A Long-Read Sequencing Approach for Direct Haplotype Phasing in Clinical Settings
Abstract
1. Introduction
2. Results
2.1. Optimization of APOE Locus Amplification
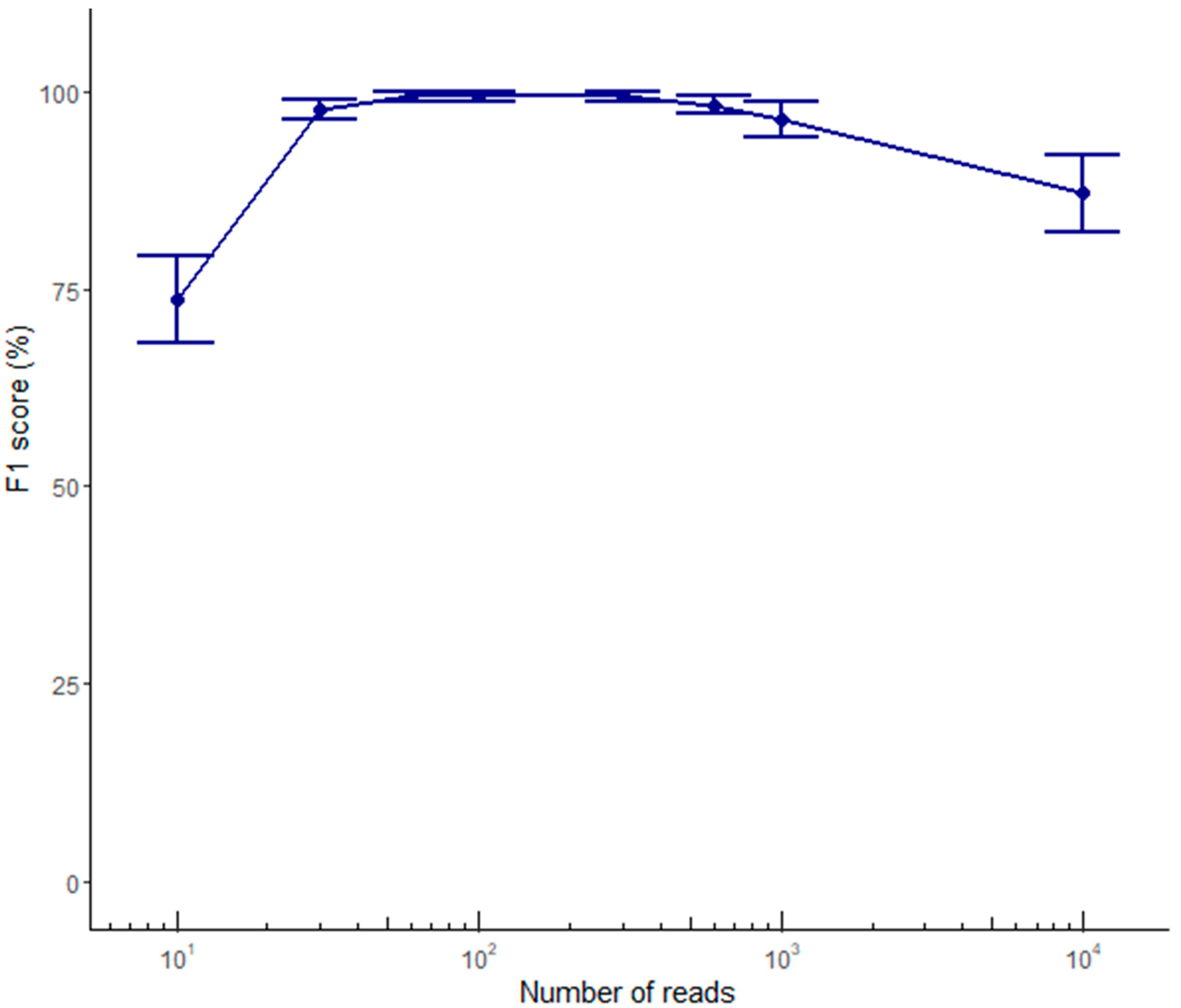
2.2. Variant Calling Performance Using ONT Data
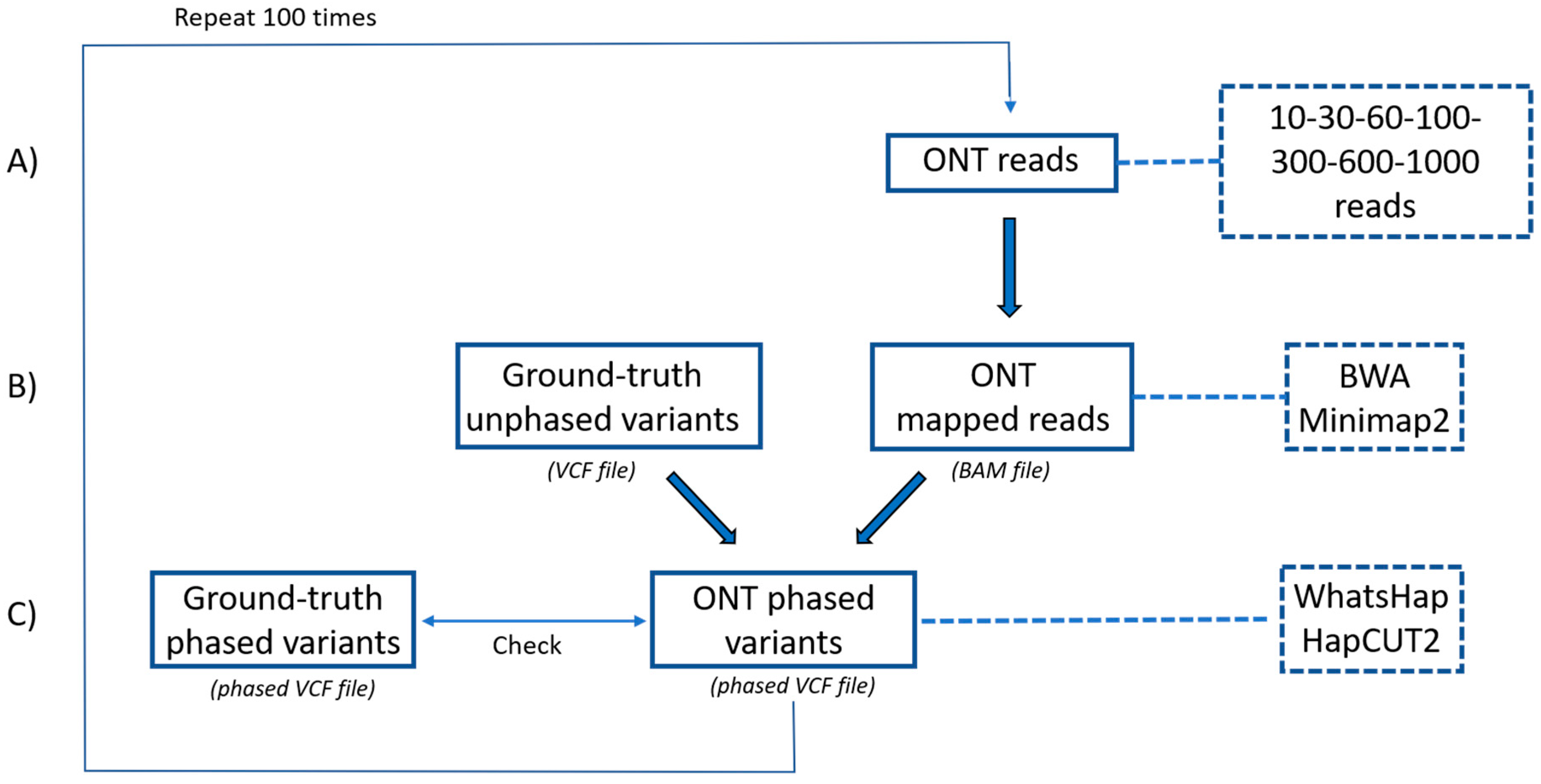
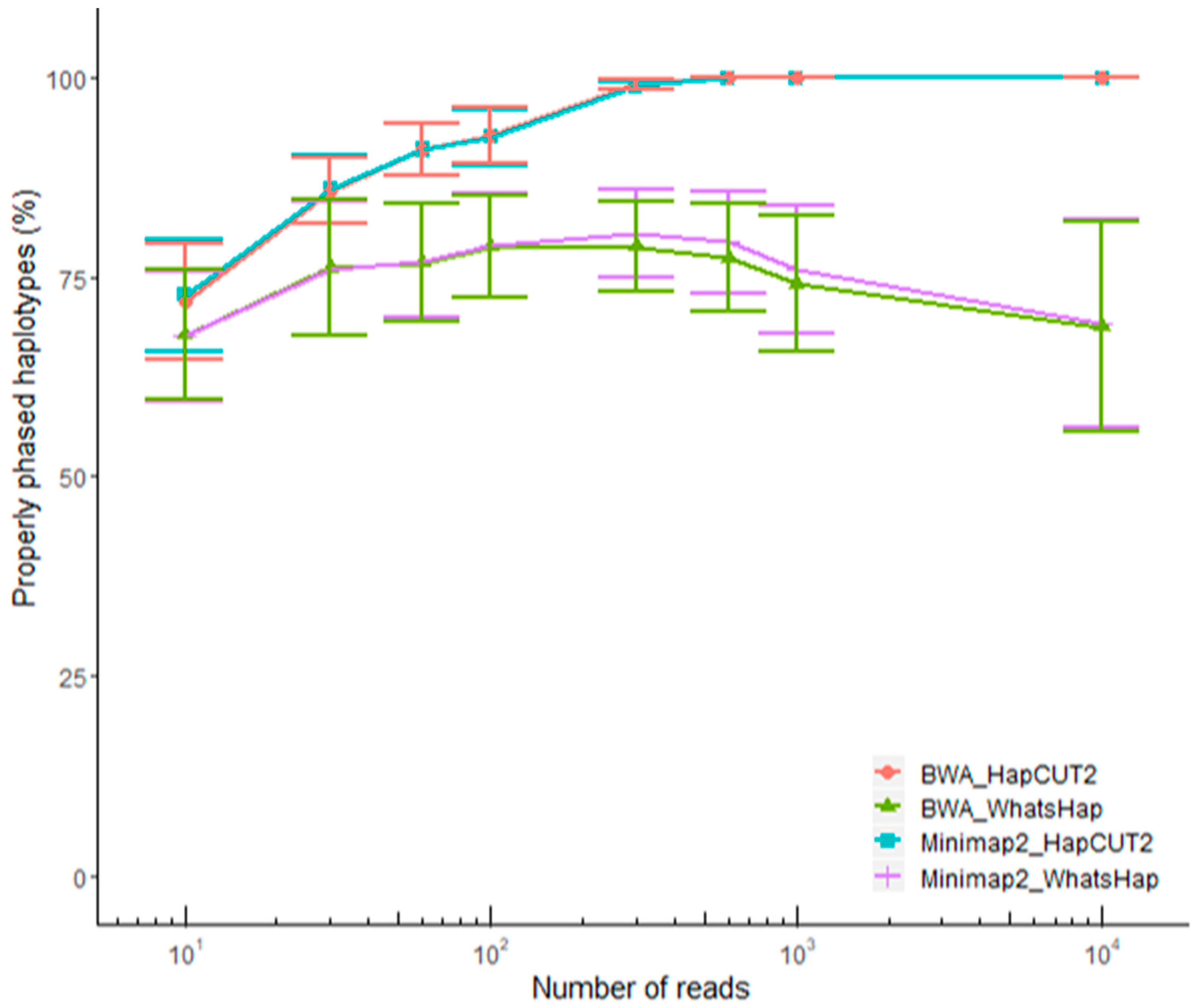
2.3. Optimization of a Haplotype Phasing Pipeline Based on ONT Data
3. Discussion
4. Materials and Methods
4.1. DNA Extraction
4.2. PCR Amplification of The Target Region
4.3. 10× Genomics Library Preparation, Sequencing, and Data Analysis
4.4. ONT Sequencing and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Scitable by Nature Education. Available online: https://www.nature.com/scitable/definition/haplotype-haplotypes-142/ (accessed on 30 November 2020).
- Allen, M.; Kachadoorian, M.; Quicksall, Z.; Zou, F.; Chai, H.S.; Younkin, C.; E Crook, J.; Pankratz, V.S.; Carrasquillo, M.M.; Krishnan, S.; et al. Association of MAPT haplotypes with Alzheimer’s disease risk and MAPT brain gene expression levels. Alzheimer’s Res. Ther. 2014, 6, 39. [Google Scholar] [CrossRef]
- Williams, M.A.; McKay, G.J.; Carson, R.; Craig, D.; Silvestri, G.; Passmore, P. Age-Related Macular Degeneration-Associated Genes in Alzheimer Disease. Am. J. Geriatr. Psychiatry 2015, 23, 1290–1296. [Google Scholar] [CrossRef]
- Lescai, F.; Chiamenti, A.M.; Codemo, A.; Pirazzini, C.; D’Agostino, G.; Ruaro, C.; Ghidoni, R.; Benussi, L.; Galimberti, D.; Esposito, F.; et al. An APOE Haplotype Associated with Decreased epsilon4 Expression Increases the Risk of Late Onset Alzheimer’s Disease. J. Alzheimer’s Dis. 2011, 24, 235–245. [Google Scholar] [CrossRef]
- Navarro, S.; Medina, P.; Mira, Y.; Estelles, A.; Villa, P.; Ferrando, F.; Vaya, A.; Bertina, R.M.; España, F. Haplotypes of the EPCR gene, prothrombin levels, and the risk of venous thrombosis in carriers of the prothrombin G20210A mutation. Haematol. 2008, 93, 885–891. [Google Scholar] [CrossRef][Green Version]
