Tn Antigen Expression Defines an Immune Cold Subset of Mismatch-Repair Deficient Colorectal Cancer
Abstract
1. Introduction
2. Results
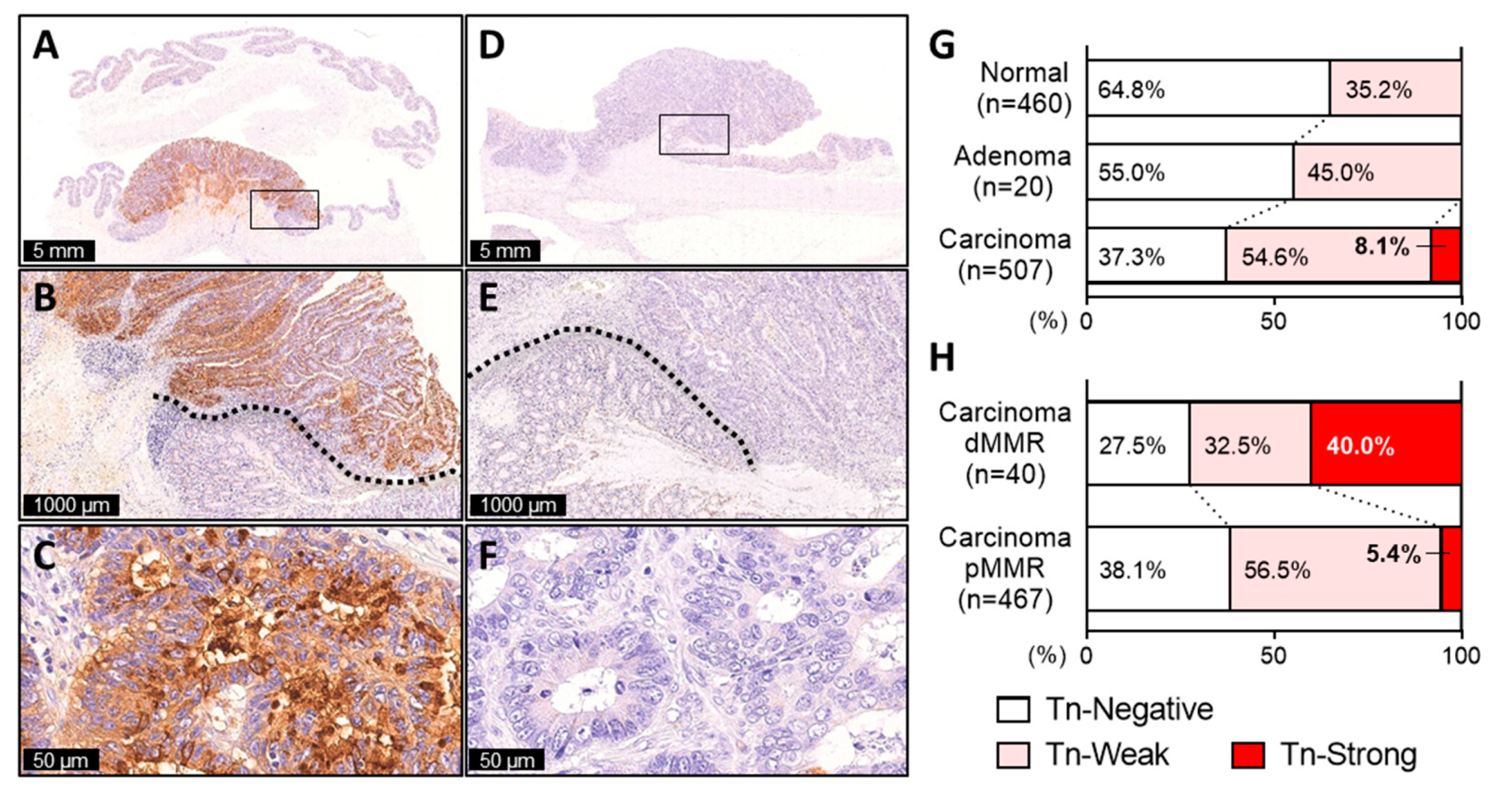
2.1. Tn Antigen Expression in CRC
2.2. A Small Subset of CRC Exhibiting Diffuse and Intense Tn Staining
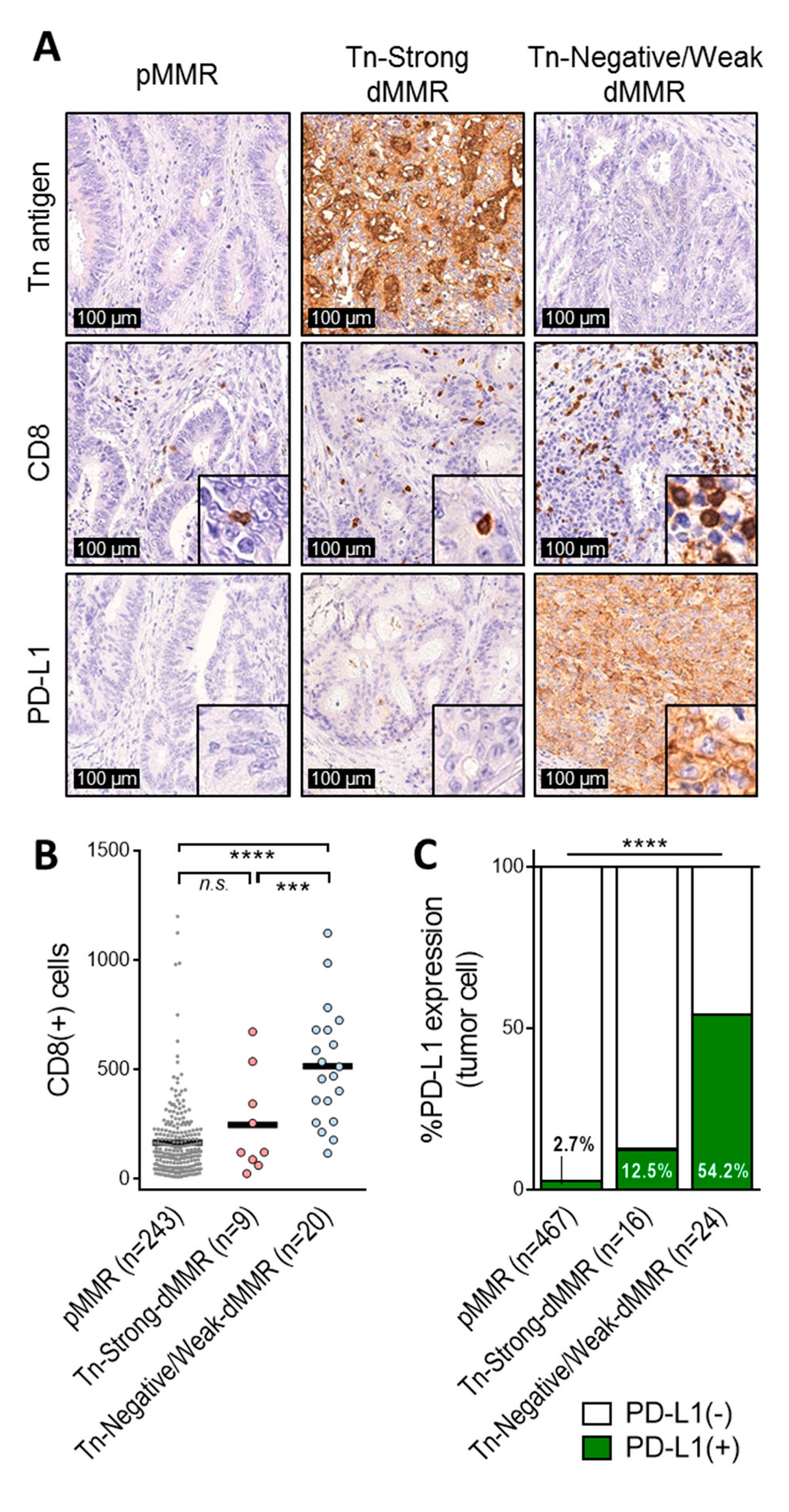
2.3. Tn-Strong dMMR Tumors Showed Immune Cold Characteristics
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. Immunohistochemistry
4.3. Assessment of Staining
4.4. Determination of MMR Status
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CRC | colorectal cancer |
| dMMR | deficient mismatch-repair |
| pMMR | proficient mismatch-repair |
| MSI-H | high-level microsatellite instability |
| MSS | microsatellite stable |
| TME | tumor microenvironment |
| TIL | tumor-infiltrating lymphocyte |
| ICI | immune checkpoint inhibitors |
| TACA | tumor-associated carbohydrate antigen |
| FFPE | formalin-fixed paraffin-embedded |
References
- Network, T.C.G.A. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Sveen, A.; Kopetz, S.; Lothe, R.A. Biomarker-guided therapy for colorectal cancer: Strength in complexity. Nat. Rev. Clin. Oncol. 2020, 17, 11–32. [Google Scholar] [CrossRef] [PubMed]
- Llosa, N.J.; Cruise, M.; Tam, A.; Wicks, E.C.; Hechenbleikner, E.M.; Taube, J.M.; Blosser, R.L.; Fan, H.; Wang, H.; Luber, B.S.; et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015, 5, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Overman, M.J.; Lonardi, S.; Wong, K.Y.M.; Lenz, H.J.; Gelsomino, F.; Aglietta, M.; Morse, M.A.; Van Cutsem, E.; McDermott, R.; Hill, A.; et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J. Clin. Oncol. 2018, 36, 773–779. [Google Scholar] [CrossRef]
- Le, D.T.; Kim, T.W.; Van Cutsem, E.; Geva, R.; Jager, D.; Hara, H.; Burge, M.; O’Neil, B.; Kavan, P.; Yoshino, T.; et al. Phase ii open-label study of pembrolizumab in treatment-refractory, microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: Keynote-164. J. Clin. Oncol. 2020, 38, 11–19. [Google Scholar] [CrossRef]
