Abstract
Muscular dystrophies are a group of more than 160 different human neuromuscular disorders characterized by a progressive deterioration of muscle mass and strength. The causes, symptoms, age of onset, severity, and progression vary depending on the exact time point of diagnosis and the entity. Congenital myopathies are rare muscle diseases mostly present at birth that result from genetic defects. There are no known cures for congenital myopathies; however, recent advances in gene therapy are promising tools in providing treatment. This review gives an overview of the mouse models used to investigate the most common muscular dystrophies and congenital myopathies with emphasis on their potentials and limitations in respect to human applications.
1. Introduction
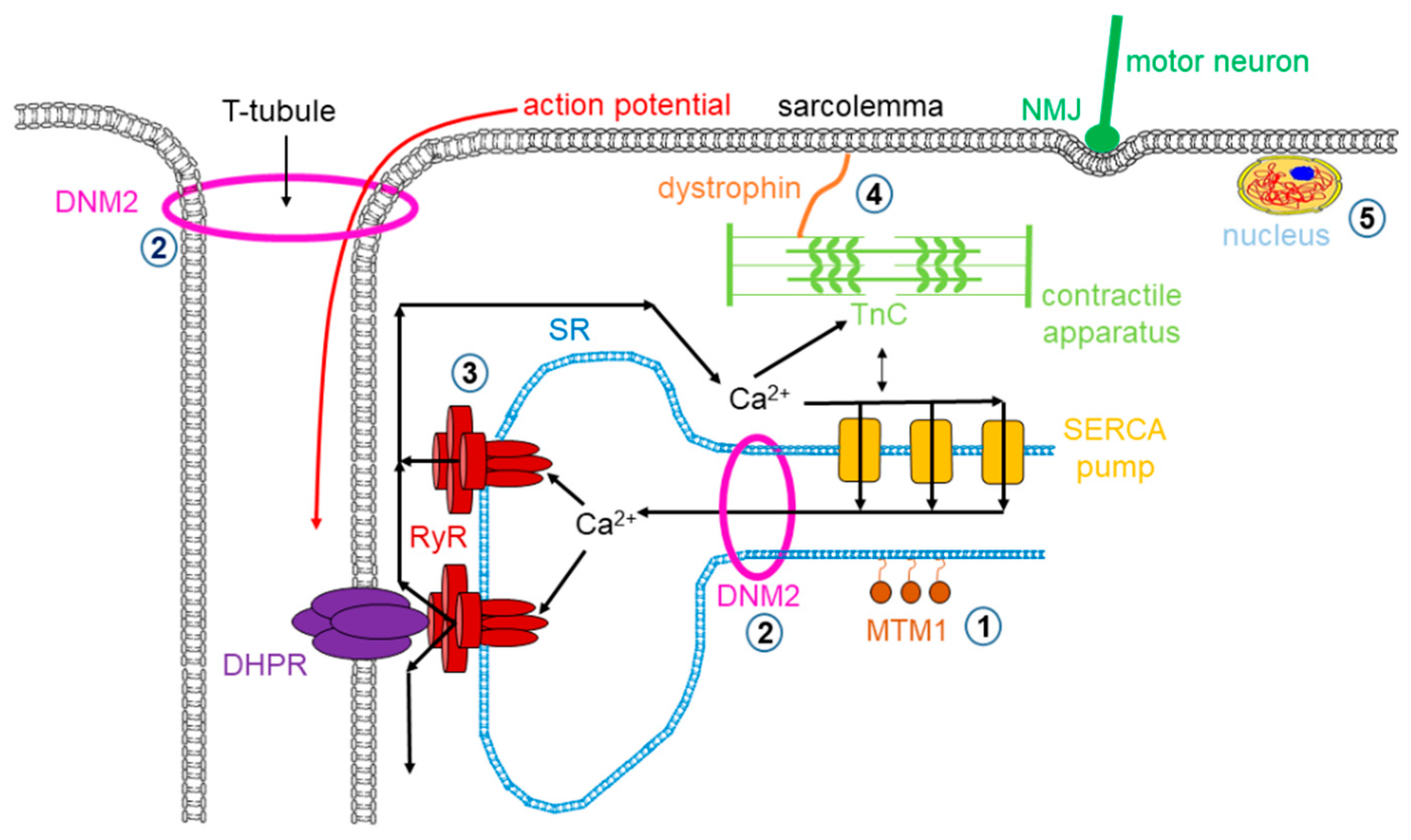
Muscle dystrophy is a muscle disease that leads to a progressive loss of muscle mass and a weakened musculoskeletal system in accordance with age of onset, severity, and the group of muscles affected. Dystrophy is an umbrella name that encompasses more than 30 genetic disorders that progress over time, leading to degeneration and weakness of the muscles. The phenotype of muscular dystrophy is an endpoint that arises from a disparate set of genetic and biochemically heterogeneous pathways. Genes associated with muscular dystrophies encode proteins of the plasma membrane (sarcolemma), terminal cisternae, extracellular matrix, and the sarcomere, as well as nuclear membrane components (Figure 1).
Figure 1.
Mutations in skeletal muscle causing muscle disorders. ➀. Altered lipid phosphatase activity of MTM1; ➁. defects in microtubule dynamics or vesicular traffic (DNM2); ➂. defective calcium release from the SR via RyRs; ➃. mutated or missing dystrophin; ➄. defects in alternative splicing due to MBLN1, CELF1 and DUX4 malfunction. (MTM1: myotubularin 1; DNM2: dynamin 2; SR: sarcoplasmic reticulum; RyR: ryanodine receptor; MBLN1: muscle blind-like; CELF: CUGBP/Elav-like factors; DUX4: homebox protein 4; DHPR: dihydropyridine receptor; NMJ: neuromuscular junction; SERCA: sarco(endo)plasmic reticulum calcium pump).
Myopathies are a diversified family of disorders characterized by pathological structure and/or the functioning of skeletal muscles. Inherited myopathies include a clinically, histopathologically, and genetically heterogeneous group of rare genetic muscle diseases that are characterized by architectural anomalies in the muscle fibers.
In the present review, our aim was to focus primarily on the mouse models used in preclinical studies of the amplest muscle disorders with emphasis on their potentials and limitations in respect to human applications. Here, we chose to elaborate exclusively on mouse models as they are easy to breed, maintain in large numbers, and genetically modify; however, one must note that there are various mammalian model systems available that are not addressed here due to length constraints. The importance of establishing similarities and differences between the human disease condition and murine animal models and the potential obstacles and limitations that arise from these differences when attempting to elucidate a prospective therapeutic strategy for muscle disorders is now generally accepted. Even though scientists have accesses to robust methods for the diagnosis and extensive characterization of disease progression along with a vast array of animal models that recapitulate well (but not entirely) muscle disorders, the available therapies are still palliative, minimizing symptoms rather than addressing the true cause of the disease.
2. Muscular Dystrophies
Muscular dystrophies (MDs) are a group of inherited disorders in which the voluntary muscles that control movement, in some instances the heart muscles and eventually the diaphragm, progressively weaken and lose their ability to maintain proper function. There are more than 30 types of MDs that vary in severity, symptoms, and causes. In recent years, the classification of MDs has been adjusted in order to correspond to the newly available information related to the primary protein dysfunctions and their localizations. As a consequence, by convention, the MDs had been classified according to the main clinical and biopsy findings, age of onset, and rate of progression into nine major forms: (1) Becker, (2) congenital, (3) Duchenne, (4) distal, (5) Emery–Dreifuss, (6) facioscapulohumeral, (7) limb–girdle, (8) myotonic, and (9) oculopharyngeal muscular dystrophy. In the present review, we tackle the most common forms of MDs in humans.