- Vymetalkova, V.P.; Soucek, P.; Kunická, T.; Jiraskova, K.; Brynychová, V.; Pardini, B.; Novosadova, V.; Polivkova, Z.; Kubáčková, K.; Kozevnikovova, R.; et al. Genotype and Haplotype Analyses of TP53 Gene in Breast Cancer Patients: Association with Risk and Clinical Outcomes. PLOS ONE 2015, 10, e0134463. [Google Scholar] [CrossRef]
- Schächter, F.; Faure-Delanef, L.; Guénot, F.; Rouger, H.; Froguel, P.; Lesueur-Ginot, L.; Cohen, D. Genetic associations with human longevity at the APOE and ACE loci. Nat. Genet. 1994, 6, 29–32. [Google Scholar] [CrossRef]
- Soerensen, M.; Dato, S.; Tan, Q.; Thinggaard, M.; Kleindorp, R.; Beekman, M.; Suchiman, H.E.D.; Jacobsen, R.; McGue, M.; Stevnsner, T.; et al. Evidence from case-control and longitudinal studies supports associations of genetic variation in APOE, CETP, and IL6 with human longevity. AGE 2012, 35, 487–500. [Google Scholar] [CrossRef]
- Ferrari, R.; Wang, Y.; Vandrovcova, J.; Guelfi, S.; Witeolar, A.; Karch, C.M.; Schork, A.J.; Fan, C.C.; Brewer, J.B.; Momeni, P.; et al. Genetic architecture of sporadic frontotemporal dementia and overlap with Alzheimer’s and Parkinson’s diseases. J. Neurol. Neurosurg. Psychiatry 2017, 88, 152–164. [Google Scholar] [CrossRef]
- Coon, K.D.; Myers, A.J.; Craig, D.W.; Webster, J.A.; Pearson, J.V.; Lince, D.H.; Zismann, V.L.; Beach, T.G.; Leung, D.; Bryden, L.; et al. A High-Density Whole-Genome Association Study Reveals That APOE Is the Major Susceptibility Gene for Sporadic Late-Onset Alzheimer’s Disease. J. Clin. Psychiatry 2007, 68, 613–618. [Google Scholar] [CrossRef]
- Corder, E.H.; Saunders, A.M.; Strittmatter, W.J.; E Schmechel, D.; Gaskell, P.C.; Small, G.W.; Roses, A.D.; Haines, J.L.; A Pericak-Vance, M. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993, 261, 921–923. [Google Scholar] [CrossRef]
- Babenko, V.N.; Afonnikov, D.A.; Ignatieva, E.V.; Klimov, A.V.; Gusev, F.E.; Rogaev, E.I. Haplotype analysis of APOE intragenic SNPs. BMC Neurosci. 2018, 19, 29–40. [Google Scholar] [CrossRef]
