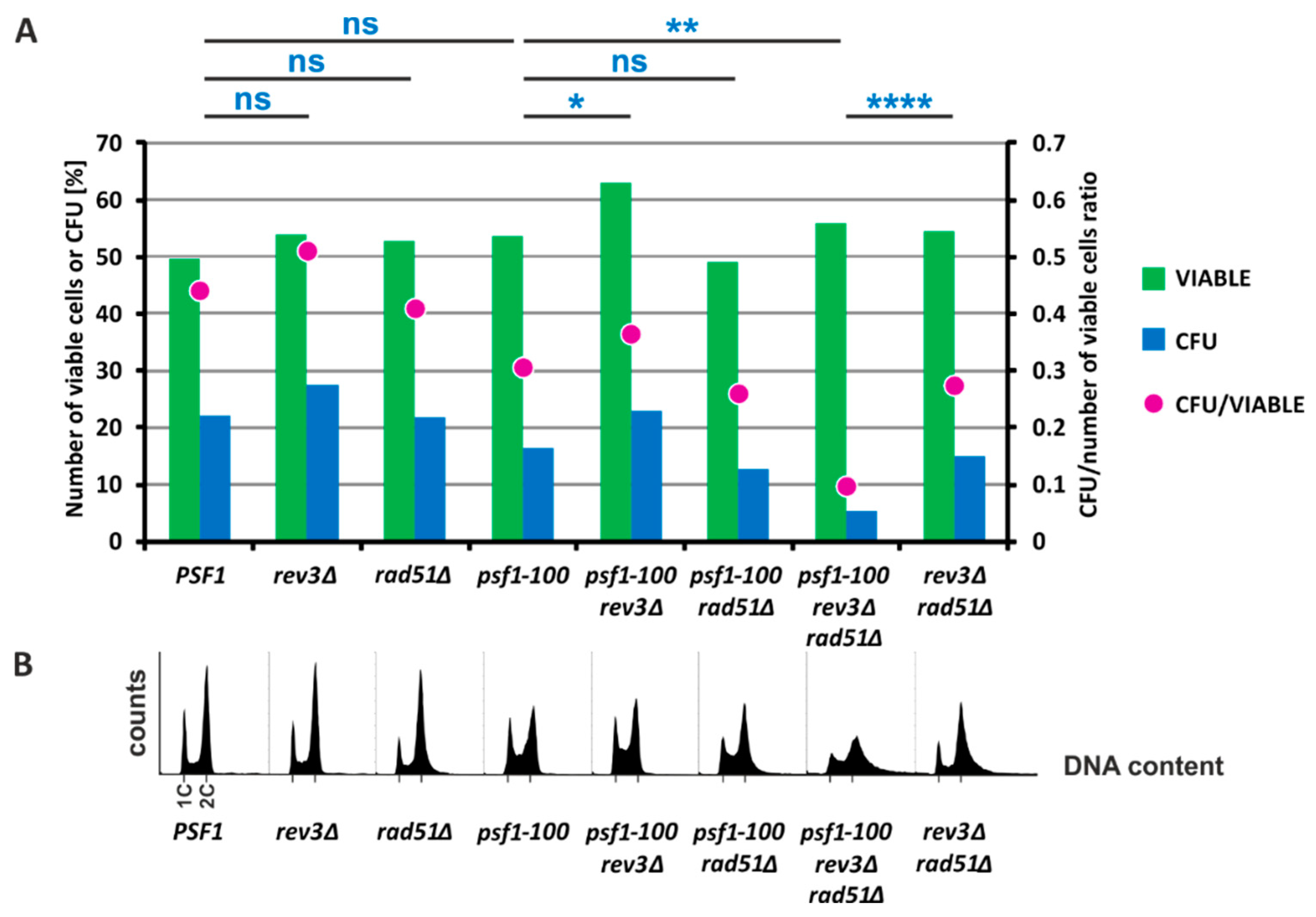
2.5. HR and Pol ζ-Dependent Synthesis Play Important Roles in psf1-100 Cells
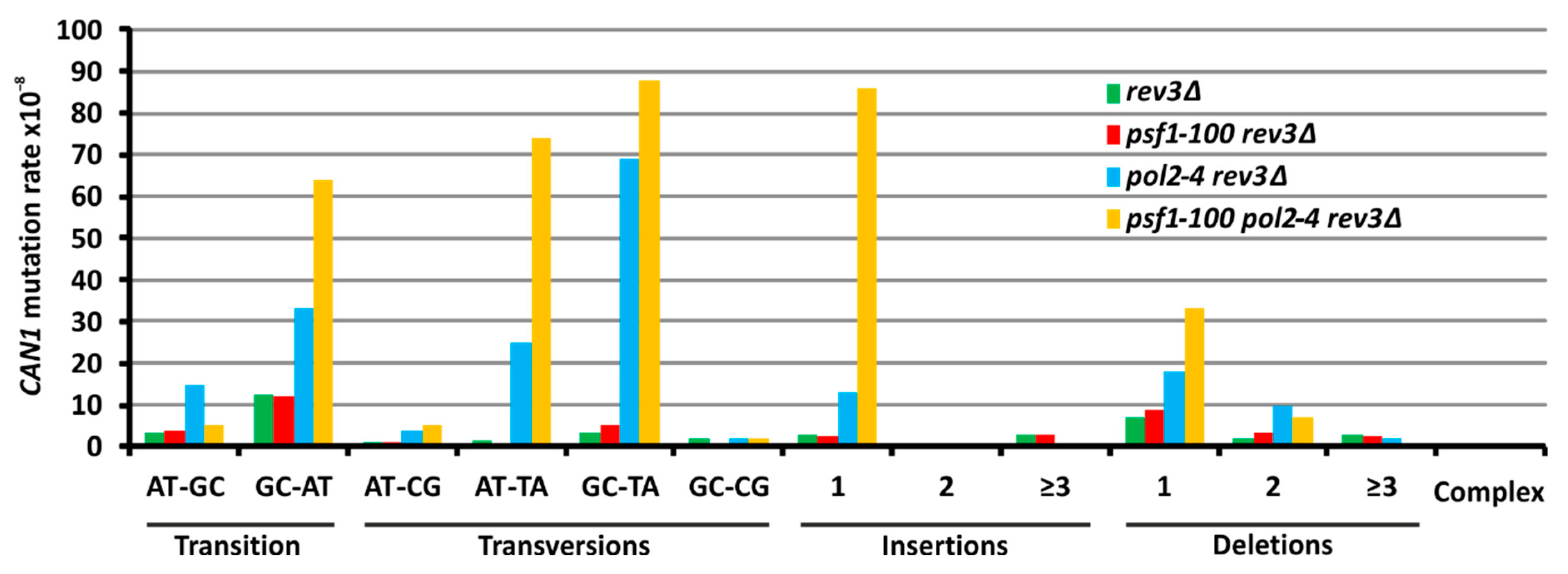
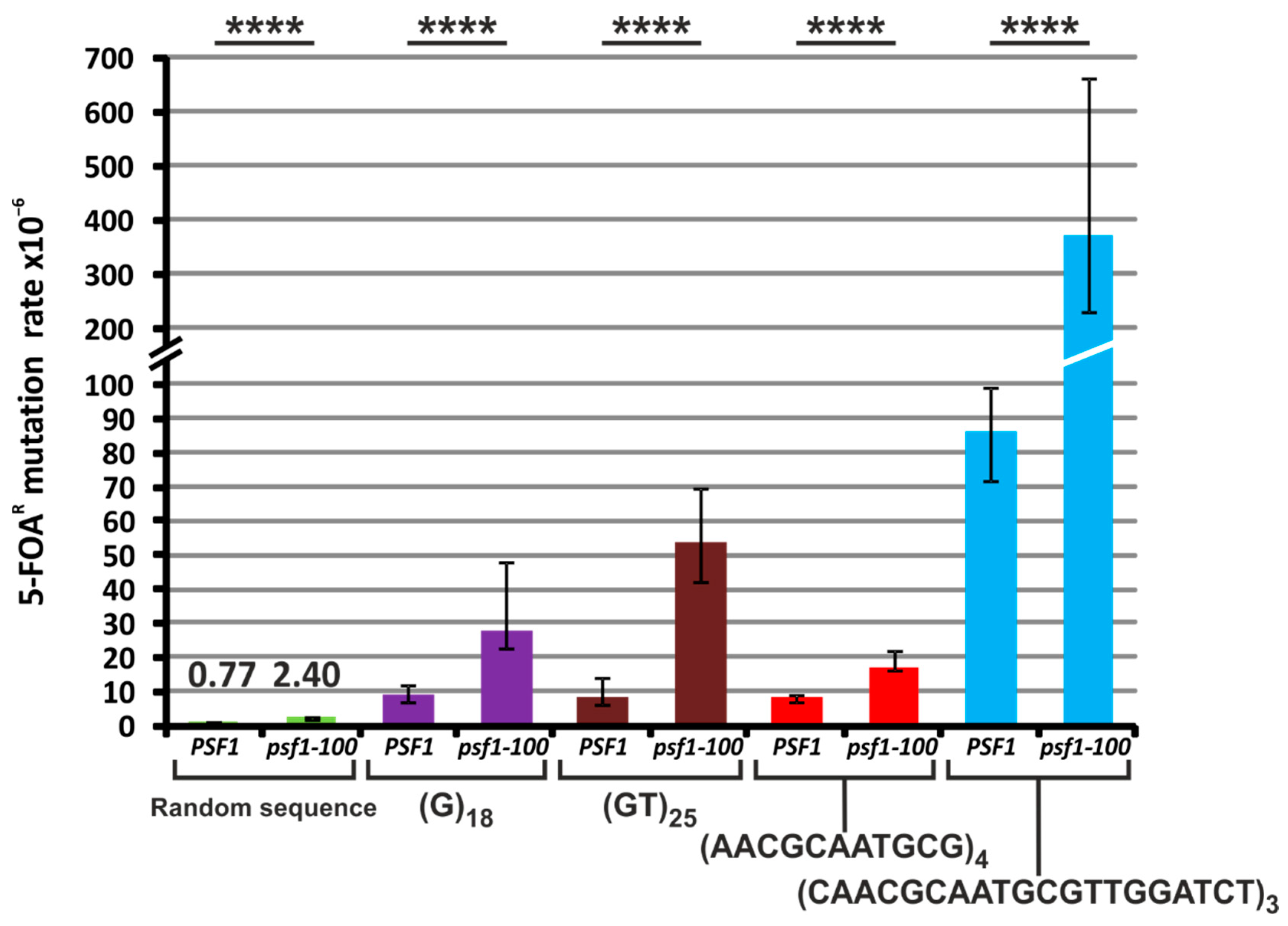
2.6. The psf1-100 Allele Facilitates Primer-Template Rearrangements, Frameshift Formation and the Instability of Repeated DNA Tracts
3. Discussion
4. Materials and Methods
4.1. Strains, Media and General Methods
4.2. Construction of Yeast Strains
4.3. Protein Purification
4.4. Pol ε-GINS In Vitro Interaction Assay
4.5. In Vitro Replication Assay
4.6. Measurement of Spontaneous Mutation Frequency at the CAN1 Locus
4.7. Calculation of Mutation Rates and Statistical Analysis
4.8. CANR Mutation Spectrum
4.9. Detection of Recombination Events
4.10. Determination of UV Radiation Sensitivity and UV-Induced Mutagenesis at the CAN1 Locus
4.11. Synchronization in G1 Phase with α-Factor
4.12. Flow Cytometry Analysis
4.13. Western Blot Analysis
4.14. Identification of Rad52, Rad51 and Rfa1 Foci by Fluorescence Microscopy
4.15. Yeast Viability Assessment
4.16. Stability of Repetitive Sequences
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BIR | Break-induced recombination |
| CMG | Cdc45, Mcm2–7, GINS complex |
| CMGE | Cdc45, Mcm2–7, GINS, DNA polymerase epsilon (Pol ε) complex |
| GINS | Psf1, Psf2, Psf3, Sld5 complex |
| HR | Homologous recombination |
| TS | Template switch |
References
- Ubhi, T.; Brown, G.W. Exploiting DNA replication stress for cancer treatment. Cancer Res. 2019, 79, 1730–1739. [Google Scholar] [CrossRef] [PubMed]
- Barnes, R.; Eckert, K. Maintenance of genome integrity: How mammalian cells orchestrate genome duplication by coordinating replicative and specialized DNA polymerases. Genes (Basel) 2017, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Kurth, I.; O’Donnell, M.E. New insights into replisome fluidity during chromosome replication. Trends Biochem. Sci. 2013, 38, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Yuan, Z.; Georgescu, R.; Li, H.; O’Donnell, M. The eukaryotic CMG helicase pumpjack and integration into the replisome. Nucleus 2016, 7, 146–154. [Google Scholar] [CrossRef][Green Version]
- Ishimi, Y. Regulation of MCM2–7 function. Genes Genet. Syst. 2018, 93, 125–133. [Google Scholar] [CrossRef]
- Li, H.; O’Donnell, M.E. The Eukaryotic CMG Helicase at the Replication Fork: Emerging Architecture Reveals an Unexpected Mechanism. BioEssays 2018, 40, 1700208. [Google Scholar] [CrossRef]
- Tercero, J.A. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 2000, 19, 2082–2093. [Google Scholar] [CrossRef]
- Aparicio, O.M.; Weinstein, D.M.; Bell, S.P. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell 1997, 91, 59–69. [Google Scholar] [CrossRef]
- Zou, L.; Stillman, B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science 1998, 280, 593–596. [Google Scholar] [CrossRef]
- Gambus, A.; Jones, R.C.; Sanchez-Diaz, A.; Kanemaki, M.; van Deursen, F.; Edmondson, R.D.; Labib, K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006, 8, 358–366. [Google Scholar] [CrossRef]
- Moyer, S.E.; Lewis, P.W.; Botchan, M.R. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 2006, 103, 10236–10241. [Google Scholar] [CrossRef] [PubMed]
- Boos, D.; Frigola, J.; Diffley, J.F.X. Activation of the replicative DNA helicase: Breaking up is hard to do. Curr. Opin. Cell Biol. 2012, 24, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Takayama, Y.; Kamimura, Y.; Okawa, M.; Muramatsu, S.; Sugino, A.; Araki, H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003, 17, 1153–1165. [Google Scholar] [CrossRef] [PubMed]
