Kisspeptin Influences the Reproductive Axis and Circulating Levels of microRNAs in Senegalese Sole
Abstract
1. Introduction
2. Results
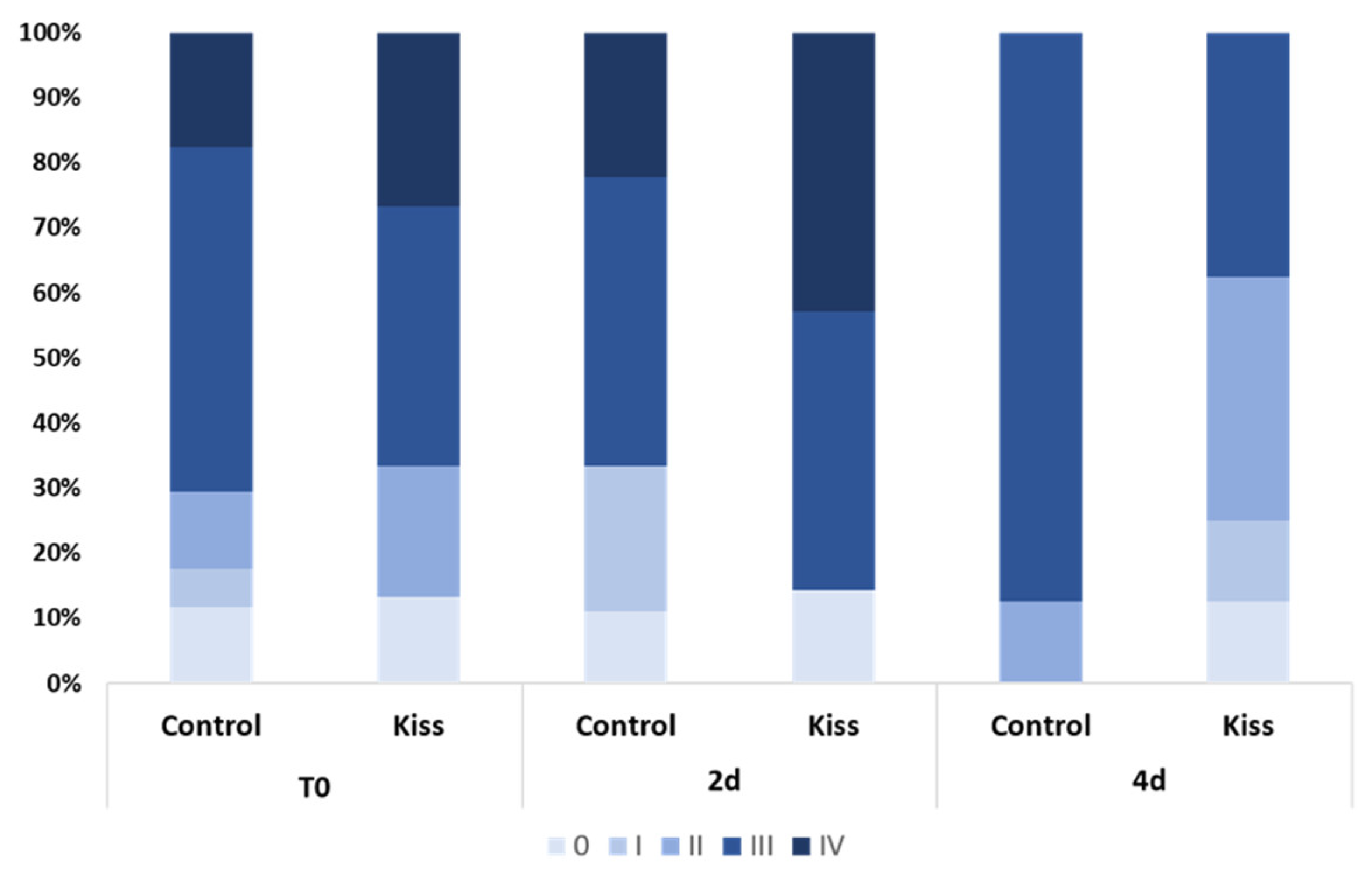
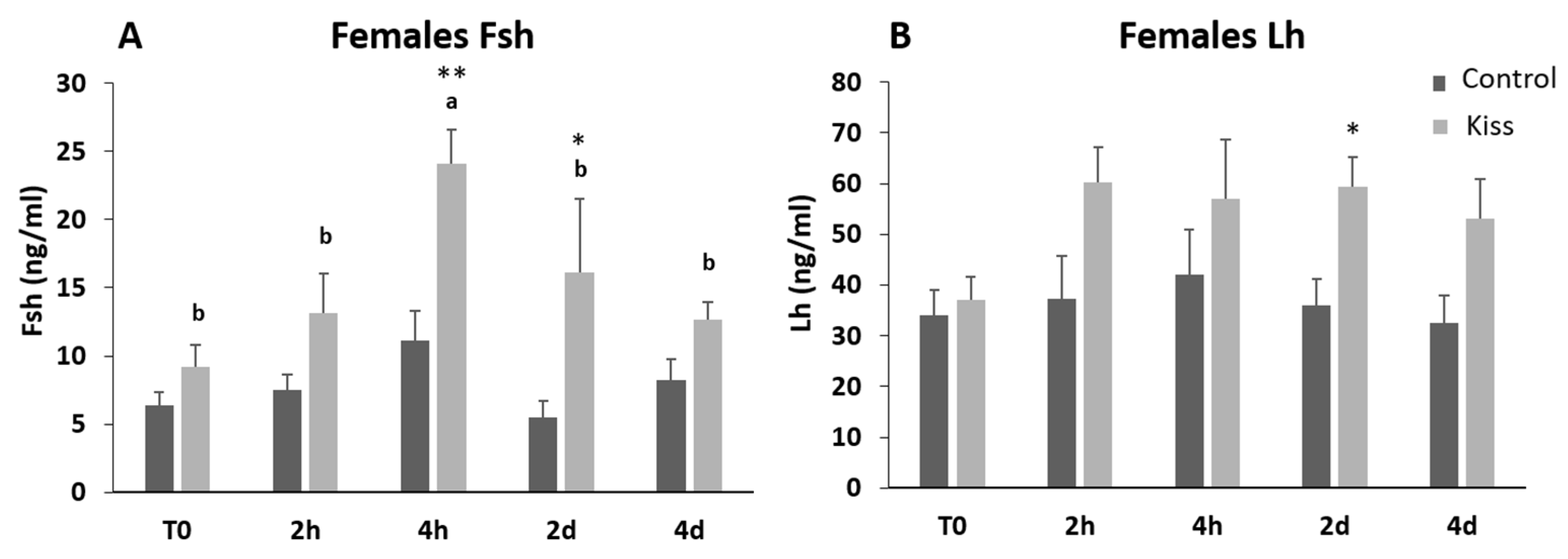
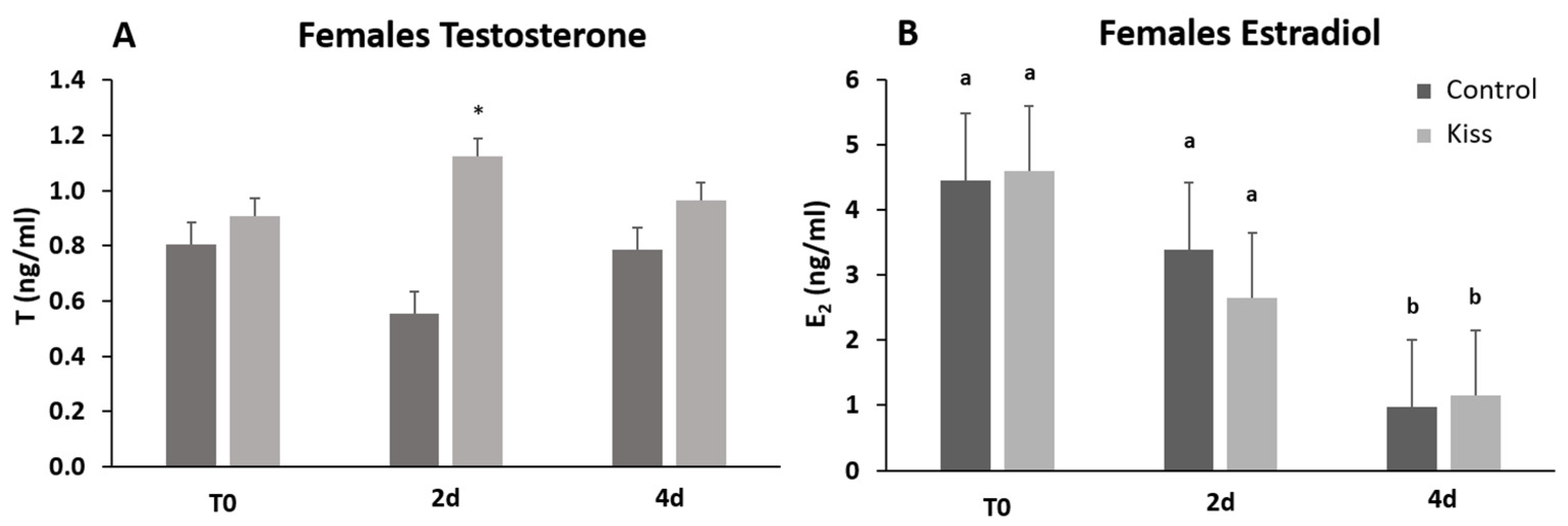
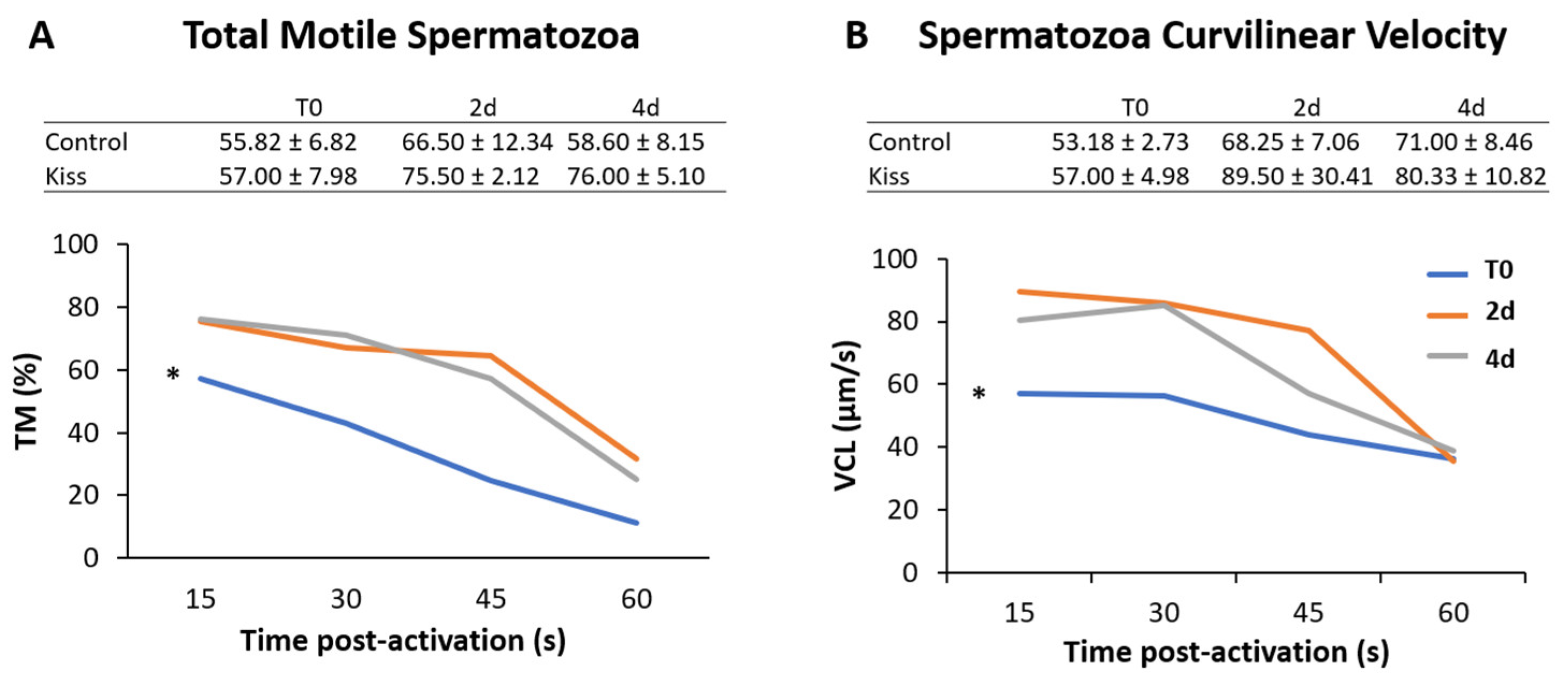
2.1. Kisspeptin Treatment Affects the Reproductive axis
2.2. Circulating sncRNA Profiles in Plasma
2.3. Bioinformatic Prediction of mRNAs Targets by Differentially Expressed (DE) miRNAs
3. Discussion
4. Materials and Methods
4.1. Animals and Housing
4.2. Experimental Design
4.3. Sperm Quality Analysis
4.4. Analysis of Hormones in Blood Plasma
4.5. microRNAs Analysis
4.5.1. Isolation, Libraries Preparation, and Sequencing
4.5.2. Data Processing and Annotations
4.5.3. mRNA Target Prediction
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Clarke, H.; Dhillo, W.S.; Jayasena, C.N. Comprehensive review on kisspeptin and its role in reproductive disorders. Endocrinol. Metab. 2015, 30, 124–141. [Google Scholar] [CrossRef]
- Mechaly, A.S.; Viñas, J.; Piferrer, F. The kisspeptin system genes in teleost fish, their structure and regulation, with particular attention to the situation in Pleuronectiformes. Gen. Comp. Endocrinol. 2013, 188, 258–268. [Google Scholar] [CrossRef]
- Jayasena, C.N.; Abbara, A.; Comninos, A.N.; Nijher, G.M.K.; Christopoulos, G.; Narayanaswamy, S.; Izzi-Engbeaya, C.; Sridharan, M.; Mason, A.J.; Warwick, J.; et al. Kisspeptin-54 triggers egg maturation in women undergoing in vitro fertilization. J. Clin. Investig. 2014, 124, 3667–3677. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, C.N.; Abbara, A.; Narayanaswamy, S.; Comninos, A.N.; Ratnasabapathy, R.; Bassett, P.; Mogford, J.T.; Malik, Z.; Calley, J.; Ghatei, M.A.; et al. Direct comparison of the effects of intravenous kisspeptin-10, kisspeptin-54 and GnRH on gonadotrophin secretion in healthy men. Hum. Reprod. 2015, 30, 1934–1941. [Google Scholar] [CrossRef] [PubMed]
- Filby, A.L.; Aerle, R.V.; Duitman, J.; Tyler, C.R. The Kisspeptin/Gonadotropin-releasing hormone pathway and molecular signaling of puberty in fish. Biol. Reprod. 2008, 78, 278–289. [Google Scholar] [CrossRef]
- Cowan, M.; Paullada-Salmerón, J.A.; López-Olmeda, J.F.; Sánchez-Vázquez, F.J.; Muñoz-Cueto, J.A. Effects of pinealectomy on the neuroendocrine reproductive system and locomotor activity in male European sea bass, Dicentrarchus labrax. Comp. Biochem. Physiol. A 2017, 207, 1–12. [Google Scholar] [CrossRef]
- Zmora, N.; Stubblefield, J.D.; Wong, T.-T.; Levavi-Sivan, B.; Millar, R.P.; Zohar, Y. Kisspeptin antagonists reveal Kisspeptin 1 and Kisspeptin 2 differential regulation of reproduction in the teleost, Morone saxatilis. Biol. Reprod. 2015, 93, 76. [Google Scholar] [CrossRef]
- Somoza, G.M.; Mechaly, A.S.; Trudeau, V.L. Kisspeptin and GnRH interactions in the reproductive brain of teleosts. Gen. Comp. Endocrinol. 2020, 298, 113568. [Google Scholar] [CrossRef]
- Felip, A.; Zanuy, S.; Pineda, R.; Pinilla, L.; Carrillo, M.; Tena-Sempere, M.; Gómez, A. Evidence for two distinct KiSS genes in non-placental vertebrates that encode kisspeptins with different gonadotropin-releasing activities in fish and mammals. Mol. Cell Endocrinol. 2009, 312, 61–71. [Google Scholar] [CrossRef]
