Staphylococcus aureus and Hyper-IgE Syndrome
Abstract
1. Introduction
2. Hyper-IgE Syndrome (HIES)
2.1. Genetics
2.2. Immunoglobulins
2.3. TLRs
2.4. STAT3
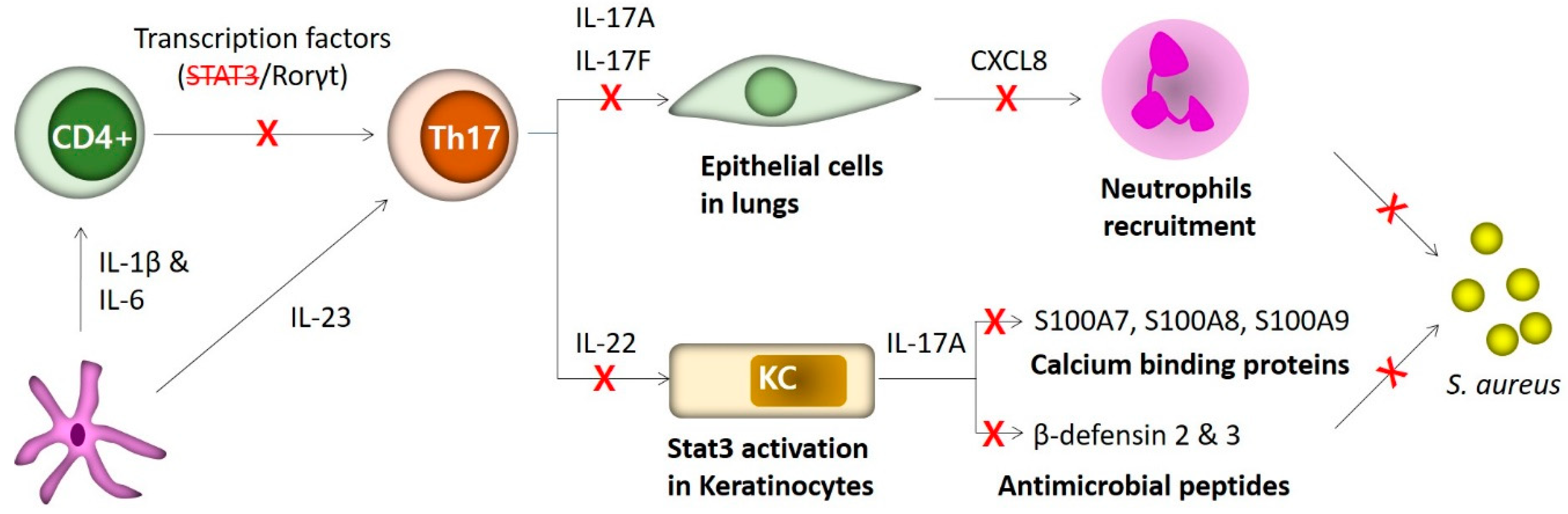
3. TH17 Cells in Staphylococcus Infections of Hyper-IgE Syndrome (HIES)
3.1. TH17 Cells
3.2. STAT3 Mutations
3.3. Molecular Mechanisms Conferring Site-Specific Susceptibility to S. aureus Infections in HIES Patients
3.4. Antimicrobial Peptides in Dermatitis of HIES Patients
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Foster, T. Immune evasion by staphylococci. Nat. Rev. Microbiol. 2005, 3, 948–958. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, H.; Melles, D.C.; Vos, M.C.; Van Leeuwen, W.; Van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- Liu, G.Y. Molecular Pathogenesis of Staphylococcus aureus Infection. Pediatr. Res. 2009, 65, 71R–77R. [Google Scholar] [CrossRef] [PubMed]
- Gordon, R.J.; Lowy, F.D. Pathogenesis of Methicillin-Resistant Staphylococcus aureus Infection. Clin. Infect. Dis. 2008, 46, S350–S359. [Google Scholar] [CrossRef] [PubMed]
- Rooijakkers, S.H.M.; Ruyken, M.; Roos, A.; Daha, M.R.; Presanis, J.S.; Sim, R.B.; Van Wamel, W.J.B.; Van Kessel, K.P.M.; Strijp, J.A.G.V. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 2005, 6, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Graves, S.F.; Kobayashi, S.D.; DeLeo, F.R. Community-associated methicillin-resistant Staphylococcus aureus immune evasion and virulence. J. Mol. Med. 2010, 88, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Rooijakkers, S.H.; Van Kessel, K.P.; Van Strijp, J.A. Staphylococcal innate immune evasion. Trends Microbiol. 2005, 13, 596–601. [Google Scholar] [CrossRef]
- Klevens, R.M.; Morrison, M.A.; Nadle, J.; Petit, S.; Gershman, K.; Ray, S.; Harrison, L.H.; Lynfield, R.; Dumyati, G.; Townes, J.M.; et al. Invasive Methicillin-Resistant Staphylococcus aureus Infections in the United States. JAMA 2007, 298, 1763–1771. [Google Scholar] [CrossRef]
- Miller, L.G.; Perdreau-Remington, F.; Rieg, G.; Mehdi, S.; Perlroth, J.; Bayer, A.S.; Tang, A.W.; Phung, T.O.; Spellberg, B. Necrotizing Fasciitis Caused by Community-Associated Methicillin-Resistant Staphylococcus aureusin Los Angeles. N. Engl. J. Med. 2005, 352, 1445–1453. [Google Scholar] [CrossRef]
- Moran, G.J.; Krishnadasan, A.; Gorwitz, R.J.; Fosheim, G.E.; McDougal, L.K.; Carey, R.B.; Talan, D.A. Methicillin-Resistant S. aureus Infections among Patients in the Emergency Department. N. Engl. J. Med. 2006, 355, 666–674. [Google Scholar] [CrossRef]
- Gould, I.M. Clinical relevance of increasing glycopeptide MICs against Staphylococcus aureus. Int. J. Antimicrob. Agents 2008, 31, 1–9. [Google Scholar] [CrossRef]