- Kanemaki, M.; Sanchez-Diaz, A.; Gambus, A.; Labib, K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 2003, 423, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Takase, Y.; Komori, Y.; Hashimoto, Y.; Arata, T.; Kamimura, Y.; Araki, H.; Takisawa, H. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev. 2003, 17, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Schwacha, A. The Mcm2–7 Complex Has In Vitro Helicase Activity. Mol. Cell 2008, 31, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Kang, M.S.; Ryu, E.; Myung, K. Eukaryotic DNA replication: Orchestrated action of multi-subunit protein complexes. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2018, 809, 58–69. [Google Scholar] [CrossRef]
- Bell, S.P.; Labib, K. Chromosome Duplication in Saccharomyces cerevisiae. Genetics 2016, 203, 1027–1067. [Google Scholar] [CrossRef]
- Lujan, S.A.; Williams, J.S.; Kunkel, T.A. DNA Polymerases Divide the Labor of Genome Replication. Trends Cell Biol. 2016, 26, 640–654. [Google Scholar] [CrossRef]
- Nick McElhinny, S.A.; Gordenin, D.A.; Stith, C.M.; Burgers, P.M.J.; Kunkel, T.A. Division of Labor at the Eukaryotic Replication Fork. Mol. Cell 2008, 30, 137–144. [Google Scholar] [CrossRef]
- Georgescu, R.E.; Langston, L.; Yao, N.Y.; Yurieva, O.; Zhang, D.; Finkelstein, J.; Agarwal, T.; O’Donnell, M.E. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat. Struct. Mol. Biol. 2014, 21, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, Y.I.; Maki, S.; Maki, H.; Kunkel, T.A. Evidence for interplay among yeast replicative DNA polymerases alpha, delta and epsilon from studies of exonuclease and polymerase active site mutations. BMC Biol. 2004, 2, 11. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Garbacz, M.A.; Lujan, S.A.; Burkholder, A.B.; Cox, P.B.; Wu, Q.; Zhou, Z.X.; Haber, J.E.; Kunkel, T.A. Evidence that DNA polymerase δ contributes to initiating leading strand DNA replication in Saccharomyces cerevisiae. Nat. Commun. 2018, 9, 858. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.X.; Lujan, S.A.; Burkholder, A.B.; Garbacz, M.A.; Kunkel, T.A. Roles for DNA polymerase δ in initiating and terminating leading strand DNA replication. Nat. Commun. 2019, 10, 3992. [Google Scholar] [CrossRef]
- Yeeles, J.T.P.; Janska, A.; Early, A.; Diffley, J.F.X. How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication. Mol. Cell 2016, 65, 105–116. [Google Scholar] [CrossRef]
- Aria, V.; Yeeles, J.T.P. Mechanism of Bidirectional Leading-Strand Synthesis Establishment at Eukaryotic DNA Replication Origins. Mol. Cell 2019, 73, 199–211. [Google Scholar] [CrossRef]
- Pursell, Z.F.; Isoz, I.; Lundström, E.-B.; Johansson, E.; Kunkel, T.A. Yeast DNA polymerase ε participates in leading-strand DNA replication. Science 2007, 317, 127–130. [Google Scholar] [CrossRef]
- Budd, M.E.; Campbell, J.L. DNA polymerases δ and ε are required for chromosomal replication in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993, 13, 496–505. [Google Scholar] [CrossRef][Green Version]
- Dmowski, M.; Rudzka, J.; Campbell, J.L.; Jonczyk, P.; Fijałkowska, I.J. Mutations in the Non-Catalytic Subunit Dpb2 of DNA Polymerase Epsilon Affect the Nrm1 Branch of the DNA Replication Checkpoint. PLOS Genet. 2017, 13, e1006572. [Google Scholar] [CrossRef]
- Dmowski, M.; Fijałkowska, I.J. Diverse roles of Dpb2, the non-catalytic subunit of DNA polymerase ε. Curr. Genet. 2017, 63, 983–987. [Google Scholar] [CrossRef]
- Dua, R.; Levy, D.L.; Campbell, J.L. Role of the putative zinc finger domain of Saccharomyces cerevisiae DNA polymerase epsilon in DNA replication and the S/M checkpoint pathway. J. Biol. Chem. 1998, 273, 30046–30055. [Google Scholar] [CrossRef] [PubMed]
- Navas, T.A.; Zhou, Z.; Elledge, S.J. DNA polymerase ε links the DNA replication machinery to the S phase checkpoint. Cell 1995, 80, 29–39. [Google Scholar] [CrossRef]