- Akazome, Y.; Kanda, S.; Okubo, K.; Oka, Y. Functional and evolutionary insights into vertebrate kisspeptin systems from studies of fish brain. J. Fish Biol. 2010, 76, 161–182. [Google Scholar] [CrossRef]
- Morais, S.; Aragão, C.; Cabrita, E.; Conceição, L.E.C.; Constenla, M.; Costas, B.; Dias, J.; Duncan, N.; Engrola, S.; Estevez, A.; et al. New developments and biological insights into the farming of Solea senegalensis reinforcing its aquaculture potential. Rev. Aquacult. 2016, 8, 227–263. [Google Scholar] [CrossRef]
- Guzmán, J.M.; Norberg, B.; Ramos, J.; Mylonas, C.C.; Mañanós, E.L. Vitellogenin, steroid plasma levels and spawning performance of cultured female Senegalese sole (Solea senegalensis). Gen. Comp. Endocrinol. 2008, 156, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Rasines, I.; Gómez, M.; Martín, I.; Rodríguez, C.; Mañanós, E.; Chereguini, O. Artificial fertilization of Senegalese sole (Solea senegalensis): Hormone therapy administration methods, timing of ovulation and viability of eggs retained in the ovarian cavity. Aquaculture 2012, 326–329, 129–135. [Google Scholar] [CrossRef]
- Carazo, I.; Chereguini, O.; Martín, I.; Huntingford, F.; Duncan, N. Reproductive ethogram and mate selection in captive wild Senegalese sole (Solea senegalensis). Span. J. Agric. Res. 2016, 14, e0401. [Google Scholar] [CrossRef]
- Chauvigné, F.; Fatsini, E.; Duncan, N.; Ollé, J.; Zanuy, S.; Gómez, A.; Cerdà, J. Plasma levels of follicle-stimulating and luteinizing hormones during the reproductive cycle of wild and cultured Senegalese sole (Solea senegalensis). Comp. Biochem. Physiol. A 2016, 191, 35–43. [Google Scholar] [CrossRef]
- Cabrita, E.; Soares, F.; Beirão, J.; García-López, A.; Martínez-Rodríguez, G.; Dinis, M.T. Endocrine and milt response of Senegalese sole, Solea senegalensis, males maintained in captivity. Theriogenology 2011, 75, 1–9. [Google Scholar] [CrossRef]
- Rasines, I.; Gómez, M.; Martín, I.; Rodríguez, C.; Mañanós, E.; Chereguini, O. Artificial fertilisation of cultured Senegalese sole (Solea senegalensis): Effects of the time of day of hormonal treatment on inducing ovulation. Aquaculture 2013, 392–395, 94–97. [Google Scholar] [CrossRef]
- Guzmán, J.M.; Ramos, J.; Mylonas, C.C.; Mañanós, E.L. Spawning performance and plasma levels of GnRHa and sex steroids in cultured female Senegalese sole (Solea senegalensis) treated with different GnRHa-delivery systems. Aquaculture 2009, 291, 200–209. [Google Scholar] [CrossRef]
- Riesco, M.F.; Oliveira, C.; Soares, F.; Gavaia, P.J.; Dinis, M.T.; Cabrita, E. Solea senegalensis sperm cryopreservation: New insights on sperm quality. PLoS ONE 2017, 12, e0186542. [Google Scholar] [CrossRef]
- Fernández, I.; Fernandes, J.M.O.; Roberto, V.P.; Kopp, M.; Oliveira, C.; Riesco, M.F.; Dias, J.; Cox, C.J.; Leonor Cancela, M.; Cabrita, E.; et al. Circulating small non-coding RNAs provide new insights into vitamin K nutrition and reproductive physiology in teleost fish. Biochim. Biophys. Acta 2019, 1863, 39–51. [Google Scholar] [CrossRef]
- Agulleiro, M.J.; Anguis, V.; Cañavate, J.P.; Martínez-Rodríguez, G.; Mylonas, C.C.; Cerdà, J. Induction of spawning of captive reared Senegal sole (Solea senegalensis) using different administration methods for gonadotropin-releasing hormone agonist. Aquaculture 2006, 257, 511–524. [Google Scholar] [CrossRef]