2.1. Dystrophinopathies
Dystrophinopathies cover a spectrum of X-linked muscle diseases ranging from mild to severe forms that include Duchenne muscular dystrophy (DMD), Becker muscular dystrophy (BMD), and DMD-associated dilated cardiomyopathy (DCM). DMD/BMD are neuromuscular genetic disorders characterized by progressive muscle degeneration, weakness, and wasting due to the alterations of a critical muscle protein called dystrophin, which is a relatively long (110 nm), rod-shaped intracellular protein localized at the cytoplasmic face of the sarcolemma in cardiac and skeletal muscles [1,2]. Dystrophin connects γ-actin of the subsarcolemmal cytoskeleton system to a complex of proteins in the surface membrane (dystrophin protein complex, DPC) and helps keep the muscle cell intact and orchestrates the transmission of force laterally across the muscle during contraction (see Figure 1). Growing evidence suggests that dystrophin also has a major role in regulating signaling pathways that activate nitric oxide (NO) production, Ca2+ entry, and the production of reactive oxygen species (ROS). The absence or reduced expression of dystrophin beside other members of the DPC complex causes dystrophinophathies.
DMD is the most common muscular dystrophy in children affecting primarily boys due to classical X-linked recessive genetics according to which males who carry the mutation express the disease while females are carriers. The incidence of DMD is approximately 1 in 3500 [3,4,5,6]. DMD symptom onset is in early childhood, usually between ages 2 and 6. The symptoms of DMD include decreased muscle size accompanied by progressive weakness and atrophy of skeletal and heart muscles. Early signs of DMD are muscle weakness of weight-bearing muscles and may include delayed ability to sit, stand, or walk, difficulties learning to speak, and general cognitive impairment. Most children with DMD use a wheelchair by their early teens. Heart and breathing problems also begin in the teen years, leading to serious life-threatening complications, and patients usually die in the third or fourth decade due to respiration or cardiac failure.
Frame-shift mutations or other genetic rearrangement in the dystrophin gene abolish protein expression that disturbs the connection between the cytoskeleton and the extracellular matrix, making muscle fibers more susceptible to contraction-induced membrane damage. As a result, the uncontrolled influx of calcium ions occurs inevitably, leading to progressive myofiber degeneration [7,8]. These pathologic processes are accompanied by chronic inflammation and fibrosis [9], as evidenced by macrophage infiltration [10]. In DMD, skeletal muscles active myofiber necrosis, and cellular infiltration can be histologically identified, furthermore regenerating myofibers containing centrally located nuclei, and a large variety of myofiber sizes are often detected. This phenotype is particularly pronounced in the diaphragm, which undergoes progressive degeneration and myofiber loss, causing an approximately 5-fold reduction in muscle isometric strength [11].
In-frame deletions often generate truncated dystrophin and result in BMD characterized by skeletal muscle weakness with milder symptoms and later onset that appear between the ages of 2 and 16 but in some cases as late as the twenties. Its incidence has been estimated to be between 1 in 30,000 male births [12,13].
Plentiful mouse models have been developed to better understand the basic molecular biology of DMD. Currently, there are nearly 60 different animal models for DMD, and the list keeps growing. For a comprehensive lineup, see Table 1 and the review by McGreevey and colleagues [14]. The TREAT-NMD Alliance (https://treat-nmd.org/research-overview/preclinical-research/) is an initiative to improve preclinical trial design and execution for the most common mouse models of DMD, spinal muscular atrophy (SMA), and congenital muscular dystrophy.

Table 1.
Mouse models of Duchenne muscular dystrophy.
With all its caveats, the most widely used animal model for DMD research is the C57BL/10ScSn-Dmdmdx/J (BL10-mdx; available from the Jackson laboratory, JL#001801) mouse in which the dystrophic phenotype arises because of a point mutation (C to T transition) in exon 23, which results in a stop codon and truncated dystrophin protein. This spontaneous mutation was discovered in the early 1980s in a colony of C57BL/10ScSn mice due to elevated serum creatine kinase (CK) and histological evidence of myopathy [15]. The mdx muscles seem more susceptible to contraction- and stretch-induced damage revealed as sarcolemmal tears [64]. Normal physiological control of calcium homeostasis is lost in mdx mice [65,66], and similar to the human condition, calcium levels are increased in myofibers isolated from mdx mice [67].
DMD is a multi-systemic condition affecting many parts of the body and resulting in atrophy of the skeletal, cardiac, and respiratory muscles. DMD disease progression in mdx mice has several distinctive phases. In the first 2 weeks, the mdx muscle is indistinguishable from that of normal mice. Between 3 and 6 weeks, it undergoes astonishing necrosis. Subsequently, the majority of skeletal muscle enters a relatively robust regeneration phase. As a hallmark of the disease, mdx limb muscles often become hypertrophic during this phase. The diaphragm is an exception, as it shows progressive deterioration, as seen in affected humans [11]. Severe dystrophic phenotypes, such as muscle wasting, scoliosis, and heart failure do not occur until mdx mice are 15 months or older [68,69,70,71,72,73]. Despite being deficient for dystrophin, mdx mice display overall minimal clinical symptoms; their lifespan is only reduced by ≈25% (vs. 75% decrease in humans) without obvious signs of dilated cardiomyopathy [14,37]. The robust skeletal muscle regeneration might explain somewhat the slowly progressive phenotype observed in mdx mice.
The mdx mouse has been crossed to several different genetic backgrounds, including the Albino, BALB/c, C3H, C57BL/6, DBA2, and FVB strains; several immune-deficient mdx strains were also engineered (see Table 1). Phenotypic variation has been observed in different backgrounds. Several other dystrophin-deficient lines (Dup2, DMD-null, Dp71-null, mdx52, and mdxβgeo) were also created using various genetic engineering techniques. The DMD-null mouse was created by deleting the entire DMD genomic region using the Cre-loxP technology [24] resulting in the ablation dystrophin isoforms expression in all tissues. Further models (mdxcv) were created by chemical mutagenesis programs by treating mice with N-ethyl-N-nitrosourea, a chemical mutagen, so that each strain carries a different point mutation [20,74]. By eliminating myogenic differentiation 1 (MyoD), a master myogenic regulator, from mdx mice, Megeney et al. obtained a MyoD/dystrophin double-mutant mouse that shows marked myopathy, dilated cardiomyopathy, and premature death [45,46,75]. Another similar approach was the generation of telomerase/mdx double-mutant mice (mTR/mdx) that show more severe muscle wasting and cardiac defects [44,76].
Two other proteins, utrophin and α7-integrin, fulfill the same function as dystrophin, and their relative expression is upregulated in mdx mice. The genetic elimination of utrophin, which is expressed along the sarcolemma in developing muscle, exhibits 80% homology and shares structural and functional motifs with dystrophin and α7-integrin; their deletion in mdx mice lead to the creation of utrophin/dystrophin and integrin/dystrophin double-knockout (dko) mice, respectively [33,34,35,46,77]. The dko mice show much more severe muscle disease symptoms (similar to or even worse than that of humans with DMD); however, they are difficult to generate and care for. Utrophin heterozygous mdx mice might represent an intermediate model between the extreme dko mice and mildly affected mdx mice [78,79].
Second mutations have been introduced to “humanize” mice (e.g., inactivation of cytidine monophosphate sialic acid hydrolase (Cmah)) and to mutate genes involved in cytoskeleton-ECM interactions (e.g. desmin and laminin); however, the introduction of a second mutation not present in human DMD turned out to produce a much more severe phenotype and complicated data interpretation [14,36,46,80].
To test if the “humanization” of telomere lengths could recapitulate the DMD disease phenotype, the mdx4cv/mTRG2 dko mice were generated, which seem to recapitulate the best of both the skeletal muscle and cardiovascular features of human DMD [44]. Nevertheless, there are still a few tenable therapies for DMD, so the need for appropriate mouse models more similar to the mdx model is emphasized. Even with an improved delivery of promising strategies such as gene editing or exon skipping, testing must be done in mice with the full spectrum of DMD pathology.