- Venderbosch, S.; Nagtegaal, I.D.; Maughan, T.S.; Smith, C.G.; Cheadle, J.P.; Fisher, D.; Kaplan, R.; Quirke, P.; Seymour, M.T.; Richman, S.D.; et al. Mismatch repair status and braf mutation status in metastatic colorectal cancer patients: A pooled analysis of the cairo, cairo2, coin, and focus studies. Clin. Cancer Res. 2014, 20, 5322–5330. [Google Scholar] [CrossRef]
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A., Jr. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361–375. [Google Scholar] [CrossRef]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (checkmate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. Pd-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Kannagi, R.; Sakuma, K.; Miyazaki, K.; Lim, K.T.; Yusa, A.; Yin, J.; Izawa, M. Altered expression of glycan genes in cancers induced by epigenetic silencing and tumor hypoxia: Clues in the ongoing search for new tumor markers. Cancer Sci. 2010, 101, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Noda, M.; Okayama, H.; Tachibana, K.; Sakamoto, W.; Saito, K.; Thar Min, A.K.; Ashizawa, M.; Nakajima, T.; Aoto, K.; Momma, T.; et al. Glycosyltransferase gene expression identifies a poor prognostic colorectal cancer subtype associated with mismatch repair deficiency and incomplete glycan synthesis. Clin. Cancer Res. 2018, 24, 4468–4481. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, L.R.; Carrascal, M.A.; Barbas, A.; Ramalho, J.S.; Novo, C.; Delannoy, P.; Videira, P.A. Challenges in antibody development against tn and sialyl-tn antigens. Biomolecules 2015, 5, 1783–1809. [Google Scholar] [CrossRef] [PubMed]
- Posey, A.D., Jr.; Schwab, R.D.; Boesteanu, A.C.; Steentoft, C.; Mandel, U.; Engels, B.; Stone, J.D.; Madsen, T.D.; Schreiber, K.; Haines, K.M.; et al. Engineered car t cells targeting the cancer-associated tn-glycoform of the membrane mucin muc1 control adenocarcinoma. Immunity 2016, 44, 1444–1454. [Google Scholar] [CrossRef] [PubMed]
- RodrIguez, E.; Schetters, S.T.T.; van Kooyk, Y. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 2018, 18, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Beckwith, D.M.; Cudic, M. Tumor-associated o-glycans of muc1: Carriers of the glyco-code and targets for cancer vaccine design. Semin. Immunol. 2020, 47, 101389. [Google Scholar] [CrossRef]
