Multi-Omics Analysis Provides Novel Insight into Immuno-Physiological Pathways and Development of Thermal Resistance in Rainbow Trout Exposed to Acute Thermal Stress
Abstract
1. Introduction
2. Results
2.1. Fish under Acute Thermal Stress
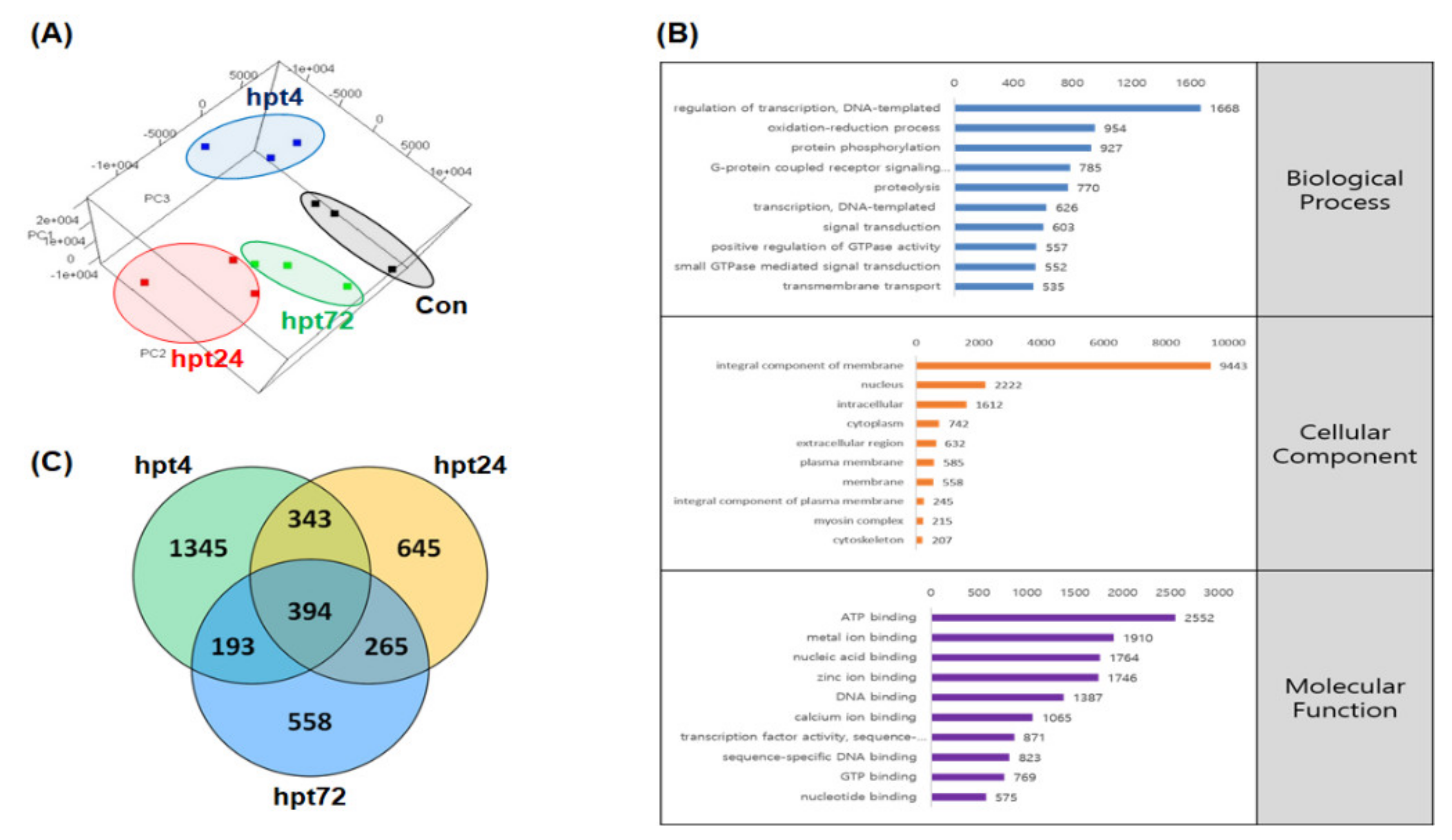
2.2. Genome-Guided Transcriptome Assembly
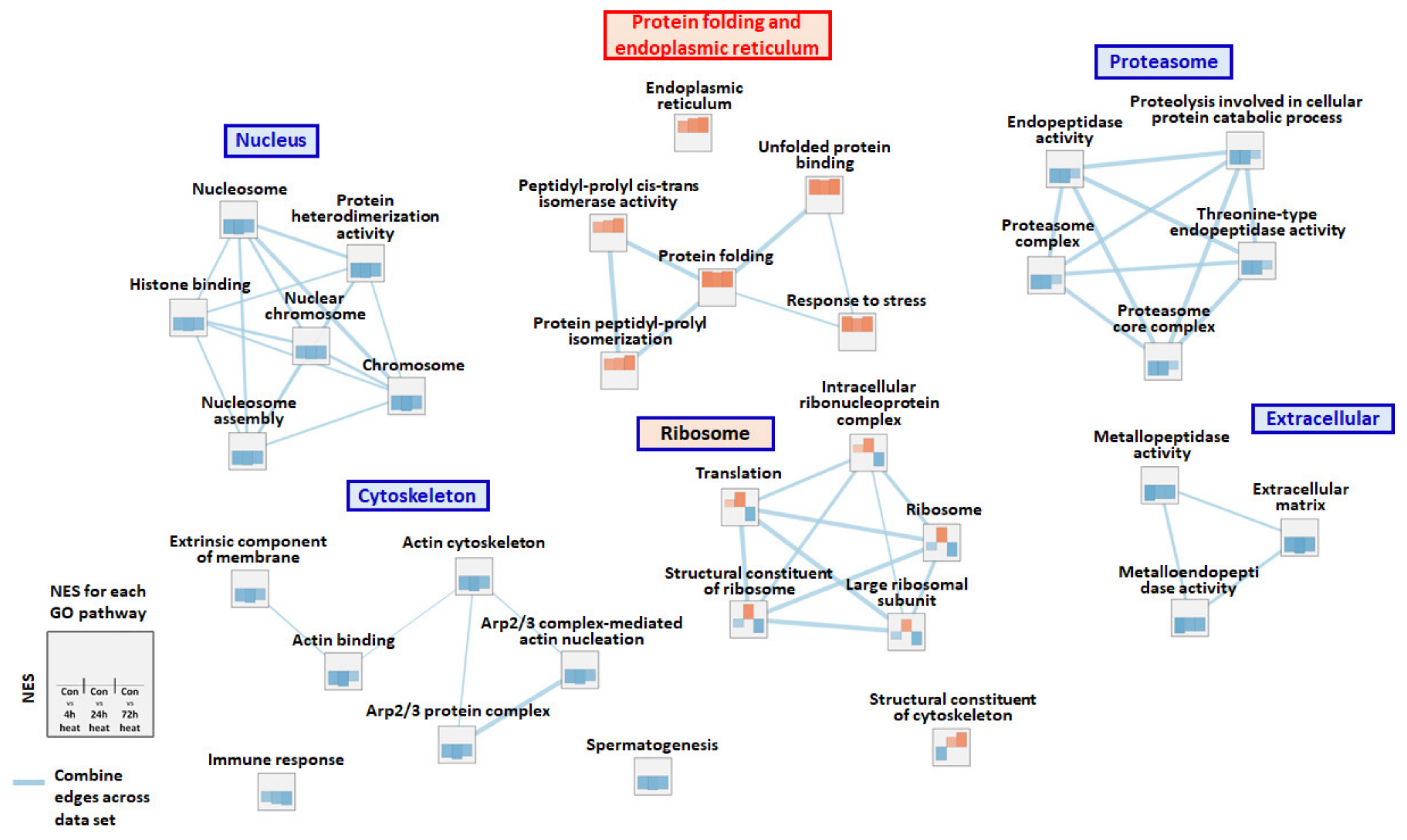
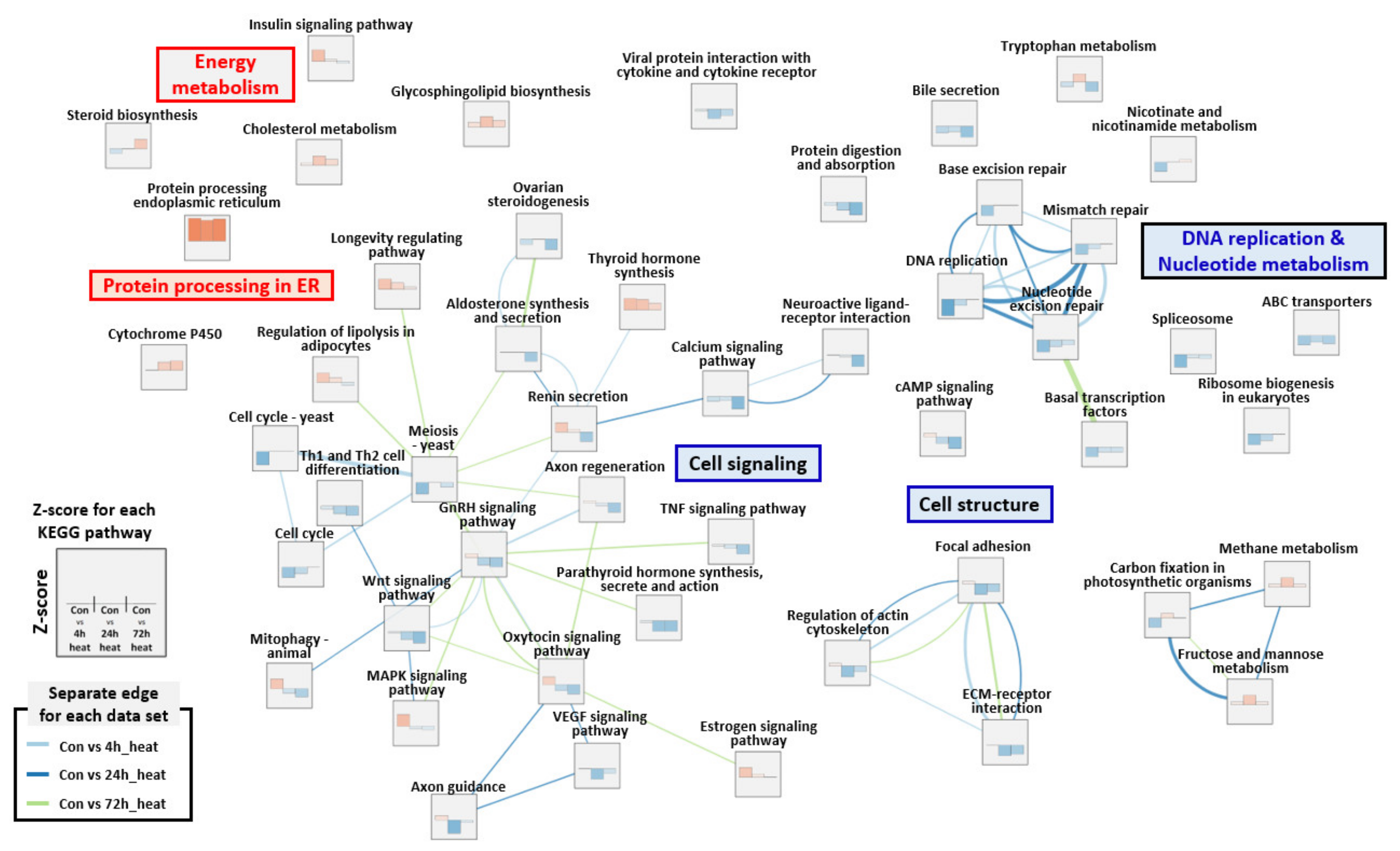
2.3. Gene Set Enrichment Analysis (GSEA)
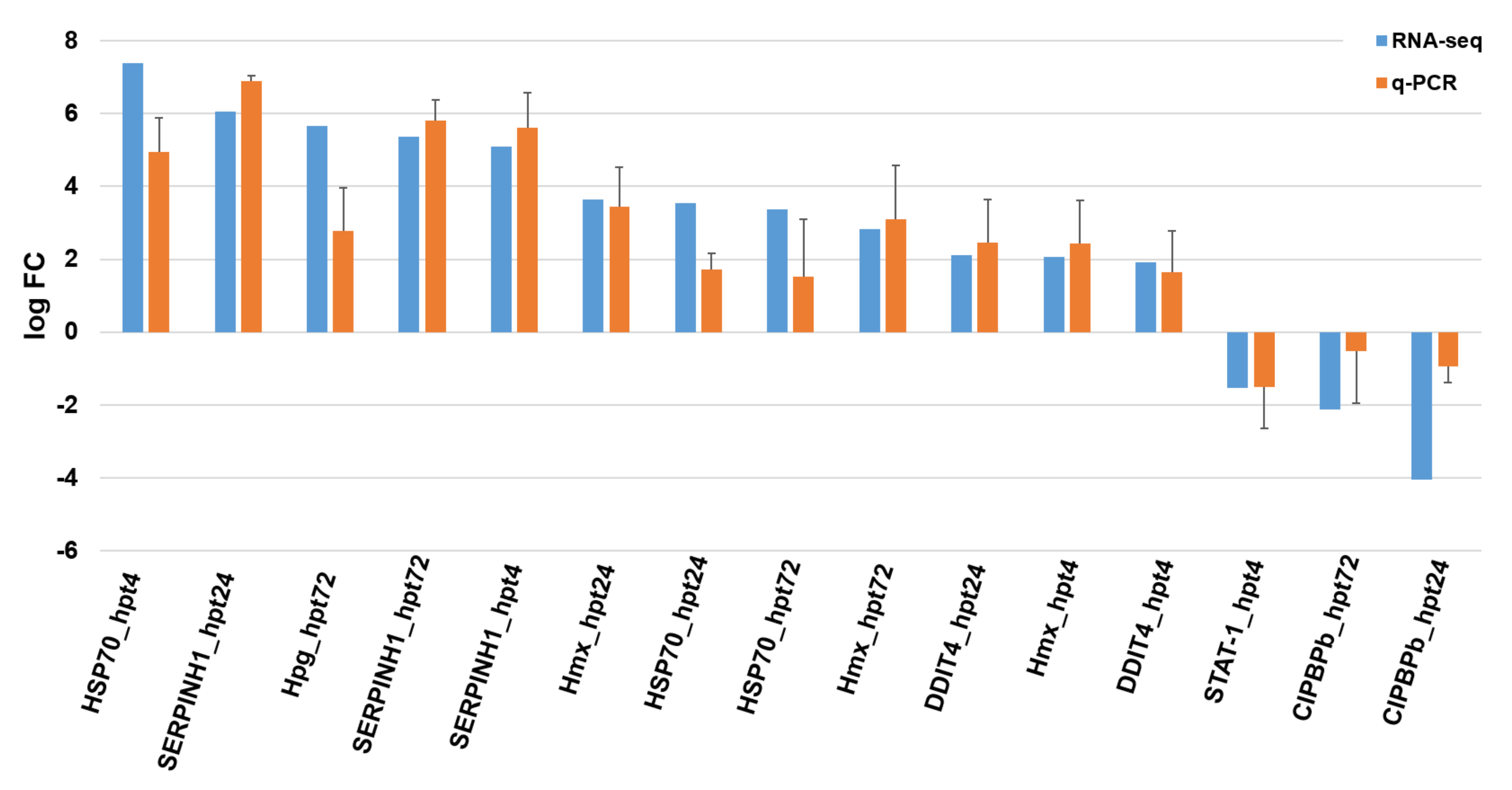
2.4. Differentially Expressed Gene (DEG) Analysis and RNA-Seq Validation
2.5. Serological Analysis
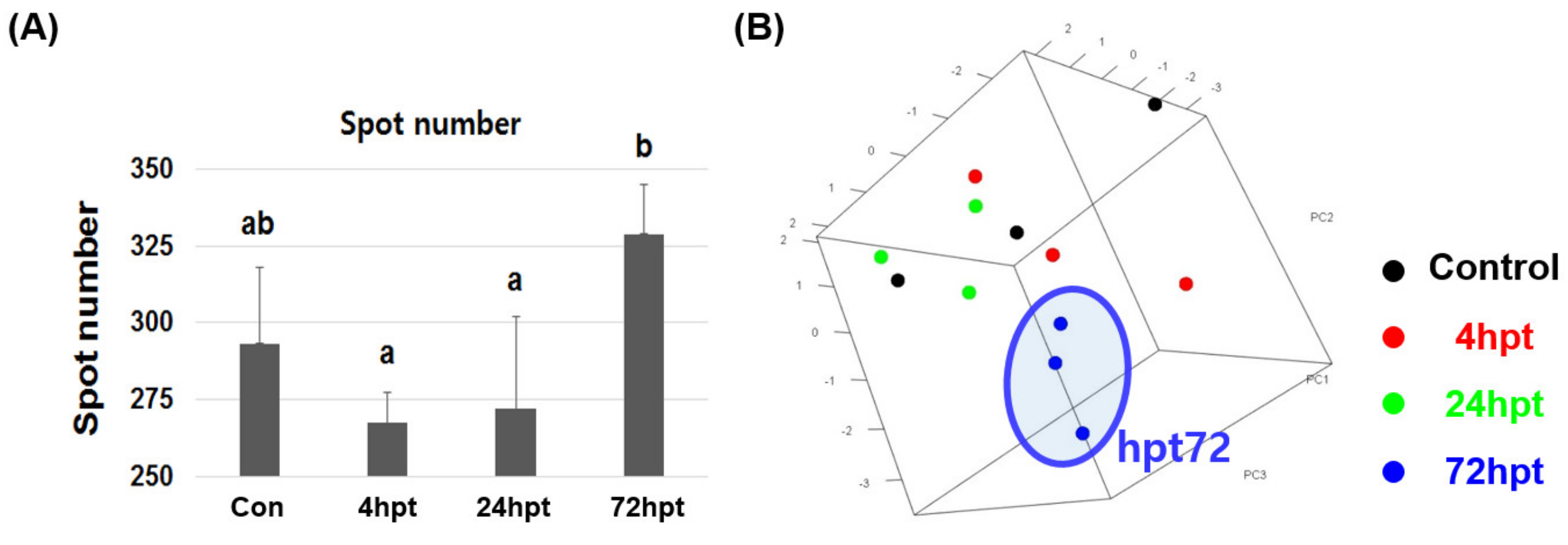
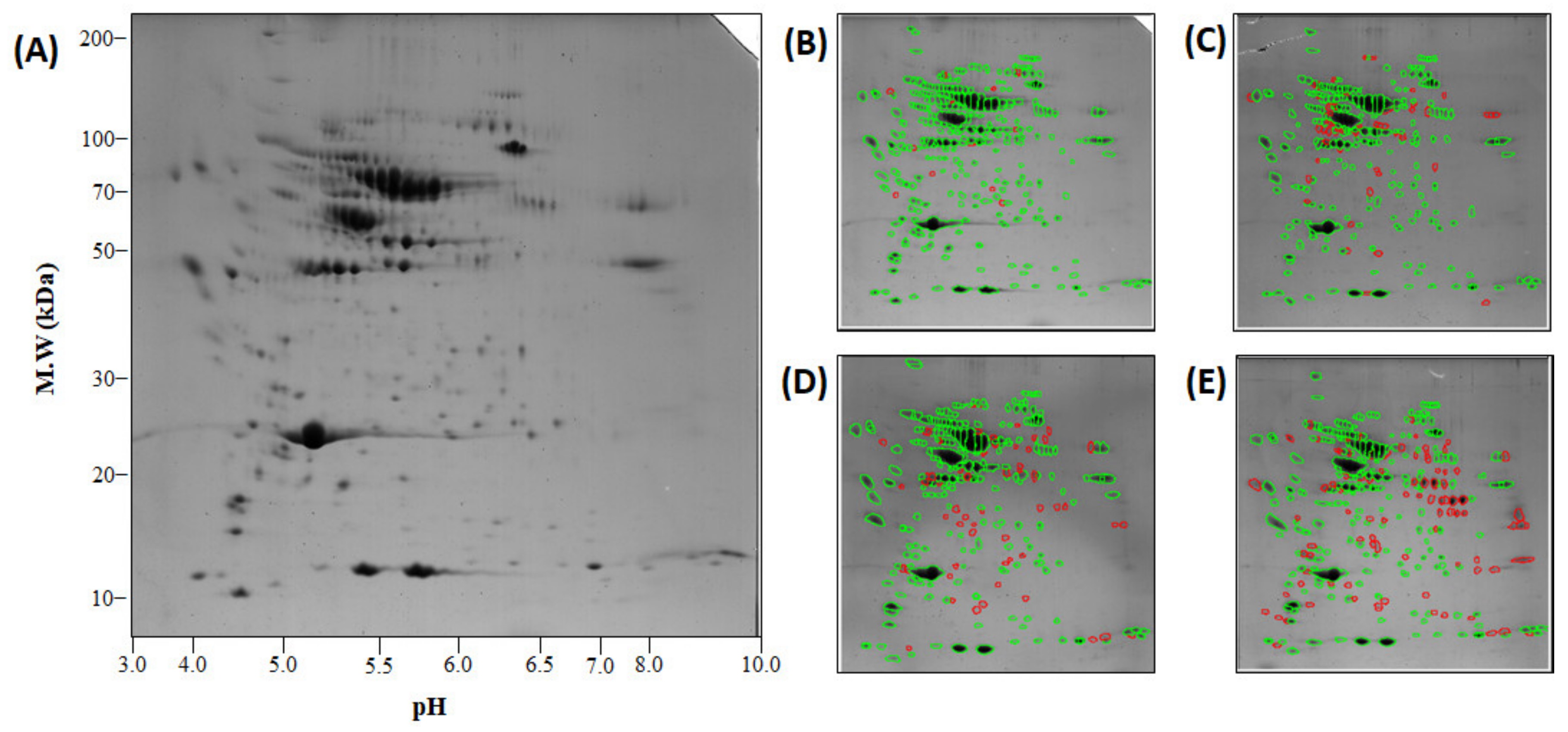
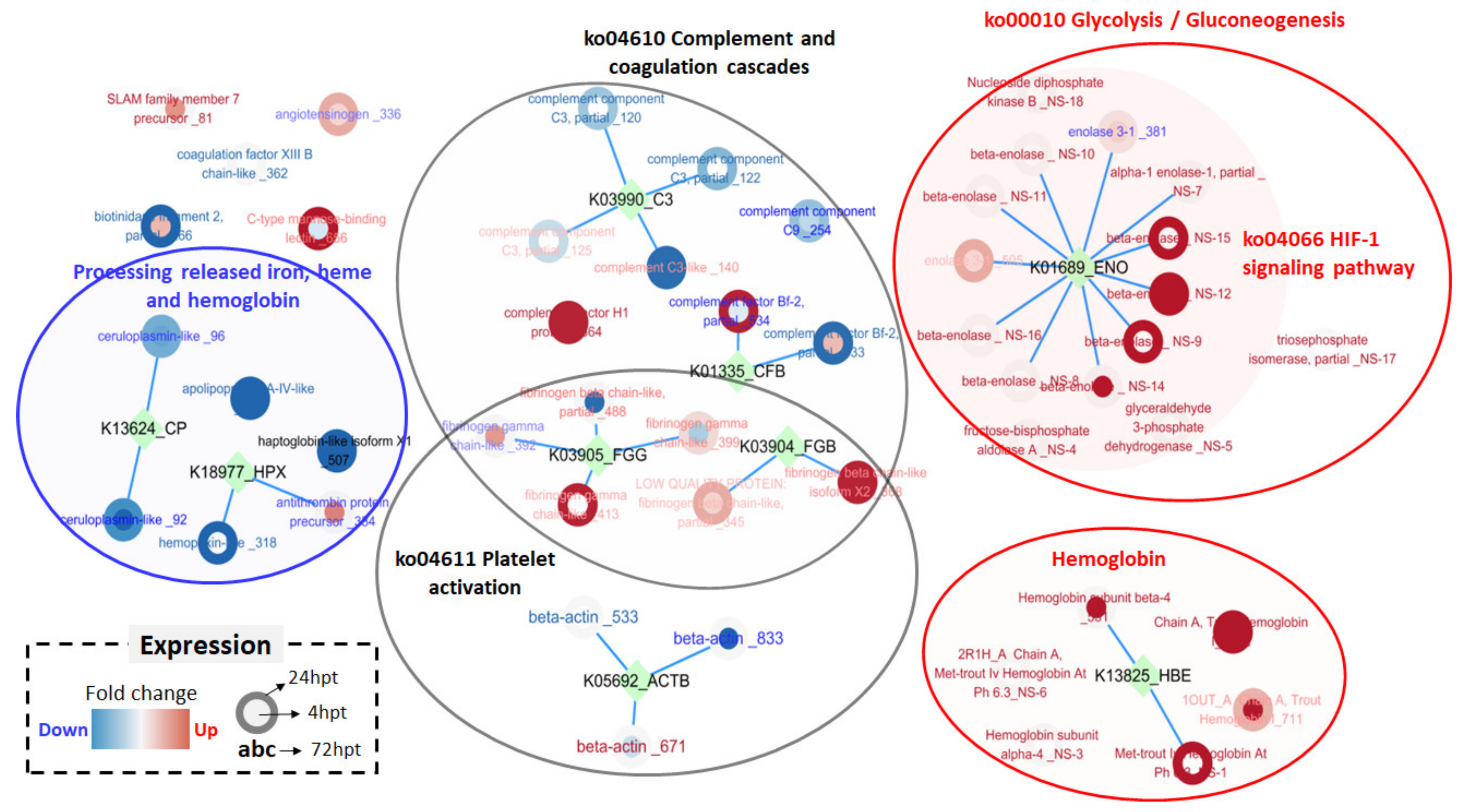
2.6. Analysis of Plasma Proteome and Differentially Expressed Proteins
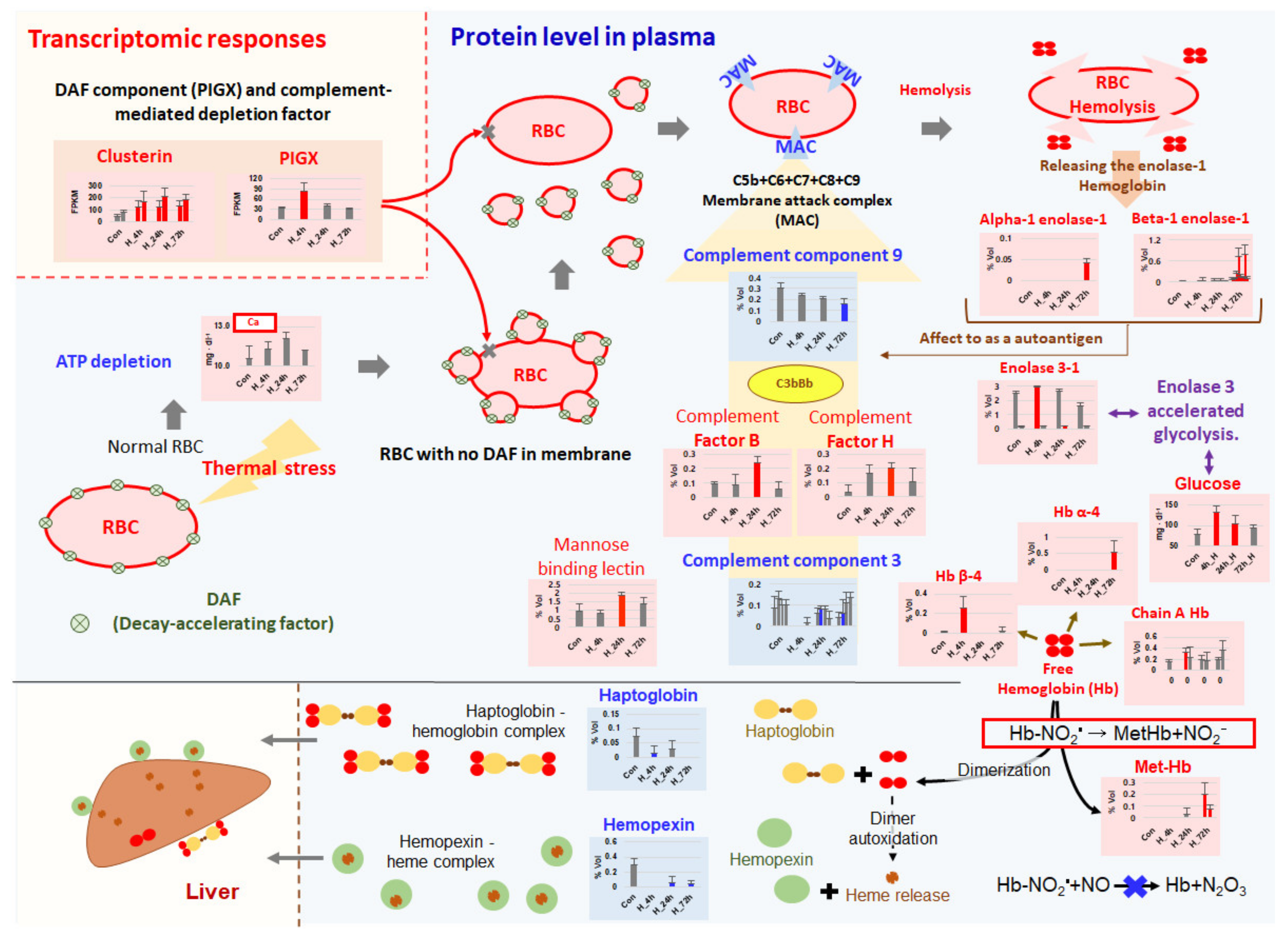
3. Discussion
3.1. DNA Replication Stress
3.2. Protein Processing in the Endoplasmic Reticulum and Glycolysis
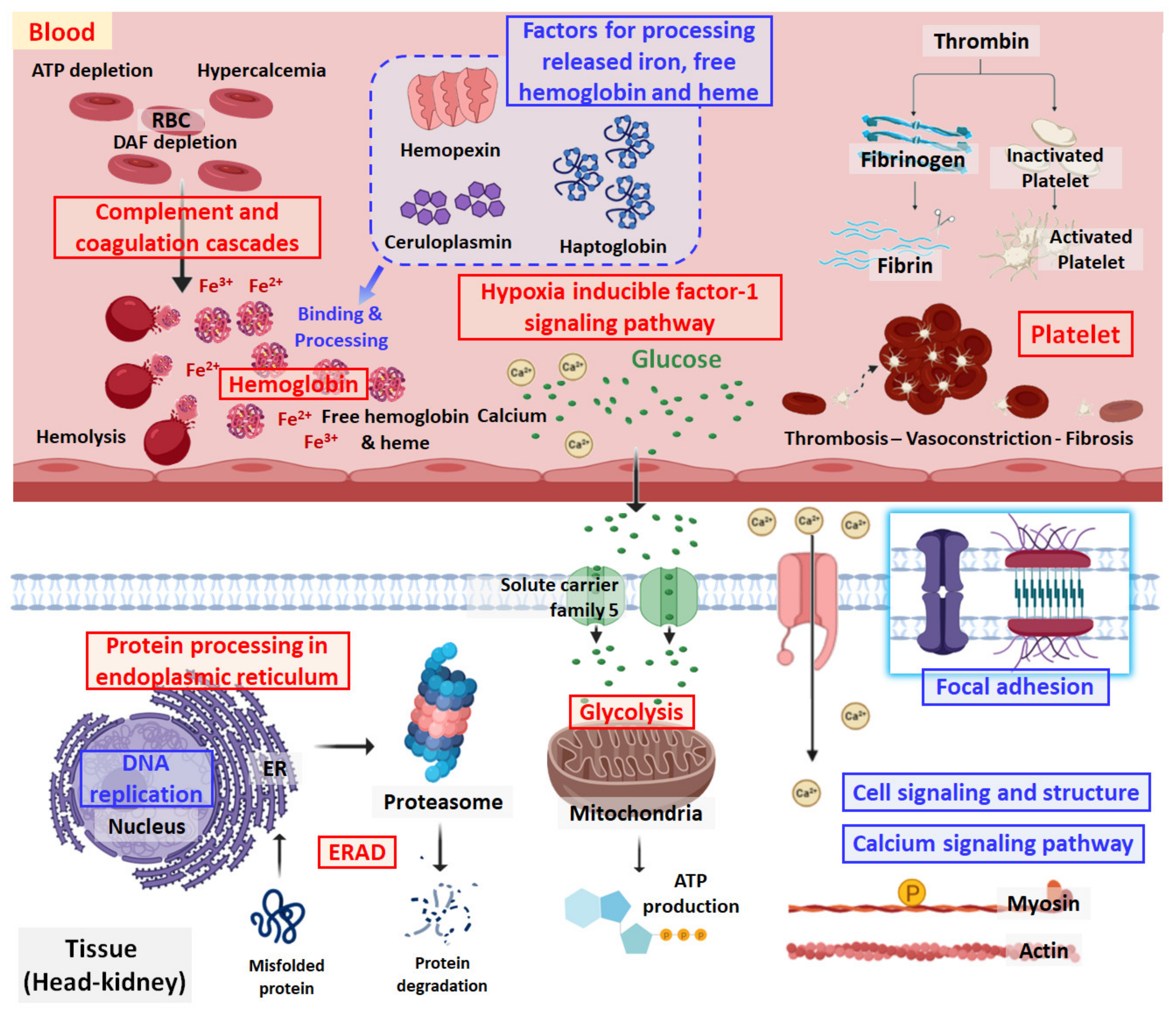
3.3. Complement-Associated Hemolysis
3.4. Removal of Cell-Free Hemoglobin
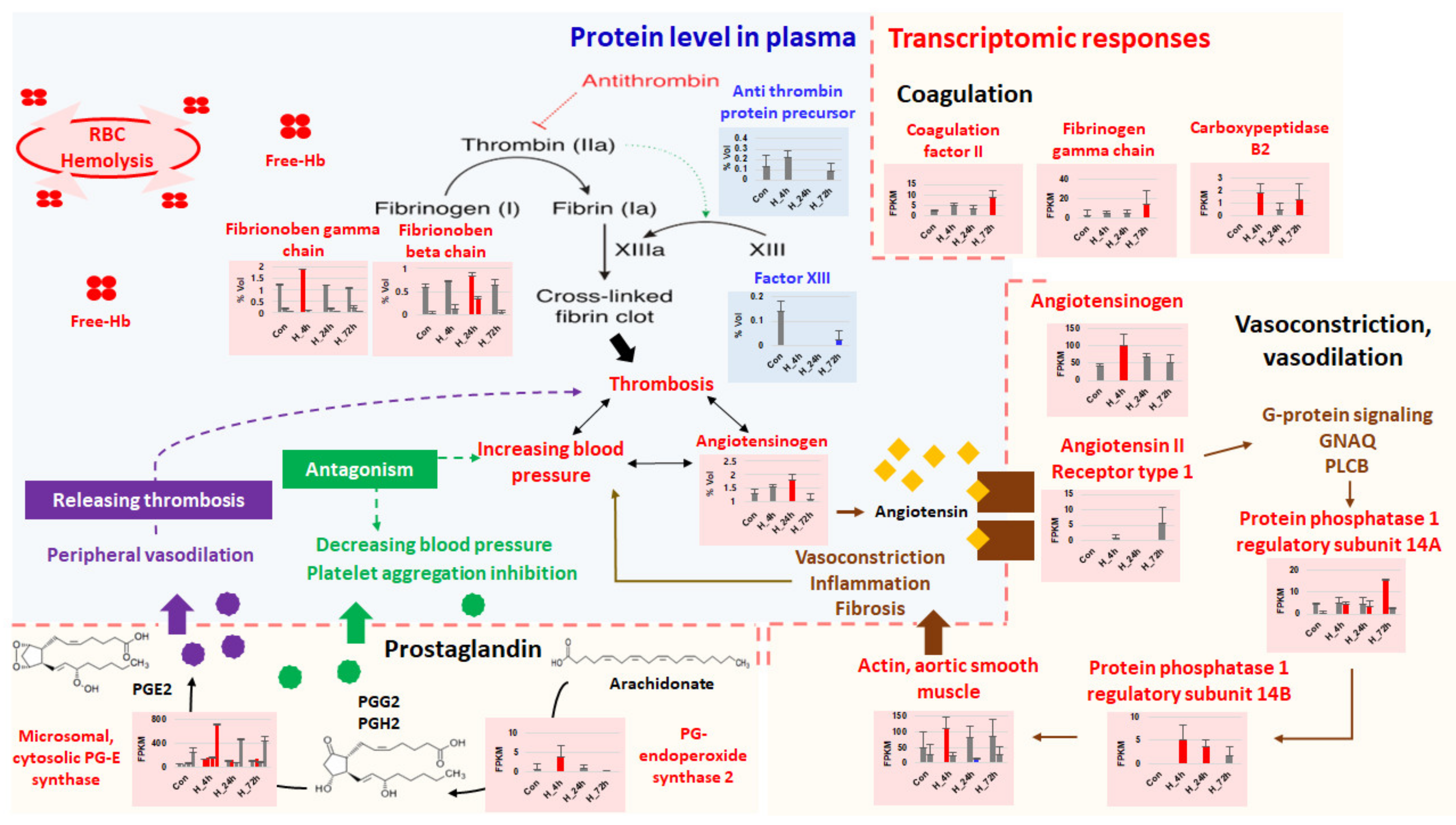
3.5. Thrombosis and Hypertension/Vasoconstriction
4. Materials and Methods
4.1. Acute Thermal Stress
4.2. RNA Extraction and RNA-seq
4.3. Genome-Guided Assembly and Annotation
4.4. Selection of Differentially Expressed Genes and Functional Annotation
4.5. Validation of RNA-seq Results Using qPCR
4.6. Analysis of the Biochemical Factors in Plasma
4.7. Two-Dimensional Electrophoresis (2DE)
4.8. Data Availability Statement
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sun, M.; Li, J.; Gai, C.; Chang, Z.; Li, J.; Zhao, F. Pharmacokinetics of Difloxacin in Olive Flounder Paralichthys olivaceus at Two Water Temperatures. J. Vet. Pharmacol. Ther. 2014, 37, 186–191. [Google Scholar] [CrossRef]
- Yue, X.; Huan, P.; Hu, Y.; Liu, B. Integrated Transcriptomic and Proteomic Analyses Reveal Potential Mechanisms Linking Thermal Stress and Depressed Disease Resistance in the Turbot Scophthalmus maximus. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Sunday, J.M.; Bates, A.E.; Dulvy, N.K. Thermal Tolerance and the Global Redistribution of Animals. Nat. Clim. Chang. 2012, 2, 686–690. [Google Scholar] [CrossRef]
- Comte, L.; Olden, J.D. Evolutionary and Environmental Determinants of Freshwater Fish Thermal Tolerance and Plasticity. Glob. Chang. Biol. 2017, 23, 728–736. [Google Scholar] [CrossRef]
- Pinsky, M.L.; Eikeset, A.M.; McCauley, D.J.; Payne, J.L.; Sunday, J.M. Greater Vulnerability to Warming of Marine Versus Terrestrial Ectotherms. Nature 2019, 569, 108–111. [Google Scholar] [CrossRef]
- Gosselin, J.L.; Anderson, J.J. Step-Patterned Survivorship Curves: Mortality and Loss of Equilibrium Responses to High Temperature and Food Restriction in Juvenile Rainbow Trout (Oncorhynchus mykiss). PLoS ONE 2020, 15, e0233699. [Google Scholar] [CrossRef]
- Roychowdhury, P.; Aftabuddin, M.; Pati, M.K. Thermal Stress Altered Growth Performance and Metabolism and Induced Anaemia and Liver Disorder in Labeo rohita. Aquacult. Res. 2020, 51, 1406–1414. [Google Scholar] [CrossRef]
- Madeira, D.; Araújo, J.; Vitorino, R.; Capelo, J.; Vinagre, C.; Diniz, M.S. Ocean Warming Alters Cellular Metabolism and Induces Mortality in Fish Early Life Stages: A Proteomic Approach. Environ. Res. 2016, 148, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Whittington, R.; Reddacliff, G. Influence of Environmental Temperature on Experimental Infection of Redfin Perch (Perca fluviatilis) and Rainbow Trout (Oncorhynchus mykiss) with Epizootic Haematopoietic Necrosis Virus, an Australian Iridovirus. Aust. Vet. J. 1995, 72, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Magnadóttir, B.; Lange, S.; Steinarsson, A.; Gudmundsdottir, S. Infection Dynamics of Two Renal Myxozoans in Hatchery Reared Fry and Juvenile Atlantic Cod Gadus morhua L. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1999, 122, 173–180. [Google Scholar]
