Yil102c-A is a Functional Homologue of the DPMII Subunit of Dolichyl Phosphate Mannose Synthase in Saccharomyces cerevisiae
Abstract
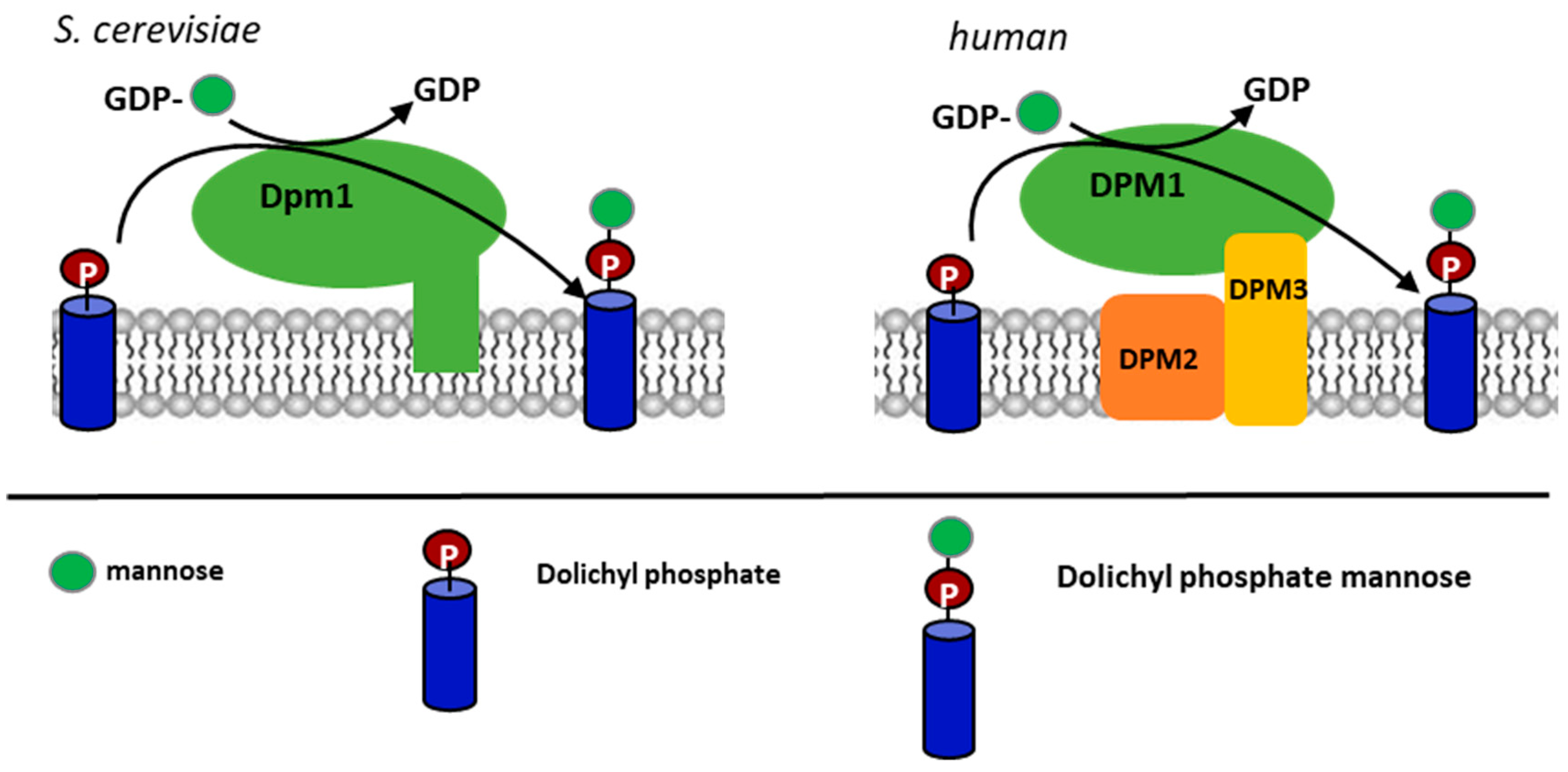
1. Introduction
2. Results
2.1. Identification and Cloning of YIL102c-A
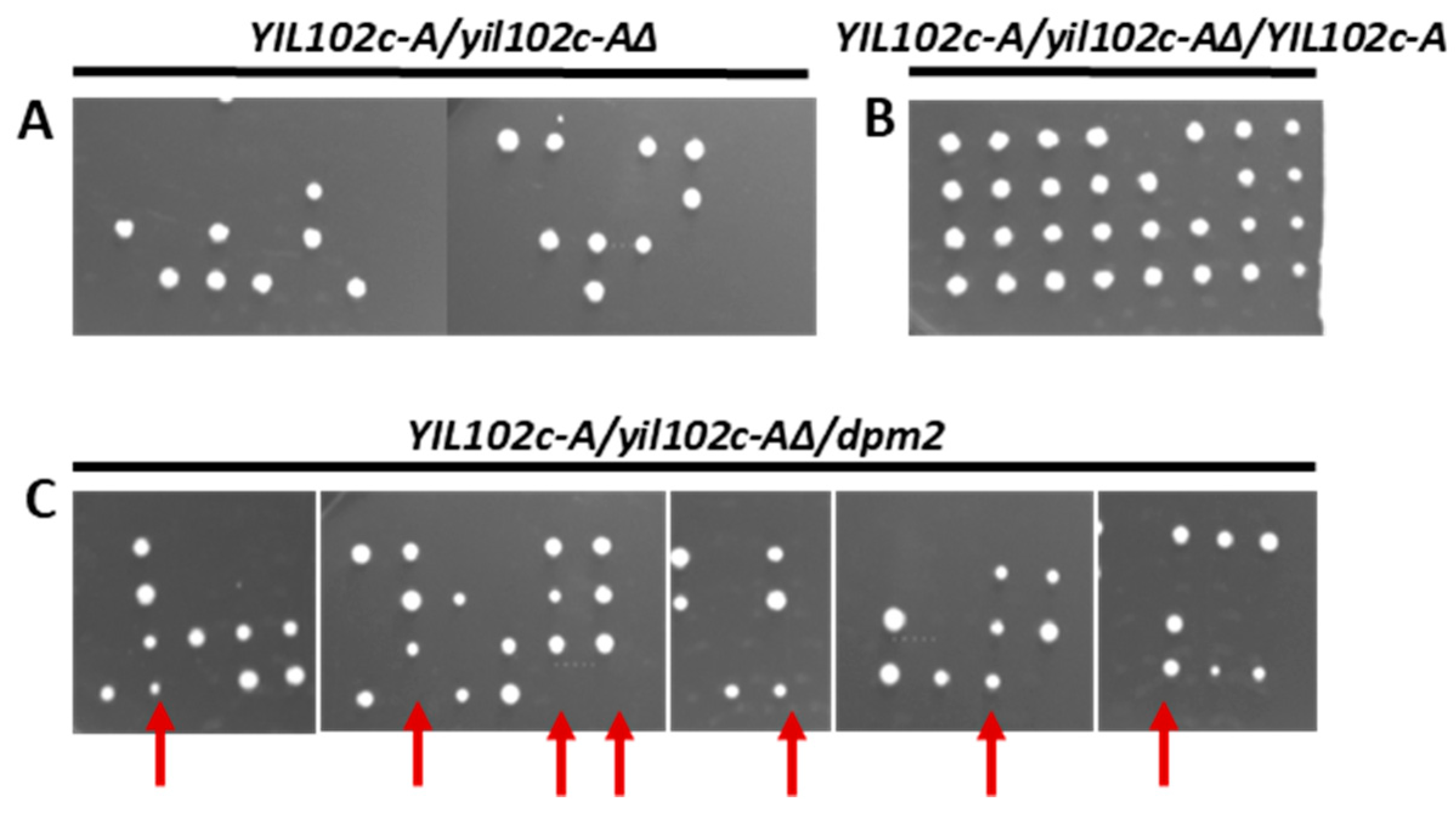
2.2. Lack of YIL102c-A is Lethal
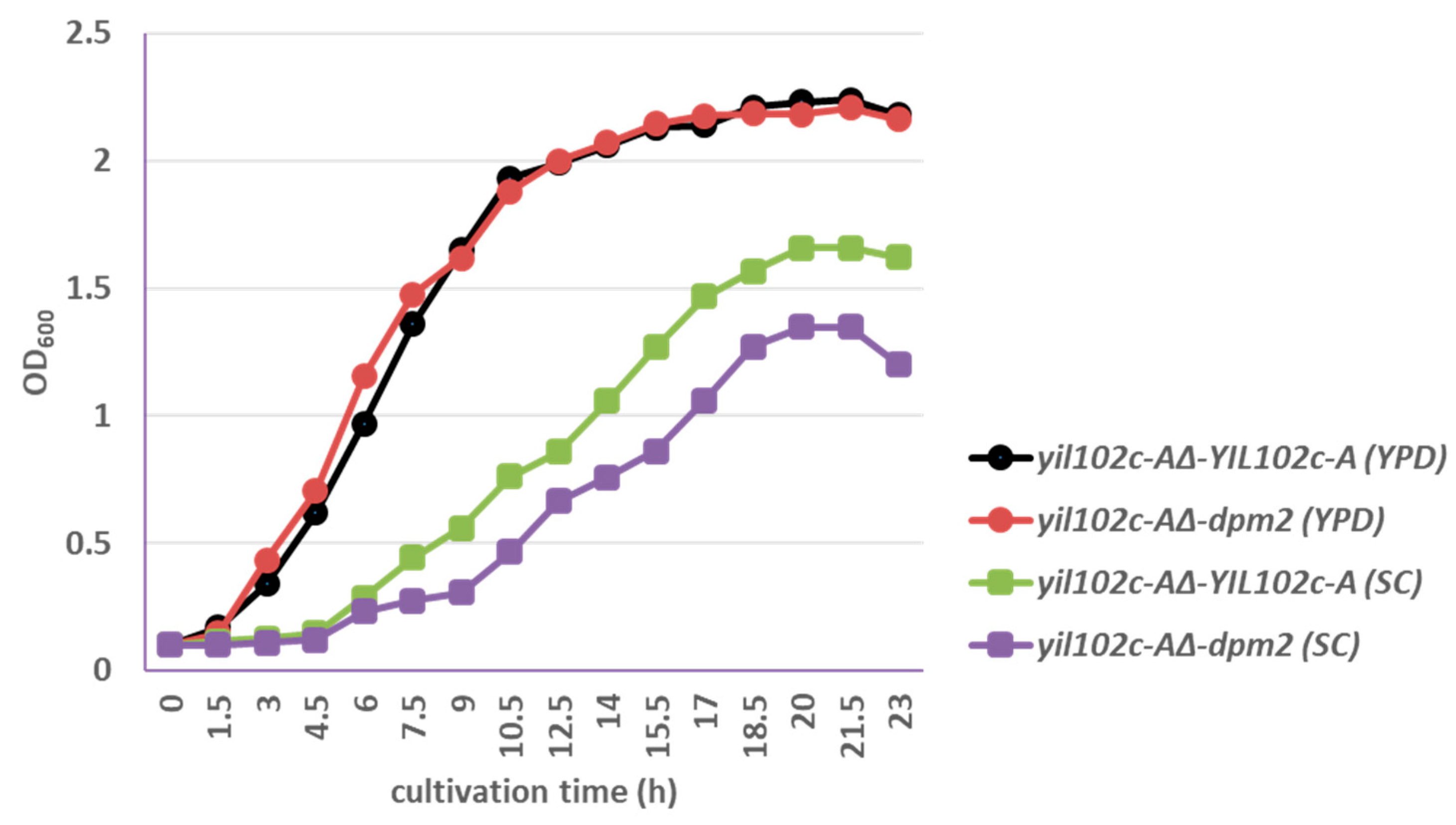
2.3. Complementation of YIL102c-A Deletion in S. cerevisiae by dpm2 Gene from Trichoderma
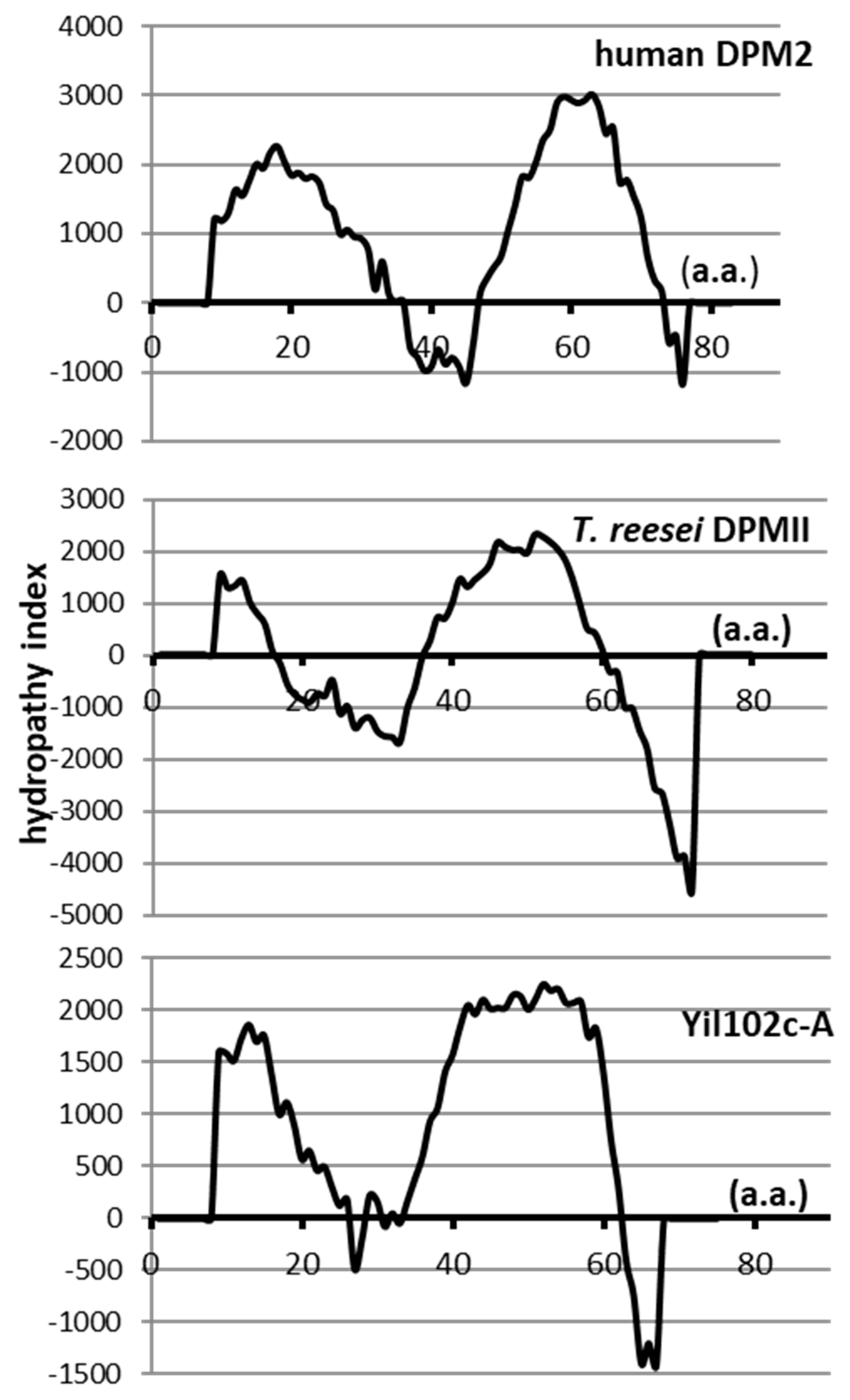
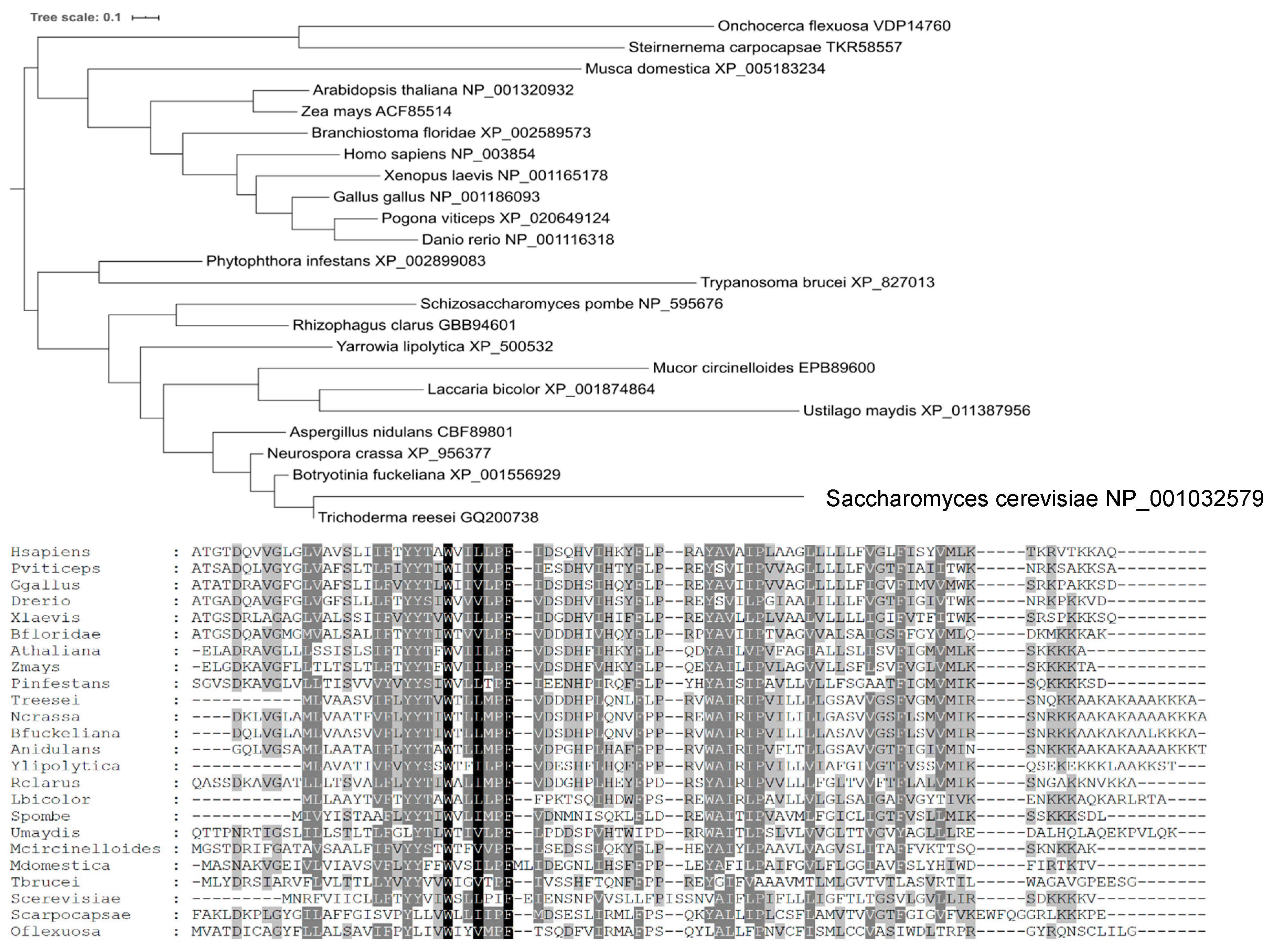
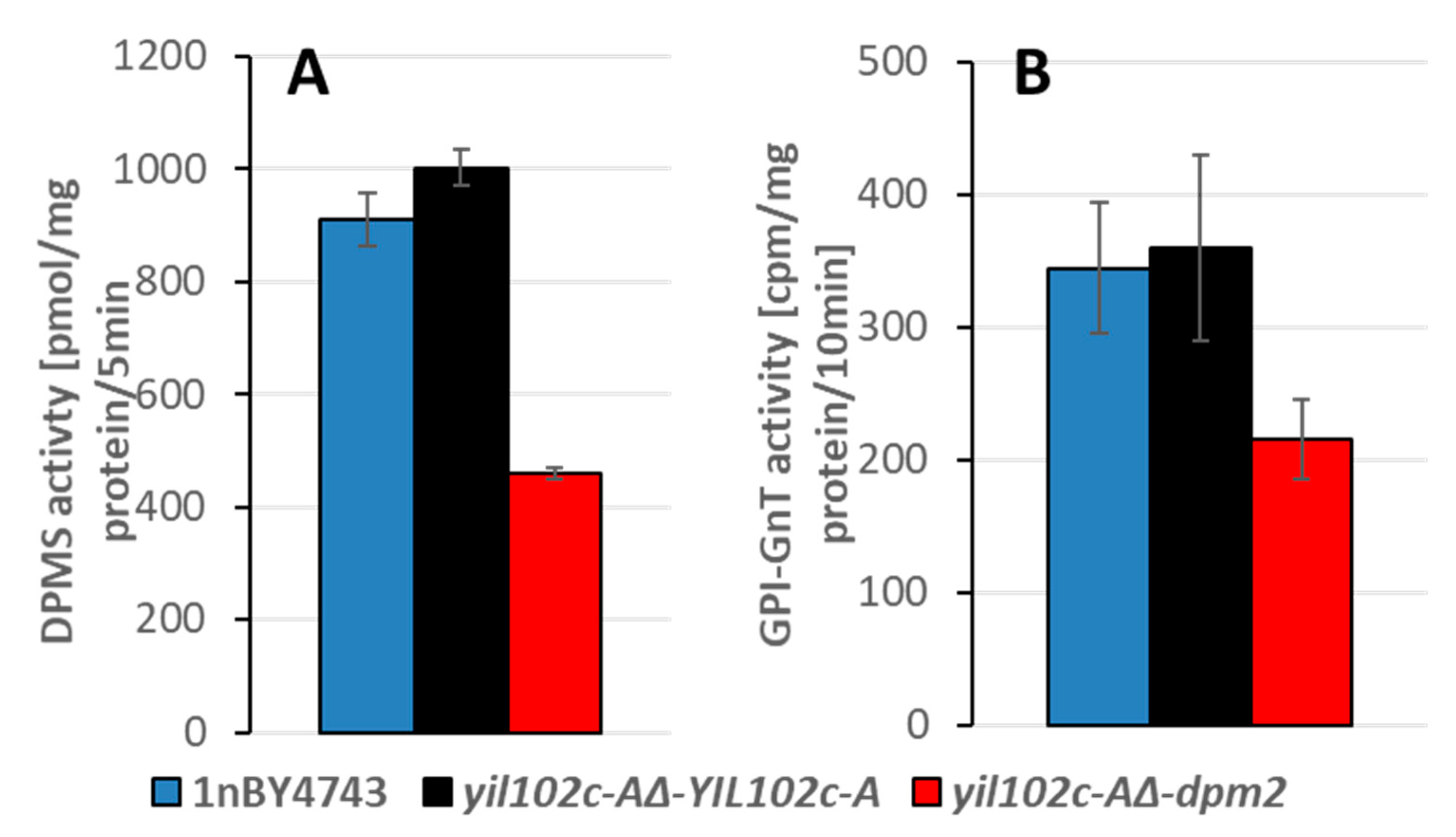
2.4. Comparison of Yil102c-A and DPMII Proteins
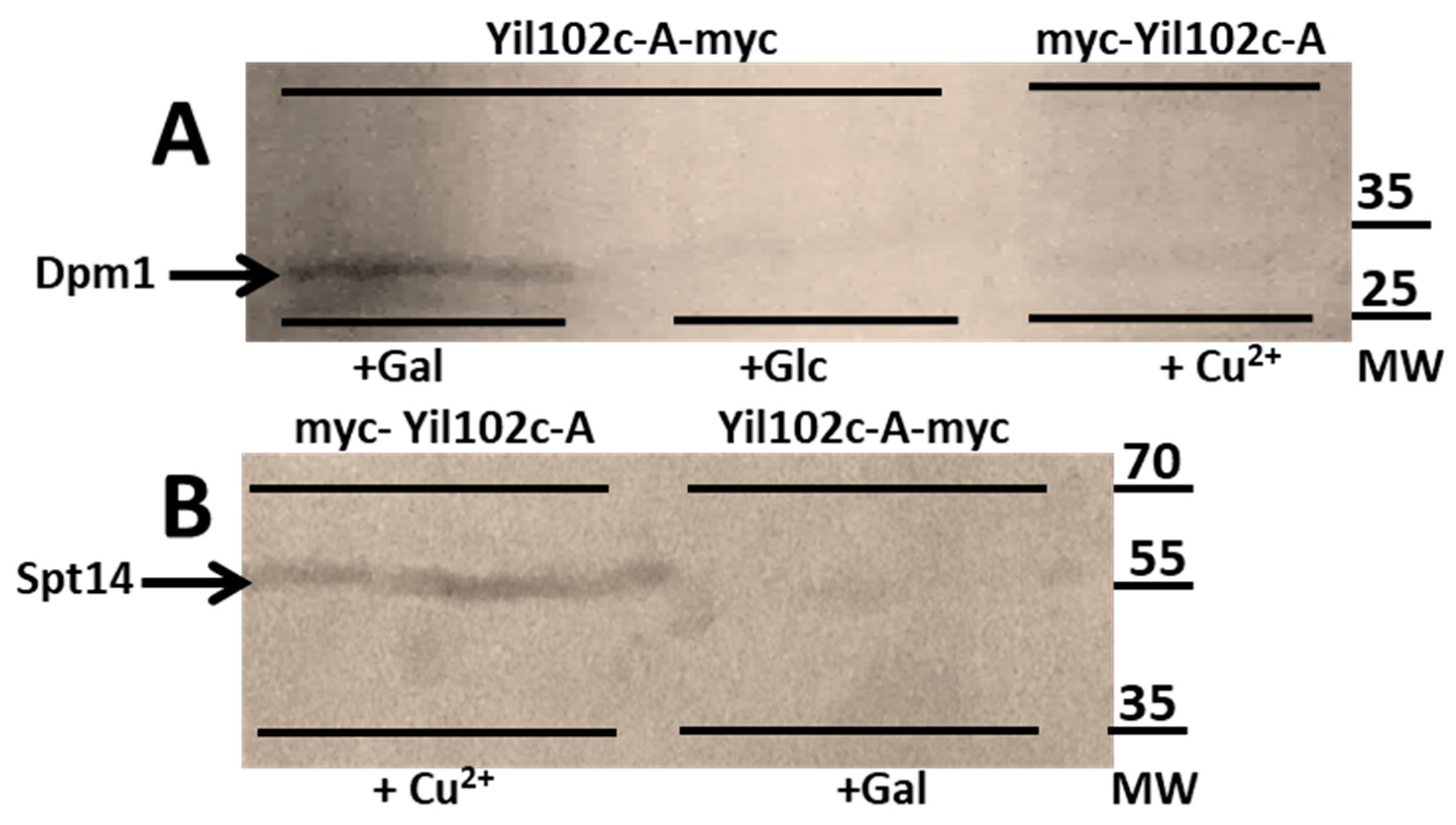
2.5. Yil102c-A Interacts with DPM1 and Spt14 (PIG-A)
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Deletion of YIL102c-A Gene in BY4743 Strain
4.3. Overexpression of YIL102c-A-myc in S. cerevisiae
4.4. Membrane Preparation
4.5. Protein Determination
4.6. Dolichyl Phosphate Mannose Synthase (EC 2.4.1.83) Activity
4.7. Glucosylphosphatidylinositol-N-Acetylglucosaminyl Transferase (GPI-GnT) (EC 2.4.1.198) Activity
4.8. Coimmunoprecipitation was Done Using Myc-Trap Agarose kit
4.9. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kornfeld, R.; Kornfeld, S. Assembly of asparagine linked oligosaccharides. Annu. Rev. Biochem. 1985, 54, 631–664. [Google Scholar] [CrossRef] [PubMed]
