Functional Identification of Serine Hydroxymethyltransferase as a Key Gene Involved in Lysostaphin Resistance and Virulence Potential of Staphylococcus aureus Strains
Abstract
1. Introduction
2. Results
2.1. Lysostaphin Resistance Pattern in ST72 Isolates
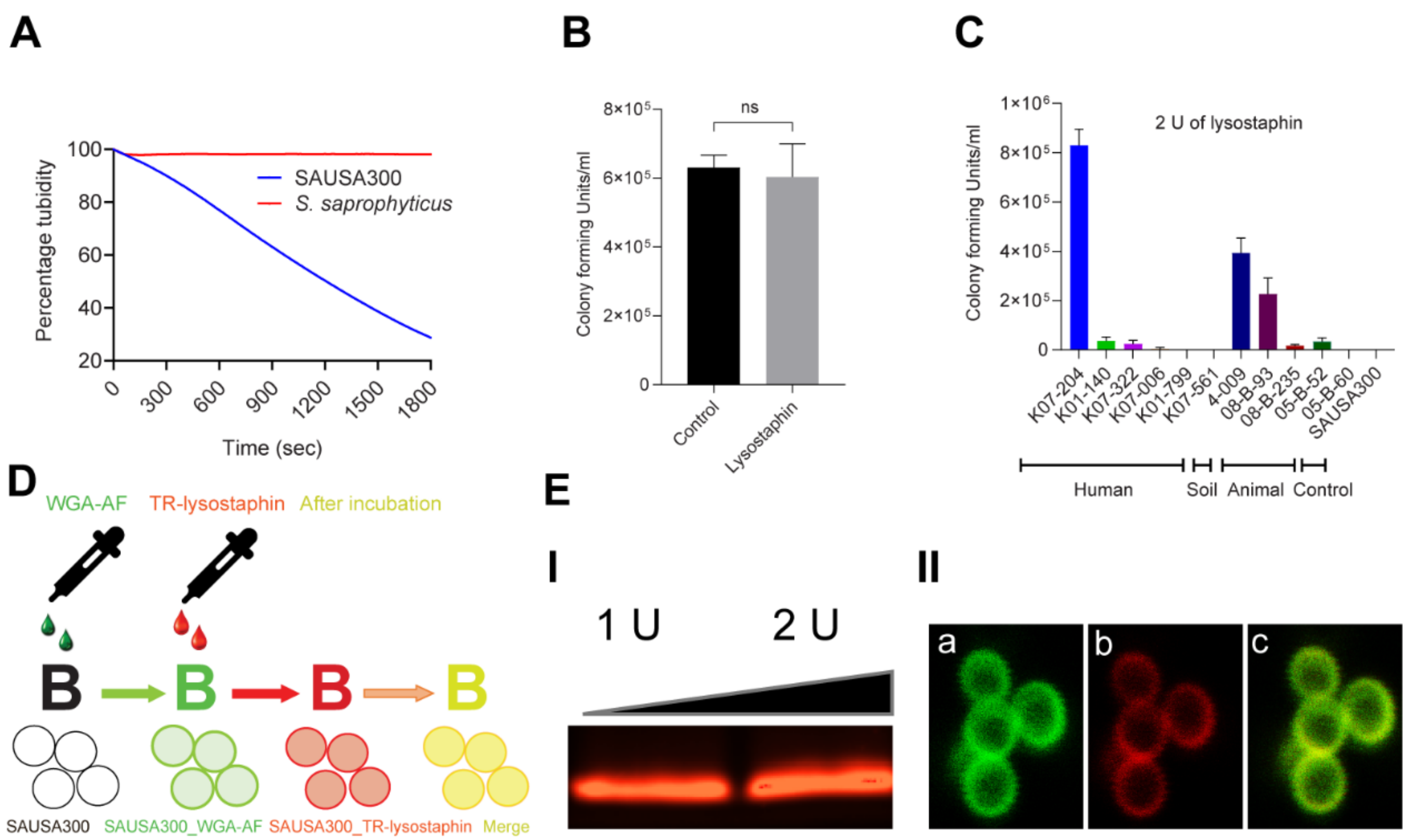
2.2. Phenotypic Assessment of the Lysostaphin Resistance in ST72 Isolates
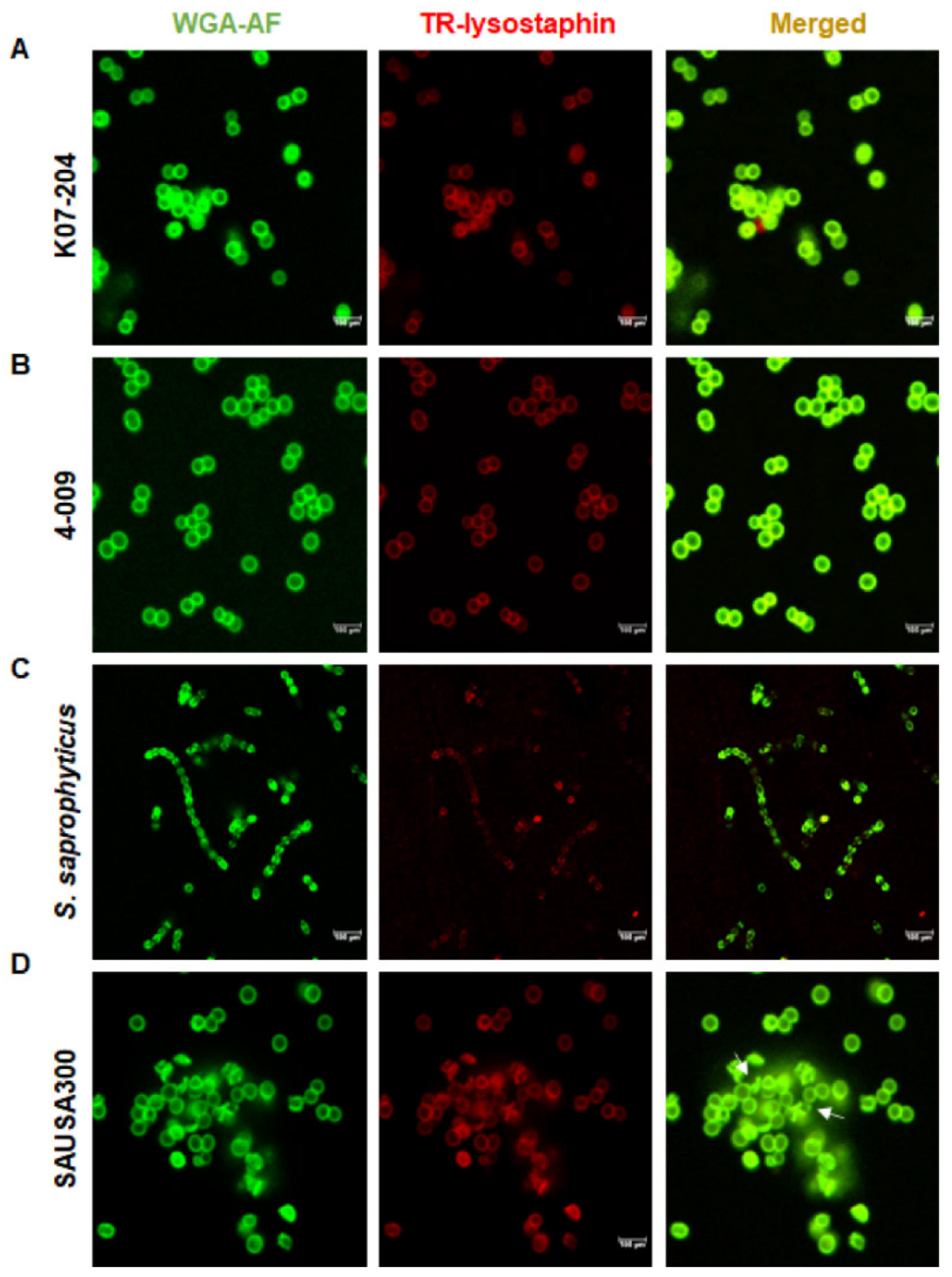
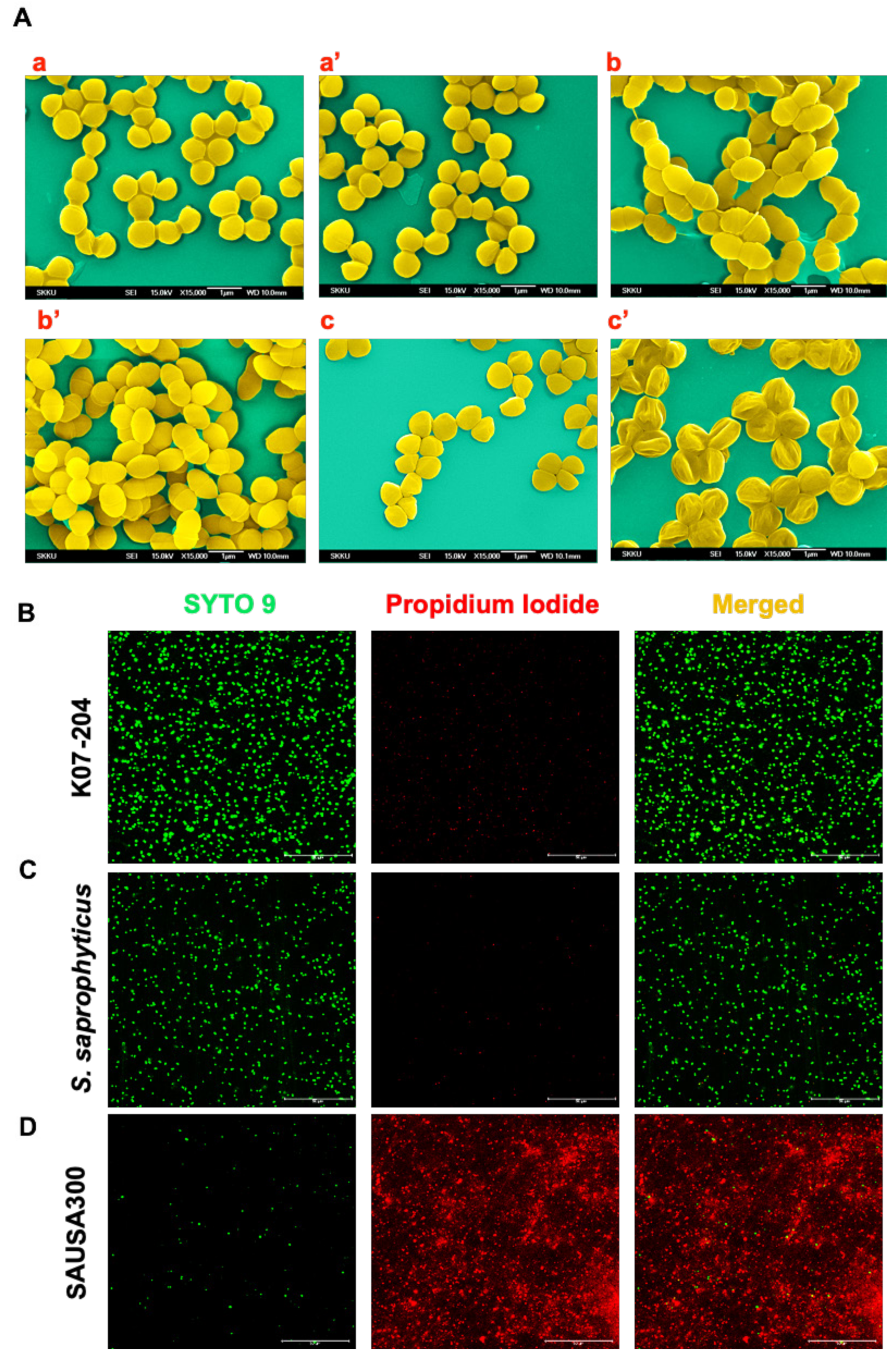
2.3. Confirmation of the Lysostaphin Resistance in ST72 Isolates
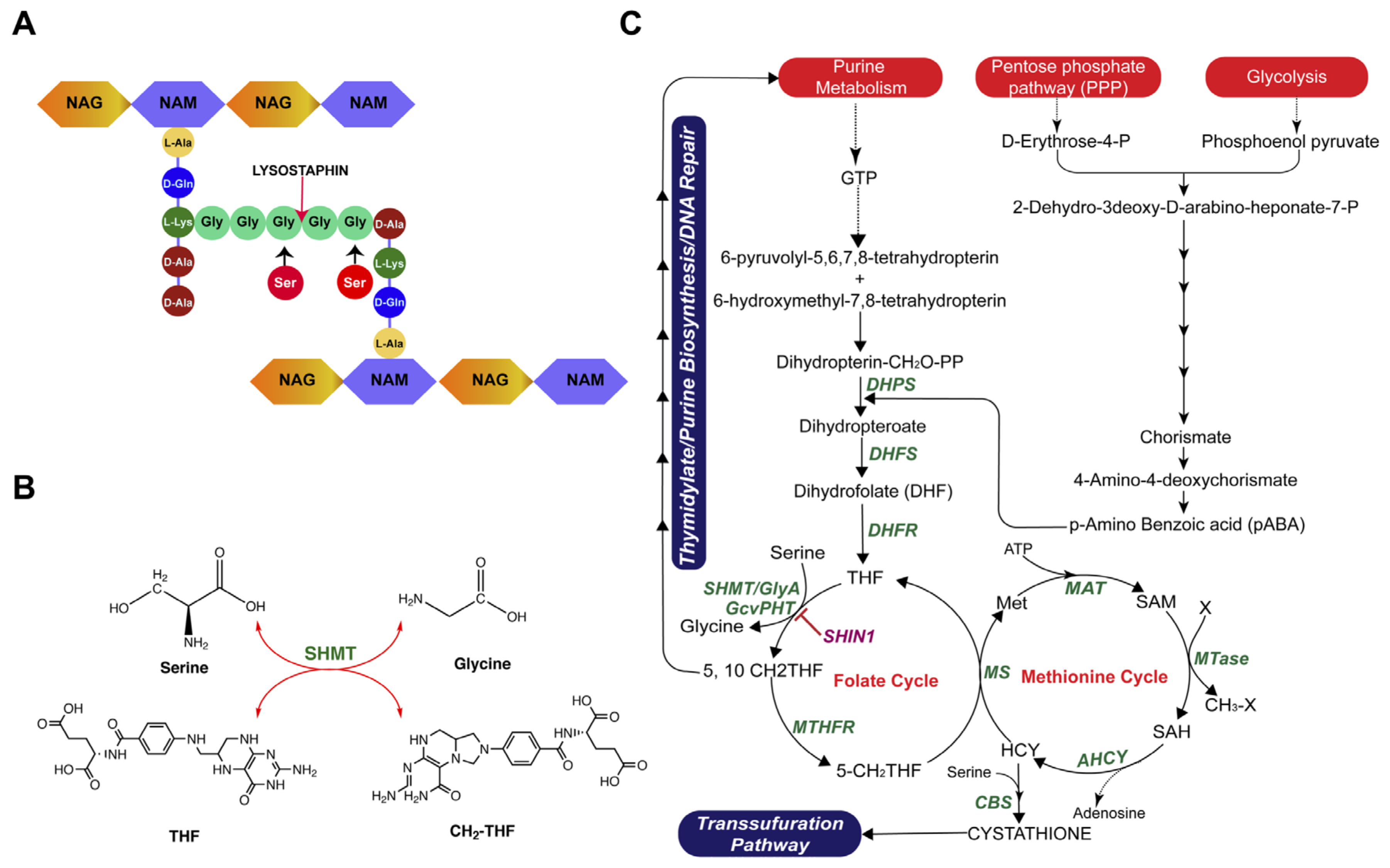