- Steinkraus, G.; White, R.; Friedrich, L. Vancomycin MIC creep in non-vancomycin-intermediate Staphylococcus aureus (VISA), vancomycin-susceptible clinical methicillin-resistant S. aureus (MRSA) blood isolates from 2001 05. J. Antimicrob. Chemother. 2007, 60, 788–794. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.S.; Fowler, V.G.; Shukla, S.K.; Rose, W.E.; Proctor, R.A. Development of a vaccine against Staphylococcus aureus invasive infections: Evidence based on human immunity, genetics and bacterial evasion mechanisms. FEMS Microbiol. Rev. 2020, 44, 123–153. [Google Scholar] [CrossRef]
- Buckley, R.H.; Wray, B.B.; Belmaker, E.Z. Extreme hyperimmunoglobulinemia E and undue susceptibility to infection. Pediatrics 1972, 49, 59–70. [Google Scholar] [PubMed]
- Davis, S.; Schaller, J.; Wedgwood, R.; Harvard, M. JOB’S SYNDROME. Lancet 1966, 287, 1013–1015. [Google Scholar] [CrossRef]
- Grimbacher, B.; Holland, S.M.; Gallin, J.I.; Greenberg, F.; Hill, S.C.; Malech, H.L.; Miller, J.A.; O’Connell, A.C.; Puck, J.M. Hyper-IgE Syndrome with Recurrent Infections—An Autosomal Dominant Multisystem Disorder. N. Engl. J. Med. 1999, 340, 692–702. [Google Scholar] [CrossRef]
- Grimbacher, B.; Dutra, A.S.; Holland, S.M.; Fischer, R.E.; Pao, M.; Gallin, J.I.; Puck, J.M. Analphoid marker chromosome in a patient with hyper-IgE syndrome, autism, and mild mental retardation. Genet. Med. 1999, 1, 213–218. [Google Scholar] [CrossRef]
- Grimbacher, B.; Schäffer, A.A.; Holland, S.M.; Davis, J.; Gallin, J.I.; Malech, H.L.; Atkinson, T.P.; Belohradsky, B.H.; Buckley, R.H.; Cossu, F.; et al. Genetic Linkage of Hyper-IgE Syndrome to Chromosome 4. Am. J. Hum. Genet. 1999, 65, 735–744. [Google Scholar] [CrossRef]
- Holland, S.; DeLeo, F.R.; Elloumi, H.Z.; Hsu, A.P.; Uzel, G.; Brodsky, N.; Freeman, A.F.; Demidowich, A.; Davis, J.; Turner, M.L.C.; et al. STAT3 Mutations in the Hyper-IgE Syndrome. N. Engl. J. Med. 2007, 357, 1608–1619. [Google Scholar] [CrossRef]
- Minegishi, Y.; Saito, M.; Metin, A.; Karasuyama, H.; Tsuchiya, S.; Tsuge, I.; Takada, H.; Hara, T.; Kawamura, N.; Ariga, T.; et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 2007, 448, 1058–1062. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Noguchi, K.; Shi, W.; Tanaka, T.; Matsumoto, M.; Yoshida, N.; Kishimoto, T.; Akira, S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 1997, 94, 3801–3804. [Google Scholar] [CrossRef]
- Sowerwine, K.J.; Holland, S.M.; Freeman, A.F. Hyper-IgE syndrome update. Ann. N. Y. Acad. Sci. 2012, 1250, 25–32. [Google Scholar] [CrossRef]
- Renner, E.D.; Rylaarsdam, S.; Aňover-Sombke, S.; Rack, A.L.; Reichenbach, J.; Carey, J.C.; Zhu, Q.; Jansson, A.F.; Barboza, J.; Schimke, L.F.; et al. Novel signal transducer and activator of transcription 3 (STAT3) mutations, reduced TH17 cell numbers, and variably defective STAT3 phosphorylation in hyper-IgE syndrome. J. Allergy Clin. Immunol. 2008, 122, 181–187. [Google Scholar] [CrossRef]
- De Beaucoudrey, L.; Puel, A.; Filipe-Santos, O.; Cobat, A.; Ghandil, P.; Chrabieh, M.; Feinberg, J.; Von Bernuth, H.; Samarina, A.; Jannière, L.; et al. Mutations in STAT3 and IL12RB1 impair the development of human IL-17–producing T cells. J. Exp. Med. 2008, 205, 1543–1550. [Google Scholar] [CrossRef]