- Piertney, S.; Oliver, M. The Evolutionary Ecology of the Major Histocompatibility Complex. Heredity 2006, 96, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Marcos-López, M.; Gale, P.; Oidtmann, B.; Peeler, E. Assessing the Impact of Climate Change on Disease Emergence in Freshwater Fish in the United Kingdom. Transbound. Emerg. Dis. 2010, 57, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, Y.; Liu, Z.; Kang, Y.; Wang, J. Transcriptomic Responses to Heat Stress in Rainbow Trout Oncorhynchus mykiss Head Kidney. Fish Shellfish Immunol. 2018, 82, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Webster, T.M.U.; Rodriguez-Barreto, D.; Martin, S.A.; Van Oosterhout, C.; Orozco-terWengel, P.; Cable, J.; Hamilton, A.; Garcia de Leaniz, C.; Consuegra, S. Contrasting Effects of Acute and Chronic Stress on the Transcriptome, Epigenome, and Immune Response of Atlantic Salmon. Epigenetics 2018, 13, 1191–1207. [Google Scholar] [CrossRef]
- Hokanson, K.E.; Kleiner, C.F.; Thorslund, T.W. Effects of Constant Temperatures and Diel Temperature Fluctuations on Specific Growth and Mortality Rates and Yield of Juvenile Rainbow Trout, Salmo gairdneri. J. Fish. Board Can. 1977, 34, 639–648. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Z.; Wang, Y.N.; Kang, Y.J.; Wang, J.F.; Shi, H.N.; Huang, J.Q.; Jiang, L. Effects of Heat Stress on Serum Cortisol, Alkaline Phosphatase Activity and Heat Shock Protein 40 and 90β mRNA Expression in Rainbow Trout Oncorhynchus mykiss. Biologia 2016, 71, 109–115. [Google Scholar] [CrossRef]
- Matthews, K.; Berg, N. Rainbow Trout Responses to Water Temperature and Dissolved Oxygen Stress in Two Southern California Stream Pools. J. Fish Biol. 1997, 50, 50–67. [Google Scholar] [CrossRef]
- Verhille, C.E.; English, K.K.; Cocherell, D.E.; Farrell, A.P.; Fangue, N.A. High Thermal Tolerance of a Rainbow Trout Population Near its Southern Range Limit Suggests Local Thermal Adjustment. Conserv. Physiol. 2016, 4, cow057. [Google Scholar] [CrossRef]
- Li, B.; Sun, S.; Zhu, J.; Yanli, S.; Wuxiao, Z.; Ge, X. Transcriptome Profiling and Histology Changes in Juvenile Blunt Snout Bream (Megalobrama amblycephala) Liver Tissue in Response to Acute Thermal Stress. Genomics 2019, 111, 242–250. [Google Scholar] [CrossRef]
- Song, M.; Zhao, J.; Wen, H.; Li, Y.; Li, J.; Li, L.; Tao, Y. The Impact of Acute Thermal Stress on the Metabolome of the Black Rockfish (Sebastes schlegelii). PLoS ONE 2019, 14, e0217133. [Google Scholar] [CrossRef]
- Akbarzadeh, A.; Günther, O.P.; Houde, A.L.; Li, S.; Ming, T.J.; Jeffries, K.M.; Hinch, S.G.; Miller, K.M. Developing Specific Molecular Biomarkers for Thermal Stress in Salmonids. BMC Genom. 2018, 19, 749. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-Omics Approaches to Disease. Genome Biol. 2017, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Hu, Y.; Zhang, X.; Wei, X.; Wang, T.; Yin, S. Integrated Application of Multi-Omics Provides Insights into Cold Stress Responses in Pufferfish Takifugu fasciatus. BMC Genom. 2019, 20, 563. [Google Scholar] [CrossRef]