- Rashidijahanabad, Z.; Huang, X. Recent advances in tumor associated carbohydrate antigen based chimeric antigen receptor t cells and bispecific antibodies for anti-cancer immunotherapy. Semin. Immunol. 2020, 47, 101390. [Google Scholar] [CrossRef]
- Freire, T.; Lo-Man, R.; Bay, S.; Leclerc, C. Tn glycosylation of the muc6 protein modulates its immunogenicity and promotes the induction of th17-biased t cell responses. J. Biol. Chem. 2011, 286, 7797–7811. [Google Scholar] [CrossRef]
- van Vliet, S.J.; Gringhuis, S.I.; Geijtenbeek, T.B.; van Kooyk, Y. Regulation of effector t cells by antigen-presenting cells via interaction of the c-type lectin mgl with cd45. Nat. Immunol. 2006, 7, 1200–1208. [Google Scholar] [CrossRef]
- van Vliet, S.J.; Bay, S.; Vuist, I.M.; Kalay, H.; Garcia-Vallejo, J.J.; Leclerc, C.; van Kooyk, Y. Mgl signaling augments tlr2-mediated responses for enhanced il-10 and tnf-alpha secretion. J. Leukoc. Biol. 2013, 94, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Cornelissen, L.A.M.; Blanas, A.; Zaal, A.; van der Horst, J.C.; Kruijssen, L.J.W.; O’Toole, T.; van Kooyk, Y.; van Vliet, S.J. Tn antigen expression contributes to an immune suppressive microenvironment and drives tumor growth in colorectal cancer. Front. Oncol. 2020, 10, 1622. [Google Scholar] [CrossRef] [PubMed]
- Schrock, A.B.; Ouyang, C.; Sandhu, J.; Sokol, E.; Jin, D.; Ross, J.S.; Miller, V.A.; Lim, D.; Amanam, I.; Chao, J.; et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in msi-high metastatic colorectal cancer. Ann. Oncol. 2019, 30, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Sahin, I.H.; Akce, M.; Alese, O.; Shaib, W.; Lesinski, G.B.; El-Rayes, B.; Wu, C. Immune checkpoint inhibitors for the treatment of msi-h/mmr-d colorectal cancer and a perspective on resistance mechanisms. Br. J. Cancer 2019, 121, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Loupakis, F.; Depetris, I.; Biason, P.; Intini, R.; Prete, A.A.; Leone, F.; Lombardi, P.; Filippi, R.; Spallanzani, A.; Cascinu, S.; et al. Prediction of benefit from checkpoint inhibitors in mismatch repair deficient metastatic colorectal cancer: Role of tumor infiltrating lymphocytes. Oncologist 2020, 25, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Tauriello, D.V.F.; Palomo-Ponce, S.; Stork, D.; Berenguer-Llergo, A.; Badia-Ramentol, J.; Iglesias, M.; Sevillano, M.; Ibiza, S.; Canellas, A.; Hernando-Momblona, X.; et al. Tgfbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018, 554, 538–543. [Google Scholar] [CrossRef]