2.4. Investigating the Existing Mechanism of Lysostaphin Resistance
2.5. Comparative Genomics Analysis of lysr K07-204 and lyss K07-561
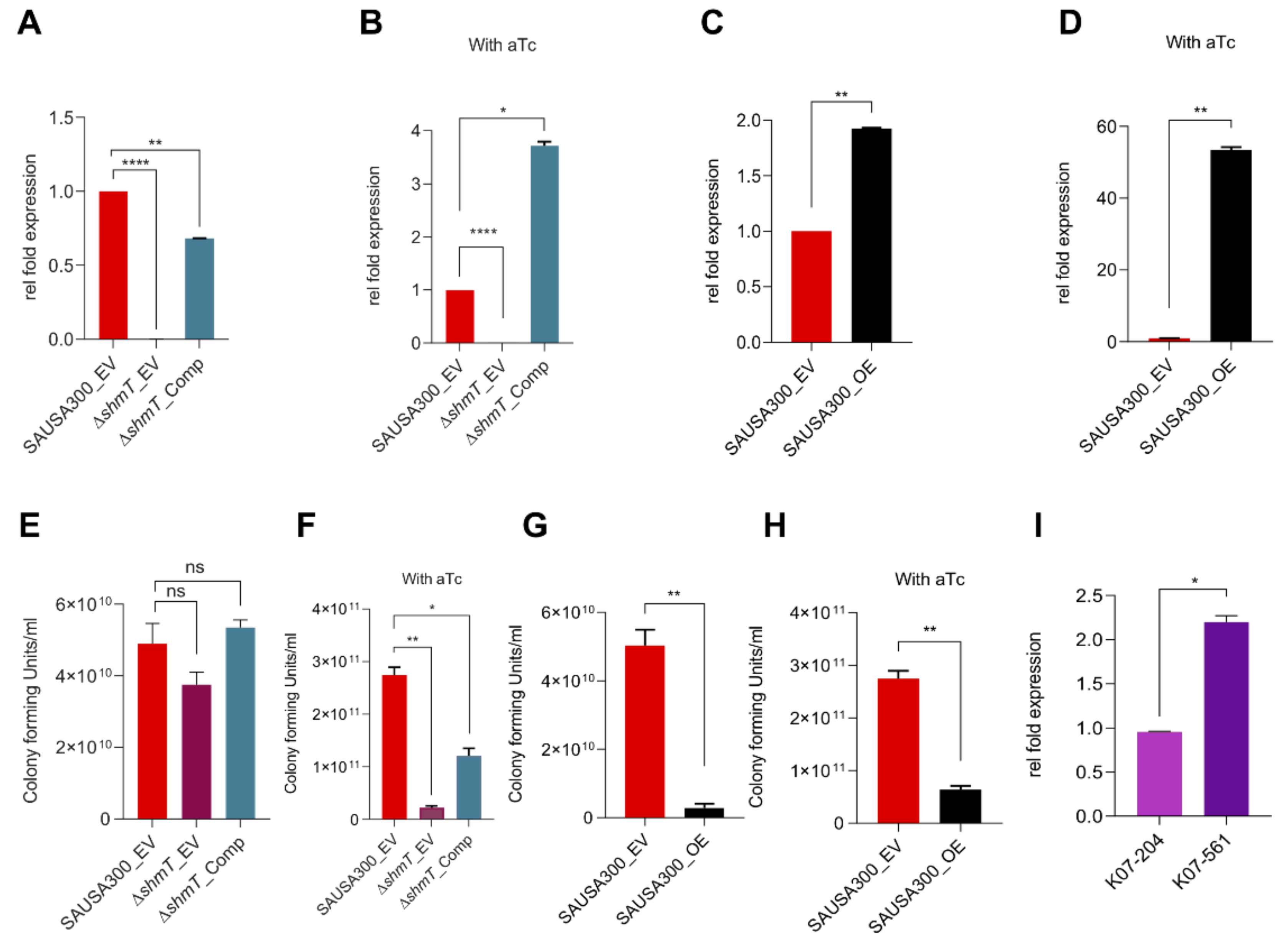
2.6. Role of shmT in Lysostaphin Resistance
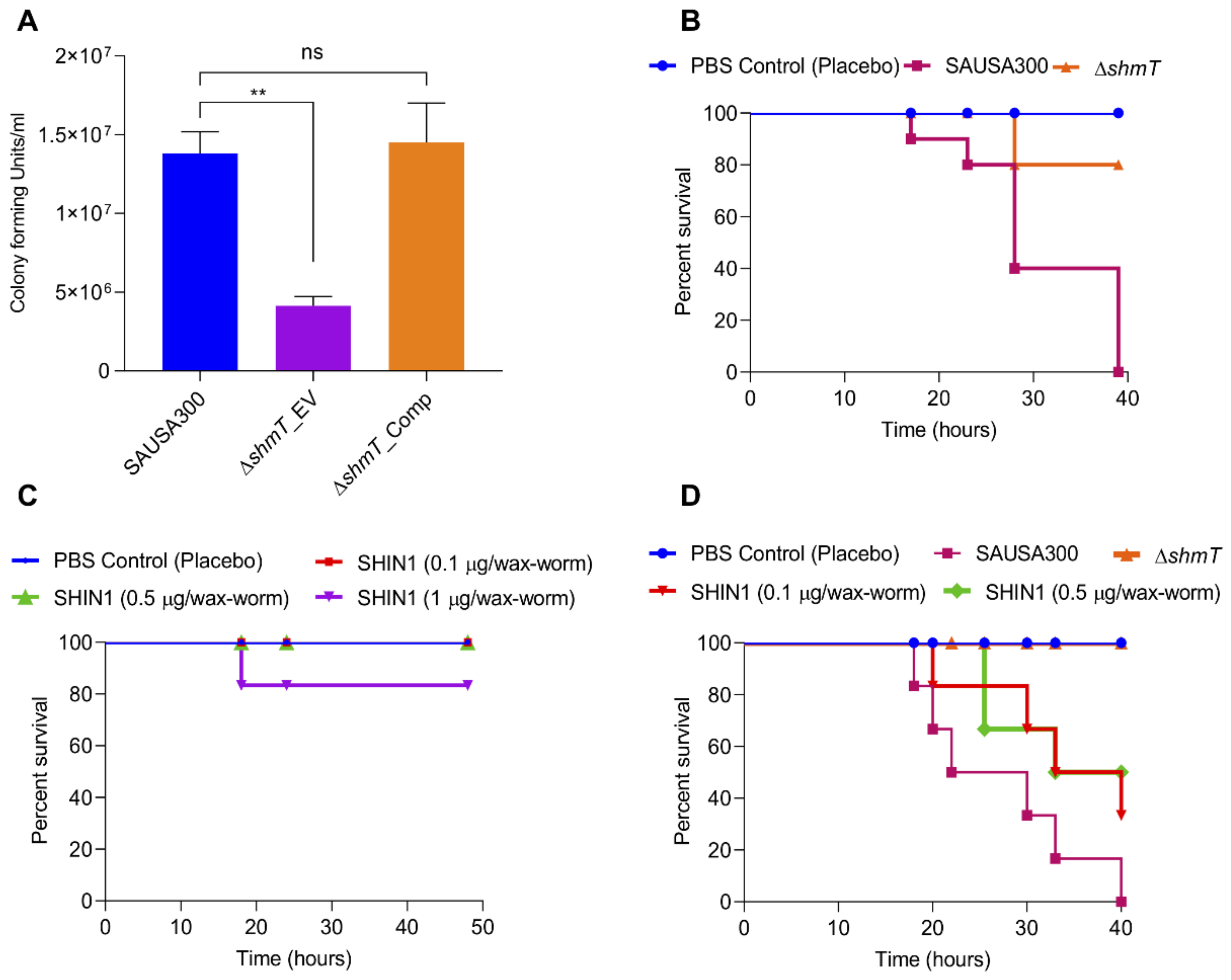
2.7. Role of shmT in Maintenance of Virulence Potential of S. aureus
3. Discussion
4. Materials and Methods
4.1. Cells, Chemicals, and Reagents
4.2. Bacterial Cell Growth Conditions
4.3. Conjugation of TR-X Succinimidyl Ester with Lysostaphin
4.4. Assessment of ST72 Response to Lysostaphin Treatment
4.5. Turbidity Reduction Assay