- Hirschberg, C.B.; Snider, M.D. Topography of glycosylation in the rough endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 1987, 56, 63–87. [Google Scholar] [CrossRef] [PubMed]
- Orlean, P. Dolichol phosphate mannose synthase is required in vivo for glycosyl phosphatidylinositol membrane anchoring, O mannosylation, and N glycosylation of protein in Saccharomyces cerevisiae. Mol. Cell Biol. 1990, 10, 5796–5805. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Inoue, N. Dissecting and manipulating the pathway for glycosylphos-phatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 2000, 4, 632–638. [Google Scholar] [CrossRef]
- Helenius, A.; Aebi, M. Intracellular functions of N-linked glycans. Science 2001, 291, 2364–2369. [Google Scholar] [CrossRef]
- Maeda, Y.; Kinoshita, T. Dolichol-phosphate mannose synthase: Structure, function and regulation. Biochim. Biophys. Acta 2008, 1780, 861–868. [Google Scholar] [CrossRef]
- Doucey, M.-A.; Hess, D.; Cacan, R.; Hofsteenge, J. Protein C-mannosylation is enzyme-catalyzed and uses dolichyl-phosphate-mannose as a precursor. Mol. Biol. Cell 1998, 9, 291–300. [Google Scholar] [CrossRef]
- Orlean, P.; Albright, C.; Robbins, P.W. Cloning and sequencing of the yeast gene for dolichol phosphate mannose synthase, an essential protein. J. Biol. Chem. 1988, 263, 17499–17507. [Google Scholar]