- Rebl, A.; Verleih, M.; Köbis, J.M.; Kühn, C.; Wimmers, K.; Köllner, B.; Goldammer, T. Transcriptome Profiling of Gill Tissue in Regionally Bred and Globally Farmed Rainbow Trout Strains Reveals Different Strategies for Coping with Thermal Stress. Mar. Biotechnol. 2013, 15, 445–460. [Google Scholar] [CrossRef]
- Kang, Y.; Liu, Z.; Shi, H.; Wang, J.; Huang, J.; Li, Y.; Li, J.; Wang, Y. Label-Free Quantification of Protein Expression in the Rainbow Trout (Oncorhynchus mykiss) in Response to Short-Term Exposure to Heat Stress. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 30, 158–168. [Google Scholar] [CrossRef]
- Andreyeva, A.Y.; Kukhareva, T.A.; Soldatov, A.A. Cellular Composition and Proliferation Levels in the Hematopoietic Tissue of Black Scorpionfish (Scorpaena porcus L.) Head Kidney and Spleen during the Spawning and Wintering Periods. Anat. Rec. 2019, 302, 1136–1143. [Google Scholar] [CrossRef]
- Lewis, J.M.; Hori, T.S.; Rise, M.L.; Walsh, P.J.; Currie, S. Transcriptome Responses to Heat Stress in the Nucleated Red Blood Cells of the Rainbow Trout (Oncorhynchus mykiss). Physiol. Genom. 2010, 42, 361–373. [Google Scholar] [CrossRef]
- Allison, D.B.; Cui, X.; Page, G.P.; Sabripour, M. Microarray Data Analysis: From Disarray to Consolidation and Consensus. Nat. Rev. Genet. 2006, 7, 55–65. [Google Scholar] [CrossRef]
- Quackenbush, J. Computational Analysis of Microarray Data. Nat. Rev. Genet. 2001, 2, 418–427. [Google Scholar] [CrossRef]
- Han, H.; Lee, S.; Lee, I. NGSEA: Network-Based Gene Set Enrichment Analysis for Interpreting Gene Expression Phenotypes with Functional Gene Sets. Mol. Cells 2019, 42, 579–588. [Google Scholar] [PubMed]
- Zeman, M.K.; Cimprich, K.A. Causes and Consequences of Replication Stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Berti, M.; Vindigni, A. Replication Stress: Getting Back on Track. Nat. Struct. Mol. Biol. 2016, 23, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Mazouzi, A.; Velimezi, G.; Loizou, J.I. DNA Replication Stress: Causes, Resolution and Disease. Exp. Cell Res. 2014, 329, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Macheret, M.; Halazonetis, T.D. DNA Replication Stress as a Hallmark of Cancer. Annu. Rev. Pathol. Mech. Dis. 2015, 10, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; D’Andrea, A.D. Regulation of DNA Cross-Link Repair by the Fanconi anemia/BRCA Pathway. Genes Dev. 2012, 26, 1393–1408. [Google Scholar] [CrossRef] [PubMed]
- Ragoonanan, V.; Hubel, A.; Aksan, A. Response of the Cell membrane–cytoskeleton Complex to Osmotic and freeze/thaw Stresses. Cryobiology 2010, 61, 335–344. [Google Scholar] [CrossRef]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.; Lamond, A.I. The Nucleolus Under Stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef]
- Vogel, J.L.; Parsell, D.A.; Lindquist, S. Heat-Shock Proteins Hsp104 and Hsp70 Reactivate mRNA Splicing After Heat Inactivation. Curr. Biol. 1995, 5, 306–317. [Google Scholar] [CrossRef][Green Version]
- Coz-Rakovac, R.; Strunjak-Perovic, I.; Hacmanjek, M.; Lipej, Z.; Sostaric, B. Blood Chemistry and Histological Properties of Wild and Cultured Sea Bass (Dicentrarchus labrax) in the North Adriatic Sea. Vet. Res. Commun. 2005, 29, 677–687. [Google Scholar] [CrossRef]
- Ji, L.; Sun, G.; Li, J.; Wang, Y.; Du, Y.; Li, X.; Liu, Y. Effect of Dietary β-Glucan on Growth, Survival and Regulation of Immune Processes in Rainbow Trout (Oncorhynchus mykiss) Infected by Aeromonas Salmonicida. Fish Shellfish Immunol. 2017, 64, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Hoseki, J.; Ushioda, R.; Nagata, K. Mechanism and Components of Endoplasmic Reticulum-Associated Degradation. J. Biochem. 2010, 147, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Keller, J.; Huang, F.A.; Markesbery, W. Decreased Levels of Proteasome Activity and Proteasome Expression in Aging Spinal Cord. Neuroscience 2000, 98, 149–156. [Google Scholar] [CrossRef]