- Ma, C.S.; Chew, G.Y.; Simpson, N.; Priyadarshi, A.; Wong, M.; Grimbacher, B.; Fulcher, D.A.; Tangye, S.G.; Cook, M.C. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J. Exp. Med. 2008, 205, 1551–1557. [Google Scholar] [CrossRef]
- Heimall, J.; Davis, J.; Shaw, P.A.; Hsu, A.P.; Gu, W.; Welch, P.; Holland, S.M.; Freeman, A.F. Paucity of genotype–phenotype correlations in STAT3 mutation positive Hyper IgE Syndrome (HIES). Clin. Immunol. 2011, 139, 75–84. [Google Scholar] [CrossRef]
- Kane, A.; Deenick, E.K.; Ma, C.S.; Cook, M.C.; Uzel, G.; Tangye, S.G. STAT3 is a central regulator of lymphocyte differentiation and function. Curr. Opin. Immunol. 2014, 28, 49–57. [Google Scholar] [CrossRef]
- Béziat, V.; Tavernier, S.J.; Chen, Y.-H.; Ma, C.S.; Materna, M.; Laurence, A.; Staal, J.; Aschenbrenner, D.; Roels, L.; Worley, L.; et al. Dominant-negative mutations in human IL6ST underlie hyper-IgE syndrome. J. Exp. Med. 2020, 217, e20191804. [Google Scholar] [CrossRef]
- Nieminen, P.; Morgan, N.V.; Fenwick, A.L.; Parmanen, S.; Veistinen, L.; Mikkola, M.L.; Van Der Spek, P.J.; Giraud, A.; Judd, L.M.; Arte, S.; et al. Inactivation of IL11 Signaling Causes Craniosynostosis, Delayed Tooth Eruption, and Supernumerary Teeth. Am. J. Hum. Genet. 2011, 89, 67–81. [Google Scholar] [CrossRef]
- Spencer, S.; Bal, S.K.; Egner, W.; Allen, H.L.; Raza, S.I.; Ma, C.A.; Gürel, M.; Zhang, Y.; Sun, G.; Sabroe, R.A.; et al. Loss of the interleukin-6 receptor causes immunodeficiency, atopy, and abnormal inflammatory responses. J. Exp. Med. 2019, 216, 1986–1998. [Google Scholar] [CrossRef]
- Kotlarz, D.; Ziętara, N.; Uzel, G.; Weidemann, T.; Braun, C.J.; Diestelhorst, J.; Krawitz, P.M.; Robinson, P.N.; Hecht, J.; Puchałka, J.; et al. Loss-of-function mutations in the IL-21 receptor gene cause a primary immunodeficiency syndrome. J. Exp. Med. 2013, 210, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.Y.M.; Geha, S. Clinical and Immunologic Aspects of the Hyperimmunoglobulin E Syndrome. Hematol. Oncol. Clin. N. Am. 1988, 2, 81–100. [Google Scholar] [CrossRef]
- Erlewyn-Lajeunesse, M.D.S. Hyperimmunoglobulin-E syndrome with recurrent infection: A review of current opinion and treatment. Pediatr. Allergy Immunol. 2000, 11, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Takada, H.; Nomura, A.; Ohga, S.; Shibata, R.; Hara, T. Distinct gene expression patterns of peripheral blood cells in hyper-IgE syndrome. Clin. Exp. Immunol. 2005, 140, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Sheerin, K.A.; Buckley, R.H. Antibody responses to protein, polysaccharide, and ΦX174 antigens in the hyperimmunoglobulinemia E (hyper-IgE) syndrome. J. Allergy Clin. Immunol. 1991, 87, 803–811. [Google Scholar] [CrossRef]