- Guzmán, J.M.; Ramos, J.; Mylonas, C.C.; Mañanós, E.L. Comparative effects of human chorionic gonadotropin (hCG) and gonadotropin-releasing hormone agonist (GnRHa) treatments on the stimulation of male Senegalese sole (Solea senegalensis) reproduction. Aquaculture 2011, 316, 121–128. [Google Scholar] [CrossRef]
- Chauvigné, F.; Ollé, J.; González, W.; Duncan, N.; Giménez, I.; Cerdà, J. Toward developing recombinant gonadotropin-based hormone therapies for increasing fertility in the flatfish Senegalese sole. PLoS ONE 2017, 12, e0174387. [Google Scholar] [CrossRef]
- Chauvigné, F.; González, W.; Ramos, S.; Ducat, C.; Duncan, N.; Giménez, I.; Cerdà, J. Seasonal- and dose-dependent effects of recombinant gonadotropins on sperm production and quality in the flatfish Solea senegalensis. Comp. Biochem. Physiol. A 2018, 225, 59–64. [Google Scholar] [CrossRef]
- Mechaly, A.S.; Viñas, J.; Piferrer, F. Gene structure analysis of kisspeptin-2 (Kiss2) in the Senegalese sole (Solea senegalensis): Characterization of two splice variants of Kiss2, and novel evidence for metabolic regulation of kisspeptin signaling in non-mammalian species. Mol. Cell Endocrinol. 2011, 339, 14–24. [Google Scholar] [CrossRef]
- Mechaly, A.S.; Viñas, J.; Piferrer, F. Sex-specific changes in the expression of kisspeptin, kisspeptin receptor, gonadotropins and gonadotropin receptors in the Senegalese sole (Solea senegalensis) during a full reproductive cycle. Comp. Biochem. Physiol. A 2012, 162, 364–371. [Google Scholar] [CrossRef]
- Collins, L.J.; Schönfeld, B.; Chen, X.S. Chapter 4-the epigenetics of non-coding RNA. In Handbook of Epigenetics; Tollefsbol, T., Ed.; Academic Press: San Diego, CA, USA, 2011; pp. 49–61. [Google Scholar]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Silva, M.; Melo, S.A. Non-coding RNAs in exosomes: New players in cancer biology. Curr. Genomics 2015, 16, 295–303. [Google Scholar] [CrossRef]
- Viereck, J.; Thum, T. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ. Res. 2017, 120, 381–399. [Google Scholar] [CrossRef]
- Virant-Klun, I.; Ståhlberg, A.; Kubista, M.; Skutella, T. MicroRNAs: From female fertility, germ cells, and stem cells to cancer in humans. Stem Cells Int. 2016, 2016, 3984937. [Google Scholar] [CrossRef]
- Robles, V.; Valcarce, D.G.; Riesco, M.F. Non-coding RNA regulation in reproduction: Their potential use as biomarkers. Noncoding RNA Res. 2019, 4, 54–62. [Google Scholar] [CrossRef]
- Gilad, S.; Meiri, E.; Yogev, Y.; Benjamin, S.; Lebanony, D.; Yerushalmi, N.; Benjamin, H.; Kushnir, M.; Cholakh, H.; Melamed, N.; et al. Serum MicroRNAs are promising novel biomarkers. PLoS ONE 2008, 3, e3148. [Google Scholar] [CrossRef]
- Rome, S. Use of miRNAs in biofluids as biomarkers in dietary and lifestyle intervention studies. Genes Nutr. 2015, 10, 1–10. [Google Scholar] [CrossRef]
- Sun, Z.; Hao, T.; Tian, J. Identification of exosomes and its signature miRNAs of male and female Cynoglossus semilaevis. Sci. Rep. 2017, 7, 860. [Google Scholar] [CrossRef]
- Ma, J.; Li, Y.; Wu, M.; Zhang, C.; Che, Y.; Li, W.; Li, X. Serum immune responses in common carp (Cyprinus carpio L.) to paraquat exposure: The traditional parameters and circulating microRNAs. Fish Shellfish Immunol. 2018, 76, 133–142. [Google Scholar] [CrossRef]