- Keller, J.N.; Gee, J.; Ding, Q. The Proteasome in Brain Aging. Ageing Res. Rev. 2002, 1, 279–293. [Google Scholar] [CrossRef]
- Goldbaum, O.; Riedel, M.; Stahnke, T.; Richter-Landsberg, C. The Small Heat Shock Protein HSP25 Protects Astrocytes Against Stress Induced by Proteasomal Inhibition. Glia 2009, 57, 1566–1577. [Google Scholar] [CrossRef]
- Lee, M.J.; Na, K.; Jeong, S.; Lim, J.; Kim, S.A.; Lee, M.; Song, S.Y.; Kim, H.; Hancock, W.S.; Paik, Y. Identification of Human Complement Factor B as a Novel Biomarker Candidate for Pancreatic Ductal Adenocarcinoma. J. Proteome Res. 2014, 13, 4878–4888. [Google Scholar] [CrossRef]
- Goldbaum, O.; Vollmer, G.; Richter-Landsberg, C. Proteasome Inhibition by MG-132 Induces Apoptotic Cell Death and Mitochondrial Dysfunction in Cultured Rat Brain Oligodendrocytes but Not in Astrocytes. Glia 2006, 53, 891–901. [Google Scholar] [CrossRef]
- Lee, D.H.; Goldberg, A.L. Proteasome Inhibitors Cause Induction of Heat Shock Proteins and Trehalose, which Together Confer Thermotolerance in Saccharomyces Cerevisiae. Mol. Cell. Biol. 1998, 18, 30–38. [Google Scholar] [CrossRef]
- Losser, M.; Damoisel, C.; Payen, D. Bench-to-Bedside Review: Glucose and Stress Conditions in the Intensive Care Unit. Crit. Care 2010, 14, 231. [Google Scholar] [CrossRef]
- Løvoll, M.; Kilvik, T.; Boshra, H.; Bøgwald, J.; Sunyer, J.O.; Dalmo, R.A. Maternal Transfer of Complement Components C3-1, C3-3, C3-4, C4, C5, C7, Bf, and Df to Offspring in Rainbow Trout (Oncorhynchus mykiss). Immunogenetics 2006, 58, 168–179. [Google Scholar] [CrossRef]
- Sunyer, J.O.; Zarkadis, I.; Sarrias, M.R.; Hansen, J.D.; Lambris, J.D. Cloning, Structure, and Function of Two Rainbow Trout Bf Molecules. J. Immunol. 1998, 161, 4106–4114. [Google Scholar] [PubMed]
- Kriebardis, A.G.; Antonelou, M.H.; Stamoulis, K.E.; Economou-Petersen, E.; Margaritis, L.H.; Papassideri, I.S. RBC-derived Vesicles during Storage: Ultrastructure, Protein Composition, Oxidation, and Signaling Components. Transfusion 2008, 48, 1943–1953. [Google Scholar] [CrossRef] [PubMed]
- Babiker, A.A.; Ronquist, G.; Nilsson, U.R.; Nilsson, B. Transfer of Prostasomal CD59 to CD59-deficient Red Blood Cells Results in Protection Against complement-mediated Hemolysis. Am. J. Reprod. Immunol. 2002, 47, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Salzer, U.; Hinterdorfer, P.; Hunger, U.; Borken, C.; Prohaska, R. Ca -Dependent Vesicle Release from Erythrocytes Involves Stomatin-Specific Lipid Rafts, Synexin (Annexin VII), and Sorcin. Blood J. Am. Soc. Hematol. 2002, 99, 2569–2577. [Google Scholar] [CrossRef]
- Butikofer, P.; Kuypers, F.A.; Xu, C.M.; Chiu, D.T.; Lubin, B. Enrichment of Two Glycosyl-Phosphatidylinositol-Anchored Proteins, Acetylcholinesterase and Decay Accelerating Factor, in Vesicles Released from Human Red Blood Cells. Blood 1989, 74, 1481–1485. [Google Scholar] [CrossRef]
- Miwa, T.; Zhou, L.; Hilliard, B.; Molina, H.; Song, W. Crry, but Not CD59 and DAF, is Indispensable for Murine Erythrocyte Protection in Vivo from Spontaneous Complement Attack. Blood 2002, 99, 3707–3716. [Google Scholar] [CrossRef]
- Vogt, A.M.; Elsässer, A.; Nef, H.; Bode, C.; Kübler, W.; Schaper, J. Increased Glycolysis as Protective Adaptation of Energy Depleted, Degenerating Human Hibernating Myocardium. Mol. Cell. Biochem. 2003, 242, 101–107. [Google Scholar] [CrossRef]