- Langston, L.D.; Zhang, D.; Yurieva, O.; Georgescu, R.E.; Finkelstein, J.; Yao, N.Y.; Indiani, C.; O’Donnell, M.E. CMG helicase and DNA polymerase ε form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc. Natl. Acad. Sci. USA 2014, 111, 15390–15395. [Google Scholar] [CrossRef] [PubMed]
- Georgescu, R.E.; Schauer, G.D.; Yao, N.Y.; Langston, L.D.; Yurieva, O.; Zhang, D.; Finkelstein, J.; O’Donnell, M.E.; O’Donnell, M.E.; O’Donnell, M.E. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. Elife 2015, 4, e04988. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Shi, Y.; Georgescu, R.E.; Yuan, Z.; Chait, B.T.; Li, H.; O’Donnell, M.E. The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol. 2015, 22, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.C.; Janska, A.; Goswami, P.; Renault, L.; Abid Ali, F.; Kotecha, A.; Diffley, J.F.X.; Costa, A. CMG–Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proc. Natl. Acad. Sci. USA 2017, 114, 201700530. [Google Scholar] [CrossRef]
- Yeeles, J.T.P.; Poli, J.; Marians, K.J.; Pasero, P. Rescuing stalled or damaged replication forks. Cold Spring Harb. Perspect. Biol. 2013, 5, a012815. [Google Scholar] [CrossRef]
- Bermudez, V.P.; Farina, A.; Raghavan, V.; Tappin, I.; Hurwitz, J. Studies on human DNA polymerase epsilon and GINS complex and their role in DNA replication. J. Biol. Chem. 2011, 286, 28963–28977. [Google Scholar] [CrossRef]
- Sengupta, S.; Van Deursen, F.; De Piccoli, G.; Labib, K. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr. Biol. 2013, 23, 543–552. [Google Scholar] [CrossRef]
- Hizume, K.; Endo, S.; Muramatsu, S.; Kobayashi, T.; Araki, H. DNA polymerase ε-dependent modulation of the pausing property of the CMG helicase at the barrier. Genes Dev. 2018, 32, 1315–1320. [Google Scholar] [CrossRef]
- Kesti, T.; Flick, K.; Keränen, S.; Syväoja, J.E.; Wittenberg, C. DNA Polymerase ε Catalytic Domains Are Dispensable for DNA Replication, DNA Repair, and Cell Viability. Mol. Cell 1999, 3, 679–685. [Google Scholar] [CrossRef]
- Dua, R.; Levy, D.L.; Campbell, J.L. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol ε and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 1999, 274, 22283–22288. [Google Scholar] [CrossRef]
- Araki, H.; Hamatake, R.K.; Johnston, L.H.; Sugino, A. DPB2, the gene encoding DNA polymerase II subunit B, is required for chromosome replication in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1991, 88, 4601–4605. [Google Scholar] [CrossRef] [PubMed]
- Ohya, T.; Maki, S.; Kawasaki, Y.; Sugino, A. Structure and function of the fourth subunit (Dpb4p) of DNA polymerase epsilon in Saccharomyces cerevisiae. Nucleic Acids Res. 2000, 28, 3846–3852. [Google Scholar] [CrossRef] [PubMed]
- Araki, H.; Hamatake, R.K.; Morrison, A.; Johnson, A.L.; Johnston, L.H.; Sugino, A. Cloning DPB3, the gene encoding the third subunit of DNA polymerase II of Saccharomyces cerevisiae. Nucleic Acids Res. 1991, 19, 4867–4872. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yuan, Z.; Georgescu, R.; Schauer, G.D.; O’Donnell, M.E.; Li, H. Structure of the polymerase ε holoenzyme and atomic model of the leading strand replisome. Nat. Commun. 2020, 11, 3156. [Google Scholar] [CrossRef]
- Goswami, P.; Abid Ali, F.; Douglas, M.E.; Locke, J.; Purkiss, A.; Janska, A.; Eickhoff, P.; Early, A.; Nans, A.; Cheung, A.M.C.; et al. Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome. Nat. Commun. 2018, 9, 5061. [Google Scholar] [CrossRef]
- Muramatsu, S.; Hirai, K.; Tak, Y.S.; Kamimura, Y.; Araki, H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol ε, and GINS in budding yeast. Genes Dev. 2010, 24, 602–612. [Google Scholar] [CrossRef]