Dysferlinopathies are caused by the lack of functional dysferlin, which is a key protein involved in membrane repair processes causing Myoshi myopathy or dysferlin-related limb girdle muscular dystrophy (LGMD R2) [81]. The dysferlin-deficient mice (dysf-/-) replicate well human dysferlinopathies, showing similarities with the human condition although with milder histopathological aspects. Due to space restrictions, we did not further elaborate on these mouse models (for a comprehensive review, see [82] and more recently [83].
2.2. Myotonic Dystrophy
Myotonic dystrophy (DM) is an autosomal dominantly inherited disorder and the most prevalent form of muscular dystrophy in adulthood. Clinical characterization of the disease was done first by Steiner in 1909. DM is a complex genetic disease with diverse symptoms affecting multiple organs, such as skeletal muscle, cardiac muscle, the endocrine and gastrointestinal system, reproductive system, and central nervous system (CNS). Symptoms range from muscle weakness and wasting both in skeletal muscle and in heart, arrhythmias, or conduction abnormalities, disorders in the function of the neuromuscular junction, neurologic impairment such as excessive daytime sleepiness and motivation deficit, insulin resistance, cataracts, and male infertility. There are two major forms of the disease: myotonic dystrophy type I (DM1 or Steiner’s disease) and myotonic dystrophy type II (DM2 or proximal myotonic myopathy), which are associated to partially similar clinical appearances but distinct genetic defects [57].
Several hypotheses have been suggested to explain the complex symptoms of DM. The genetic background responsible for classic myotonic dystrophy documented by Steiner was discovered in 1992. An expansion of a CTG trinucleotide repeat in the 3′ untranslated region of the dystrophia myotonica protein kinase gene (DMPK) has been identified, which is a mutation that has been transcribed into RNA but not translated into protein [57]. Based on DMPK haploinsufficiency theory, the expanded repeats inhibit DMPK mRNA or protein production, which is in agreement with observation in DM1 patient muscle and cell cultures demonstrating a decreased expression of DMPK mRNA and protein [84]. On the other hand, DMPK-knockout mice did not display myotonia but rather mild myopathy [85]. Although DMPK haploinsufficiency alone is not sufficient to explain the features of DM1, the CTG repeats might influence the expression of neighboring genes as well. The haploinsufficiency of SIX5 and of other adjacent genes such as myotonic dystrophy gene with tryptophan and aspartic acid (WD) repeats, DMWD [86], and the FCGRT gene, encoding the Immunoglobulin G Fc Fragment Receptor and Transporter, has also been suggested to contribute to DM1 pathogenesis [87]. Indeed, Six5 knockout mice develop cataracts [88,89] but without any muscular deficiency. The next concept was the RNA gain-of-function hypothesis assuming that the mutant RNA transcribed from the expanded allele is capable of inducing symptoms of the disease. The HSALR transgenic mouse model (among others) confirms this theory [90]. In HSALR mice 250 CTG repeats were expressed in the 3′ end of the human skeletal α-actin gene that implied myotonia and muscle degeneration characteristic in DM1 without a multisystem phenotype.
DM2 was identified in 1998 with a different genetic mutation from that of DM1 [91]. In 2001, DM2 was reported as a result of CCTG repeats within intron 1 of the nucleic acid-binding protein (CNBP) gene (known also as zinc finger 9 gene, ZNF9) [92]. In both types of DM, there is a nucleotide repeat expansion; however, completely different genes are affected. Nevertheless, DM1 and DM2 have similar symptoms bringing up the idea of a common pathogenic mechanism. One candidate is a process through interaction with RNA-binding proteins. The transcripts with nucleotide repeat expansions can accumulate in the nucleus (see Figure 1) and form RNA aggregates/foci interfering with protein families such as the muscleblind-like (MBNL), CUGBP/Elav-like factors (CELF), and the RNA binding Fox (RBFOX) being the most important splicing regulators in skeletal muscle [93,94,95].
MBNL1 is sequestered on the expanded CUG repeats producing a loss of function, while CELF1 is upregulated due to the activation of protein kinase C, leading to its stabilization. These processes result in irregular splicing profiles of MBNL1- and CELF1-regulated transcripts in adult skeletal muscle and heart, or even during embryonic to adult switch in the splicing pattern. MBNL1 (Mbnl1ΔE3/ΔE3) knockout mice with targeted deletion of MBNL1 exon 3—where an RNA-binding motif is located—underpin this model, since these animals reproduce several features of DM including muscle, eye, and RNA splicing disorders (alternative splicing regulation in the brain is slightly affected, it depends mainly on the loss of MBNL2) [96]. Moreover, the adeno-associated virus-mediated overexpression of MBNL1 in HSALR mice is able to lessen the myotonia [24]. Verification also arises from the tissue-specific induction of CELF1 overexpression in adult mouse skeletal muscle, where muscle impairment detected in DM1 has been reproduced [97], while CELF1 overexpression in the heart leads to cardiac abnormalities similar to DM1 [98] (the role of CELF1 in DM2 is not clarified). These phenotypes were related to miss-splicing. These animal models imply that the overexpression of toxic CTG or CCTG repeats, depletion of MBNL1, or overexpression of CELF1 would all eventuate in similar splicing alterations, which initiate downstream signaling pathways resulting in the phenotype and/or molecular background of myotonic dystrophies.
The modified splicing apparatus can affect other genes in diverse signal transduction pathways leading to disrupted protein synthesis or the presence of different protein isoforms and the modified localization of proteins. Focusing on muscle, an aberrant regulation of RNA-binding proteins causes splicing alterations in the voltage-gated chloride channel 1 (CLCN1) transcript resulting in myotonia with delayed muscle relaxation in skeletal muscle cells [57,99]. Alternative splicing defects of BIN1 (bridging integrator 1), a lipid-binding protein responsible for the biogenesis of the transverse (T) tubules, has also been associated with muscle weakness. The inactive form of BIN1 causes damages of the excitation–contraction coupling (ECC) [100]. Another protein concerned is the calcium channel CaV1.1. Mis-splicing of the CACNA1S gene contributes to muscle weakness. Alternative splicing of the genes encoding ryanodine receptor 1 (RyR1) and SERCA1 expression are also altered modifying contractility of the muscles [101]. Altered splicing of several other genes encoding structural proteins has also been described in DM, as a few examples: DTNA (encoding dystrobrevin-α), MYOM1 (encoding myomesin1), NEB (encoding nebulin), TNNT3 (encoding fast troponin T3), DMD (encoding dystrophin), MTMR1 (encoding myotubularin-related protein 1), and CAPN3 (encoding intracellular protease Calpain 3). Atypical splicing of the insulin receptor (IR) may take part in the formation of insulin resistance. The genes mentioned above represent just a few examples of the more than 30 miss regulated splicing identified in DM patient’s tissue samples or of the more than 60 aberrant splicing described in mice tissues [102].
According to the previously listed conditions, molecular, and genetic variations, an “overall DM” model system should meet several requirements. At this moment, none of the available mouse models recapitulates all aspects of DM (for a comprehensive list of these mouse models, see Table 2). At the same time, the mouse models generated so far provided us with a significant tool in understanding the disease mechanism. There are different approaches in the various models. The inactivation of DM genes, the overexpression of toxic CTG/CTGG repeats, the induced alterations in splicing through MBNL1 inactivation, or CELF1 overexpression have resulted in a transgenic mouse model that was suitable for the examination of different aspects of the disease. Despite the growing number of already identified transcripts and the increased amount of data on altered pathways, the precise mechanism of the DMs is poorly understood. The available and the new mouse models to be established in the future can help scientists to discover a disease-modifying therapy.

Table 2.
Mouse models of myotonic dystrophies.
2.3. Facioscapulohumeral Dystrophy
Facioscapulohumeral dystrophy (FSHD) also known as Landouzy–Dejerine syndrome is the 3rd most common autosomal dominant form of muscular dystrophy after DMD and DM. Its prevalence is 1:8500 to 15,000, and males are more often symptomatic compared to females [123]. The disease tends to progress slowly with periods of rapid deterioration, and it affects the face, shoulder blades, and upper arms muscles, leading to difficulty chewing or swallowing and slanted shoulders. Currently, there is no cure for FSHD, as no pharmaceuticals have proven effective for alleviating the disease course. Prognosis is variable, but most people with the disease have a normal lifespan.