- Berger, M.; Kirkpatrick, C.H.; Goldsmith, P.K.; Gallin, J.I. IgE antibodies to Staphylococcus aureus and Candida albicans in patients with the syndrome of hyperimmunoglobulin E and recurrent infections. J. Immunol. 1980, 125, 2437–2443. [Google Scholar] [PubMed]
- Dreskin, S.C.; Goldsmith, P.K.; Gallin, J.I. Immunoglobulins in the hyperimmunoglobulin E and recurrent infection (Job’s) syndrome. Deficiency of anti-Staphylococcus aureus immunoglobulin A. J. Clin. Investig. 1985, 75, 26–34. [Google Scholar] [CrossRef]
- Pène, J.; Gauchat, J.-F.; Lécart, S.; Drouet, E.; Guglielmi, P.; Boulay, V.; Delwail, A.; Foster, D.; Lecron, J.-C.; Yssel, H. Cutting edge: IL-21 is a switch factor for the production of IgG1 and IgG3 by human B cells. J. Immunol. 2004, 172, 5154–5157. [Google Scholar] [CrossRef]
- Avery, D.T.; Deenick, E.K.; Ma, C.S.; Suryani, S.; Simpson, N.; Chew, G.Y.; Chan, T.D.; Palendira, U.; Bustamante, J.; Boisson-Dupuis, S.; et al. B cell–intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J. Exp. Med. 2010, 207, 155–171. [Google Scholar] [CrossRef]
- Suto, A.; Nakajima, H.; Hirose, K.; Suzuki, K.; Kagami, S.-I.; Seto, Y.; Hoshimoto, A.; Saito, Y.; Foster, D.C.; Iwamoto, I. Interleukin 21 prevents antigen-induced IgE production by inhibiting germ line Cε transcription of IL-4–stimulated B cells. Blood 2002, 100, 4565–4573. [Google Scholar] [CrossRef]
- Wood, N.; Bourque, K.; Donaldson, D.D.; Collins, M.; Vercelli, D.; Goldman, S.J.; Kasaian, M.T. IL-21 effects on human IgE production in response to IL-4 or IL-13. Cell. Immunol. 2004, 231, 133–145. [Google Scholar] [CrossRef]
- Van De Veen, W.; Krätz, C.E.; McKenzie, C.I.; Aui, P.M.; Neumann, J.; Van Noesel, C.J.M.; Wirz, O.F.; Hagl, B.; Kröner, C.; Spielberger, B.D.; et al. Impaired memory B-cell development and antibody maturation with a skewing toward IgE in patients with STAT3 hyper-IgE syndrome. Allergy 2019, 74, 2394–2405. [Google Scholar] [CrossRef]
- Stentzel, S.; Hagl, B.; Abel, F.; Kahl, B.C.; Rack-Hoch, A.; Bröker, B.M.; Renner, E.D. Reduced Immunoglobulin (Ig) G Response to Staphylococcus aureus in STAT3 Hyper-IgE Syndrome. Clin. Infect. Dis. 2017, 64, 1279–1282. [Google Scholar] [CrossRef]
- Hawn, T.R.; Ozinsky, A.; Williams, L.M.; Rodrigues, S.; Clark, A.; Pham, U.; Hill, H.R.; Ochs, H.; Aderem, A.; Liles, W.C. Hyper-IgE Syndrome Is Not Associated with Defects in Several Candidate Toll-Like Receptor Pathway Genes. Hum. Immunol. 2005, 66, 842–847. [Google Scholar] [CrossRef]
- Renner, E.D.; Pawlita, I.; Hoffmann, F.; Hornung, V.; Hartl, D.; Albert, M.; Jansson, A.; Endres, S.; Hartmann, G.; Belohradsky, B.H.; et al. No Indication for a Defect in Toll-Like Receptor Signaling in Patients with Hyper-IgE Syndrome. J. Clin. Immunol. 2005, 25, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Minegishi, Y.; Saito, M.; Morio, T.; Watanabe, K.; Agematsu, K.; Tsuchiya, S.; Takada, H.; Hara, T.; Kawamura, N.; Ariga, T.; et al. Human Tyrosine Kinase 2 Deficiency Reveals Its Requisite Roles in Multiple Cytokine Signals Involved in Innate and Acquired Immunity. Immunity 2006, 25, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-M.; McAleer, J.P.; Zheng, M.; Pociask, D.A.; Kaplan, M.H.; Qin, S.; Reinhart, T.A.; Kolls, J.K. Innate Stat3-mediated induction of the antimicrobial protein Reg3γ is required for host defense against MRSA pneumonia. J. Exp. Med. 2013, 210, 551–561. [Google Scholar] [CrossRef]