- Xiao, H.; Gao, Y.; Liu, L.; Li, Y. Association between polymorphisms in the promoter region of the apolipoprotein E (APOE) gene and Alzheimer’s disease: A meta-analysis. EXCLI J. 2017, 16, 921–938. [Google Scholar]
- Deelen, J.; Beekman, M.; Uh, H.-W.; Helmer, Q.; Kuningas, M.; Christiansen, L.; Kremer, D.; Van Der Breggen, R.; Suchiman, H.E.D.; Lakenberg, N.; et al. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell 2011, 10, 686–698. [Google Scholar] [CrossRef]
- Deelen, J.; Beekman, M.; Uh, H.-W.; Broer, L.; Ayers, K.L.; Tan, Q.; Kamatani, Y.; Bennet, A.M.; Tamm, R.; Trompet, S.; et al. Genome-wide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Hum. Mol. Genet. 2014, 23, 4420–4432. [Google Scholar] [CrossRef]
- Nebel, A.; Kleindorp, R.; Caliebe, A.; Nothnagel, M.; Blanché, H.; Junge, O.; Wittig, M.; Ellinghaus, D.; Flachsbart, F.; Wichmann, H.-E.; et al. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mech. Ageing Dev. 2011, 132, 324–330. [Google Scholar] [CrossRef]
- Lin, R.; Zhang, Y.; Yan, D.; Liao, X.; Gong, G.; Hu, J.; Fu, Y.; Cai, W. Association of common variants in TOMM40/APOE/APOC1 region with human longevity in a Chinese population. J. Hum. Genet. 2016, 61, 323–328. [Google Scholar] [CrossRef]
- Snyder, M.W.; Adey, A.; Kitzman, J.O.; Shendure, J. Haplotype-resolved genome sequencing: Experimental methods and applications. Nat. Rev. Genet. 2015, 16, 344–358. [Google Scholar] [CrossRef]
- Huang, M.; Tu, J.; Lu, Z. Recent Advances in Experimental Whole Genome Haplotyping Methods. Int. J. Mol. Sci. 2017, 18, 1944. [Google Scholar] [CrossRef]
- Porubský, D.; Sanders, A.D.; Van Wietmarschen, N.; Falconer, E.; Hills, M.; Spierings, D.C.; Bevova, M.R.; Guryev, V.; Lansdorp, P.M. Direct chromosome-length haplotyping by single-cell sequencing. Genome Res. 2016, 26, 1565–1574. [Google Scholar] [CrossRef]
- Putnam, N.H.; O’Connell, B.L.; Stites, J.C.; Rice, B.J.; Blanchette, M.; Calef, R.; Troll, C.J.; Fields, A.; Hartley, P.D.; Sugnet, C.W.; et al. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 2016, 26, 342–350. [Google Scholar] [CrossRef]