- Endo, E.; Okayama, H.; Saito, K.; Nakajima, S.; Yamada, L.; Ujiie, D.; Kase, K.; Fujita, S.; Endo, H.; Sakamoto, W.; et al. A tgfbeta-dependent stromal subset underlies immune checkpoint inhibitor efficacy in DNA mismatch repair-deficient/microsatellite instability-high colorectal cancer. Mol. Cancer Res. 2020, 18, 1402–1413. [Google Scholar] [CrossRef]
- Janssen, E.; Subtil, B.; de la Jara Ortiz, F.; Verheul, H.M.W.; Tauriello, D.V.F. Combinatorial immunotherapies for metastatic colorectal cancer. Cancers 2020, 12, 1875. [Google Scholar] [CrossRef]
- Grasso, C.S.; Giannakis, M.; Wells, D.K.; Hamada, T.; Mu, X.J.; Quist, M.; Nowak, J.A.; Nishihara, R.; Qian, Z.R.; Inamura, K.; et al. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov. 2018, 8, 730–749. [Google Scholar] [CrossRef]
- Sveen, A.; Johannessen, B.; Tengs, T.; Danielsen, S.A.; Eilertsen, I.A.; Lind, G.E.; Berg, K.C.G.; Leithe, E.; Meza-Zepeda, L.A.; Domingo, E.; et al. Multilevel genomics of colorectal cancers with microsatellite instability-clinical impact of jak1 mutations and consensus molecular subtype 1. Genome Med. 2017, 9, 46. [Google Scholar] [CrossRef]
- Chia, J.; Goh, G.; Bard, F. Short o-galnac glycans: Regulation and role in tumor development and clinical perspectives. Biochim. Biophys. Acta 2016, 1860, 1623–1639. [Google Scholar] [CrossRef]
- Sun, X.; Ju, T.; Cummings, R.D. Differential expression of cosmc, t-synthase and mucins in tn-positive colorectal cancers. BMC Cancer 2018, 18, 827. [Google Scholar] [CrossRef] [PubMed]
- Itzkowitz, S.H.; Yuan, M.; Montgomery, C.K.; Kjeldsen, T.; Takahashi, H.K.; Bigbee, W.L.; Kim, Y.S. Expression of tn, sialosyl-tn, and t antigens in human colon cancer. Cancer Res. 1989, 49, 197–204. [Google Scholar] [PubMed]