- Gov, N.; Safran, S. Red Blood Cell Membrane Fluctuations and Shape Controlled by ATP-Induced Cytoskeletal Defects. Biophys. J. 2005, 88, 1859–1874. [Google Scholar] [CrossRef]
- Ashida, H.; Hong, Y.; Murakami, Y.; Shishioh, N.; Sugimoto, N.; Kim, Y.U.; Maeda, Y.; Kinoshita, T. Mammalian PIG-X and Yeast Pbn1p are the Essential Components of Glycosylphosphatidylinositol-Mannosyltransferase I. Mol. Biol. Cell 2005, 16, 1439–1448. [Google Scholar] [CrossRef]
- Londou, A.; Mikrou, A.; Zarkadis, I.K. Cloning and Characterization of Two Clusterin Isoforms in Rainbow Trout. Mol. Immunol. 2008, 45, 470–478. [Google Scholar] [CrossRef]
- De Córdoba, S.R.; De Jorge, E.G. Translational mini-review Series on Complement Factor H: Genetics and Disease Associations of Human Complement Factor H. Clin. Exp. Immunol. 2008, 151, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fibach, E.; Dana, M. Oxidative Stress in Paroxysmal Nocturnal Hemoglobinuria and Other Conditions of Complement-Mediated Hemolysis. Free Radic. Biol. Med. 2015, 88, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Amer, J.; Zelig, O.; Fibach, E. Oxidative Status of Red Blood Cells, Neutrophils, and Platelets in Paroxysmal Nocturnal Hemoglobinuria. Exp. Hematol. 2008, 36, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Nagababu, E.; Chrest, F.J.; Rifkind, J.M. Hydrogen-Peroxide-Induced Heme Degradation in Red Blood Cells: The Protective Roles of Catalase and Glutathione Peroxidase. Biochim. Biophys. Acta 2003, 1620, 211–217. [Google Scholar] [CrossRef]
- Bunn, H.F.; Jandl, J.H. Exchange of Heme among Hemoglobins and between Hemoglobin and Albumin. J. Biol. Chem. 1968, 243, 465–475. [Google Scholar]
- Balla, G.; Vercellotti, G.M.; Muller-Eberhard, U.; Eaton, J.; Jacob, H.S. Exposure of Endothelial Cells to Free Heme Potentiates Damage Mediated by Granulocytes and Toxic Oxygen Species. Lab. Invest. 1991, 64, 648–655. [Google Scholar]
- Kumar, S.; Bandyopadhyay, U. Free Heme Toxicity and its Detoxification Systems in Human. Toxicol. Lett. 2005, 157, 175–188. [Google Scholar] [CrossRef]
- Nagy, E.; Eaton, J.W.; Jeney, V.; Soares, M.P.; Varga, Z.; Galajda, Z.; Szentmiklósi, J.; Méhes, G.; Csonka, T.; Smith, A. Red Cells, Hemoglobin, Heme, Iron, and Atherogenesis. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1347–1353. [Google Scholar] [CrossRef]
- Jeney, V.; Balla, G.; Balla, J. Red Blood Cell, Hemoglobin and Heme in the Progression of Atherosclerosis. Front. Physiol. 2014, 5, 379. [Google Scholar] [CrossRef]
- Smith, A.; McCulloh, R.J. Hemopexin and Haptoglobin: Allies against Heme Toxicity from Hemoglobin Not Contenders. Front. Physiol. 2015, 6, 187. [Google Scholar] [CrossRef]
- Delanghe, J.R.; Langlois, M.R. Hemopexin: A Review of Biological Aspects and the Role in Laboratory Medicine. Clin. Chim. Acta 2001, 312, 13–23. [Google Scholar] [CrossRef]
- Yang, W.S.; Stockwell, B.R. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016, 26, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Hou, W.; Song, X.; Yu, Y.; Huang, J.; Sun, X.; Kang, R.; Tang, D. Ferroptosis: Process and Function. Cell Death Differ. 2016, 23, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Barnham, K.J.; Bush, A.I. Metals in Alzheimer’s and Parkinson’s Diseases. Curr. Opin. Chem. Biol. 2008, 12, 222–228. [Google Scholar] [CrossRef]