- Minegishi, Y.; Saito, M.; Nagasawa, M.; Takada, H.; Hara, T.; Tsuchiya, S.; Agematsu, K.; Yamada, M.; Kawamura, N.; Ariga, T.; et al. Molecular explanation for the contradiction between systemic Th17 defect and localized bacterial infection in hyper-IgE syndrome. J. Exp. Med. 2009, 206, 1291–1301. [Google Scholar] [CrossRef] [PubMed]
- Aujla, S.J.; Chan, Y.R.; Husain, S.; Kreindler, J.L.; Dubin, P.J.; Pilewski, J.M.; Myerburg, M.M.; Mason, C.A.; Iwakura, Y.; Kolls, J.K.; et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat. Med. 2008, 14, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.S.; Cho, J.S. Immunity against Staphylococcus aureus cutaneous infections. Nat. Rev. Immunol. 2011, 11, 505–518. [Google Scholar] [CrossRef]
- Farmand, S.; Kremer, B.; Häffner, M.; Pütsep, K.; Bergman, P.; Sundin, M.; Ritterbusch, H.; Seidl, M.; Follo, M.; Henneke, P.; et al. Eosinophilia and reduced STAT3 signaling affect neutrophil cell death in autosomal-dominant Hyper-IgE syndrome. Eur. J. Immunol. 2018, 48, 1975–1988. [Google Scholar] [CrossRef] [PubMed]
- Myles, I.A.; Anderson, E.D.; Earland, N.J.; Zarember, K.A.; Sastalla, I.; Williams, K.W.; Gough, P.; Moore, I.N.; Ganesan, S.; Fowler, C.J.; et al. TNF overproduction impairs epithelial staphylococcal response in hyper IgE syndrome. J. Clin. Investig. 2018, 128, 3595–3604. [Google Scholar] [CrossRef] [PubMed]
- Dong, C. TH17 cells in development: An updated view of their molecular identity and genetic programming. Nat. Rev. Immunol. 2008, 8, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.I.; Zhou, L.; Littman, D.R. Transcriptional regulation of Th17 cell differentiation. Semin. Immunol. 2007, 19, 409–417. [Google Scholar] [CrossRef]
- Chen, Z.; Laurence, A.; O’Shea, J.J. Signal transduction pathways and transcriptional regulation in the control of Th17 differentiation. Semin. Immunol. 2007, 19, 400–408. [Google Scholar] [CrossRef]
- Ishigame, H.; Kakuta, S.; Sudo, K.; Nakae, S.; Sasakawa, C.; Iwakura, Y.; Nagai, T.; Kadoki, M.; Nambu, A.; Komiyama, Y.; et al. Differential Roles of Interleukin-17A and -17F in Host Defense against Mucoepithelial Bacterial Infection and Allergic Responses. Immunity 2009, 30, 108–119. [Google Scholar] [CrossRef]
- Kolls, J.K.; Lindén, A. Interleukin-17 Family Members and Inflammation. Immunity 2004, 21, 467–476. [Google Scholar] [CrossRef]
- Toy, D.; Kugler, D.; Wolfson, M.; Bos, T.V.; Gurgel, J.; Derry, J.; Tocker, J.; Peschon, J. Cutting Edge: Interleukin 17 Signals through a Heteromeric Receptor Complex. J. Immunol. 2006, 177, 36–39. [Google Scholar] [CrossRef]