- García-López, Á.; Fernández-Pasquier, V.; Couto, E.; Canario, A.V.M.; Sarasquete, C.; Martínez-Rodríguez, G. Testicular development and plasma sex steroid levels in cultured male Senegalese sole, Solea senegalensis Kaup. Gen. Comp. Endocrinol. 2006, 147, 343–351. [Google Scholar] [CrossRef]
- Beck, B.H.; Fuller, S.A.; Peatman, E.; McEntire, M.E.; Darwish, A.; Freeman, D.W. Chronic exogenous kisspeptin administration accelerates gonadal development in basses of the genus Morone. Comp. Biochem. Physiol. A 2012, 162, 265–273. [Google Scholar] [CrossRef]
- Nocillado, J.N.; Zohar, Y.; Biran, J.; Levavi-Sivan, B.; Elizur, A. Chronic kisspeptin administration stimulated gonadal development in pre-pubertal male yellowtail kingfish (Seriola lalandi; Perciformes) during the breeding and non-breeding season. Gen. Comp. Endocrinol. 2013, 191, 168–176. [Google Scholar] [CrossRef]
- Selvaraj, S.; Ohga, H.; Nyuji, M.; Kitano, H.; Nagano, N.; Yamaguchi, A.; Matsuyama, M. Subcutaneous administration of Kiss1 pentadecapeptide accelerates spermatogenesis in prepubertal male chub mackerel (Scomber japonicus). Comp. Biochem. Physiol. A 2013, 166, 228–236. [Google Scholar] [CrossRef]
- Zmora, N.; Stubblefield, J.; Zulperi, Z.; Biran, J.; Levavi-Sivan, B.; Muñoz-Cueto, J.A.; Zohar, Y. Differential and gonad stage-dependent roles of kisspeptin1 and kisspeptin2 in reproduction in the modern teleosts, Morone Species. Biol. Reprod. 2012, 86, 177. [Google Scholar] [CrossRef]
- Ohga, H.; Selvaraj, S.; Matsuyama, M. The roles of kisspeptin system in the reproductive physiology of fish, with special reference to chub mackerel studies as main axis. Front. Endocrinol. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Beato, S.; Toledo-Solís, F.J.; Fernández, I. Vitamin K in vertebrates’ reproduction: Further puzzling pieces of evidence from teleost fish species. Biomolecules 2020, 10, 1303. [Google Scholar] [CrossRef]
- Kirilov, M.; Clarkson, J.; Liu, X.; Roa, J.; Campos, P.; Porteous, R.; Schütz, G.; Herbison, A.E. Dependence of fertility on kisspeptin-Gpr54 signaling at the GnRH neuron. Nat. Commun. 2013, 4, 2492. [Google Scholar] [CrossRef]
- Tang, H.; Liu, Y.; Luo, D.; Ogawa, S.; Yin, Y.; Li, S.; Zhang, Y.; Hu, W.; Parhar, I.S.; Lin, H.; et al. The kiss/kissr systems are dispensable for zebrafish reproduction: Evidence from gene knockout studies. Endocrinology 2015, 156, 589–599. [Google Scholar] [CrossRef]
- Liu, H.; Todd, E.V.; Lokman, P.M.; Lamm, M.S.; Godwin, J.R.; Gemmell, N.J. Sexual plasticity: A fishy tale. Mol. Reprod. Dev. 2017, 84, 171–194. [Google Scholar] [CrossRef]
- Nakajo, M.; Kanda, S.; Karigo, T.; Takahashi, A.; Akazome, Y.; Uenoyama, Y.; Kobayashi, M.; Oka, Y. Evolutionally conserved function of kisspeptin neuronal system is nonreproductive regulation as revealed by nonmammalian study. Endocrinology 2017, 159, 163–183. [Google Scholar] [CrossRef]
- Zhao, Y.; Lin, M.-C.A.; Mock, A.; Yang, M.; Wayne, N.L. Kisspeptins modulate the biology of multiple populations of gonadotropin-releasing hormone neurons during embryogenesis and adulthood in Zebrafish (Danio rerio). PLoS ONE 2014, 9, e104330. [Google Scholar] [CrossRef][Green Version]
- Gu, Y.; Zhang, L.; Chen, X. Differential expression analysis of Paralichthys olivaceus microRNAs in adult ovary and testis by deep sequencing. Gen. Comp. Endocrinol. 2014, 204, 181–184. [Google Scholar] [CrossRef]