- Lieberman-Aiden, E.; Van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef]
- Zheng, G.X.; Lau, B.T.; Schnall-Levin, M.; Jarosz, M.; Bell, J.M.; Hindson, C.M.; Kyriazopoulou-Panagiotopoulou, S.; Masquelier, D.A.; Merrill, L.; Terry, J.M.; et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol. 2016, 34, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; A Sasani, T.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T.; et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018, 36, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Porubsky, D.; Garg, S.; Sanders, A.D.; Korbel, J.O.; Guryev, V.; Lansdorp, P.M.; Marschall, T. Dense and accurate whole-chromosome haplotyping of individual genomes. Nat. Commun. 2017, 8, 1293. [Google Scholar] [CrossRef] [PubMed]
- Rang, F.J.; Kloosterman, W.P.; De Ridder, J. From squiggle to basepair: Computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Laver, T.W.; Caswell, R.C.; Moore, K.A.; Poschmann, J.; Johnson, M.B.; Owens, M.M.; Ellard, S.; Paszkiewicz, K.H.; Weedon, M.N. Pitfalls of haplotype phasing from amplicon-based long-read sequencing. Sci. Rep. 2016, 6, 21746. [Google Scholar] [CrossRef]
- Edge, P.; Bafna, V.; Bansal, V. HapCUT2: Robust and accurate haplotype assembly for diverse sequencing technologies. Genome Res. 2017, 27, 801–812. [Google Scholar] [CrossRef]
- Patterson, M.; Marschall, T.; Pisanti, N.; Van Iersel, L.; Stougie, L.; Klau, G.W.; Hu, Y.-J. WhatsHap: Weighted Haplotype Assembly for Future-Generation Sequencing Reads. J. Comput. Biol. 2015, 22, 498–509. [Google Scholar] [CrossRef]
- Kuleshov, V. Probabilistic single-individual haplotyping. Bioinform. 2014, 30, i379–i385. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinform. 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- 10x Genomics. Discontinuation of Linked-Reads. Available online: https://www.10xgenomics.com/products/linked-reads (accessed on 30 November 2020).