- Grabowska, E.; Wronska, U.; Denkiewicz, M.; Jaszczur, M.; Respondek, A.; Alabrudzinska, M.; Suski, C.; Makiela-Dzbenska, K.; Jonczyk, P.; Fijalkowska, I.J. Proper functioning of the GINS complex is important for the fidelity of DNA replication in yeast. Mol. Microbiol. 2014, 92, 659–680. [Google Scholar] [CrossRef]
- Handa, T.; Kanke, M.; Takahashi, T.S.; Nakagawa, T.; Masukata, H. DNA polymerization-independent functions of DNA polymerase epsilon in assembly and progression of the replisome in fission yeast. Mol. Biol. Cell 2012, 23, 3240–3253. [Google Scholar] [CrossRef]
- Nelson, J.R.; Lawrence, C.W.; Hinkle, D.C. Thymine-Thymine Dimer Bypass by Yeast DNA Polymerase ζ. Science (80-) 1996, 272, 1646–1649. [Google Scholar] [CrossRef] [PubMed]
- Baranovskiy, A.G.; Lada, A.G.; Siebler, H.M.; Zhang, Y.; Pavlov, Y.I.; Tahirov, T.H. DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J. Biol. Chem. 2012, 287, 17281–17287. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.E.; Prakash, L.; Prakash, S. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc. Natl. Acad. Sci. USA 2012, 109, 12455–12460. [Google Scholar] [CrossRef] [PubMed]
- Makarova, A.V.; Stodola, J.L.; Burgers, P.M. A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012, 40, 11618–11626. [Google Scholar] [CrossRef]
- Vaisman, A.; Woodgate, R. Translesion DNA polymerases in eukaryotes: What makes them tick? Crit. Rev. Biochem. Mol. Biol. 2017, 52, 274–303. [Google Scholar] [CrossRef]
- Martin, S.K.; Wood, R.D. DNA polymerase ζ in DNA replication and repair. Nucleic Acids Res. 2019, 47, 8348–8361. [Google Scholar] [CrossRef]
- Kochenova, O.V.; Daee, D.L.; Mertz, T.M.; Shcherbakova, P.V. DNA Polymerase ζ-Dependent Lesion Bypass in Saccharomyces cerevisiae Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA. PLoS Genet. 2015, 11, e1005110. [Google Scholar] [CrossRef]
- Lemontt, J.F. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics 1971, 68, 21–33. [Google Scholar]
- Quah, S.K.; Von Borstel, R.C.; Hastings, P.J. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics 1980, 96, 819–839. [Google Scholar]
- Northam, M.R.; Robinson, H.A.; Kochenova, O.V.; Shcherbakova, P.V. Participation of DNA polymerase ζ in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics 2010, 184, 27–42. [Google Scholar] [CrossRef]
- Aksenova, A.; Volkov, K.; Maceluch, J.; Pursell, Z.F.; Rogozin, I.B.; Kunkel, T.A.; Pavlov, Y.I.; Johansson, E. Mismatch repair-independent increase in spontaneous mutagenesis in yeast lacking non-essential subunits of DNA polymerase epsilon. PLoS Genet. 2010, 6, e1001209. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.R.; Nguyen, H.D.; Wang, X.; Bielinsky, A.K. Mcm10 deficiency causes defective-replisome-induced mutagenesis and a dependency on error-free postreplicative repair. Cell Cycle 2014, 13, 1737–1748. [Google Scholar] [CrossRef] [PubMed]
- Garbacz, M.; Araki, H.; Flis, K.; Bebenek, A.; Zawada, A.E.; Jonczyk, P.; Makiela-Dzbenska, K.; Fijalkowska, I.J. Fidelity consequences of the impaired interaction between DNA polymerase epsilon and the GINS complex. DNA Repair 2015, 29, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Kraszewska, J.; Garbacz, M.; Jonczyk, P.; Fijalkowska, I.J.; Jaszczur, M. Defect of Dpb2p, a noncatalytic subunit of DNA polymerase ε, promotes error prone replication of undamaged chromosomal DNA in Saccharomyces cerevisiae. Mutat. Res. 2012, 737, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Szwajczak, E.; Fijalkowska, I.J.; Suski, C. The CysB motif of Rev3p involved in the formation of the four-subunit DNA polymerase ζ is required for defective-replisome-induced mutagenesis. Mol. Microbiol. 2017, 106, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Northam, M.R.; Garg, P.; Baitin, D.M.; Burgers, P.M.J.; Shcherbakova, P.V. A novel function of DNA polymerase ζ regulated by PCNA. EMBO J. 2006, 25, 4316–4325. [Google Scholar] [CrossRef]