- Haselbeck, A.; Tanner, W. Purification of GDP mannose: Dolichyl phosphate-O-D-mannosyl transferase. Eur. J. Biochem. 1989, 181, 663–668. [Google Scholar] [CrossRef]
- Mazhari-Tabrizi, R.; Eckert, V.; Blank, M.; Muller, R.; Mumberg, D.; Funk, M.; Schwarz, R.T. Cloning and functional expression of glycosyltransferases from parasitic protozoans by heterologous complementation in yeast: The dolichol phosphate mannose synthase from Trypanosoma brucei. Biochem. J. 1996, 316, 853–858. [Google Scholar] [CrossRef]
- Zimmerman, J.W.; Specht, C.A.; Cazares, B.X.; Robbins, P.W. The isolation of a Dol-P-Man synthase from Ustilago maydis that functions in Saccharomyces cerevisiae. Yeast 1996, 12, 765–771. [Google Scholar] [CrossRef]
- Colussi, P.A.; Taron, C.H.; Mach, J.C.; Orlean, P. Human and Saccharomyces cerevisiae dolichol phosphate mannose synthases represent two classes of the enzyme, but both function in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 1997, 94, 7873–7878. [Google Scholar] [CrossRef] [PubMed]
- Kruszewska, J.S.; Saloheimo, M.; Migdalski, A.; Orlean, P.; Penttila, M.; Palamarczyk, G. Dolichol phosphate mannose synthase from the filamentous fungus Trichoderma reesei. Glycobiology 2000, 10, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Tanaka, S.; Hino, J.; Kangawa, K.; Kinoshita, T. Human dolichol-phosphate-mannose synthase consists of three subunits, DPM1, DPM2 and DPM3. EMBO J. 2000, 11, 2475–2482. [Google Scholar] [CrossRef]