- He, P.; Wei, P.; Chen, X.; Lin, Y.; Peng, J. Identification and characterization of microRNAs in the gonad of Trachinotus ovatus using Solexa sequencing. Comp. Biochem. Physiol. D 2019, 30, 312–320. [Google Scholar] [CrossRef]
- McEwen, T.J.; Yao, Q.; Yun, S.; Lee, C.Y.; Bennett, K.L. Small RNA in situ hybridization in Caenorhabditis elegans, combined with RNA-seq, identifies germline-enriched microRNAs. Develop. Biol. 2016, 418, 248–257. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Farlora, R.; Valenzuela-Miranda, D.; Alarcón-Matus, P.; Gallardo-Escárate, C. Identification of microRNAs associated with sexual maturity in rainbow trout brain and testis through small RNA deep sequencing. Mol. Reprod. Dev. 2015, 82, 651–662. [Google Scholar] [CrossRef]
- Presslauer, C.; Tilahun Bizuayehu, T.; Kopp, M.; Fernandes, J.M.O.; Babiak, I. Dynamics of miRNA transcriptome during gonadal development of zebrafish. Sci. Rep. 2017, 7, 43850. [Google Scholar] [CrossRef] [PubMed]
- Petrelli, A.; Carollo, R.; Cargnelutti, M.; Iovino, F.; Callari, M.; Cimino, D.; Todaro, M.; Mangiapane, L.R.; Giammona, A.; Cordova, A.; et al. Correction: By promoting cell differentiation, miR-100 sensitizes basal-like breast cancer stem cells to hormonal therapy. Oncotarget 2019, 10, 5003–5004. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Jiang, S.; Xiong, Y.; Fu, H.; Zhang, W.; Wang, Y.; Gong, Y.; Jin, S.; Wu, Y. Integrated analysis of differentially expressed microRNAs and mRNAs to screen miRNAs and genes related to reproduction in Macrobrachium nipponense. 3 Biotechnol. 2019, 9, 327. [Google Scholar] [CrossRef]
- Li, Y.; Fang, Y.; Liu, Y.; Yang, X. MicroRNAs in ovarian function and disorders. J. Ovarian Res. 2015, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- Sirotkin, A.V.; Ovcharenko, D.; Grossmann, R.; Lauková, M.; Mlyncek, M. Identification of microRNAs controlling human ovarian cell steroidogenesis via a genome-scale screen. J. Cell Physiol. 2009, 219, 415–420. [Google Scholar] [CrossRef]
- Fichtlscherer, S.; Zeiher, A.M.; Dimmeler, S. Circulating microRNAs: Biomarkers or mediators of cardiovascular diseases? Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2383–2390. [Google Scholar] [CrossRef]
- Xia, J.H.; He, X.P.; Bai, Z.Y.; Yue, G.H. Identification and characterization of 63 MicroRNAs in the Asian seabass Lates calcarifer. PLoS ONE 2011, 6, e17537. [Google Scholar] [CrossRef]
- Bizuayehu, T.T.; Babiak, I. MicroRNA in Teleost Fish. Genome Biol. Evol. 2014, 6, 1911–1937. [Google Scholar] [CrossRef]
- Goll, M.G.; Halpern, M.E. Chapter 5-DNA methylation in zebrafish. In Progress in Molecular Biology and Translational Science; Cheng, X., Blumenthal, R.M., Eds.; Academic Press: Cambdrige, MA, USA, 2011; Volume 101, pp. 193–218. [Google Scholar]
- Rai, K.; Jafri, I.F.; Chidester, S.; James, S.R.; Karpf, A.R.; Cairns, B.R.; Jones, D.A. Dnmt3 and G9a cooperate for tissue-specific development in zebrafish. J. Biol. Chem. 2010, 285, 4110–4121. [Google Scholar] [CrossRef]
- Campos, C.; Valente, L.M.P.; Conceição, L.E.C.; Engrola, S.; Fernandes, J.M.O. Temperature affects methylation of the myogenin putative promoter, its expression and muscle cellularity in Senegalese sole larvae. Epigenetics 2013, 8, 389–397. [Google Scholar] [CrossRef]