The FSHD is a very complex disease with primate-specific genetic and epigenetic components. It is caused by the epigenetic de-repression of the double homebox protein 4 (DUX4) retrogene on chromosome 4, in the 4q35 region that leads to a gain-of-function disease [124]. DUX4 is expressed in early human development, while in mature tissues, it is suppressed. In FSHD, DUX4 is inadequately turned off, which can be due to several different mutations. The mutation termed “D4Z4 contraction” defines the FSHD type 1 (FSHD1), making up 95% of all FSHD cases, whereas the disease caused by other mutations is classified as FSHD2 or contraction independent.
There are a few FSHD1 mouse models available for preclinical efficiency testing prior to human clinical trials, but due to the unusual nature of the disease locus, these models will not recapitulate accurately the genetic and pathophysiological spectrum of the human condition, and overall, these models remain sub-optimal in assessing therapeutic efficacy (Table 3). The most significant hurdle that is impossible to overcome is that the D4Z4 macrosatellite encoding the toxic DUX4 retrogene is specific to primates, which impedes the possibility of working with a natural model of the disease [125]. Several xenograft models were developed in which skeletal muscle tissue from FSHD patients or muscle precursor cells were transplanted into the mouse muscle (see Table 3). There are currently no mouse models for FSHD2.

Table 3.
Mouse models of facioscapulohumeral muscular dystrophy.
3. Myopathies
Myopathies contain a wide range of skeletal muscle disorders characterized by the irregular structure or muscle function. Myopathic patients show decreased physical activity without any disruption of sensory or autonomic function. Almost in all myopathies, symptoms affect proximal muscles bilaterally. Considering that a lot of myopathies cause progressive deterioration in the daily activity of patients, supportive therapy is often needed to overcome the physical and psychological effects of these diseases. Congenital myopathies are in a clinically, histopathologically, and genetically diverse group of rare hereditary skeletal muscle diseases that are characterized by structural abnormalities in the muscle fibers. They are subdivided into five subgroups: (1) congenital fiber-type disproportion myopathy; (2) centronuclear myopathies; (3) nemaline myopathies; (4) core myopathies; and (5) myosin storage (hyaline body) myopathy. The unusually broad genetic and clinical heterogeneity of these diseases stimulates for more expanded research on animal models. Furthermore, because of the lack of useful therapies, further studies are required to find candidates to cure patients with different types of myopathies.
3.1. Core Myophaties
Core myopathies are classified into congenital myopathies with variable clinical appearance, but they are usually associated with decreased muscle tone, pronounced muscle weakness and skeletal malformation; interestingly, symptoms do not or slowly progress with age.
3.1.1. Multi Minicore Disease
The neuromuscular illness multi-minicore disease (MmD) is characterized by multiple, amorphous cores seen on muscle biopsy and clinical features of a congenital myopathy. “Minicore” means that as a result of reduced or depleted oxidative activity, multiple core structures are visible in the muscle fiber [140,141]. MmD cores have a few or no mitochondria along with multiple internally placed nuclei, and type 1 fiber dominance is characteristic in the affected muscles (see Figure 1).
Several forms of MmD have been identified. Among others, there are (1) the classic (responsible for ≈75% of all cases), (2) the progressive, (3) the antenatal, and (4) the ophthalmoplegic as the most common forms. The classic MmD has typical orthopedic disorders such as kyphoscoliosis [142] with respiratory abnormalities [143,144]. Genetic heterogeneity is responsible for the clinical variability. Recessive mutation of the ryanodine receptor gene can give a wide range of clinical features consisting of external ophthalmoplegia and distal weakness [145]. Recessive mutations in SELENON gene encoding selenoprotein N (SEPN1) [146,147] result in the classic phenotype, with spinal rigidity, respiratory impairment, and early scoliosis as typical characteristics. A severe form of MmD with bad prognosis can develop as a result of mutations in MYH7 gene encoding myosin heavy chain beta (MHC-β) isoform with cardiac involvement [148]. Mutations in MEGF10 encoding multiple epidermal growth factor-like domains protein 10 causes MmD with serious weakness, respiratory impairment, and scoliosis [149]. Mutations in CACNA1S (Voltage-Gated Calcium Channel Subunit Alpha1 S) or in SCN4A (Voltage-Gated Sodium Channel Alpha Subunit 4) have also been associated with MmD [150]. Mutations in TTN gene encoding the titin sarcomere component affect the Ig domain of the proximal I-band and can cause a congenital titinopathy, which manifests as an early onset of MmD without affecting the heart [151]. Rare MmD diseases with atypical cores caused by the autosomal-dominant CCDC78 (coiled-coil domain containing 78) mutations are diagnosed also as a centronuclear myopathy [152]. Last but not least, MmD can also be caused by mutations in ACTA1 [153], ACTN2 [154], and FXR1 [155], encouraging the creation of different non-ryanodine core myopathy mouse models to better understand these rare muscle disorders. As a result, various transgenic animal models have been developed to identify the disease progression mechanisms for some mutations in order to explain genotype–phenotype correlations. Without claiming completeness, the most studied core myopathy mutations in mouse models causing MmD are summarized in Table 4.

Table 4.
Mouse models for multi-minicore disease and central core disease.
The most severe disorders of core myopathy are due to decreased RyR1 expression. In 1994, Takekura and colleagues developed a homozygous RyR1-/- mice from the RyR1-knockout RyR1skrrm1 and RyR1tmAlle strain, in which the anchoring cytoplasmic ‘foot’ domain is missing. Homozygous RyR1-/- mice die already perinatal due to respiratory failure and skeletal abnormalities. Interestingly, heterozygous RyR1skrrm1/+ and RyRtmAlle/+ mice do not have any apparent pathological irregularities [156,157]. This seems to correlate with human pathology; namely RyR1-associated core disorders caused by autosomal-dominant mutations or bi-allelic recessive RyR1 loss, rather than heterozygous loss [161].
In a recent study by Elbaz et al. (2019) a new mouse model was developed carrying heterozygous recessive RyR1 mutations isogenic with those identified in severely affected MmD patients (see Table 4). The authors came to the conclusion that the bi-allelic RyR1 p.A4329D mutation is responsible for a milder phenotype than its mono-allelic variant, causing changes in the biochemical properties and physiological functions, namely by focusing on the slowly twitch while sparing the fast twitch muscles [162].
As mentioned above, recessive mutations in SELENON gene encoding selenoprotein N (SEPN) result in an important part of the classical MmD. Since SEPN regulates Ca2+ levels in the ER/SR (sarcoplasmic reticulum) via SERCA2 activation [159], in Selenon1-/- mice, myofibers have excessive oxidative/nitrosative stress and abnormal Ca2+ handling because of the dysfunctional ER-stress response and inhibited SERCA2 activity. Moreover, according to the experiments of Castets et al. (2011), in Selenon1-/- mice, ER stress and high cytosolic Ca2+ levels caused impaired muscle regeneration deficiencies because of the reduced satellite cell numbers [163].
To the best of our knowledge, at the time of writing this review, there are no models for the extremely rare MYH7, for TTN and for ACTA1 mutations, which cause a subset of MmD with cardiac involvement [148,153,164].
3.1.2. Central Core Disease
Central core disease (CCD) is a subgroup of core myopathies, an autosomal inherited muscle disorder, characterized by core-like lesions in myofibers [165,166]. RyR1 mutations are found in the background of the majority of cases (Figure 1); these missense substitutions mostly are identified in three hotspots: (1) in the N-terminal between C35 and R614, (2) central between D2129 and R2458, and (3) C-terminal regions between I3916 and G4942 in the amino acid sequence of RyR1 [167]. Several studies proved that CCD mutations enhance the sensitivity of RyR1, resulting in a gain of function. This alteration influences the ECC and Ca2+ homeostasis via leaky RyR1 channels and altered EC uncoupling mechanisms. In the case of CCD, either mechanism could explain why muscle weakness was observed in patients with the disease [68,168]. A decreased threshold for channel activation and impaired coupling between DHPR and RyR1 may be responsible for the observed pathological symptoms.