- Zembek, P.; Perlińska-Lenart, U.; Rawa, K.; Górka-Nieć, W.; Palamarczyk, G.; Kruszewska, J.S. Cloning and functional analysis of the dpm2 and dpm3 genes from Trichoderma reesei expressed in a Saccharomyces cerevisiae dpm1∆ mutant strain. Biol. Chem. 2011, 392, 517–527. [Google Scholar] [CrossRef]
- Tomita, S.; Inoue, N.; Maeda, Y.; Ohishi, K.; Takeda, J.; Kinoshita, T. A homologue of Saccharomyces cerevisiae Dpm1p is not sufficient for synthesis of dolichol-phosphate-mannose in mammalian cells. J. Biol. Chem. 1998, 273, 9249–9254. [Google Scholar] [CrossRef]
- Ashida, H.; Maeda, Y.; Kinoshita, T. DPM1, the catalytic subunit of dolichol-phosphate mannose synthase, is tethered to and stabilized on the endoplasmic reticulum membrane by DPM3. J. Biol. Chem. 2006, 181, 896–904. [Google Scholar] [CrossRef]
- Watanabe, R.; Murakami, Y.; Marmor, M.D.; Inoue, N.; Maeda, Y.; Hino, J.; Kangawa, K.; Julius, M.; Kinoshita, T. Initial enzyme for glycosylphosphatidylinositol biosynthesis requires PIG-P and is regulated by DPM2. EMBO J. 2000, 19, 4402–4411. [Google Scholar] [CrossRef]
- Orlean, P.; Menon, A.K. GPI anchoring of protein in yeast and mammalian cells, or: How we learned to stop worrying and love glycophospholipids. J. Lipid Res. 2007, 48, 993–1011. [Google Scholar] [CrossRef]