- Wright, W.D.; Shah, S.S.; Heyer, W.D. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10524–10535. [Google Scholar] [CrossRef]
- Sun, Y.; McCorvie, T.J.; Yates, L.A.; Zhang, X. Structural basis of homologous recombination. Cell. Mol. Life Sci. 2020, 77, 3–18. [Google Scholar] [CrossRef]
- Syeda, A.H.; Hawkins, M.; McGlynn, P. Recombination and Replication. Cold Spring Harb. Perspect. Biol. 2014, 6, 1–14. [Google Scholar] [CrossRef]
- Piazza, A.; Heyer, W.D. Moving forward one step back at a time: Reversibility during homologous recombination. Curr. Genet. 2019, 65, 1333–1340. [Google Scholar] [CrossRef]
- Barkley, L.R.; Song, I.Y.; Zou, Y.; Vaziri, C. Reduced expression of GINS complex members induces hallmarks of pre-malignancy in primary untransformed human cells. Cell Cycle 2009, 8, 1577–1588. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cottineau, J.; Kottemann, M.C.; Lach, F.P.; Kang, Y.H.; Vély, F.; Deenick, E.K.; Lazarov, T.; Gineau, L.; Wang, Y.; Farina, A.; et al. Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency. J. Clin. Investig. 2017, 127, 1991–2006. [Google Scholar] [CrossRef] [PubMed]
- Homma, T.; Freire, B.; Honjo Kawahira, R.; Dauber, A.; Funari, M.; Lerario, A.; Nishi, M.; Albuquerque, E.; Vasques, G.; Collett-Solberg, P.; et al. Genetic Disorders in Prenatal Onset Syndromic Short Stature. J. Pediatr. 2019, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, I.; Miyamoto, M.; Shibata, T.; Akashi-tanaka, S.; Kinoshita, T.; Mogushi, K.; Oda, K.; Ueno, M.; Takakura, N.; Mizushima, H.; et al. Up-regulation of PSF1 promotes the growth of breast cancer cells. Genes Cells 2010, 15, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhou, Q.-Z.; Zhang, X.-L.; Wang, Z.-Z.; Wang, P. Identification of Hub Genes Using Co-Expression Network Analysis in Breast Cancer as a Tool to Predict Different Stages. Med. Sci. Monit. 2019, 8873–8890. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Cui, D.; Kawano, H.; Yoshitomi, C.; Nobuyuki, S.; Eiji, T. Induced expression of GINS complex is an essential step for reactivation of quiescent stem-like tumor cells within the peri- necrotic niche in human glioblastoma. J. Cancer Res. Clin. Oncol. 2019, 145, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Toda, H.; Seki, N.; Kurozumi, S.; Shinden, Y.; Yamada, Y. RNA-sequence-based microRNA expression signature in breast cancer: Tumor-suppressive miR-101-5p regulates molecular pathogenesis. Mol. Oncol. 2020, 14, 426–446. [Google Scholar] [CrossRef]
- Seo, Y.-S.; Kang, Y.-H. The Human Replicative Helicase, the CMG Complex, as a Target for Anti-cancer Therapy. Front. Mol. Biosci. 2018, 5, 26. [Google Scholar] [CrossRef]
- Reed, D.R.; Alexandrow, M.G. Myc and the Replicative CMG Helicase: The Creation and Destruction of Cancer: Myc Over-Activation of CMG Helicases Drives Tumorigenesis and Creates a Vulnerability in CMGs for Therapeutic Intervention. BioEssays 2020, 42, 1900218. [Google Scholar] [CrossRef]
- Yeeles, J.T.P.; Deegan, T.D.; Janska, A.; Early, A.; Diffley, J.F.X. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [Google Scholar] [CrossRef]
- Maréchal, A.; Zou, L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015, 25, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Bhat, K.P.; Cortez, D. RPA and RAD51: Fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 2018, 25, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Lisby, M.; Barlow, J.H.; Burgess, R.C.; Rothstein, R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 2004, 118, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Lisby, M.; Rothstein, R.; Mortensen, U.H. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 2001, 98, 8276–8282. [Google Scholar] [CrossRef]