- Firmino, J.; Carballo, C.; Armesto, P.; Campinho, M.A.; Power, D.M.; Manchado, M. Phylogeny, expression patterns and regulation of DNA Methyltransferases in early development of the flatfish, Solea senegalensis. BMC Dev. Biol. 2017, 17, 11. [Google Scholar] [CrossRef]
- Dawkins, J.L.; Hulme, D.J.; Brahmbhatt, S.B.; Auer-Grumbach, M.; Nicholson, G.A. Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause hereditary sensory neuropathy type I. Nat. Genet. 2001, 27, 309–312. [Google Scholar] [CrossRef]
- Han, G.; Gupta, S.D.; Gable, K.; Niranjanakumari, S.; Moitra, P.; Eichler, F.; Brown, R.H., Jr.; Harmon, J.M.; Dunn, T.M. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc. Natl. Acad. Sci. USA 2009, 106, 8186–8191. [Google Scholar] [CrossRef]
- Crabos, M.; Yamakado, T.; Heizmann, C.W.; Cerletti, N.; Bühler, F.R.; Erne, P. The calcium binding protein tropomyosin in human platelets and cardiac tissue: Elevation in hypertensive cardiac hypertrophy. Eur. J. Clin. Investig. 1991, 21, 472–478. [Google Scholar] [CrossRef]
- Kumar, N.M.; Gilula, N.B. The gap junction communication channel. Cell 1996, 84, 381–388. [Google Scholar] [CrossRef]
- Francis, R.J.B.; Lo, C.W. Primordial germ cell deficiency in the connexin 43 knockout mouse arises from apoptosis associated with abnormal p53 activation. Development 2006, 133, 3451–3460. [Google Scholar] [CrossRef]
- Kidder, G.M.; Vanderhyden, B.C. Bidirectional communication between oocytes and follicle cells: Ensuring oocyte developmental competence. Can. J. Physiol. Pharmacol. 2010, 88, 399–413. [Google Scholar] [CrossRef]
- Cerdá, J.L.; Petrino, T.R.; Wallace, R.A. Functional heterologous gap junctions in Fundulus ovarian follicles maintain meiotic arrest and permit hydration during oocyte maturation. Dev. Biol. 1993, 160, 228–235. [Google Scholar] [CrossRef]
- Zhang, L.; Feng, T.; Spicer, L.J. The role of tight junction proteins in ovarian follicular development and ovarian cancer. Reproduction 2018, 155, R183. [Google Scholar] [CrossRef]
- Hewitt, K.J.; Agarwal, R.; Morin, P.J. The claudin gene family: Expression in normal and neoplastic tissues. BMC Cancer 2006, 6, 186. [Google Scholar] [CrossRef]
- Pramod, A.B.; Foster, J.; Carvelli, L.; Henry, L.K. SLC6 transporters: Structure, function, regulation, disease association and therapeutics. Mol. Aspects Med. 2013, 34, 197–219. [Google Scholar] [CrossRef]
- Cerdà, J.; Subhedar, N.; Reich, G.; Wallace, R.A.; Selman, K. Oocyte sensitivity to serotonergic regulation during the follicular cycle of the teleost Fundulus heteroclitus. Biol. Reprod. 1998, 59, 53–61. [Google Scholar] [CrossRef]
- Beny-Shefer, Y.; Zilkha, N.; Lavi-Avnon, Y.; Bezalel, N.; Rogachev, I.; Brandis, A.; Dayan, M.; Kimchi, T. Nucleus accumbens dopamine signaling regulates sexual preference for females in male mice. Cell Rep. 2017, 21, 3079–3088. [Google Scholar] [CrossRef] [PubMed]
- Vacher, C.; Mañanos, E.L.; Breton, B.; Marmignon, M.H.; Saligaut, C. Modulation of pituitary dopamine D1 or D2 receptors and secretion of follicle stimulating hormone and luteinizing hormone during the annual reproductive cycle of female rainbow trout. J. Neuroendocrinol. 2000, 12, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Riesco, M.F.; Valcarce, D.G.; Martínez-Vázquez, J.M.; Martín, I.; Calderón-García, A.Á.; Gonzalez-Nunez, V.; Robles, V. Male reproductive dysfunction in Solea senegalensis: New insights into an unsolved question. Reprod. Fertil. Dev. 2019, 31, 1104–1115. [Google Scholar] [CrossRef]