Transgenic animals for CCD have been developed in order to study the altered protein function and pathological consequence. Malignant hyperthermia (MH) mutations in RyR1 might also cause CCD. The development of cores was formerly studied in two MH/CCD mouse lines: RyR1Y522S/+ and RyR1R163C/+. The Y522S and R163C mutations result in CCD in humans. Based on comprehensive studies, cores could not be detected in the RyR1R163C/+ mice [169,170]. However, RyR1Y522S/+ mouse showed progressive core development, and the localized regions containing damaged mitochondria were associated with disrupted sarcomeres and T-tubules, which could explain why cores were identified in patients with RyR1 mutations [171].
The RyR1T4826I/T4826I knock-in mouse showed elevated resting Ca2+ levels; the mutant aged male mice had core myopathy-like features in the soleus muscle, including Z-line disorientation and impaired sarcomere organization [172]. Zvaritch and colleagues (2007) employed a knock-in mouse line expressing the EC-uncoupling RyR1 mutation, I4895T, which corresponds to one of the most common I4898T CCD mutations in humans, resulting in an EC-uncoupled phenotype due to the impaired function of RyR1. The heterozygous mutation causes severe clinical appearance in human patients. Ile-4898 is located in a highly conserved GGIG4899 motif, forming the selectivity filter of the Ca2+ release channel. Based on in vitro functional studies, all amino acid substitutions at position 4898 negatively influences Ca2+ release channel conductance. RyR1I4895T/I4895T mice die perinatally because of paralyzed respiratory muscles [173]. Intact RyR1 Ca2+ release units and maintained SR Ca2+ content was detected in these mice; however, myofiber cultures showed disrupted RyR1-mediated Ca2+ release. Heterozygous RyR1I4898T/+ mice are born and despite exhibiting hypotonia and respiratory distress, they survive and do not show apparent skeletal deformities. On the other hand, mice have progressive congenital myopathy related to muscle weakness with age. These mice have been described to also develop cores, minicores, and rods [160,161]. The disease is slowly progressive, insufficient contractility was observed at the age of 2 months in fast and slow twitch muscles. Many mice show different degrees of impaired motor function at the age of 8 months. A combination of WT and mutant subunits randomly influences RyR1 functionality in a RyR1 tetramer that is proposed to contribute to phenotypic variability in RyR1-related disorders [161].
RyR1I4898T/+ mice (Table 4) are offered as the most appropriate genetically and phenotypically valid model of a RyR1-related congenital myopathy [160].
3.2. Centronuclear Myopathies
Centronuclear myopathy (CNM) is a general term for the family of rare genetic skeletal muscle diseases caused by a mutation in a definite gene. These disorders show muscle weakness ranging from mild to serious. Symptoms often start at birth in the serious forms of the myopathy, but they can also appear at any point during life, even though the onset in adulthood is rare. The name of CNM originates from the centrally located nucleus of the muscle fiber (normally located at the periphery). There are multiple genetic forms of CNM along with an X-linked form known as myotubular myopathy (XLMTM) caused by mutations in the myotubularin (MTM1) gene. There are a few autosomal forms as well, usually linked to three different genes: dynamin 2 (DNM2), bridging integrator 1 (BIN1), and RyR1 that have been identified to cause autosomal forms of CNM (Table 5).

Table 5.
Mouse models for centronuclear myopathies.
3.2.1. MTM1
X-linked myotubular myopathy (XLMTM) is a rare (1:50,000) congenital disease of skeletal muscle affecting only males [182]. XLMTM shares a general pathological trait in skeletal muscle, which is hypotrophic myofibers having centrally located nuclei [183,184,185]. The cause of the disease is mutations in the MTM1 gene encoding the universal phosphatase myotubularin, which plays a role in the phosphatidylinositol 3-kinase pathway to regulate intracellular vesicular transport and membrane trafficking [186,187,188,189,190]. MTM1 has effects on both types of phosphatidylinositol–phosphate [PtdIns(3,5)P2 or PtdIns(3)P] [186,191,192,193]. To date, more than 200 loss-of-function mutations of the MTM1 gene have been found in myotubular myopathy patients [194,195,196].
XLMTM patients are classified into three groups (mild, intermediate, or severe phenotype) based on remaining ventilator capacity [197]. Most of the patients have the severe phenotype showing serious hypotonia and a lack of spontaneous breathing at birth [198,199]. These newborns usually die within the first months of life. Long-term survivors in this group need continuous ventilation support [198,200]. Patients with mild and intermediate phenotypes can breathe independently at least a few hours daily [197,200].
Studies performed on mouse models lacking myotubularin have shown that skeletal muscle is the prime tissue affected in the pathogenesis of myotubular myopathy, and the protein is necessary for proper muscle development and the normal distribution of myofibrillar organelles. MTM1 knockout (KO) mice evolve centronuclear myopathy, starting at around one month after birth, showing dynamic muscle weakness that critically decreases lifespan to a maximum of 2–3 months [174].
Al Qusairi and colleagues reported that ECC of skeletal muscle is the major target of myotubular myopathy [201]. Using MTM1 KO mice, they showed that myopathic muscle fibers have abnormal longitudinally oriented T-tubules and a decreased number of triads (the structure formed by a T tubule with a sarcoplasmic reticulum (SR) on both sides; Figure 1). As a consequence, depolarizations evoked a calcium release from the SR that is strongly decreased while the SR calcium content and the removal of Ca2+ from the myoplasm were unaffected. These changes were accompanied with the 3-fold reduction in the level of RyR1. The authors hypothesized that the abnormal SR Ca2+ release causes the failure of muscle function in MTM1 KO mice.
Further investigation of MTM1 KO mice revealed the fact that the blockade of phosphatidylinositol 3-kinase (PtdIns 3-kinase) activity restores the defected Ca2+ release from the SR in isolated muscle fibers and increases the mobility and extends the lifespan of these mice [202]. The same group showed that the muscle fibers of MTM1 KO mice exhibit spontaneous elementary Ca2+ release events (sparks) with 30 times higher frequency than control fibers at resting conditions. These sparks occur at locations in the fibers where RyR1s lack the control of the voltage sensor DHPR because of the disrupted T-tubule membrane [203].
In the meantime, another murine model of XLMTM was developed by introducing a c.205C > T base change in MTM1 exon 4 [175]. These mice have a longer lifespan than MTM1 KO mice and show a milder MTM phenotype; however, they still present significant muscle weakness and atrophy. The genetic defect of these mice has a human counterpart.
To date, no effective treatment exists for XLMTM patients; however, other potentially usable therapeutic targets were suggested according to animal studies. The same authors who developed the MTM1 KO mouse proved that one intramuscular inoculation of myotubularin expressing adeno-associated virus (AAV) in MTM1 KO mice reverted the pathological phenotype in the injected muscle. The myotubularin replacement substantially corrected mitochondria and nuclei positioning in myofibers. These positive changes greatly increased muscle volume and force [204]. Later, they suggested enzyme replacement therapy, since myotubularin is a cytoplasmic enzyme, it does not have mannosylation, and it circulates in the blood. By delivering myotubularin in a fusion protein form, researchers were able to improve the structure and function of MTM1 KO muscle [205].
Dowling and colleagues showed that abnormal neuromuscular junction (NMJ) signal transmission is a crucial and likely manageable aspect of the MTM1 disease pathogenesis. An acetylcholinesterase inhibitor treatment significantly improved the fatigability and treadmill performance in MTM1 KO mice [206]. However, this type of treatment has not been tried in human therapy yet.