- Orntoft, T.F.; Harving, N.; Langkilde, N.C. O-linked mucin-type glycoproteins in normal and malignant colon mucosa: Lack of t-antigen expression and accumulation of tn and sialosyl-tn antigens in carcinomas. Int. J. Cancer 1990, 45, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Itzkowitz, S.H.; Bloom, E.J.; Lau, T.S.; Kim, Y.S. Mucin associated tn and sialosyl-tn antigen expression in colorectal polyps. Gut 1992, 33, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Oshikiri, T.; Miyamoto, M.; Morita, T.; Fujita, M.; Miyasaka, Y.; Senmaru, N.; Yamada, H.; Takahashi, T.; Horita, S.; Kondo, S. Tumor-associated antigen recognized by the 22-1-1 monoclonal antibody encourages colorectal cancer progression under the scanty cd8+ t cells. Clin. Cancer Res. 2006, 12, 411–416. [Google Scholar] [CrossRef][Green Version]
- Jiang, Y.; Liu, Z.; Xu, F.; Dong, X.; Cheng, Y.; Hu, Y.; Gao, T.; Liu, J.; Yang, L.; Jia, X.; et al. Aberrant o-glycosylation contributes to tumorigenesis in human colorectal cancer. J. Cell Mol. Med. 2018, 22, 4875–4885. [Google Scholar] [CrossRef]
- Pages, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Cheever, M.A.; Allison, J.P.; Ferris, A.S.; Finn, O.J.; Hastings, B.M.; Hecht, T.T.; Mellman, I.; Prindiville, S.A.; Viner, J.L.; Weiner, L.M.; et al. The prioritization of cancer antigens: A national cancer institute pilot project for the acceleration of translational research. Clin. Cancer Res. 2009, 15, 5323–5337. [Google Scholar] [CrossRef]
- Scheid, E.; Major, P.; Bergeron, A.; Finn, O.J.; Salter, R.D.; Eady, R.; Yassine-Diab, B.; Favre, D.; Peretz, Y.; Landry, C.; et al. Tn-muc1 dc vaccination of rhesus macaques and a phase i/ii trial in patients with nonmetastatic castrate-resistant prostate cancer. Cancer Immunol. Res. 2016, 4, 881–892. [Google Scholar] [CrossRef]
- Amedei, A.; Asadzadeh, F.; Papi, F.; Vannucchi, M.G.; Ferrucci, V.; Bermejo, I.A.; Fragai, M.; De Almeida, C.V.; Cerofolini, L.; Giuntini, S.; et al. A structurally simple vaccine candidate reduces progression and dissemination of triple-negative breast cancer. iScience 2020, 23, 101250. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, P.; Artaud, C.; Bay, S.; Ganneau, C.; Campone, M.; Delaloge, S.; Gourmelon, C.; Loirat, D.; Medioni, J.; Pein, F.; et al. The fully synthetic glycopeptide mag-tn3 therapeutic vaccine induces tumor-specific cytotoxic antibodies in breast cancer patients. Cancer Immunol. Immunother. 