- Gilpatrick, T.; Lee, I.; Graham, J.E.; Raimondeau, E.; Bowen, R.; Heron, A.; Downs, B.; Sukumar, S.; Sedlazeck, F.J.; Timp, W. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat. Biotechnol. 2020, 38, 433–438. [Google Scholar] [CrossRef]
- Edge, P.; Bansal, V. Longshot enables accurate variant calling in diploid genomes from single-molecule long read sequencing. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Luo, R.; Wong, C.-L.; Wong, Y.-S.; Tang, C.-I.; Liu, C.-M.; Leung, C.-M.; Lam, T.-W. Exploring the limit of using a deep neural network on pileup data for germline variant calling. Nat. Mach. Intell. 2020, 2, 220–227. [Google Scholar] [CrossRef]
- Ammar, R.; Paton, T.A.; Torti, D.; Shlien, A.; Bader, G.D. Long read nanopore sequencing for detection of HLA and CYP2D6 variants and haplotypes. F1000Research 2015, 4, 17. [Google Scholar] [CrossRef]
- Stancu, M.C.; Van Roosmalen, M.J.; Renkens, I.; Nieboer, M.M.; Middelkamp, S.; De Ligt, J.; Pregno, G.; Giachino, D.; Mandrile, G.; Valle-Inclan, J.E.; et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Leija-Salazar, M.; Sedlazeck, F.; Toffoli, M.; Mullin, S.; Mokretar, K.; Athanasopoulou, M.; Donald, A.; Sharma, R.; Hughes, D.; Schapira, A.H.V.; et al. Detection of GBA missense mutations and other variants using the Oxford Nanopore MinION. Mol. Genet. Genomic. Med. 2019, 7, 3. [Google Scholar] [CrossRef]
- Maestri, S.; Cosentino, E.; Paterno, M.; Freitag, H.; Garces, J.M.; Marcolungo, L.; Alfano, M.; Njunjić, I.; Schilthuizen, M.; Slik, F.J.; et al. A Rapid and Accurate MinION-Based Workflow for Tracking Species Biodiversity in the Field. Genes 2019, 10, 468. [Google Scholar] [CrossRef]
- Menegon, M.; Cantaloni, C.; Rodriguez-Prieto, A.; Centomo, C.; Abdelfattah, A.; Rossato, M.; Bernardi, M.; Xumerle, L.; Loader, S.; Delledonne, M. On site DNA barcoding by nanopore sequencing. PLoS ONE 2017, 12, e0184741. [Google Scholar] [CrossRef]
- Knot, I.E.; Zouganelis, G.D.; Weedall, G.D.; Wich, S.A.; Rae, R. DNA Barcoding of Nematodes Using the MinION. Front. Ecol. Evol. 2020, 8, 8. [Google Scholar] [CrossRef]
- Karst, S.; Ziels, R.; Kirkegaard, R.; Sørensen, E.; McDonald, D.; Zhu, Q.; Knight, R.; Albertsen, M. Enabling high-accuracy long-read amplicon sequences using unique molecular identifiers with Nanopore or PacBio sequencing. bioRxiv 2020. Available online: https://www.biorxiv.org/content/10.1101/645903v3.full (accessed on 30 November 2020).
- Wick, R.R.; Judd, L.M.; E Holt, K. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019, 20, 1–10. [Google Scholar] [CrossRef]
- Vereecke, N.; Bokma, J.; Haesebrouck, F.; Nauwynck, H.; Boyen, F.; Pardon, B.; Theuns, S. High quality genome assemblies of Mycoplasma bovis using a taxon-specific Bonito basecaller for MinION and Flongle long-read nanopore sequencing. BMC Bioinform. 2020, 21, 1–16. [Google Scholar] [CrossRef]
- Tytgat, O.; Gansemans, Y.; Weymaere, J.; Rubben, K.; Deforce, D.; Van Nieuwerburgh, F. Nanopore Sequencing of a Forensic STR Multiplex Reveals Loci Suitable for Single-Contributor STR Profiling. Genes 2020, 11, 381. [Google Scholar] [CrossRef]
- Gabrieli, T.; Sharim, H.; Michaeli, Y.; Ebenstein, Y. Cas9-Assisted Targeting of CHromosome segments (CATCH) for targeted nanopore sequencing and optical genome mapping. BioRxiv 2017. Available online: https://www.biorxiv.org/content/10.1101/110163v3 (accessed on 30 November 2020).