Inhibition of the phosphoinositide 3-kinase PIK3C2B improved the motor function and prolonged lifespan of the MTM1-deficient mice [207]. Another study on double KO mice demonstrated that the reduction of dynamin 2 (DNM2) expression in MTM1–/y mice was enough to decrease the early XLMTM lethality as well as most hallmarks of the disease; it also increased the lifespan of mice [176]. A systemic application of DNM2 antisense oligonucleotides in MTM1-KO mice was shown to prevent the development of muscle myopathy by reducing the DNM2 protein level [208]. In addition, this type of treatment in severely affected mice reversed the muscle pathology within 2 weeks.
Recently, two studies presented beneficial effects of long-term tamoxifen treatment, which increased the lifespan of MTM1 KO mice by improving the overall motor function [209,210]. Tamoxifen, a selective estrogen receptor modulator used in breast cancer therapy, eliminated successfully the molecular, histological, and functional hallmarks of XLMTM. Tamoxifen is the first long-term used and safe drug with a promising therapeutic potential for XLMTM patients.
A fresh study introduced a miR-199a-1-MTM1 dko mice model [177]. This research group demonstrated an upregulation in the expression of the intragenic microRNA miR-199a-1 and DNM2 as a host gene in XLCNM skeletal muscle. The dko mice displayed longer lifespans and improved muscle histology and strength. Their results suggest that this microRNA is a potential target in therapies to manage XLCNM.
3.2.2. Dynamin2
DNM2-related myopathies are the consequence of a missense mutation in the dynamin 2 (DNM2) gene, leading to an autosomal congenital dominant disease. Some cases of DNM2-related CNM may occur spontaneously (sporadically) with no previous family history of the disorder (i.e., new mutations) [211]. The encoded protein is universally expressed and associated to membrane trafficking and endocytosis, and it plays a role in centrosome cohesion and actin assembly. This large GTPase protein has five functionally distinguishable domains: the N-terminal domain is the GTPase; the middle domain (MD); the domain homologues to pleckstrin (PH); the GTPase effector domain; and it ends in an arginine and proline-rich domain at the C-terminal (PRD) [212]. The foremost found patients suffering in DNM2-related autosomal dominant CNM showed a slowly progressing muscle weakness, and the disease affected mainly distal muscles with onset in early adulthood. Shortly afterwards, four new mutations in the DNM2 gene were found in children presenting neonatal hypotonia manifested in weak suckling as well as lower limb and facial muscle weakness [213]. The number of DNM2 mutations increased when new missense mutations in the PH domain in the C-terminal region were found [36]. These genetic failures were associated with a very severe clinical phenotype present from infancy but also in adults. To date, a little over more than 100 human mutations have already been reported in DNM2 gene with different onset and phenotypes [214], and from these, 35 human mutations of the DNM2 gene have been identified associated to CNM. The only common characteristic is the morphological hallmarks: hypotrophic fibers with centralized nuclei. In the past few years, several new human mutations of dynamin2 were identified (i.e., p.G359D in the middle domain by Chen et al., 2018). It should be noted that patients with mutations of DNM2 often present a disorder of the peripheral nerve (Charcot–Marie–Tooth disease).
The first murine model of DNM2 mutation was generated by Durieux and colleagues (2010, KI-DNM2R465W). This knock-in (KI) heterozygous mouse model mimicked the most common mutation in the DNM2 human gene known at that time. KI mice showed progressively developing muscle weakness from 3 weeks of age, and atrophy developed at around 2 months of age. The membrane trafficking was severely altered, and a high level structural disorganization of muscle fibers (Figure 1) was accounted as the main mechanism of the disease [214]. A modified intracellular Ca2+ homeostasis was also reported: the resting intracellular [Ca2+], the sarcolemmal calcium permeability, and the releasable SR Ca2+ content was increased in muscle fibers from KI-DNM2R465W mice [215,216]. This mouse model was further investigated in detail, and the density of the calcium current through DHPRs and the rate of voltage-activated SR calcium release were found to be reduced. Fibers from the mutant KI mice produced elusive spontaneous Ca2+ release events under resting condition, which were not present in control animals [217].
Another research group developed a double KO mouse model lacking miR-133a-1 and miR-133a-2 showing progressive CNM [179]. The myopathy was accompanied with mitochondrial dysfunction, which can be attributed partly due to the upregulation of DNM2 [179]. These mice showed T-tubule disorganization, leading to impaired EC coupling functions.
As previously mentioned, the reduction of DNM2 expression improved the lifespan of XLMTM animals (see the subsection on MTM1) [176]. The same research group proved also that the DNM2 modulation can be used as a therapeutic application for patients with BIN1 defects. BIN1-related CNM is caused by mutations to the amphiphysin 2 (BIN1) genes and is inherited as an autosomal recessive condition. BIN1 and DNM2 are ubiquitous proteins involved in membrane remodeling. Cowling and colleagues generated BIN1 and DNM2 double KO mice that survived at least one and half years and had maintained muscle force and a normally organized structure of muscle fibers. The authors have hypothesized that DNM2 and BIN1 regulate muscle maturation and work through a common pathway, and they depicted BIN1 as negatively regulating DNM2. It was shown that lowering the level of DNM2 after birth could be sufficient to turn back the decline of muscle functions and progression of XLMTM [180].
If we take into account the fact that the total elimination of DNM2 is lethal at embryonic stages in mice, but the heterozygous KO mice are viable with unaffected muscle function, a potential therapeutic approach can be the reduction of the expression of the mutant allele without affecting the wild-type allele [176]. This concept was established by developing allele-specific siRNA sequences to specifically reduce the human and murine DNM2–mRNA containing the p.R465W mutation [218]. The technique resulted in a promising functional restoration of muscle function in mice.
Following the same train of thought, another research group used a single intramuscular injection of adeno-associated virus-shRNA against DNM2 in a knock-in mouse harboring the p.R465W mutation. Five weeks post injection, the fiber size distribution and muscle mass were improved [219]. The authors established also a systemic treatment by using intraperitoneal injections of antisense oligonucleotides against DNM2 weekly for 5 weeks. This treatment was similarly successful in minimizing pathological symptoms in DNM2R465W/+ mice.
CRISPR/Cas9 technology for genome editing is a recently emerging elegant technique. This was used in a study investigating an allele-specific correction or inactivation of a heterozygous mutation in the DNM2 gene. DNM2R465W/+ murine myoblasts showed less hallmarks of the disease after CRISPR/Cas9 correction of the dominant point mutation [220].
A fresh study targeting the S619L missense mutation successfully used DNM2 reduction with antisense oligonucleotides. Histological, force, and locomotor defects were partially or fully rescued just after 3 weeks of treatment in mice [181].
4. Malignant Hyperthermia
One of the most severe emergency situations that may occur in the operating room is caused by malignant hyperthermia (MH) susceptibility (MHS) of the patient. MH syndrome is an idiosyncratic reaction to volatile anesthetics such as halothane, isoflurane, desflurane, sevoflurane, and the depolarizing muscle relaxant succinylcholine. Symptoms include general muscle contracture, which leads to a rapid increase of the body temperature (1 °C/5 min), lactic acidosis, and hyperkalemia. These symptoms are likely to be fatal unless the patient is immediately treated with the muscle relaxant dantrolene and the body is cooled down [221,222].