- Galano, A.; Medina, M.E.; Tan, D.X.; Reiter, R.J. Melatonin and its Metabolites as Copper Chelating Agents and their Role in Inhibiting Oxidative Stress: A Physicochemical Analysis. J. Pineal Res. 2015, 58, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Waggoner, D.J.; Bartnikas, T.B.; Gitlin, J.D. The Role of Copper in Neurodegenerative Disease. Neurobiol. Dis. 1999, 6, 221–230. [Google Scholar] [CrossRef]
- Wolf, T.L.; Kotun, J.; Meador-Woodruff, J.H. Plasma Copper, Iron, Ceruloplasmin and Ferroxidase Activity in Schizophrenia. Schizophr. Res. 2006, 86, 167–171. [Google Scholar] [CrossRef]
- Yuan, D.S.; Stearman, R.; Dancis, A.; Dunn, T.; Beeler, T.; Klausner, R.D. The Menkes/Wilson Disease Gene Homologue in Yeast Provides Copper to a Ceruloplasmin-Like Oxidase Required for Iron Uptake. Proc. Natl. Acad. Sci. USA 1995, 92, 2632–2636. [Google Scholar] [CrossRef]
- Arnal, N.; Cristalli, D.O.; de Alaniz, M.J.; Marra, C.A. Clinical Utility of Copper, Ceruloplasmin, and Metallothionein Plasma Determinations in Human Neurodegenerative Patients and their First-Degree Relatives. Brain Res. 2010, 1319, 118–130. [Google Scholar] [CrossRef]
- Attieh, Z.K.; Mukhopadhyay, C.K.; Seshadri, V.; Tripoulas, N.A.; Fox, P.L. Ceruloplasmin Ferroxidase Activity Stimulates Cellular Iron Uptake by a Trivalent Cation-Specific Transport Mechanism. J. Biol. Chem. 1999, 274, 1116–1123. [Google Scholar] [CrossRef]
- Shiva, S.; Wang, X.; Ringwood, L.A.; Xu, X.; Yuditskaya, S.; Annavajjhala, V.; Miyajima, H.; Hogg, N.; Harris, Z.L.; Gladwin, M.T. Ceruloplasmin is a NO Oxidase and Nitrite Synthase that Determines Endocrine NO Homeostasis. Nat. Chem. Biol. 2006, 2, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Samuel, T.K.; Gitlin, J.D. Copper and Nitric Oxide Meet in the Plasma. Nat. Chem. Biol. 2006, 2, 452–453. [Google Scholar] [CrossRef] [PubMed]
- Gladwin, M.T.; Kim-Shapiro, D.B. The Functional Nitrite Reductase Activity of the Heme-Globins. Blood J. Am. Soc. Hematol. 2008, 112, 2636–2647. [Google Scholar] [CrossRef] [PubMed]
- Mannucci, P. Recent Progress in the Pathophysiology of Fibrinogen. Eur. Heart J. 1995, 16, 25–30. [Google Scholar] [CrossRef]
- Mateos-Cáceres, P.J.; García-Méndez, A.; Farré, A.L.; Macaya, C.; Núñez, A.; Gómez, J.; Alonso-Orgaz, S.; Carrasco, C.; Burgos, M.E.; de Andrés, R. Proteomic Analysis of Plasma from Patients during an Acute Coronary Syndrome. J. Am. Coll. Cardiol. 2004, 44, 1578–1583. [Google Scholar] [CrossRef]
- Mosesson, M. Fibrinogen and Fibrin Structure and Functions. J. Thromb. Haemost. 2005, 3, 1894–1904. [Google Scholar] [CrossRef]
- Machlus, K.R.; Cardenas, J.C.; Church, F.C.; Wolberg, A.S. Causal Relationship between Hyperfibrinogenemia, Thrombosis, and Resistance to Thrombolysis in Mice. Blood J. Am. Soc. Hematol. 2011, 117, 4953–4963. [Google Scholar] [CrossRef]
- Davalos, D.; Akassoglou, K. Fibrinogen as a Key Regulator of Inflammation in Disease. Semin. Immunopathol. 2012, 34, 43–62. [Google Scholar] [CrossRef]
- Brown, N.J.; Vaughan, D.E. Prothrombotic Effects of Angiotensin. Adv. Intern. Med. 2000, 45, 419–429. [Google Scholar]
- Leung, P.; Wong, T.; Lam, S.; Chan, H.; Wong, P. Testicular Hormonal Regulation of the Renin-Angiotensin System in the Rat Epididymis. Life Sci. 2000, 66, 1317–1324. [Google Scholar] [CrossRef]
- Kobori, H.; Harrison-Bernard, L.M.; Navar, L.G. Expression of Angiotensinogen mRNA and Protein in Angiotensin II-Dependent Hypertension. J. Am. Soc. Nephrol. 2001, 12, 431–439. [Google Scholar] [PubMed]