- Zheng, Y.; Valdez, P.A.; Danilenko, D.M.; Hu, Y.; Sa, S.M.; Gong, Q.; Abbas, A.R.; Modrusan, Z.; Ghilardi, N.; De Sauvage, F.J.; et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 2008, 14, 282–289. [Google Scholar] [CrossRef]
- Huang, W.; Na, L.; Fidel, P.L.; Schwarzenberger, P. Requirement of Interleukin-17A for Systemic Anti–Candida albicansHost Defense in Mice. J. Infect. Dis. 2004, 190, 624–631. [Google Scholar] [CrossRef]
- Saito, M.; Nagasawa, M.; Takada, H.; Hara, T.; Tsuchiya, S.; Agematsu, K.; Yamada, M.; Kawamura, N.; Ariga, T.; Tsuge, I.; et al. Defective IL-10 signaling in hyper-IgE syndrome results in impaired generation of tolerogenic dendritic cells and induced regulatory T cells. J. Exp. Med. 2011, 208, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Minegishi, Y.; Karasuyama, H. Defects in Jak-STAT-mediated cytokine signals cause hyper-IgE syndrome: Lessons from a primary immunodeficiency. Int. Immunol. 2008, 21, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Minegishi, Y.; Karasuyama, H. Genetic origins of hyper-IgE syndrome. Curr. Allergy Asthma Rep. 2008, 8, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Grimbacher, B.; Holland, S.M.; Puck, J.M. Hyper-IgE syndromes. Immunol. Rev. 2005, 203, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Minegishi, Y.; Karasuyama, H. Hyperimmunoglobulin E syndrome and tyrosine kinase 2 deficiency. Curr. Opin. Allergy Clin. Immunol. 2007, 7, 506–509. [Google Scholar] [CrossRef]
- Milner, J.D.; Brenchley, J.M.; Davis, J.; Hsu, A.; Asher, A.I.; O’Shea, J.; Holland, S.M.; Paul, W.E.; Douek, D.C.; Laurence, A.; et al. Impaired TH17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 2008, 452, 773–776. [Google Scholar] [CrossRef]
- Happel, K.I.; Dubin, P.J.; Zheng, M.; Ghilardi, N.; Lockhart, C.; Quinton, L.J.; Odden, A.R.; Shellito, J.E.; Bagby, G.J.; Nelson, S.; et al. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J. Exp. Med. 2005, 202, 761–769. [Google Scholar] [CrossRef]
- Winkelstein, J.A.; Marino, M.C.; Buckley, R.H.; Foster, C.B.; Chanock, S.J.; Dickler, H.; Johnston, R.B.; Boyle, J.; Curnutte, J.; Gallin, J.I.; et al. Chronic Granulomatous Disease: Report on a National Registry of 368 Patients. Medicine 2000, 79, 155–169. [Google Scholar] [CrossRef]
- Kolls, J.K.; McCray, P.B.; Chan, Y.R. Cytokine-mediated regulation of antimicrobial proteins. Nat. Rev. Immunol. 2008, 8, 829–835. [Google Scholar] [CrossRef]
- Happel, K.I.; Zheng, M.; Young, E.; Quinton, L.J.; Lockhart, E.; Ramsay, A.J.; Shellito, J.E.; Schurr, J.R.; Bagby, G.J.; Nelson, S.; et al. Cutting Edge: Roles of Toll-Like Receptor 4 and IL-23 in IL-17 Expression in Response to Klebsiella pneumoniae Infection. J. Immunol. 2003, 170, 4432–4436. [Google Scholar] [CrossRef]