- Kinoshita, T.; Inoue, N.; Murakami, Y. Glycosylphosphatidylinositol-N-Acetylglucosaminyltransferase (GPI-GlcNAc Transferase): A complex comprised of PIGA, PIGC, PIGH, PIGQ, PIGP, PIGY and DPM2. In Handbook of Glycosyltransferases and Related Genes; Taniguchi, N., Honke, K., Fukuda, M., Narimatsu, H., Yamaguchi, Y., Angata, T., Eds.; Springer: Tokyo, Japan, 2014; Chapter 106; pp. 1193–1208. [Google Scholar]
- Hofmann, K.; Stoffel, W. TMbase—A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 1993, 374, 166. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Beck, P.J.; Orlean, P.; Albright, C.; Robbins, P.W.; Gething, M.J.; Sambrook, J.F. The Saccharomyces cerevisiae DPM1 gene encoding dolichol-phosphate-mannose synthase is able to complement a glycosylation-defective mammalian cell line. Mol. Cell. Biol. 1990, 10, 4612–4622. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, D.K.; Carrasquillo, E.A.; Hughey, P.; Schutzbach, J.S.; Martinez, J.A.; Baksi, K. In vitro phosphorylation by cAMP-dependent protein kinase up-regulates recombinant Saccharomyces cerevisiae mannosylphosphodolichol synthase. J. Biol. Chem. 2005, 280, 417–4181. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, D.K.; Aponte, E.; DaSilve, J.J. Low expression of lipid-linked oligosachcride due to a functionally altered Dol-P-Man synthase reduces protein glycosylation in cAMP-dependent protein kinase deficient Chinese hamster ovary cells. Glycoconj. J. 2004, 21, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, D.K.; Kousvelari, E.E.; Baum, B.J. cAMP-mediated protein phosphorylation of microsomal membranes increases mannosylphosphodolichol synthase activity. Proc. Natl. Acad. Sci. USA 1987, 84, 6389–6393. [Google Scholar] [CrossRef] [PubMed]
- Kruszewska, J.; Palamarczyk, G.; Kubicek, C.P. Mannosyl phospholdolichol synthase from Trichoderma reesei is activated by protein kinase dependent phosphorylation in vivo. FEMS Lett. 1991, 80, 81–86. [Google Scholar] [CrossRef]
- Gryz, E.; Perlinska-Lenart, U.; Gawarecka, K.; Jozwiak, A.; Piłsyk, S.; Jemiola-Rzeminska, M.; Bernat, P.; Muszewska, A.; Steczkiewicz, K.; Ginalski, K.; et al. Poly-saturated dolichols from filamentous fungi modulate activity of dolichol-dependent glycosyltransferase and physical properties of membranes. Int. J. Mol Sci. 2019, 20, 3043. [Google Scholar] [CrossRef] [PubMed]
- Sobering, A.K.; Watanabe, R.; Romeo, M.J.; Yan, B.C.; Specht, C.A.; Orlean, P.; Reizman, H.; Levin, D.E. Yeast Ras regulates the complex that catalyzes the first step in GPI-anchor biosynthesis at the ER. Cell 2004, 117, 637–648. [Google Scholar] [CrossRef]