- Lang, M.; Gihr, G.; Gawaz, M.; Müller, I. Hemostasis in Danio rerio: Is the Zebrafish a Useful Model for Platelet Research? J. Thromb. Haemost. 2010, 8, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Widlansky, M.E.; Gokce, N.; Keaney, J.F.; Vita, J.A. The Clinical Implications of Endothelial Dysfunction. J. Am. Coll. Cardiol. 2003, 42, 1149–1160. [Google Scholar] [CrossRef]
- Sreenarasimhaiah, J. Diagnosis and Management of Intestinal Ischaemic Disorders. BMJ 2003, 326, 1372–1376. [Google Scholar] [CrossRef]
- Adlbrecht, C.; Bonderman, D.; Plass, C.; Jakowitsch, J.; Beran, G.; Sperker, W.; Siostrzonek, P.; Glogar, D.; Maurer, G.; Lang, I.M. Active Endothelin is an Important Vasoconstrictor in Acute Coronary Thrombi. Thromb. Haemost. 2007, 97, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Herve, P.; Humbert, M.; Sitbon, O.; Parent, F.; Nunes, H.; Legal, C.; Garcia, G.; Simonneau, G. Pathobiology of Pulmonary Hypertension. the Role of Platelets and Thrombosis. Clin. Chest Med. 2001, 22, 451–458. [Google Scholar] [CrossRef]
- Philipose, S.; Konya, V.; Sreckovic, I.; Marsche, G.; Lippe, I.T.; Peskar, B.A.; Heinemann, A.; Schuligoi, R. The Prostaglandin E2 Receptor EP4 is Expressed by Human Platelets and Potently Inhibits Platelet Aggregation and Thrombus Formation. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 2416–2423. [Google Scholar] [CrossRef]
- Spina, A.P. Thermal Ecology of Juvenile Steelhead in a Warm-Water Environment. Environ. Biol. Fishes 2007, 80, 23–34. [Google Scholar] [CrossRef]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noël, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A. The Rainbow Trout Genome Provides Novel Insights into Evolution After Whole-Genome Duplication in Vertebrates. Nat. Commun. 2014, 5, 1–10. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L. WEGO: A Web Tool for Plotting GO Annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment Map: A Network-Based Method for Gene-Set Enrichment Visualization and Interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef] [PubMed]
- Cline, M.S.; Smoot, M.; Cerami, E.; Kuchinsky, A.; Landys, N.; Workman, C.; Christmas, R.; Avila-Campilo, I.; Creech, M.; Gross, B. Integration of Biological Networks and Gene Expression Data using Cytoscape. Nat. Protoc. 2007, 2, 2366. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E. PGC-1α-Responsive Genes Involved in Oxidative Phosphorylation are Coordinately Downregulated in Human Diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Subramanian, A.; Kuehn, H.; Gould, J.; Tamayo, P.; Mesirov, J.P. GSEA-P: A Desktop Application for Gene Set Enrichment Analysis. Bioinformatics 2007, 23, 3251–3253. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.J.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: How to use the entry view. In Plant Bioinformatics; Springer: Berlin/Heidelberg, Germany, 2016; pp. 23–54. [Google Scholar]
- Nolan, D.J.; Ginsberg, M.; Israely, E.; Palikuqi, B.; Poulos, M.G.; James, D.; Ding, B.; Schachterle, W.; Liu, Y.; Rosenwaks, Z. Molecular Signatures of Tissue-Specific Microvascular Endothelial Cell Heterogeneity in Organ Maintenance and Regeneration. Dev. Cell 2013, 26, 204–219. [Google Scholar] [CrossRef]
- Gravel, A.; Vijayan, M.M. Non-Steroidal Anti-Inflammatory Drugs Disrupt the Heat Shock Response in Rainbow Trout. Aquat. Toxicol. 2007, 81, 197–206. [Google Scholar] [CrossRef]
- Tan, E.; Wongwarangkana, C.; Kinoshita, S.; Suzuki, Y.; Oshima, K.; Hattori, M.; Ineno, T.; Tamaki, K.; Kera, A.; Muto, K. Global Gene Expression Analysis of Gill Tissues from Normal and Thermally Selected Strains of Rainbow Trout. Fish. Sci. 2012, 78, 1041–1049. [Google Scholar] [CrossRef]