- Madsen, E.B.; Höijer, I.; Kvist, T.; Ameur, A.; Mikkelsen, M.J. Xdrop: Targeted sequencing of long DNA molecules from low input samples using droplet sorting. Hum. Mutat. 2020, 41, 1671–1679. [Google Scholar] [CrossRef]
- Mantere, T.; Kersten, S.; Hoischen, A. Long-Read Sequencing Emerging in Medical Genetics. Front. Genet. 2019, 10, 426. [Google Scholar] [CrossRef]
- Zhao, H.; Sun, Z.; Wang, J.; Huang, H.; Kocher, J.-P.; Wang, L. CrossMap: A versatile tool for coordinate conversion between genome assemblies. Bioinform. 2014, 30, 1006–1007. [Google Scholar] [CrossRef]
- Van Der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinform. 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
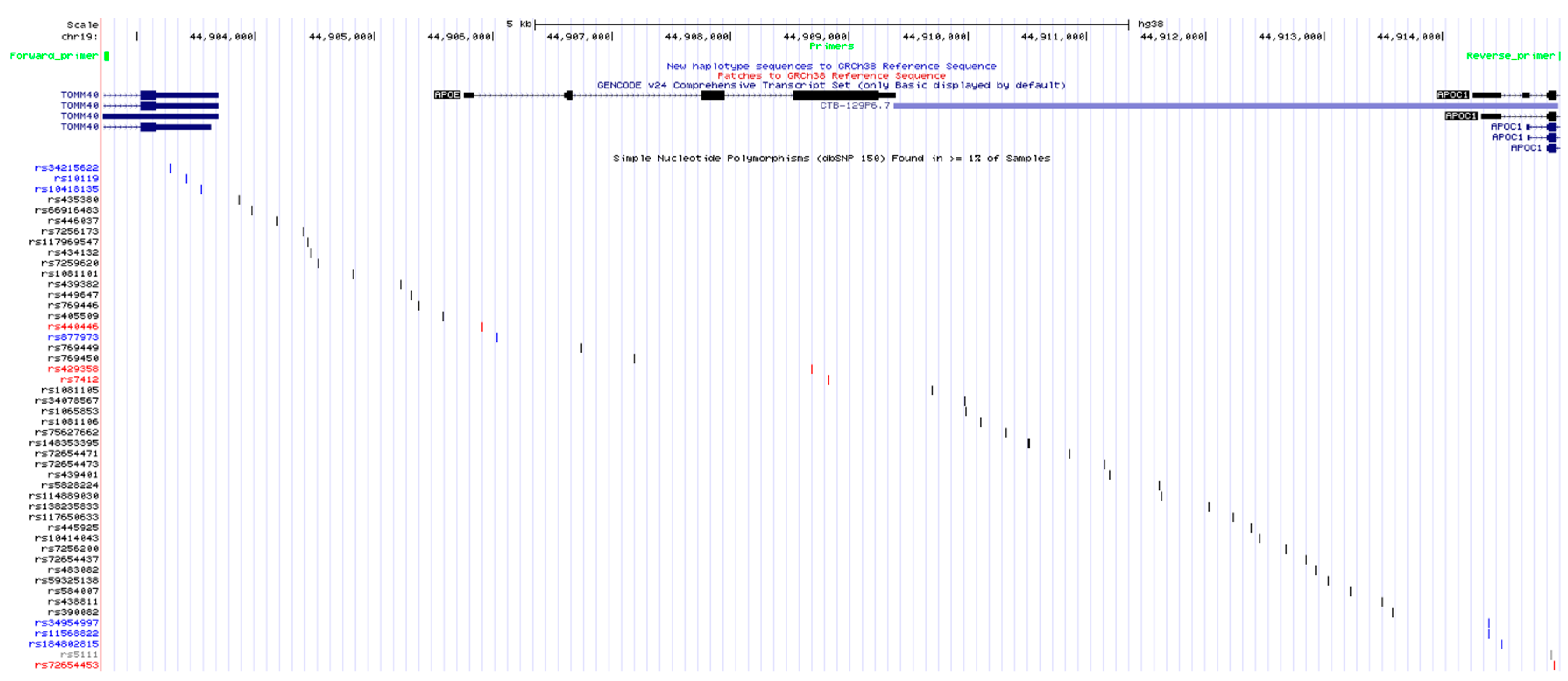
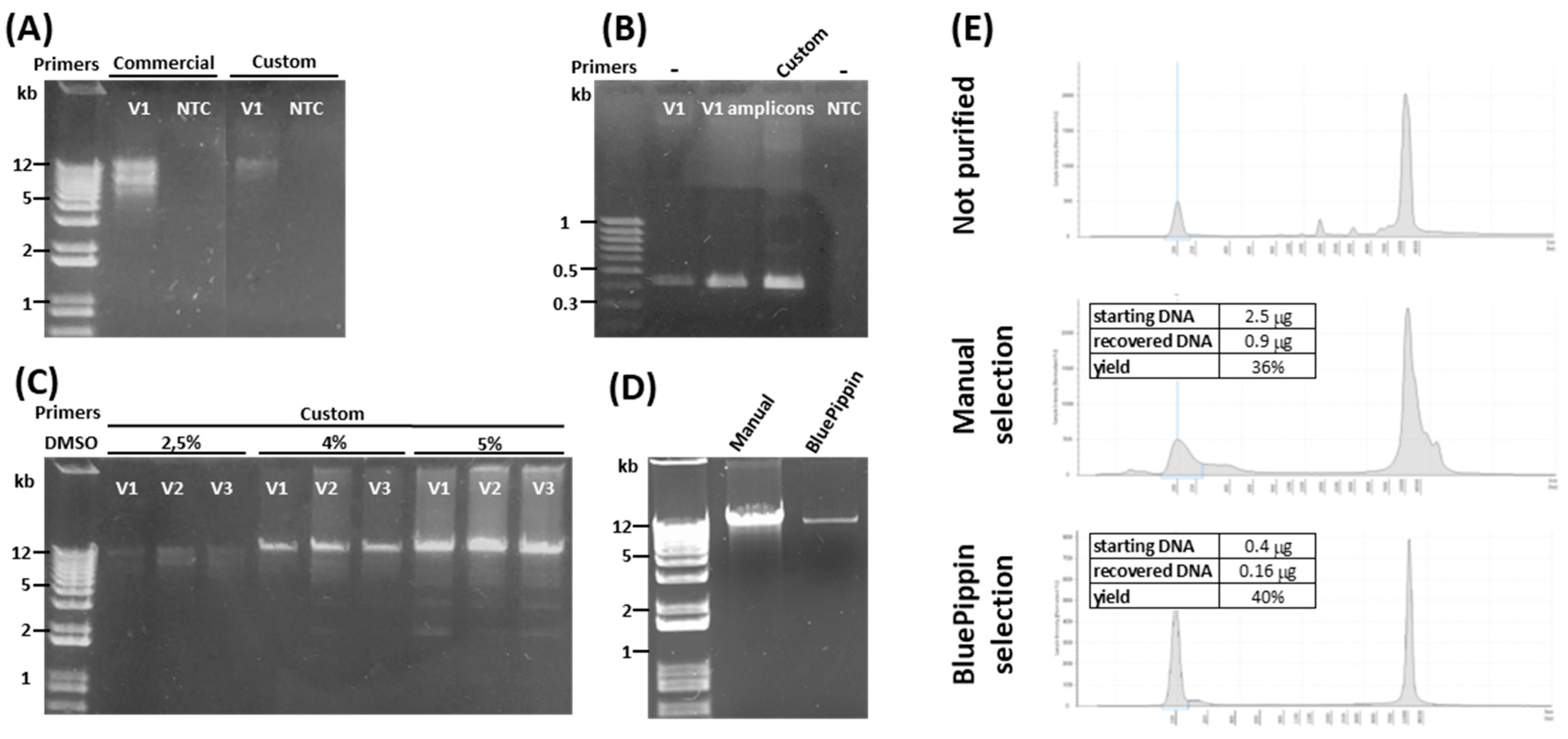

| dbSNP ID | Pos. | Ref. | Alt. |
|---|---|---|---|
| rs893292251 | chr19:44902764 | C | T |
| rs34215622 | chr19:44903281 | C | CG |
| - | chr19:44903416 | G | A |
| - | chr19:44904398 | C | T |
| rs7259620 | chr19:44904531 | G | A,C |
| rs449647 | chr19:44905307 | A | T |
| rs769446 | chr19:44905371 | T | C |
| rs405509 | chr19:44905579 | T | G |
| - | chr19:44905832 | C | T |
| rs440446 | chr19:44905910 | C | G |
| - | chr19:44906337 | G | A |
| rs769450 | chr19:44907187 | G | A |
| rs429358 | chr19:44908684 | T | C |
| rs7412 | chr19:44908822 | C | T |
| rs747519137 | chr19:44909521 | CT | C |
| - | chr19:44909967 | TG | T |
| rs1065853 | chr19:44909976 | G | C,T |
| rs75627662 | chr19:44910319 | C | T |
| - | chr19:44910393 | A | C |
| - | chr19:44910397 | T | C |
| - | chr19:44910405 | A | C |
| - | chr19:44910410 | A | C |
| rs72654469 | chr19:44910678 | T | C |
| rs72654473 | chr19:44911142 | C | A |
| rs439401 | chr19:44911194 | T | C |
| rs5828224 | chr19:44911609 | AT | A |
| rs445925 | chr19:44912383 | G | A,C |
| rs483082 | chr19:44912921 | G | T |
| rs59325138 | chr19:44913034 | C | T |
| rs584007 | chr19:44913221 | A | G |