The prevalence of MH crisis ranges from 1:5000 to 1:50,000 anesthesia. In the past 40 years—since dantrolene sodium must be available in all operating rooms—the mortality of MH dropped from over 80% to less than 5% [223,224]. Fortunately, animal models since the 1970s made great progress in understanding the pathophysiology and clinical manifestation of MH. The most widely used experimental animals were pigs from certain pig breeds, such as Pietrain, Landrace, Yorkshire, and Poland China, which were affected by MHS. The genetical cause that accounts for the syndrome was discovered by MacLennon’s group, who identified a common mutation (R615C) in the gene encoding the skeletal muscle type RyR (Figure 1), which was responsible for the porcine MHS phenotype, suggesting that a RyR1 mutation is linked to human MHS, too [225,226]. Since then, more than 200 MHS mutations have been identified in the human gene [227]. They are clustered in three mutation hotspots (N-terminal, aa 35–614; central, aa 2163–2458; C-terminal, aa 4550–4940) [228]. Mutations are believed to destabilize the closed-state conformation of the Ca2+ release channel (i.e., RyR1); therefore, all these MH-susceptible RyRs share a common, overactive, gain-of-function phenotype [229]. Hypersensitive gating has been demonstrated in response to major RyR agonists (such as caffeine and ATP), and most importantly for Ca2+ [230,231]. This feature creates a low, unsafe stimulation threshold for halothane, resulting in uncontrolled Ca2+ release and a consequent contracture of resting muscles when exposed to therapeutic concentrations of volatile anesthetics. In addition, diminished inhibition by Mg2+ has also been demonstrated, which may contribute to the pathogenesis, too [232,233]. Although the porcine model was extremely useful in preclinical studies of the RyR inhibitor dantrolene [233,234], this model has many disadvantages [235,236,237,238,239,240,241]. For example, R615C is a recessive mutation in pigs. Moreover, R615C represents only 2% of all human mutations. Apparently, the detailed understanding of the pathomechanism of MH required genetically modified mouse models covering all three hotspots. To date, four MHS RyR1 knock-in muse genotypes are available: Y524S, R163C, G2435R, and T4826I [169,172,230,242,243,244,245]. All these mice reproduce a typical MHS phenotype, displaying whole body contractions and elevated core temperatures in response to therapeutic concentrations of halothane or isoflurane. Similar to pigs, MHS mice exhibit heat-stress-induced MH episodes. Common features of muscle fibers or myotubes include elevated resting intracellular Ca2+ concentration and increased susceptibility to caffeine- and heat-induced contractures in vitro.
Y524S was the first murine model of human MHS (Y522S). Homozygous mice show severe skeletal and muscular abnormalities and die at the early stage of intrauterine life (17th day) or soon after birth. Heterozygous mice are viable and reproductive [169,242,243]. R163C (also R163C in human) is a dominant heterozygous mutation with no phenotype until exposed to a trigger agent. Homozygous are not viable at birth [230,244]. The mouse carrying the mutation G2435R is the model for the most common human MHS mutation G2434R. G2434R mutation has been found in 16% of families tested. Both homozygous and heterozygous mice are viable and fertile, although some homozygous males died spontaneously [245].
KI mice heterozygous or homozygous for T4826I RyR both survive, although homozygous animals were more sensitive to halothane and heat stress. In addition, males were reported to be more susceptible to MH trigger agents than females [172].
In summary, all four MHS RyR KI mouse strains summarized in Table 6 accurately mimic the patient’s phenotype and provide invaluable tools to investigate the detailed pathomechanism of MH and will be useful in the future to discover new, potential trigger agents [246,247].

Table 6.
Mouse models for malignant hyperthermia.
5. Conclusions
Although skeletal muscle disorders represent rare diseases and affect only a smaller portion of the population, the disability imposed on the affected person as well as the necessity to care for these individuals carries a significant economic burden for the society and the healthcare system. This justifies the need for appropriate animal models in developing new therapies and testing the proposed interventions to understand the nature of the given disorders.
The present review summarizes the most commonly used mouse models for a subset of muscle disorders with the highest prevalence in the human population; these mouse models provide important insights into causal gene relationships, have forged our understanding of molecular mechanisms and disease pathogenesis, and have driven progress toward a cure for muscle disorders. Most of the skeletal human diseases have the afferent mouse models, whose human relevance is still pending as these models are limited in their presentation of the human pathologies; however, there are promising results based on the recent advances achieved via elegant approaches such as gene editing or alternative splicing. Unfortunately, only a fraction of muscle disorders possess effective therapy at this moment, so finding and/or establishing an appropriate animal model is an important step toward the direction of understanding the complex pathomechanism of the disease and ultimately develop effective therapies.
For the existing mouse models summarized in Table 1, Table 2, Table 3, Table 4, Table 5 and Table 6, in most cases, therapies have been tested, although some of these have been not implemented in human treatment; nevertheless. They represent promising approaches that should eventually reach clinical trials. Yet, one has to acknowledge the obvious differences between the two species as well as the difficulties in targeting genetic therapies in human patients.
Taken altogether, we hope that this summary will help and encourage the scientific community to continue the search for proper animal models and therapies for muscle disorders.
Author Contributions
M.S., N.D., J.F., P.S., J.A., and B.D. wrote the manuscript. M.S., L.S., and Z.É.M. prepared the tables. M.S. and P.S. prepared the figures. M.S., B.D., and L.C. took care of conceptualization, editing and review of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The research was financed by the Thematic Excellence Programme of the Ministry for Innovation and Technology in Hungary (ED_18-1-2019-0028), within the framework of the Space Sciences thematic programme of the University of Debrecen. Furthermore, this work was supported by NKFIH PD-128370 grant of the Hungarian National Research, Development and Innovation Office (Hungary) and the GINOP-2.3.2-15-2016-00044 project.
Acknowledgments
The authors would like to thank Judit Boczán (University of Debrecen) for diligently reading the manuscript and providing feedback from the standpoint of a practicing neurologist.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hoffman, E.P.; Brown, R.H.; Kunkel, L.M. Dystrophin: The protein product of the duchenne muscular dystrophy locus. Cell 1987, 51, 919–928. [Google Scholar] [CrossRef]
- Ahn, A.H.; Kunkel, L.M. The structural and functional diversity of dystrophin. Nat. Genet. 1993, 3, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Emery, A.E.H.; Muntoni, F. Duchenne muscular dystrophy. Hum. Genet. 2004, 115, 529. [Google Scholar] [CrossRef]
- Ryder, S.; Leadley, R.M.; Armstrong, N.; Westwood, M.; De Kock, S.; Butt, T.; Jain, M.; Kleijnen, J. The burden, epidemiology, costs and treatment for Duchenne muscular dystrophy: An evidence review. Orphanet J. Rare Dis. 2017, 12, 1–21. [Google Scholar] [CrossRef]