4.6. Evaluation of Lysostaphin Binding to the Cell Wall of ST72 Isolates
4.7. Assessment of Lysostaphin Endopeptidase Activity on ST72 Isolates Using SEM
4.8. Live/Dead Staining
4.9. Evaluation of Presence/Absence and Mutational Analysis of Known Genes Responsible for Lysostaphin Resistance
4.10. Comparative Genomics-Based Metabolic Pathway Modeling
4.11. Functional Genomics of the shmT Gene from K07-204 and Cloning into pRMC2 Vector
4.12. Quantitative RT-PCR for Expression Analysis of shmT
4.13. Evaluation of Virulence in ST72 Isolates
- In vitro infection experiment. To assess the role of the shmT gene in the virulence of S. aureus, an in vitro infection experiment was performed using HEK293 and RAW264.7 mouse macrophage cell lines. Mammalian cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum in a 5% carbon dioxide humidified incubator at 37 °C. Cells were split in 6-well plates (1.0 × 106 cells/well) and incubated for 24 h. After washing with 1× PBS, bacterial cells were provided with invasion medium (DMEM without FBS) before 2 h of infection. The RAW264.7 and HEK293 cells were infected with SAUSA300 strains at moi 10 (multiplicity of infection 10) for 30 min as described previously [32]. Extracellular cells were killed using lysostaphin (5 U) and gentamicin (400 µg/mL). Cells were washed three times with 1× PBS to remove residual antibacterial agents. Cells were collected by trypsinization and collected by centrifugation. The cell pellet was washed once again with 1× PBS and treated with 0.04% Triton-X100 to break open the mammalian cells to recover the intracellular bacterial cells. Intracellular bacterial cells were diluted in 1× PBS and the bacterial count was assessed by dilution plating of 100 µl on TSA plates for enumeration of CFU.