2020, 69, 703–716. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Marada, V.; Cai, Q.; Kizerwetter, M.; He, Y.; Wolf, S.P.; Schreiber, K.; Clausen, H.; Schreiber, H.; Kranz, D.M. Structure-guided engineering of the affinity and specificity of cars against tn-glycopeptides. Proc. Natl. Acad. Sci. USA 2020, 117, 15148–15159. [Google Scholar] [CrossRef] [PubMed]
- Titov, A.; Valiullina, A.; Zmievskaya, E.; Zaikova, E.; Petukhov, A.; Miftakhova, R.; Bulatov, E.; Rizvanov, A. Advancing car t-cell therapy for solid tumors: Lessons learned from lymphoma treatment. Cancers 2020, 12, 125. [Google Scholar] [CrossRef]
- Ashizawa, M.; Okayama, H.; Ishigame, T.; Thar Min, A.K.; Saito, K.; Ujiie, D.; Murakami, Y.; Kikuchi, T.; Nakayama, Y.; Noda, M.; et al. Mirna-148a-3p regulates immunosuppression in DNA mismatch repair-deficient colorectal cancer by targeting pd-l1. Mol. Cancer Res. 2019, 17, 1403–1413. [Google Scholar]
| Total (n = 507) | Tn-Negative/Weak | Tn-Strong | p-Value | ||
|---|---|---|---|---|---|
| n = 466 (91.9%) | n = 41 (8.1%) | ||||
| Age | 0.233 | ||||
| Mean ± SD | 68.4 ± 11.6 | 68.2 ± 11.4 | 70.5 ± 13.3 | ||
| Gender | 0.315 | ||||
| Male | 319 | 290 (62.2%) | 29 (70.7%) | ||
| Female | 188 | 176 (37.8%) | 12 (29.3%) | ||
| Location | 0.004 | ||||
| Proximal colon | 193 | 168 (36.1%) | 25 (61.0%) | ||
| Distal colon | 133 | 126 (27.0%) | 7 (17.1%) | ||
| Rectum | 181 | 172 (36.9%) | 9 (22.0%) | ||
| Tumor differentiation | <0.0001 | ||||
| Well-Moderately | 478 | 447 (95.9%) | 31 (75.6%) | ||
| Poorly | 29 | 19 (4.1%) | 10 (24.4%) | ||
| Histology | <0.0001 | ||||
| Non-mucinous | 482 | 451 (96.8%) | 31 (75.6%) | ||
| Mucinous | 25 | 15 (3.2%) | 10 (24.4%) | ||
| Tumor invasion | 0.050 | ||||
| Tis | 32 | 30 (6.4%) | 2 (4.9%) | ||
| T1 | 61 | 59 (12.7%) | 2 (4.9%) | ||
| T2 | 73 | 69 (14.8%) | 4 (9.8%) | ||
| T3 | 194 | 177 (38.0%) | 17 (41.5%) | ||
| T4 | 147 | 131 (28.1%) | 16 (39.0%) | ||
| Lymphatic invasion | 0.858 | ||||
| Absent | 147 | 136 (29.2%) | 11 (26.8%) | ||
| Present | 360 | 330 (70.8%) | 30 (73.2%) | ||
| Venous invasion | 1.000 | ||||
| Absent | 129 | 119 (25.5%) | 10 (24.4%) | ||
| Present | 378 | 347 (74.5%) | 31 (75.6%) | ||