- Worby, C.A.; Dixon, J.E. Sorting out the cellular functions of sorting nexins. Nat. Rev. Mol. Cell Biol. 2002, 3, 919–931. [Google Scholar] [CrossRef]
- Ling, K.-H.; Hewitt, C.A.; Tan, K.-L.; Cheah, P.-S.; Vidyadaran, S.; Lai, M.-I.; Lee, H.-C.; Simpson, K.; Hyde, L.; Pritchard, M.A.; et al. Functional transcriptome analysis of the postnatal brain of the Ts1Cje mouse model for Down syndrome reveals global disruption of interferon-related molecular networks. BMC Genomics 2014, 15, 624. [Google Scholar] [CrossRef]
- Webber, E.; Li, L.; Chin, L.-S. Hypertonia-associated protein Trak1 is a novel regulator of endosome-to-lysosome trafficking. J. Mol. Biol. 2008, 382, 638–651. [Google Scholar] [CrossRef][Green Version]
- Brickley, K.; Stephenson, F.A. Trafficking kinesin protein (TRAK)-mediated transport of mitochondria in axons of hippocampal neurons. J. Biol. Chem. 2011, 286, 18079–18092. [Google Scholar] [CrossRef] [PubMed]
- García-López, A.; Anguis, V.; Couto, E.; Canario, A.V.M.; Cañavate, J.P.; Sarasquete, C.; Martínez-Rodríguez, G. Non-invasive assessment of reproductive status and cycle of sex steorid levels in a captive wild broosdstock of senegalese sole Solea senagalensis (Kaup). Aquaculture 2006, 254, 583–593. [Google Scholar] [CrossRef]
- Chauvigné, F.; Verdura, S.; Mazón, M.J.; Boj, M.; Zanuy, S.; Gómez, A.; Cerdà, J. Development of a flatfish-specific enzyme-linked immunosorbent assay for Fsh using a recombinant chimeric gonadotropin. Gen. Comp. Endocrinol. 2015, 221, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 3. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Mullany, L.E.; Herrick, J.S.; Wolff, R.K.; Slattery, M.L. MicroRNA seed region length impact on target messenger RNA expression and survival in colorectal cancer. PLoS ONE 2016, 11, e0154177. [Google Scholar] [CrossRef]
- Yartseva, V.; Takacs, C.M.; Vejnar, C.E.; Lee, M.T.; Giraldez, A.J. RESA identifies mRNA-regulatory sequences at high resolution. Nat. Methods 2017, 14, 201–207. [Google Scholar] [CrossRef]
- Zar, J.H. Biostatistical Analysis, 4th ed.; Prentice-Hall Inc.: Upper Saddle River, NJ, USA, 1999. [Google Scholar]
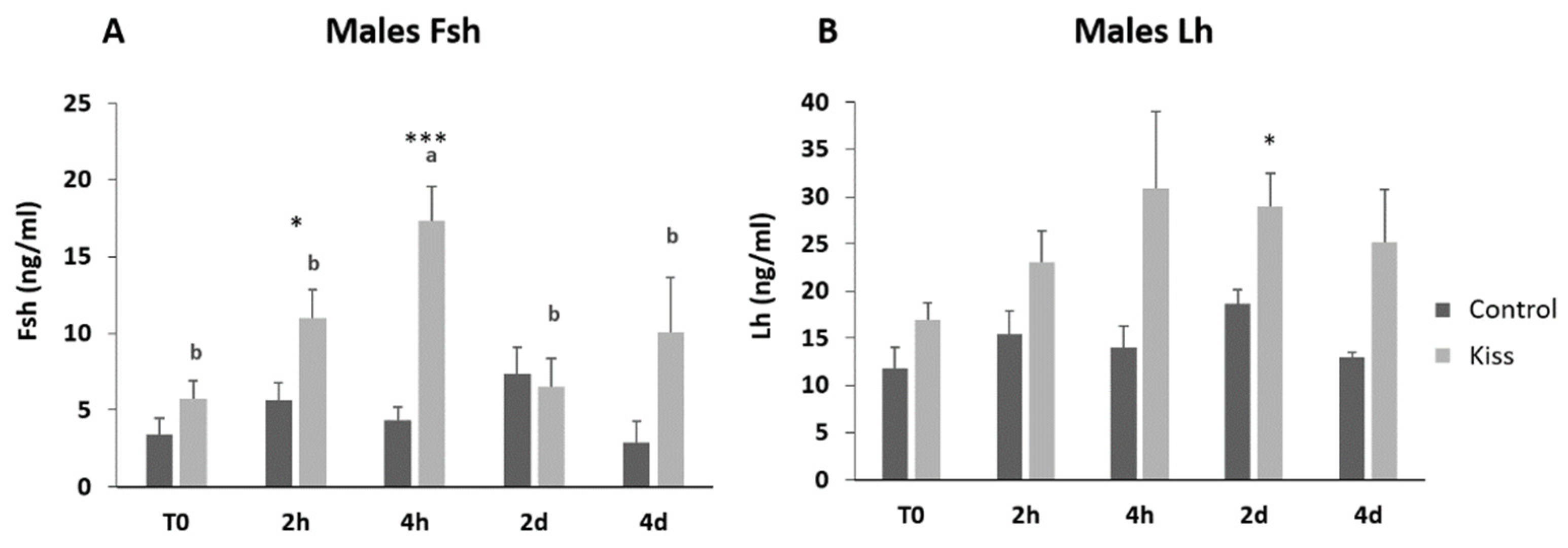
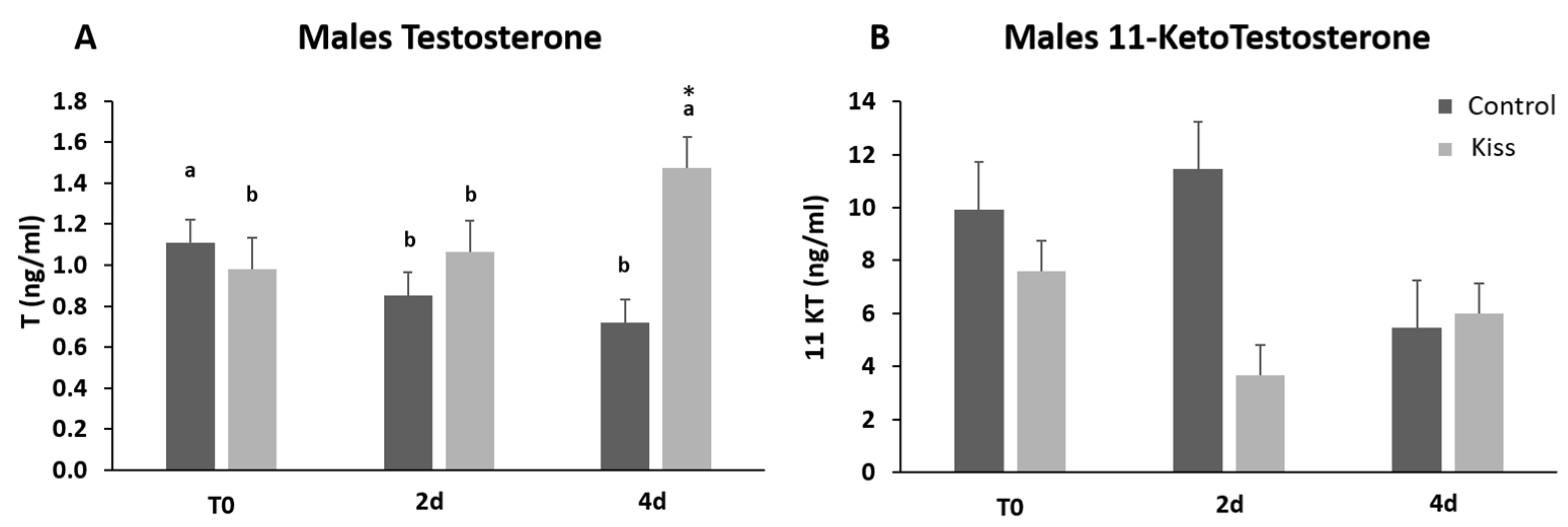
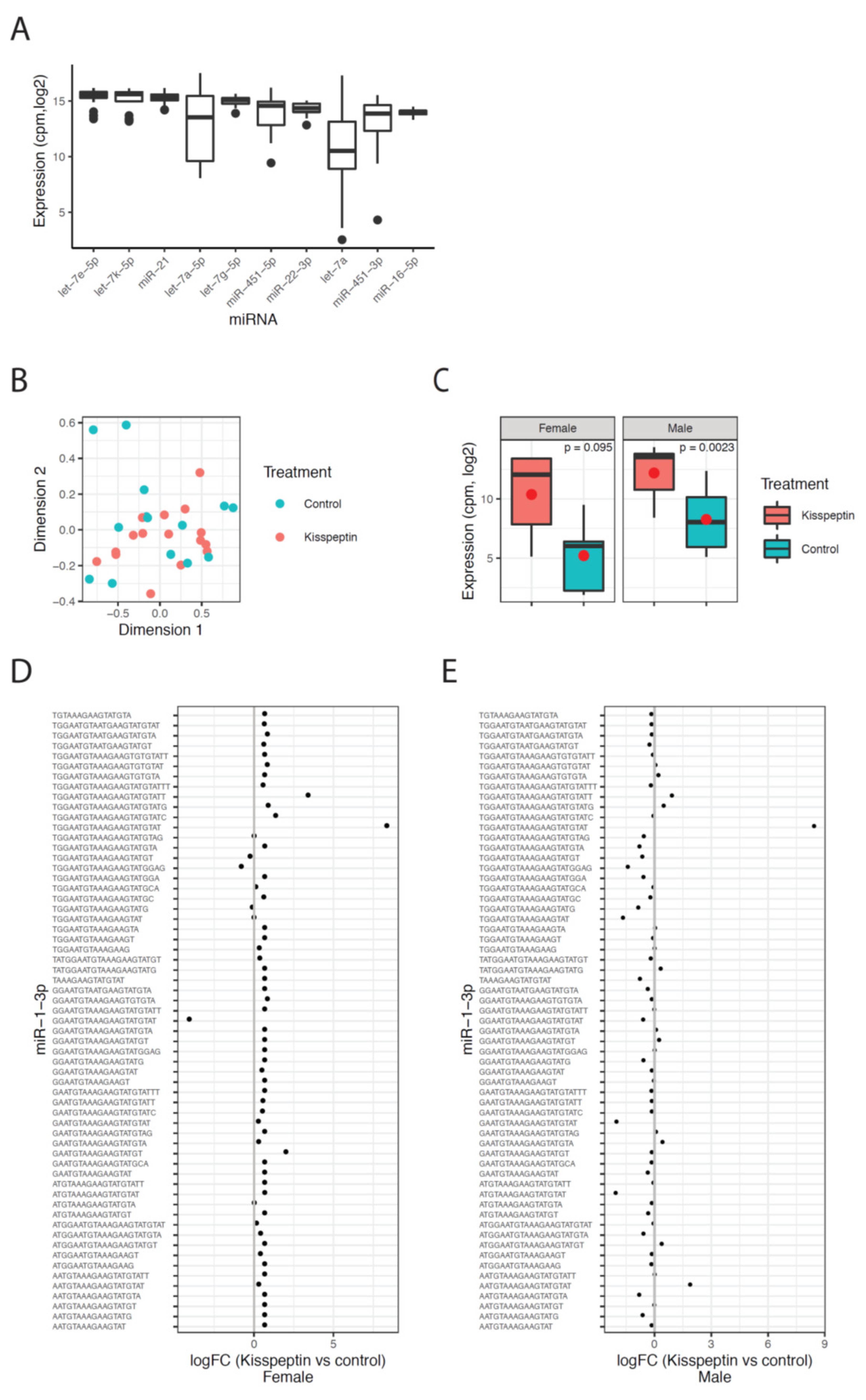
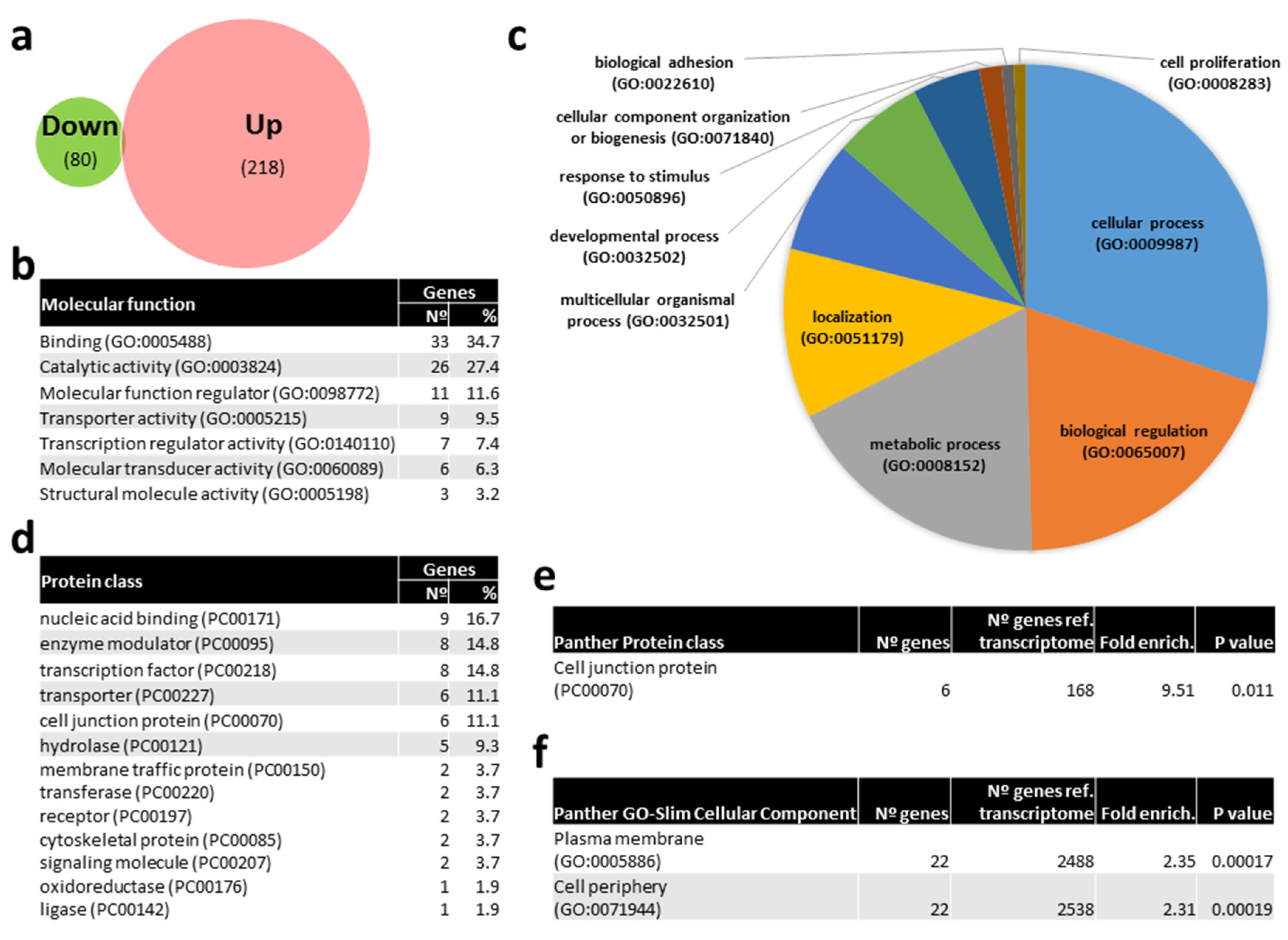
| Sampling | Treatment | Live Cells (%) |
|---|---|---|
| T0 | Kiss | 88.55 ± 12.77 |
| Control | 90.47 ± 4.81 | |
| 2 d | Kiss | 84.87 ± 6.40 |
| Control | 89.32 ± 2.96 | |
| 4 d | Kiss | 89.75 ± 4.26 |
| Control | 88.53 ± 1.35 |
| miRNA | Sequence | Log2 Fold Change | Average Expression | Adjusted p-Value |
|---|---|---|---|---|
| let-7e | TGAGGTAGTTGGTTGT | 6.295 | 2.352 | 0.00939 |
| miR-199a-3p | AGTAGTCTGCACATTGGTT | 7.973 | 3.232 | 0.00165 |
| miR-100-5p | AACCCGTAGATCCGAACTTGTG | −9.335 | 4.472 | 0.00006 |
| Kisspeptintreated | Sample ID | Sex | Sampling Point | Raw Reads | Trimmed | Trimmed % | microRNA | microRNA (%) |
| A1-1_S1 | Female | 2 d | 453,229 | 208,921 | 46 | 47,725 | 11 | |
| A1-2_S2 | Female | 2 d | 500,647 | 237,779 | 47 | 91,581 | 18 | |