- Barbera, M.A.; Petes, T.D. Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2006, 103, 12819–12824. [Google Scholar] [CrossRef]
- Branzei, D.; Szakal, B. Building up and breaking down: Mechanisms controlling recombination during replication. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 381–394. [Google Scholar] [CrossRef]
- Kramara, J.; Osia, B.; Malkova, A. Break-Induced Replication: The Where, The Why, and The How. Trends Genet. 2018, 34, 518–531. [Google Scholar] [CrossRef]
- Skoneczna, A.; Krol, K.; Skoneczny, M. How Do Yeast and Other Fungi Recognize and Respond to Genome Perturbations? In Stress Response Mechanisms in Fungi; Springer Nature: Cham, Switzerland, 2018; pp. 87–130. ISBN 9783030006839. [Google Scholar]
- Boiteux, S.; Jinks-Robertson, S. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics 2013, 193, 1025–1064. [Google Scholar] [CrossRef]
- Saugar, I.; Ortiz-Bazán, M.Á.; Tercero, J.A. Tolerating DNA damage during eukaryotic chromosome replication. Exp. Cell Res. 2014, 329, 170–177. [Google Scholar] [CrossRef]
- Buzovetsky, O.; Kwon, Y.; Pham, N.T.; Kim, C.; Ira, G.; Sung, P.; Xiong, Y. Role of the Pif1-PCNA Complex in Pol δ-Dependent Strand Displacement DNA Synthesis and Break-Induced Replication. Cell Rep. 2017, 21, 1707–1714. [Google Scholar] [CrossRef]
- Wilson, M.A.; Kwon, Y.; Xu, Y.; Chung, W.H.; Chi, P.; Niu, H.; Mayle, R.; Chen, X.; Malkova, A.; Sung, P.; et al. Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature 2013, 502, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Lydeard, J.R.; Jain, S.; Yamaguchi, M.; Haber, J.E. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 2007, 448, 820–823. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.H.; Obert, R.; Burgers, P.M.J.; Kunkel, T.A.; Resnick, M.A.; Gordenin, D.A. The 3′→5′ exonuclease of DNA polymerase δ can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl. Acad. Sci. USA 2001, 98, 5122–5127. [Google Scholar] [CrossRef] [PubMed]
- Karras, G.I.; Jentsch, S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 2010, 141, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Enserink, J.M.; Kolodner, R.D. An overview of Cdk1-controlled targets and processes. Cell Div. 2010, 5, 11. [Google Scholar] [CrossRef]
- Tanaka, S.; Araki, H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010371. [Google Scholar] [CrossRef]
- Amon, A.; Tyers, M.; Futcher, B.; Nasmyth, K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 1993, 74, 993–1007. [Google Scholar] [CrossRef]
- Deere, D.; Shen, J.; Vesey, G.; Bell, P.; Bissinger, P.; Veal, D. Flow cytometry and cell sorting for yeast viability assessment and cell selection. Yeast 1998, 14, 147–160. [Google Scholar] [CrossRef]
- Pavla, V.; Renata, L.; Ivana, M.; Jiri, H. The Stationary-Phase Cells of Saccharomyces cerevisiae Display Dynamic Actin Filaments Required for Processes Extending Chronological Life Span. Mol. Cell. Biol. 2015, 35, 3892–3908. [Google Scholar] [CrossRef]
- Streisinger, G.; Okada, Y.; Emrich, J.; Newton, J.; Tsugita, A.; Terzaghi, E.; Inouye, M. Frameshift mutations and the genetic code. Cold Spring Harb. Symp. Quant. Biol. 1966, 31, 77–84. [Google Scholar] [CrossRef]
- Morrison, A.; Sugino, A. The 3′ → 5′ exonucleases of both DNA polymerases δ and ε participate in correcting errors of DNA replication in Saccharomyces cerevisiae. MGG Mol. Gen. Genet. 1994, 242, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Tóth, G.; Gáspári, Z.; Jurka, J. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Res. 2000, 10, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Wren, J.D.; Forgacs, E.; Fondon, J.W.; Pertsemlidis, A.; Cheng, S.Y.; Gallardo, T.; Williams, R.S.; Shohet, R.V.; Minna, J.D.; Garner, H.R. Repeat Polymorphisms within Gene Regions: Phenotypic and Evolutionary Implications. Am. J. Hum. Genet. 2000, 67, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Paulson, H. Repeat expansion diseases. In Handbook of Clinical Neurology; Geschwind, D.H., Paulson, H.L., Klein, C.B.T., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 147, pp. 105–123. ISBN 0072-9752. [Google Scholar]