| Lymph node metastasis | 0.395 | ||||
| Absent | 316 | 288 (61.8%) | 28 (68.3%) | ||
| Present | 188 | 176 (37.8%) | 12 (29.3%) | ||
| Not available | 3 | 2 (0.4%) | 1 (2.4%) | ||
| Distant metastasis | 0.822 | ||||
| Absent | 428 | 394 (84.5%) | 34 (82.9%) | ||
| Present | 79 | 72 (15.5%) | 7 (17.1%) | ||
| Stage | 0.544 | ||||
| 0 | 31 | 29 (6.2%) | 2 (4.9%) | ||
| I | 111 | 106 (22.7%) | 5 (12.2%) | ||
| II | 153 | 135 (29.0%) | 18 (43.9%) | ||
| III | 133 | 124 (26.6%) | 9 (22.0%) | ||
| IV | 79 | 72 (15.5%) | 7 (17.1%) | ||
| PD-L1 expression on tumor cells | 0.487 | ||||
| Negative | 479 | 441 (94.6%) | 38 (92.7%) | ||
| Positive | 28 | 25 (5.4%) | 3 (7.3%) | ||
| MMR status | <0.0001 | ||||
| pMMR | 467 | 442 (94.8%) | 25 (61.0%) | ||
| dMMR | 40 | 24 (5.2%) | 16 (39.0%) | ||
| CD8+ cells | 0.432 | ||||
| Mean ± SD | 194.7 ± 201.2 | 197.6 ± 204.1 | 163.1 ± 167.1 | ||
| CD4+ cells | 0.432 | ||||
| Mean ± SD | 96.0 ± 96.5 | 97.4 ± 99.1 | 80.8 ± 61.3 | ||
| Foxp3+ cells | 0.967 | ||||
| Mean ± SD | 386.9 ± 221.6 | 386.7 ± 225.7 | 388.5 ± 187.9 | ||
| Tn-Negative/Weak dMMR | Tn-Strong dMMR | p-Value | ||
|---|---|---|---|---|
| n = 24 (60.0%) | n = 16 (40.0%) | |||
| Age | 0.101 | |||
| Mean ± SD | 63.4 ± 15.8 | 71.6 ± 13.7 | ||
| Gender | 1.000 | |||
| Male | 11 (45.8%) | 8 (50.0%) | ||
| Female | 13 (54.2%) | 8 (50.0%) | ||
| Location | 0.062 | |||
| Proximal colon | 15 (62.5%) | 14 (87.5%) | ||
| Distal colon | 4 (16.7%) | 2 (12.5%) | ||
| Rectum | 5 (20.8%) | 0 (0.0%) | ||
| Tumor differentiation | 0.729 | |||
| Well-Moderately | 16 (66.7%) | 12 (75.0%) | ||
| Poorly | 8 (33.3%) | 4 (25.0%) | ||
| Histology | 1.000 | |||
| Non-mucinous | 21 (87.5%) | 14 (87.5%) | ||
| Mucinous | 3 (12.5%) | 2 (12.5%) | ||
| Tumor invasion | 0.916 | |||
| T1 | 0 (0.0%) | 1 (6.3%) | ||
| T2 | 7 (29.2%) | 2 (12.5%) | ||
| T3 | 10 (41.7%) | 10 (62.5%) | ||
| T4 | 7 (29.2%) | 3 (18.8%) | ||
| Lymphatic invasion | 0.729 | |||
| Absent | 8 (33.3%) | 4 (25.0%) | ||
| Present | 16 (66.7%) | 12 (75.0%) | ||
| Venous invasion | 1.000 | |||
| Absent | 5 (20.8%) | 4 (25.0%) | ||
| Present | 19 (79.2%) | 12 (75.0%) | ||
| Lymph node metastasis | 1.000 | |||
| Absent | 17 (70.8%) | 10 (62.5%) | ||