- Ballesteros, N.A.; Saint-Jean, S.S.R.; Encinas, P.A.; Perez-Prieto, S.I.; Coll, J.M. Oral Immunization of Rainbow Trout to Infectious Pancreatic Necrosis Virus (Ipnv) Induces Different Immune Gene Expression Profiles in Head Kidney and Pyloric Ceca. Fish Shellfish Immunol. 2012, 33, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.; Abós, B.; Pignatelli, J.; von Gersdorff Jørgensen, L.; Granja, A.G.; Buchmann, K.; Tafalla, C. Early Immune Responses in Rainbow Trout Liver upon Viral Hemorrhagic Septicemia Virus (VHSV) Infection. PLoS ONE 2014, 9, e111084. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.H.; Yun, S.Y.; Lim, J.W.; Kim, H.; Kim, K.H. Proteome Analysis of Rat Pancreatic Acinar Cells: Implication for cerulein-induced Acute Pancreatitis. Proteomics 2003, 3, 2446–2453. [Google Scholar] [CrossRef] [PubMed]
- Paik, Y.; Overall, C.M.; Deutsch, E.W.; Hancock, W.S.; Omenn, G.S. Progress in the Chromosome-Centric Human Proteome Project as highlighted in the annual special issue IV. J. Proteome Res. 2016, 15, 3945–3950. [Google Scholar] [CrossRef]
| Con | 4 hpt | 24 hpt | 72 hpt | |
|---|---|---|---|---|
| GOT (U·L−1) | 556 ± 159 a | 640 ± 167 a | 632 ± 53 a | 4811 ± 1213 b |
| GPT (U·L−1) | 31 ± 11 a | 39 ± 24 a | 44 ± 6 a | 1128 ± 303 b |
| ALP (U·dL−1) | 171 ± 135 | 186 ± 78 | 300 ± 103 | 180 ± 13 |
| BUN (mg·dL−1) | 1.93 ± 0.32 | 2.30 ± 0.40 | 2.10 ± 0.46 | 1.97 ± 0.12 |
| GLU (mg·dL−1) | 79 ± 13 a | 133 ± 17 b | 105 ± 19 ab | 85 ± 7 a |
| TCHO (mg·dL−1) | 268 ± 113 | 288 ± 71 | 324 ± 96 | 230 ± 19 |
| TP (g·dL−1) | 4.00 ± 0.61 | 4.40 ± 0.26 | 4.53 ± 0.32 | 3.90 ± 0.17 |
| LDH (U·L−1) | 2012 ± 378 a | 3679 ± 234 a | 3635 ± 1052 a | 12,501 ± 1283 b |
| Ca (mg·dL−1) | 10.6 ± 0.9 | 11.3 ± 0.6 | 12.1 ± 0.5 | 11.2 ± 0 |
| Major Biological Pathway | Thermal Stress | ||
|---|---|---|---|
| Acute (In This Study) | Chronic (Huang et al. [13]) | ||
| Protein processing in endoplasmic reticulum (ER) | Up | Up | |
| Glycolysis activation | Up | Up | |
| Complement associated hemolysis | Up | N.C. | |
| Processing the hemoglobin | Up | N.C. | |
| Thrombosis | Up | N.C. | |
| Hypertension/Vasoconstriction | Up | N.C. | |
| DNA replication | Down | N.C. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roh, H.; Kim, A.; Kim, N.; Lee, Y.; Kim, D.-H. Multi-Omics Analysis Provides Novel Insight into Immuno-Physiological Pathways and Development of Thermal Resistance in Rainbow Trout Exposed to Acute Thermal Stress. Int. J. Mol. Sci. 2020, 21, 9198. https://doi.org/10.3390/ijms21239198
Roh H, Kim A, Kim N, Lee Y, Kim D-H. Multi-Omics Analysis Provides Novel Insight into Immuno-Physiological Pathways and Development of Thermal Resistance in Rainbow Trout Exposed to Acute Thermal Stress. International Journal of Molecular Sciences. 2020; 21(23):9198. https://doi.org/10.3390/ijms21239198
Chicago/Turabian StyleRoh, HyeongJin, Ahran Kim, Nameun Kim, Yoonhang Lee, and Do-Hyung Kim. 2020. "Multi-Omics Analysis Provides Novel Insight into Immuno-Physiological Pathways and Development of Thermal Resistance in Rainbow Trout Exposed to Acute Thermal Stress" International Journal of Molecular Sciences 21, no. 23: 9198. https://doi.org/10.3390/ijms21239198
APA StyleRoh, H., Kim, A., Kim, N., Lee, Y., & Kim, D.-H. (2020). Multi-Omics Analysis Provides Novel Insight into Immuno-Physiological Pathways and Development of Thermal Resistance in Rainbow Trout Exposed to Acute Thermal Stress. International Journal of Molecular Sciences, 21(23), 9198. https://doi.org/10.3390/ijms21239198