- Galleria mellonella infection model. Wild-type SAUSA300 and its isogenic shmT knockout strains were grown overnight in TSB media under orbital shaking (200 rpm) culture conditions at 37 °C. The overnight grown culture was reinoculated at 100-fold dilution in TSB for 6 h. Bacterial cells were collected by centrifugation (3220× g, 4 °C) and washed once with 1 × PBS. The cell number was maintained by adjusting the optical density (OD600 = 1) in 1 × PBS. G. mellonella were utilized within a week of their receipt. Each group of control or treatment contained 10 wax-worms. Wax-worms were injected with 2.0 ×105 cells (20 µL) in the last left posterior leg using a 0.3 mL syringe (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) and incubated at 37 °C for observation. In each experiment, a group of worms was kept as control and injected with 20 µl 1 × PBS. Worms were observed for survival at different time intervals and the experiment was terminated within 20 h.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| SHMT | Serine hydroxymethyltransferase |
| SHIN1 | Serine hydroxymethyltransferase inhibitor 1 |
References
- Nahrgang, S.; Nolte, E.; Rechel, B. The role of public health organizations in addressing antimicrobial resistance in Europe. Eur. J. Public Health 2018, 28, 169–170. [Google Scholar] [CrossRef]
- Monserrat-Martinez, A.; Gambin, Y.; Sierecki, E. Thinking outside the bug: Molecular targets and strategies to overcome antibiotic resistance. Int. J. Mol. Sci. 2019, 20, 1255. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Bouvet, C.; Gjoni, S.; Zenelaj, B.; Lipsky, B.A.; Hakko, E.; Uckay, I. Staphylococcus aureus soft tissue infection may increase the risk of subsequent staphylococcal soft tissue infections. Int. J. Infect. Dis. 2017, 60, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.; Song, P.I.; Seo, C.H.; Cheong, H.; Park, Y. Colonization and infection of the skin by S. aureus: Immune system evasion and the response to cationic antimicrobial peptides. Int. J. Mol. Sci. 2014, 15, 8753–8772. [Google Scholar] [PubMed]
- Onyango, L.A.; Alreshidi, M.M. Adaptive metabolism in staphylococci: Survival and persistence in environmental and clinical settings. J. Pathog. 2018, 2018, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Haaber, J.; Penades, J.R.; Ingmer, H. Transfer of Antibiotic resistance in Staphylococcus aureus. Trends Microbiol. 2017, 25, 893–905. [Google Scholar] [CrossRef]
- Novick, R.P.; Christie, G.E.; Penades, J.R. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 2010, 8, 541–551. [Google Scholar] [CrossRef]
- Chambers, H.F.; Deleo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar]
- Mun, Y.S.; Hwang, Y.J. Novel spa and multi-locus sequence types (MLST) of Staphylococcus aureus samples isolated from clinical specimens in Korea. Antibiotics 2019, 8, 202. [Google Scholar] [CrossRef]
- Park, S.Y.; Chung, D.R.; Yoo, J.R.; Baek, J.Y.; Kim, S.H.; Ha, Y.E.; Kang, C.I.; Peck, K.R.; Lee, N.Y.; Song, J.H. Sequence type 72 community-associated meticillin-resistant Staphylococcus aureus emerged as a predominant clone of nasal colonization in newly admitted patients. J. Hosp. Infect. 2016, 93, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep learning approach to antibiotic discovery. Cell 2020, 181, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Hussain, A.; Shakeel, F.; Ahsan, M.J.; Alshehri, S.; Webster, T.J.; Lal, U.R. Recent insights on nanomedicine for augmented infection control. Int. J. Nanomed. 2019, 14, 2301–2325. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.; De Bei, O.; Bettati, S.; Campanini, B.; Kovachka, S.; Gianquinto, E.; Spyrakis, F.; Ronda, L. Iron metabolism at the interface between host and pathogen: From nutritional immunity to antibacterial development. Int. J. Mol. Sci. 2020, 21, 2145. [Google Scholar] [CrossRef] [PubMed]
- Appelbaum, P.C. The emergence of vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2006, 12, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Lee, J.; Kim, M. The association between Staphylococcus aureus nasal colonization and symptomatic infection in children in Korea where ST72 is the major genotype: A prospective observational study. Medicine 2017, 96, e7838. [Google Scholar] [CrossRef]
- Schindler, C.A.; Schuhardt, V.T. Lysostaphin: A new bacteriolytic agent for the Staphylococcus. Proc. Natl. Acad. Sci. USA 1964, 51, 414–421. [Google Scholar] [CrossRef]
- Heath, L.S.; Heath, H.E.; Sloan, G.L. Plasmid-encoded lysostaphin endopeptidase gene of Staphylococcus simulans biovar Staphylolyticus. FEMS Microbiol. Lett. 1987, 44, 129–133. [Google Scholar] [CrossRef]
- Browder, H.P.; Zygmunt, W.A.; Young, J.R.; Tavormina, P.A. Lysostaphin: Enzymatic mode of action. Biochem. Biophys. Res. Commun. 1965, 19, 383–389. [Google Scholar] [CrossRef]
- Iversen, O.J.; Grov, A. Studies on lysostaphin. Separation and characterization of three enzymes. Eur. J. Biochem. 1973, 38, 293–300. [Google Scholar] [CrossRef]