| Present | 7 (29.2%) | 5 (31.3%) | ||
| Not available | 0 (0.0%) | 1 (6.3%) | ||
| Distant metastasis | 0.553 | |||
| Absent | 23 (95.8%) | 14 (87.5%) | ||
| Present | 1 (4.2%) | 2 (12.5%) | ||
| Stage | 0.471 | |||
| I | 6 (25.0%) | 3 (18.8%) | ||
| II | 11 (45.8%) | 7 (43.8%) | ||
| III | 6 (25.0%) | 4 (25.0%) | ||
| IV | 1 (4.2%) | 2 (12.5%) | ||
| PD-L1 expression on tumor cells | 0.010 | |||
| Negative | 11 (45.8%) | 14 (87.5%) | ||
| Positive | 13 (54.2%) | 2 (12.5%) | ||
| CD8+ cells | 0.014 | |||
| Mean ± SD | 515.1 ± 265.5 | 247.3 ± 228.0 | ||
| CD4+ cells | 0.443 | |||
| Mean ± SD | 134.1 ± 114.9 | 101.8 ± 68.8 | ||
| Foxp3+ cells | 0.129 | |||
| Mean ± SD | 456.7 ± 214.4 | 353.3 ± 189.6 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsumoto, T.; Okayama, H.; Nakajima, S.; Saito, K.; Nakano, H.; Endo, E.; Kase, K.; Ito, M.; Yamauchi, N.; Yamada, L.; et al. Tn Antigen Expression Defines an Immune Cold Subset of Mismatch-Repair Deficient Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 9081. https://doi.org/10.3390/ijms21239081
Matsumoto T, Okayama H, Nakajima S, Saito K, Nakano H, Endo E, Kase K, Ito M, Yamauchi N, Yamada L, et al. Tn Antigen Expression Defines an Immune Cold Subset of Mismatch-Repair Deficient Colorectal Cancer. International Journal of Molecular Sciences. 2020; 21(23):9081. https://doi.org/10.3390/ijms21239081
Chicago/Turabian StyleMatsumoto, Takuro, Hirokazu Okayama, Shotaro Nakajima, Katsuharu Saito, Hiroshi Nakano, Eisei Endo, Koji Kase, Misato Ito, Naoto Yamauchi, Leo Yamada, and et al. 2020. "Tn Antigen Expression Defines an Immune Cold Subset of Mismatch-Repair Deficient Colorectal Cancer" International Journal of Molecular Sciences 21, no. 23: 9081. https://doi.org/10.3390/ijms21239081
APA StyleMatsumoto, T., Okayama, H., Nakajima, S., Saito, K., Nakano, H., Endo, E., Kase, K., Ito, M., Yamauchi, N., Yamada, L., Kanke, Y., Onozawa, H., Fujita, S., Sakamoto, W., Saito, M., Saze, Z., Momma, T., Mimura, K., & Kono, K. (2020). Tn Antigen Expression Defines an Immune Cold Subset of Mismatch-Repair Deficient Colorectal Cancer. International Journal of Molecular Sciences, 21(23), 9081. https://doi.org/10.3390/ijms21239081