| rs438811 | chr19:44913484 | C | T |
| rs390082 | chr19:44913574 | T | A,C,G |
| rs72654445 | chr19:44913943 | G | A |
| - | chr19:44914318 | A | T |
| rs34954997 | chr19:44914381 | C | CTTCG |
| Sample Name | Homozygous Variants | Heterozygous Variants | Heterozygous Phased Variants | |||
|---|---|---|---|---|---|---|
| SNVs | Indels | SNVs | Indels | SNVs | Indels | |
| NA12878 | 0 | 0 | 10 | 1 | 10 | 1 |
| NA12892 | 0 | 0 | 4 | 0 | 4 | 0 |
| V1 | 6 | 3 | 11 | 1 | 11 | 1 |
| V2 | 4 | 0 | 6 | 2 | 6 | 1 |
| V3 | 4 | 1 | 4 | 1 | 4 | 1 |
| Sample Name | Number of Reads | Reads Mean Length | Number of PASS Reads (%) | PASS Reads Mean Length (bp) | Number of PASS Reads Aligned | Number of PASS Reads Covering the Whole Region |
|---|---|---|---|---|---|---|
| NA12878 | 28,251 | 11,871 | 24,335 (86%) | 12,046 | 24,330 (99.99%) | 23,799 (97.82%) |
| NA12892 | 27,410 | 11,710 | 23,541 (86%) | 12,046 | 23,541 (100%) | 23,022 (97.80%) |
| V1 | 39,993 | 11,552 | 33,489 (84%) | 12,042 | 33,486 (99.99%) | 32,941 (98.37%) |
| V2 | 38,501 | 11,690 | 33,373 (87%) | 12,044 | 33,371 (99.99%) | 32,755 (98.15%) |
| V3 | 32,084 | 11,588 | 27,165 (85%) | 12,046 | 27,162 (99.99%) | 26,673 (98.20%) |
| NA12878 | NA12892 | V1 | V2 | V3 | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNVs | Indels | SNVs | Indels | SNVs | Indels | SNVs | Indels | SNVs | Indels | |||||||||||||||||||||
| Number of Reads | TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP | TP | FN | FP |
| 10 | 10 | 0 | 9 | 0 | 1 | 1 | 4 | 0 | 3 | 0 | 0 | 0 | 16 | 2 | 0 | 0 | 3 | 3 | 9 | 1 | 10 | 0 | 2 | 2 | 6 | 2 | 3 | 0 | 2 | 3 |
| 30 | 10 | 0 | 0 | 0 | 1 | 1 | 4 | 0 | 0 | 0 | 0 | 3 | 16 | 2 | 0 | 1 | 2 | 2 | 9 | 1 | 0 | 0 | 2 | 4 | 8 | 0 | 0 | 0 | 2 | 2 |
| 60 | 10 | 0 | 0 | 0 | 1 | 2 | 4 | 0 | 0 | 0 | 0 | 3 | 17 | 1 | 0 | 0 | 3 | 2 | 10 | 0 | 0 | 0 | 2 | 2 | 8 | 0 | 0 | 1 | 1 | 2 |
| 100 | 10 | 0 | 0 | 0 | 1 | 2 | 4 | 0 | 0 | 0 | 0 | 2 | 17 | 1 | 0 | 0 | 3 | 2 | 10 | 0 | 0 | 0 | 2 | 2 | 8 | 0 | 0 | 0 | 2 | 2 |