- Mitkowski, P.; Jagielska, E.; Nowak, E.; Bujnicki, J.M.; Stefaniak, F.; Niedzialek, D.; Bochtler, M.; Sabala, I. Structural bases of peptidoglycan recognition by lysostaphin SH3b domain. Sci. Rep. 2019, 9, 5965. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Schneewind, O. Target cell specificity of a bacteriocin molecule: A C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 1996, 15, 4789–4797. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.A.; Kusuma, C.; Mond, J.J.; Kokai-Kun, J.F. Lysostaphin disrupts Staphylococcus aureus and Staphylococcus epidermidis biofilms on artificial surfaces. Antimicrob. Agents Chemother. 2003, 47, 3407–3414. [Google Scholar] [CrossRef] [PubMed]
- Climo, M.W.; Patron, R.L.; Goldstein, B.P.; Archer, G.L. Lysostaphin treatment of experimental methicillin-resistant Staphylococcus aureus aortic valve endocarditis. Antimicrob. Agents Chemother. 1998, 42, 1355–1360. [Google Scholar] [CrossRef] [PubMed]
- Patron, R.L.; Climo, M.W.; Goldstein, B.P.; Archer, G.L. Lysostaphin treatment of experimental aortic valve endocarditis caused by a Staphylococcus aureus isolate with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 1999, 43, 1754–1755. [Google Scholar] [CrossRef]
- Kumar, J.K. Lysostaphin: An antistaphylococcal agent. Appl. Microbiol. Biotechnol. 2008, 80, 555–561. [Google Scholar] [CrossRef]
- Ko, K.S.; Lim, S.K.; Jung, S.C.; Yoon, J.M.; Choi, J.Y.; Song, J.H. Sequence type 72 meticillin-resistant Staphylococcus aureus isolates from humans, raw meat and soil in South Korea. J. Med. Microbiol. 2011, 60, 442–445. [Google Scholar] [CrossRef][Green Version]
- Tschierske, M.; Mori, C.; Rohrer, S.; Ehlert, K.; Shaw, K.J.; Berger-Bachi, B. Identification of three additional femAB-like open reading frames in Staphylococcus aureus. FEMS Microbiol. Lett. 1999, 171, 97–102. [Google Scholar] [CrossRef]
- Berger-Bachi, B.; Tschierske, M. Role of fem factors in methicillin resistance. Drug Resist. Updates 1998, 1, 325–335. [Google Scholar] [CrossRef]
- Sharif, S.; Singh, M.; Kim, S.J.; Schaefer, J. Staphylococcus aureus peptidoglycan tertiary structure from carbon-13 spin diffusion. J. Am. Chem. Soc. 2009, 131, 7023–7030. [Google Scholar] [CrossRef]
- Grundling, A.; Schneewind, O. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J. Bacteriol. 2006, 188, 2463–2472. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Chaurasia, A.K.; Batool, N.; Ko, K.S.; Kim, K.K. Alternative enzyme protection assay to overcome the drawbacks of the gentamicin protection assay for measuring Entry and intracellular survival of staphylococci. Infect. Immun. 2019, 87, e00119-19. [Google Scholar] [CrossRef] [PubMed]
- Gunn, B.A.; Singleton, F.L.; Peele, E.R.; Colwell, R.R.; Keiser, J.K.; Kapper, C.O. Comparison of methods for identifying Staphylococcus and Micrococcus spp. J. Clin. Microbiol. 1981, 14, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Schuhardt, V.T.; Huber, T.W.; Pope, L.M. Electron microscopy and viability of lysostaphin-induced staphylococcal spheroplasts, protoplast-like bodies, and protoplasts. J. Bacteriol. 1969, 97, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Ehlert, K.; Tschierske, M.; Mori, C.; Schroder, W.; Berger-Bachi, B. Site-specific serine incorporation by Lif and Epr into positions 3 and 5 of the staphylococcal peptidoglycan interpeptide bridge. J. Bacteriol. 2000, 182, 2635–2638. [Google Scholar] [CrossRef]
- Thumm, G.; Gotz, F. Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol. Microbiol. 1997, 23, 1251–1265. [Google Scholar] [CrossRef]
- Dehart, H.P.; Heath, H.E.; Heath, L.S.; Leblanc, P.A.; Sloan, G.L. The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl. Environ. Microbiol. 1995, 61, 2811. [Google Scholar] [CrossRef]
- Villa, T.G.; Veiga-Crespo, P. Enzybiotics: Antibiotic Enzymes as Drugs and Therapeutics; John and Wiley and Sons: Hoboken, NJ, USA, 2010; p. 284. [Google Scholar]
- Maidhof, H.; Reinicke, B.; Blumel, P.; Bergerbachi, B.; Labischinski, H. FemA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Bacteriol. 1991, 173, 3507–3513. [Google Scholar] [CrossRef]
- Grundling, A.; Missiakas, D.M.; Schneewind, O. Staphylococcus aureus mutants with increased lysostaphin resistance. J. Bacteriol. 2006, 188, 6286–6297. [Google Scholar] [CrossRef]
- Angelaccio, S. Extremophilic SHMTs: From structure to biotechnology. BioMed Res. Int. 2013, 2013. [Google Scholar] [CrossRef]
- Ducker, G.S.; Ghergurovich, J.M.; Mainolfi, N.; Suri, V.; Jeong, S.K.; Li, S.H.-J.; Friedman, A.; Manfredi, M.G.; Gitai, Z.; Kim, H. Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 2017, 114, 11404–11409. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Villa, D.; Aguilar, M.R.; Rojo, L. Folic acid antagonists: Antimicrobial and immunomodulating mechanisms and applications. Int. J. Mol. Sci. 2019, 20, 4996. [Google Scholar] [CrossRef] [PubMed]
- Corbin, J.M.; Ruiz-Echevarría, M.J. One-carbon metabolism in prostate cancer: The role of androgen signaling. Int. J. Mol. Sci. 2016, 17, 1208. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Katayama, Y.; Matsuo, M.; Sasaki, T.; Morimoto, Y.; Sekiguchi, A.; Baba, T. Multi-drug-resistant Staphylococcus aureus and future chemotherapy. J. Infect. Chemother. 2014, 20, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Joo, E.J.; Choi, J.Y.; Chung, D.R.; Song, J.H.; Ko, K.S. Characteristics of the community-genotype sequence type 72 methicillin-resistant Staphylococcus aureus isolates that underlie their persistence in hospitals. J. Microbiol. 2016, 54, 445–450. [Google Scholar] [CrossRef]