- Sherman, F. Getting started with yeast. In Guide to Yeast Genetics and Molecular Biology; Gutherie, C., Fink, G.R., Eds.; Academic Press: Cambridge, MA, USA, 1991; Volume 194, pp. 3–21. [Google Scholar]
- Bahler, J.; Wu, J.Q.; Longtine, M.S.; Shah, N.G.; McKenzie, A., 3rd; Steever, A.B.; Wach, A.; Philippsen, P.; Pringle, J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 1998, 14, 943–951. [Google Scholar] [CrossRef]
- Ellison, M.J.; Hochstrasser, M. Epitope-tagged ubiquitin. J. Biol. Chem. 1991, 266, 21150–21157. [Google Scholar]
- Perlińska-Lenart, U.; Orłowski, J.; Laudy, E.A.; Zdebska, E.; Palamarczyk, G.; Kruszewska, J.S. Glycoprotein hypersecretion alters the cell wall in Trichoderma reesei strains expressing the Saccharomyces cerevisiae dolichylphosphate mannose synthase gene. Appl. Environm. Microbiol. 2006, 72, 7778–7784. [Google Scholar] [CrossRef][Green Version]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [PubMed]
- Kruszewska, J.; Messner, R.; Kubicek, C.P.; Palamarczyk, G. O-glycosylation of proteins by membrane fractions of Trichoderma reesei QM9414. J. Gen. Microbiol. 1989, 135, 301–307. [Google Scholar]