- Henderson, S.T.; Petes, T.D. Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992, 12, 2749–2757. [Google Scholar] [CrossRef][Green Version]
- Sia, E.A.; Kokoska, R.J.; Dominska, M.; Greenwell, P.; Petes, T.D. Microsatellite instability in yeast: Dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 1997, 17, 2851–2858. [Google Scholar] [CrossRef]
- Wierdl, M.; Dominska, M.; Petes, T.D. Microsatellite Instability in Yeast: Dependence on the Length of the Microsatellite. Genetics 1997, 779, 769–779. [Google Scholar]
- Jedrychowska, M.; Denkiewicz-Kruk, M.; Alabrudzinska, M.; Skoneczna, A.; Jonczyk, P.; Dmowski, M.; Fijalkowska, I.J. Defects in the GINS complex increase the instability of repetitive sequences via a recombination-dependent mechanism. PLoS Genet. 2019, 15, e1008494. [Google Scholar] [CrossRef]
- Boeke, J.D.; LaCroute, F.; Fink, G.R. A positive selection for mutants lacking 5′ phosphate decarboxylase activity in yeast: 5 fluoro-orotic acid resistance. Mol. Gen. Genet. 1984, 197, 345–346. [Google Scholar] [CrossRef]
- Jaszczur, M.; Flis, K.; Rudzka, J.; Kraszewska, J.; Budd, M.E.; Polaczek, P.; Campbell, J.L.; Jonczyk, P.; Fijalkowska, I.J. Dpb2p, a noncatalytic subunit of DNA polymerase ε, contributes to the fidelity of DNA replication in Saccharomyces cerevisiae. Genetics 2008, 178, 633–647. [Google Scholar] [CrossRef]
- Zhou, L.; Sun, X.; Liu, C.; Wu, Q.; Tai, M.; Wei, J.; Lei, L.; Meng, F.; Qu, K.; Xu, J. Overexpression of PSF1 is correlated with poor prognosis in hepatocellular carcinoma patients. Int. J. Biol. Markers 2015, 30, e56–e64. [Google Scholar] [CrossRef]
- Hogg, M.; Johansson, E. DNA Polymerase ε in The Eukaryotic Replisome: A Guide to Protein Structure and Function; MacNeill, S., Ed.; Springer Science+Business Media: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Masumoto, H.; Muramatsu, S.; Kamimura, Y.; Araki, H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 2002, 415, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Umemori, T.; Hirai, K.; Muramatsu, S.; Kamimura, Y.; Araki, H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 2007, 445, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Zegerman, P.; Diffley, J.F.X. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 2007, 445, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Tak, Y.-S.; Araki, H. The role of CDK in the initiation step of DNA replication in eukaryotes. Cell Div. 2007, 2, 16. [Google Scholar] [CrossRef]
- Gadgil, R.; Barthelemy, J.; Lewis, T.; Leffak, M. Replication stalling and DNA microsatellite instability. Biophys. Chem. 2017, 225, 38–48. [Google Scholar] [CrossRef]
- Sogo, J.M.; Lopes, M.; Foiani, M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 2002, 297, 599–602. [Google Scholar] [CrossRef]
- González-Prieto, R.; Muñoz-Cabello, A.M.; Cabello-Lobato, M.J.; Prado, F. Rad51 replication fork recruitment is required for DNA damage tolerance. EMBO J. 2013, 32, 1307–1321. [Google Scholar] [CrossRef]
- Malacaria, E.; Pugliese, G.M.; Honda, M.; Marabitti, V.; Aiello, F.A.; Spies, M.; Franchitto, A.; Pichierri, P. Rad52 prevents excessive replication fork reversal and protects from nascent strand degradation. Nat. Commun. 2019, 10, 1412. [Google Scholar] [CrossRef]