- Senok, A.; Ehricht, R.; Monecke, S.; Al-Saedan, R.; Somily, A. Molecular characterization of methicillin-resistant Staphylococcus aureus in nosocomial infections in a tertiary-care facility: Emergence of new clonal complexes in Saudi Arabia. New Microbes New Infect. 2016, 14, 13–18. [Google Scholar] [CrossRef]
- Read, A.F.; Woods, R.J. Antibiotic resistance management. Evol. Med. Public Health 2014, 2014, 147. [Google Scholar] [CrossRef]
- Davies, J.E. Origins, acquisition and dissemination of antibiotic resistance determinants. Ciba Found. Symp. 1997, 207, 15–27. [Google Scholar]
- Martinez, J.L.; Baquero, F. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 2000, 44, 1771–1777. [Google Scholar] [CrossRef]
- Rollin, G.; Tan, X.; Tros, F.; Dupuis, M.; Nassif, X.; Charbit, A.; Coureuil, M. Intracellular survival of Staphylococcus aureus in endothelial cells: A matter of growth or persistence. Front. Microbiol. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Pai, A.; Sudhakar, G.; Kamath, V. Enzybiotics a review. Int. J. Pharm. Res. 2013, 3, 69–71. [Google Scholar]
- Dajcs, J.J.; Hume, E.B.; Moreau, J.M.; Caballero, A.R.; Cannon, B.M.; O’Callaghan, R.J. Lysostaphin treatment of methicillin-resistant Staphylococcus aureus keratitis in the rabbit. Investig. Ophthalmol. Vis. Sci. 2000, 41, 1432–1437. [Google Scholar]
- Shah, A.; Mond, J.; Walsh, S. Lysostaphin-coated catheters eradicate Staphylococccus aureus challenge and block surface colonization. Antimicrob. Agents Chemother. 2004, 48, 2704–2707. [Google Scholar] [CrossRef]
- Stranden, A.M.; Ehlert, K.; Labischinski, H.; Berger-Bachi, B. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 1997, 179, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Sugai, M.; Fujiwara, T.; Ohta, K.; Komatsuzawa, H.; Ohara, M.; Suginaka, H. Epr, which encodes glycylglycine endopeptidase resistance, is homologous to femAB and affects serine content of peptidoglycan cross bridges in Staphylococcus capitis and Staphylococcus aureus. J. Bacteriol. 1997, 179, 4311–4318. [Google Scholar] [CrossRef]
- Dahal, N.; Abdelhamed, H.; Lu, J.; Karsi, A.; Lawrence, M.L. Tricarboxylic acid cycle and one-carbon metabolism pathways are important in Edwardsiella ictaluri virulence. PLoS ONE 2013, 8, e65973. [Google Scholar] [CrossRef]
- Chen, Y.; Chatterjee, S.S.; Porcella, S.F.; Yu, Y.S.; Otto, M. Complete genome sequence of a Panton-Valentine leukocidin-negative community-associated methicillin-resistant Staphylococcus aureus strain of sequence type 72 from Korea. PLoS ONE 2013, 8, e72803. [Google Scholar] [CrossRef]
- Batool, N.; Ko, K.S.; Chaurasia, A.K.; Kim, K.K. Draft genome sequences of lysostaphin-resistant (K07-204) and lysostaphin-susceptible (K07-561) Staphylococcus aureus sequence type 72 isolated patients in South Korea. Microbiol. Resour. Announc. 2020, 9, e010557-010520. [Google Scholar]
- Jiang, W.; Xia, B.; Liu, Z. A serine hydroxymethyltransferase from marine bacterium Shewanella algae: Isolation, purification, characterization and L-serine production. Microbiol. Res. 2013, 168, 477–484. [Google Scholar] [CrossRef]
- Chaurasia, A.K.; Thorat, N.D.; Tandon, A.; Kim, J.-H.; Park, S.H.; Kim, K.K. Coupling of radiofrequency with magnetic nanoparticles treatment as an alternative physical antibacterial strategy against multiple drug resistant bacteria. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
| Genes | CN1 | K07-204 | K07-561 | SAUSA300 | ||||
|---|---|---|---|---|---|---|---|---|
| Presence | Mutation | Presence | Mutation | Presence | Mutation | Presence | Mutation | |
| femA | √ | − | √ | − | √ | − | √ | − |
| femB | √ | − | √ | − | √ | − | √ | − |
| femX (fmhB) | √ | − | √ | − | √ | − | √ | +2 * |
| fmhC | √ | − | √ | − | √ | − | √ | − |
| lyrA | √ | − | √ | − | √ | − | √ | − |
| epr | X | X | X | X | ||||
| lss | X | X | X | X | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batool, N.; Ko, K.S.; Chaurasia, A.K.; Kim, K.K. Functional Identification of Serine Hydroxymethyltransferase as a Key Gene Involved in Lysostaphin Resistance and Virulence Potential of Staphylococcus aureus Strains. Int. J. Mol. Sci. 2020, 21, 9135. https://doi.org/10.3390/ijms21239135
Batool N, Ko KS, Chaurasia AK, Kim KK. Functional Identification of Serine Hydroxymethyltransferase as a Key Gene Involved in Lysostaphin Resistance and Virulence Potential of Staphylococcus aureus Strains. International Journal of Molecular Sciences. 2020; 21(23):9135. https://doi.org/10.3390/ijms21239135
Chicago/Turabian StyleBatool, Nayab, Kwan Soo Ko, Akhilesh Kumar Chaurasia, and Kyeong Kyu Kim. 2020. "Functional Identification of Serine Hydroxymethyltransferase as a Key Gene Involved in Lysostaphin Resistance and Virulence Potential of Staphylococcus aureus Strains" International Journal of Molecular Sciences 21, no. 23: 9135. https://doi.org/10.3390/ijms21239135
APA StyleBatool, N., Ko, K. S., Chaurasia, A. K., & Kim, K. K. (2020). Functional Identification of Serine Hydroxymethyltransferase as a Key Gene Involved in Lysostaphin Resistance and Virulence Potential of Staphylococcus aureus Strains. International Journal of Molecular Sciences, 21(23), 9135. https://doi.org/10.3390/ijms21239135