| A1-3_S3 | Female | 2 d | 567,594 | 288,477 | 51 | 129,980 | 23 | |
| A1-4_S4 | Female | 2 d | 475,441 | 209,746 | 44 | 57,941 | 12 | |
| A1-11-9_S5 | Female | 2 d | 448,350 | 219,818 | 49 | 25,468 | 6 | |
| A1-6_S11 | Male | 2 d | 409,534 | 199,896 | 49 | 57,262 | 14 | |
| A1-7_S12 | Male | 2 d | 438,943 | 207,974 | 47 | 57,138 | 13 | |
| A1-10_S13 | Male | 2 d | 497,364 | 252,675 | 51 | 63,137 | 13 | |
| A1-12_S14 | Male | 2 d | 500,206 | 222,774 | 45 | 48,918 | 10 | |
| A1-13_S15 | Male | 2 d | 431,890 | 200,621 | 46 | 35,429 | 8 | |
| A1-11-12_S16 | Male | 2 d | 448,502 | 218,492 | 49 | 157,656 | 35 | |
| A4-3_S26 | Male | 4 d | 509,277 | 258,979 | 51 | 139,680 | 27 | |
| A4-8_S27 | Male | 4 d | 395,813 | 180,599 | 46 | 41,197 | 10 | |
| A4-9_S28 | Male | 4 d | 505,458 | 256,239 | 51 | 104,044 | 21 | |
| A4-10_S29 | Male | 4 d | 498,680 | 233,040 | 47 | 49,651 | 10 | |
| A4-17_S30 | Male | 4 d | 463,867 | 223,033 | 48 | 85,875 | 19 | |
| Control untreated | A2-2_S6 | Female | 2 d | 461,160 | 227,736 | 49 | 191,846 | 42 |
| A2-5_S7 | Female | 2 d | 491,359 | 243,485 | 50 | 163,635 | 33 | |
| A2-7_S8 | Female | 2 d | 415,844 | 192,516 | 46 | 124,752 | 30 | |
| A2-9_S9 | Female | 2 d | 475,556 | 211,070 | 44 | 84,252 | 18 | |
| A2-10_S10 | Female | 2 d | 443,098 | 218,571 | 49 | 52,960 | 12 | |
| A2-11_S17 | Male | 2 d | 496,908 | 245,995 | 50 | 39,267 | 8 | |
| A2-12_S18 | Male | 2 d | 488801 | 233,186 | 48 | 65,358 | 13 | |
| A2-14_S19 | Male | 2 d | 549,711 | 227,754 | 41 | 29,918 | 5 | |
| A2-16_S20 | Male | 2 d | 502,982 | 226,236 | 45 | 41,948 | 8 | |
| A3-2_S21 | Male | 4 d | 445,074 | 219,334 | 49 | 47,514 | 11 | |
| A3-5_S22 | Male | 4 d | 500,306 | 248,426 | 50 | 93,047 | 19 | |
| A3-6_S23 | Male | 4 d | 464,452 | 219,739 | 47 | 98,906 | 21 | |
| A3-9_S24 | Male | 4 d | 470,313 | 232,473 | 49 | 133,423 | 28 | |
| A3-11_S25 | Male | 4 d | 488,090 | 247,258 | 51 | 31,354 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, C.C.V.; Fatsini, E.; Fernández, I.; Anjos, C.; Chauvigné, F.; Cerdà, J.; Mjelle, R.; Fernandes, J.M.O.; Cabrita, E. Kisspeptin Influences the Reproductive Axis and Circulating Levels of microRNAs in Senegalese Sole. Int. J. Mol. Sci. 2020, 21, 9051. https://doi.org/10.3390/ijms21239051
Oliveira CCV, Fatsini E, Fernández I, Anjos C, Chauvigné F, Cerdà J, Mjelle R, Fernandes JMO, Cabrita E. Kisspeptin Influences the Reproductive Axis and Circulating Levels of microRNAs in Senegalese Sole. International Journal of Molecular Sciences. 2020; 21(23):9051. https://doi.org/10.3390/ijms21239051
Chicago/Turabian StyleOliveira, Catarina C. V., Elvira Fatsini, Ignacio Fernández, Catarina Anjos, François Chauvigné, Joan Cerdà, Robin Mjelle, Jorge M. O. Fernandes, and Elsa Cabrita. 2020. "Kisspeptin Influences the Reproductive Axis and Circulating Levels of microRNAs in Senegalese Sole" International Journal of Molecular Sciences 21, no. 23: 9051. https://doi.org/10.3390/ijms21239051
APA StyleOliveira, C. C. V., Fatsini, E., Fernández, I., Anjos, C., Chauvigné, F., Cerdà, J., Mjelle, R., Fernandes, J. M. O., & Cabrita, E. (2020). Kisspeptin Influences the Reproductive Axis and Circulating Levels of microRNAs in Senegalese Sole. International Journal of Molecular Sciences, 21(23), 9051. https://doi.org/10.3390/ijms21239051