- Ait Saada, A.; Lambert, S.A.E.; Carr, A.M. Preserving replication fork integrity and competence via the homologous recombination pathway. DNA Repair 2018, 71, 135–147. [Google Scholar] [CrossRef]
- Halas, A.; Krawczyk, M.; Sledziewska-Gojska, E. PCNA SUMOylation protects against PCNA polyubiquitination-mediated, Rad59-dependent, spontaneous, intrachromosomal gene conversion. Mutat. Res. Mol. Mech. Mutagen. 2016, 791–792, 10–18. [Google Scholar] [CrossRef]
- Pavlov, Y.I.; Newlon, C.S.; Kunkel, T.A. Yeast origins establish a strand bias for replicational mutagenesis. Mol. Cell 2002, 10, 207–213. [Google Scholar] [CrossRef]
- Amberg, D.C.; Burke, D.J.; Strathern, J.N. Methods in Yeast Genetics. A Cold Spring Harbor Laboratory Course Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2005. [Google Scholar]
- Gietz, R.D.; Woods, R.A. Transformation of Yeast by the Lithium Acetate/Single-Stranded Carrier DNA/PEG Method; Methods in Microbiology; Elsevier: Amsterdam, The Netherlands, 1998; Volume 26, ISBN 9780125215268. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Komata, M.; Bando, M.; Araki, H.; Shirahige, K. The direct binding of Mrc1, a checkpoint mediator, to Mcm6, a replication helicase, is essential for the replication checkpoint against methyl methanesulfonate-induced stress. Mol. Cell. Biol. 2009, 29, 5008–5019. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.W. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 1991, 88, 7160–7164. [Google Scholar] [CrossRef] [PubMed]
- Lea, D.E.; Coulson, C.A. The distribution of the numbers of mutants in bacterial populations. Genetics 1947, 49, 264–285. [Google Scholar] [CrossRef]
- Knop, M.; Siegers, K.; Pereira, G.; Zachariae, W.; Winsor, B.; Nasmyth, K.; Schiebel, E. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast 1999, 15, 963–972. [Google Scholar] [CrossRef]
- Torres-Rosell, J.; Sunjevaric, I.; De Piccoli, G.; Sacher, M.; Eckert-Boulet, N.; Reid, R.; Jentsch, S.; Rothstein, R.; Aragón, L.; Lisby, M. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2007, 9, 923–931. [Google Scholar] [CrossRef]

| Genotype | CAN1 Mutation Rate | p-Value 2 | ||
|---|---|---|---|---|
| × 10−8 | 95% Confidence Intervals | Relative 1 | ||
| WT | 176.0 | 148.4–247.0 | 1.0 | |
| G2-REV3 | 232.5 | 216.7–246.1 | 1.5 | 0.059951 |
| rev3Δ | 95.3 | 88.4–117.6 | 0.6 | 0.000855 |
| psf1-100 | 331.9 | 299.8–415.1 | 2.0 | |
| psf1-100 G2-REV3 | 308.6 | 301.4–382.3 | 2.0 | 0.783131 |
| psf1-100 rev3Δ | 76.5 | 69.5–87.8 | 0.5 | 0.000017 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Denkiewicz-Kruk, M.; Jedrychowska, M.; Endo, S.; Araki, H.; Jonczyk, P.; Dmowski, M.; Fijalkowska, I.J. Recombination and Pol ζ Rescue Defective DNA Replication upon Impaired CMG Helicase—Pol ε Interaction. Int. J. Mol. Sci. 2020, 21, 9484. https://doi.org/10.3390/ijms21249484
Denkiewicz-Kruk M, Jedrychowska M, Endo S, Araki H, Jonczyk P, Dmowski M, Fijalkowska IJ. Recombination and Pol ζ Rescue Defective DNA Replication upon Impaired CMG Helicase—Pol ε Interaction. International Journal of Molecular Sciences. 2020; 21(24):9484. https://doi.org/10.3390/ijms21249484
Chicago/Turabian StyleDenkiewicz-Kruk, Milena, Malgorzata Jedrychowska, Shizuko Endo, Hiroyuki Araki, Piotr Jonczyk, Michal Dmowski, and Iwona J. Fijalkowska. 2020. "Recombination and Pol ζ Rescue Defective DNA Replication upon Impaired CMG Helicase—Pol ε Interaction" International Journal of Molecular Sciences 21, no. 24: 9484. https://doi.org/10.3390/ijms21249484
APA StyleDenkiewicz-Kruk, M., Jedrychowska, M., Endo, S., Araki, H., Jonczyk, P., Dmowski, M., & Fijalkowska, I. J. (2020). Recombination and Pol ζ Rescue Defective DNA Replication upon Impaired CMG Helicase—Pol ε Interaction. International Journal of Molecular Sciences, 21(24), 9484. https://doi.org/10.3390/ijms21249484