- Altschul, S.F.; Lipman, D.J. Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. USA 1990, 87, 5509–5513. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011, 39, W475–W478. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piłsyk, S.; Perlinska-Lenart, U.; Janik, A.; Gryz, E.; Ajchler-Adamska, M.; Kruszewska, J.S. Yil102c-A is a Functional Homologue of the DPMII Subunit of Dolichyl Phosphate Mannose Synthase in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2020, 21, 8938. https://doi.org/10.3390/ijms21238938
Piłsyk S, Perlinska-Lenart U, Janik A, Gryz E, Ajchler-Adamska M, Kruszewska JS. Yil102c-A is a Functional Homologue of the DPMII Subunit of Dolichyl Phosphate Mannose Synthase in Saccharomyces cerevisiae. International Journal of Molecular Sciences. 2020; 21(23):8938. https://doi.org/10.3390/ijms21238938
Chicago/Turabian StylePiłsyk, Sebastian, Urszula Perlinska-Lenart, Anna Janik, Elżbieta Gryz, Marta Ajchler-Adamska, and Joanna S. Kruszewska. 2020. "Yil102c-A is a Functional Homologue of the DPMII Subunit of Dolichyl Phosphate Mannose Synthase in Saccharomyces cerevisiae" International Journal of Molecular Sciences 21, no. 23: 8938. https://doi.org/10.3390/ijms21238938
APA StylePiłsyk, S., Perlinska-Lenart, U., Janik, A., Gryz, E., Ajchler-Adamska, M., & Kruszewska, J. S. (2020). Yil102c-A is a Functional Homologue of the DPMII Subunit of Dolichyl Phosphate Mannose Synthase in Saccharomyces cerevisiae. International Journal of Molecular Sciences, 21(23), 8938. https://doi.org/10.3390/ijms21238938