| 300 | 10 | 0 | 0 | 0 | 1 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 17 | 1 | 0 | 0 | 3 | 2 | 10 | 0 | 0 | 0 | 2 | 0 | 8 | 0 | 0 | 0 | 2 | 0 |
| 600 | 10 | 0 | 0 | 0 | 1 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 17 | 1 | 0 | 0 | 3 | 2 | 9 | 1 | 0 | 0 | 2 | 0 | 8 | 0 | 0 | 0 | 2 | 0 |
| 1000 | 10 | 0 | 0 | 0 | 1 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 16 | 2 | 0 | 0 | 3 | 1 | 8 | 2 | 0 | 0 | 2 | 0 | 8 | 0 | 0 | 0 | 2 | 1 |
| 10000 | 8 | 2 | 0 | 0 | 1 | 1 | 4 | 0 | 0 | 0 | 0 | 1 | 10 | 8 | 0 | 0 | 3 | 1 | 7 | 3 | 0 | 0 | 2 | 3 | 7 | 1 | 0 | 0 | 2 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maestri, S.; Maturo, M.G.; Cosentino, E.; Marcolungo, L.; Iadarola, B.; Fortunati, E.; Rossato, M.; Delledonne, M. A Long-Read Sequencing Approach for Direct Haplotype Phasing in Clinical Settings. Int. J. Mol. Sci. 2020, 21, 9177. https://doi.org/10.3390/ijms21239177
Maestri S, Maturo MG, Cosentino E, Marcolungo L, Iadarola B, Fortunati E, Rossato M, Delledonne M. A Long-Read Sequencing Approach for Direct Haplotype Phasing in Clinical Settings. International Journal of Molecular Sciences. 2020; 21(23):9177. https://doi.org/10.3390/ijms21239177
Chicago/Turabian StyleMaestri, Simone, Maria Giovanna Maturo, Emanuela Cosentino, Luca Marcolungo, Barbara Iadarola, Elisabetta Fortunati, Marzia Rossato, and Massimo Delledonne. 2020. "A Long-Read Sequencing Approach for Direct Haplotype Phasing in Clinical Settings" International Journal of Molecular Sciences 21, no. 23: 9177. https://doi.org/10.3390/ijms21239177
APA StyleMaestri, S., Maturo, M. G., Cosentino, E., Marcolungo, L., Iadarola, B., Fortunati, E., Rossato, M., & Delledonne, M. (2020). A Long-Read Sequencing Approach for Direct Haplotype Phasing in Clinical Settings. International Journal of Molecular Sciences, 21(23), 9177. https://doi.org/10.3390/ijms21239177