- Chen, Z.; Tato, C.M.; Muul, L.M.; Laurence, A.; O’Shea, J.J. Distinct regulation of interleukin-17 in human T helper lymphocytes. Arthritis Rheum. 2007, 56, 2936–2946. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.-Y.; Chen, Y.; Thai, P.; Wachi, S.; Huang, F.; Kim, C.; Harper, R.W.; Wu, R. IL-17 Markedly Up-Regulates β-Defensin-2 Expression in Human Airway Epithelium via JAK and NF-κB Signaling Pathways. J. Immunol. 2004, 173, 3482–3491. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Thai, P.; Zhao, Y.-H.; Ho, Y.-S.; DeSouza, M.M.; Wu, R. Stimulation of Airway Mucin Gene Expression by Interleukin (IL)-17 through IL-6 Paracrine/Autocrine Loop. J. Biol. Chem. 2003, 278, 17036–17043. [Google Scholar] [CrossRef] [PubMed]
- Kolls, J.K.; Kanaly, S.T.; Ramsay, A.J. Interleukin-17. Am. J. Respir. Cell Mol. Biol. 2003, 28, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Wolk, K.; Kunz, S.; Witte, E.; Friedrich, M.; Asadullah, K.; Sabat, R. IL-22 Increases the Innate Immunity of Tissues. Immunity 2004, 21, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Gallo, R.L.; Hooper, L.V. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 2012, 12, 503–516. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Nakamura, Y.; Núñez, G. Role of the microbiota in skin immunity and atopic dermatitis. Allergol. Int. 2017, 66, 539–544. [Google Scholar] [CrossRef]
- Brown, K.L.; Hancock, R.E. Cationic host defense (antimicrobial) peptides. Curr. Opin. Immunol. 2006, 18, 24–30. [Google Scholar] [CrossRef]
- Ong, P.Y.; Ohtake, T.; Brandt, C.; Strickland, I.; Boguniewicz, M.; Ganz, T.; Gallo, R.L.; Leung, D.Y. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N. Engl. J. Med. 2002, 347, 1151–1160. [Google Scholar] [CrossRef]
- Paterson, M.J.; Caldera, J.R.; Liu, G.Y.; Underhill, D.M.; Nguyen, C.; Sharma, P.; Castro, A.M.; Kolar, S.L.; Tsai, C.-M.; Limon, J.J.; et al. Harnessing antifungal immunity in pursuit of a Staphylococcus aureus vaccine strategy. PLOS Pathog. 2020, 16, e1008733. [Google Scholar] [CrossRef]
- Sanchez, M.; Kolar, S.L.; Martins, G.A.; Liu, G.Y.; Müller, S.; Reyes, C.N.; Wolf, A.J.; Ogawa, C.; Singhania, R.; De Carvalho, D.D.; et al. O-Acetylation of Peptidoglycan Limits Helper T Cell Priming and Permits Staphylococcus aureus Reinfection. Cell Host Microbe 2017, 22, 543–551. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, B.; Liu, G.Y. Staphylococcus aureus and Hyper-IgE Syndrome. Int. J. Mol. Sci. 2020, 21, 9152. https://doi.org/10.3390/ijms21239152
Park B, Liu GY. Staphylococcus aureus and Hyper-IgE Syndrome. International Journal of Molecular Sciences. 2020; 21(23):9152. https://doi.org/10.3390/ijms21239152
Chicago/Turabian StylePark, Bonggoo, and George Y. Liu. 2020. "Staphylococcus aureus and Hyper-IgE Syndrome" International Journal of Molecular Sciences 21, no. 23: 9152. https://doi.org/10.3390/ijms21239152
APA StylePark, B., & Liu, G. Y. (2020). Staphylococcus aureus and Hyper-IgE Syndrome. International Journal of Molecular Sciences, 21(23), 9152. https://doi.org/10.3390/ijms21239152