- Moat, S.J.; Bradley, D.M.; Salmon, R.; Clarke, A.; Hartley, L. Newborn bloodspot screening for Duchenne Muscular Dystrophy: 21 years experience in Wales (UK). Eur. J. Hum. Genet. 2013, 21, 1049–1053. [Google Scholar] [CrossRef]
- Romitti, P.A.; Zhu, Y.; Puzhankara, S.; James, K.A.; Nabukera, S.K.; Zamba, G.K.D.; Ciafaloni, E.; Cunniff, C.; Druschel, C.M.; Mathews, K.D.; et al. Prevalence of Duchenne and Becker Muscular Dystrophies in the United States. Pediatrics 2015, 135, 513–521. [Google Scholar] [CrossRef]
- Gissel, H. The role of Ca2+ in muscle cell damage. Ann. N. Y. Acad. Sci. 2005, 1066, 166–180. [Google Scholar] [CrossRef]
- Klingler, W.; Jurkat-Rott, K.; Lehmann-Horn, F.; Schleip, R. The role of fibrosis in Duchenne muscular dystrophy. Acta Myol. 2012, 31, 184–195. [Google Scholar]
- Wallace, G.Q.; McNally, E.M. Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies. Annu. Rev. Physiol. 2009, 71, 37–57. [Google Scholar] [CrossRef]
- Grounds, M.D.; Radley, H.G.; Lynch, G.S.; Nagaraju, K.; De Luca, A. Towards developing standard operating procedures for pre-clinical testing in the mdx mouse model of Duchenne muscular dystrophy. Neurobiol. Dis. 2008, 31, 1–19. [Google Scholar] [CrossRef]
- Stedman, H.H.; Sweeney, H.L.; Shrager, J.B.; Maguire, H.C.; Panettieri, R.A.; Petrof, B.; Narusawa, M.; Leferovich, J.M.; Sladky, J.T.; Kelly, A.M. The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy. Nature 1991, 352, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Beggs, A.H.; Hoffman, E.P.; Snyder, J.R.; Arahata, K.; Specht, L.; Shapiro, F.; Angelini, C.; Sugita, H.; Kunkel, L.M. Exploring the molecular basis for variability among patients with Becker muscular dystrophy: Dystrophin gene and protein studies. Am. J. Hum. Genet. 1991, 49, 54–67. [Google Scholar] [PubMed]
- Deburgrave, N.; Daoud, F.; Llense, S.; Barbot, J.C.; Récan, D.; Peccate, C.; Burghes, A.H.M.; Béroud, C.; Garcia, L.; Kaplan, J.-C.; et al. Protein- and mRNA-Based Phenotype–Genotype Correlations in DMD/BMD With Point Mutations and Molecular Basis for BMD With Nonsense and Frameshift Mutations in the DMD Gene. Hum Mutat. 2007, 28, 183–195. [Google Scholar] [CrossRef] [PubMed]
- McGreevy, J.W.; Hakim, C.H.; McIntosh, M.A.; Duan, D. Animal models of Duchenne muscular dystrophy: From basic mechanisms to gene therapy. DMM Dis. Model. Mech. 2015, 8, 195–213. [Google Scholar] [CrossRef]
- Bulfield, G.; Siller, W.G.; Wight, P.A.L.; Moore, K.J. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc. Natl. Acad. Sci. USA 1984, 81, 1189–1192. [Google Scholar] [CrossRef]
- Krivov, L.I.; Stenina, M.A.; Yarygin, V.N.; Polyakov, A.V.; Savchuk, V.I.; Obrubov, S.A.; Komarova, N.V. A new Genetic variant of MDX mice: Study of the phenotype. Bull. Exp. Biol. Med. 2009, 147, 625–629. [Google Scholar] [CrossRef]
- Schmidt, W.M.; Uddin, M.H.; Dysek, S.; Moser-Thier, K.; Pirker, C.; Höger, H.; Ambros, I.M.; Ambros, P.F.; Berger, W.; Bittner, R.E. DNA damage, somatic aneuploidy, and malignant sarcoma susceptibility in muscular dystrophies. PLoS Genet. 2011, 7, 1–17. [Google Scholar] [CrossRef]
- Fukada, S.I.; Morikawa, D.; Yamamoto, Y.; Yoshida, T.; Sumie, N.; Yamaguchi, M.; Ito, T.; Miyagoe-Suzuki, Y.; Takeda, S.; Tsujikawa, K.; et al. Genetic background affects properties of satellite cells and mdx phenotypes. Am. J. Pathol. 2010, 176, 2414–2424. [Google Scholar] [CrossRef]
- Wasala, N.B.; Zhang, K.; Wasala, L.P.; Hakim, C.H.; Duan, D. The FVB Background Does Not Dramatically Alter the Dystrophic Phenotype of Mdx Mice. PloS Curr. 2015, 7, 1–17. [Google Scholar] [CrossRef]
- Chapman, V.M.; Miller, D.R.; Armstrong, D.; Caskey, C.T. Recovery of induced mutations for X chromosome-linked muscular dystrophy in mice. Proc. Natl. Acad. Sci. USA 1989, 86, 1292–1296. [Google Scholar] [CrossRef]
- Aigner, B.; Rathkolb, B.; Klaften, M.; Sedlmeier, R.; Klempt, M.; Wagner, S.; Michel, D.; Mayer, U.; Klopstock, T.; Hrabé De Angelis, M.; et al. Generation of N-ethyl-N-nitrosourea-induced mouse mutants with deviations in plasma enzyme activities as novel organ-specific disease models. Exp. Physiol. 2009, 94, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Araki, E.; Nakamura, K.; Nakao, K.; Kameya, S.; Kobayashi, O.; Nonaka, I.; Kobayashi, T.; Katsuki, M. Targeted disruption of exon 52 in the mouse dystrophin gene induced muscle degeneration similar to that observed in duchenne muscular dystrophy. Biochem. Biophys. Res. Commun. 1997, 238, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Wertz, K.; Füchtbauer, E.M. Dmd(mdx-βgeo): A new allele for the mouse dystrophin gene. Dev. Dyn. 1998, 212, 229–241. [Google Scholar] [CrossRef]
- Kudoh, H.; Ikeda, H.; Kakitani, M.; Ueda, A.; Hayasaka, M.; Tomizuka, K.; Hanaoka, K. A new model mouse for Duchenne muscular dystrophy produced by 2.4 Mb deletion of dystrophin gene using Cre-loxP recombination system. Biochem. Biophys. Res. Commun. 2005, 328, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Sarig, R.; Mezger-Lallemand, V.; Gitelman, I.; Davis, C.; Fuchs, O.; Yaffe, D.; Nudel, U. Targeted inactivation of Dp71, the major non-muscle product of the DMD gene: Differential activity of the Dp71 promoter during development. Hum. Mol. Genet. 1999, 8, 1–10. [Google Scholar] [CrossRef]
- Fort, P.E.; Darche, M.; Sahel, J.A.; Rendon, A.; Tadayoni, R. Lack of dystrophin protein Dp71 results in progressive cataract formation due to loss of fiber cell organization. Mol. Vis. 2014, 20, 1480–1490. [Google Scholar]
- Wein, N.; Vulin, A.; Falzarano, M.S.; Szigyarto, C.A.K.; Maiti, B.; Findlay, A.; Heller, K.N.; Uhlén, M.; Bakthavachalu, B.; Messina, S.; et al. Translation from a DMD exon 5 IRES results in a functional dystrophin isoform that attenuates dystrophinopathy in humans and mice. Nat. Med. 2014, 20, 992–1000. [Google Scholar] [CrossRef]
- Arpke, R.W.; Darabi, R.; Mader, T.L.; Zhang, Y.; Toyama, A.; Lonetree, C.L.; Nash, N.; Lowe, D.A.; Perlingeiro, R.C.R.; Kyba, M. A new immuno-, dystrophin-deficient model, the NSG-mdx4Cv mouse, provides evidence for functional improvement following allogeneic satellite cell transplantation. Stem Cells 2013, 31, 1611–1620. [Google Scholar] [CrossRef]
- Bencze, M.; Negroni, E.; Vallese, D.; Yacoubyoussef, H.; Chaouch, S.; Wolff, A.; Aamiri, A.; Di Santo, J.P.; Chazaud, B.; Butler-Browne, G.; et al. Proinflammatory macrophages enhance the regenerative capacity of human myoblasts by modifying their kinetics of proliferation and differentiation. Mol. Ther. 2012, 20, 2168–2179. [Google Scholar] [CrossRef]
- Vallese, D.; Negroni, E.; Duguez, S.; Ferry, A.; Trollet, C.; Aamiri, A.; Vosshenrich, C.A.; Füchtbauer, E.M.; Di Santo, J.P.; Vitiello, L.; et al. The Rag2 - Il2rb - Dmd - Mouse: A novel dystrophic and immunodeficient model to assess innovating therapeutic strategies for muscular dystrophies. Mol. Ther. 2013, 21, 1950–1957. [Google Scholar] [CrossRef]
- Farini, A.; Meregalli, M.; Belicchi, M.; Battistelli, M.; Parolini, D.; D’Antona, G.; Gavina, M.; Ottoboni, L.; Constantin, G.; Bottinelli, R.; et al. T and B lymphocyte depletion has a marked effect on the fibrosis of dystrophic skeletal muscles in the scid/mdx mouse. J. Pathol. 2007, 213, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Walsh, S.; Nygren, J.; Pontén, A.; Jovinge, S. Myogenic reprogramming of bone marrow derived cells in a W41Dmdmdx deficient mouse model. PLoS ONE 2011, 6, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Rooney, J.E.; Welser, J.V.; Dechert, M.A.; Flintoff-Dye, N.L.; Kaufman, S.J.; Burkin, D.J. Severe muscular dystrophy in mice that lack dystrophin and α7 integrin. J. Cell Sci. 2006, 119, 2185–2195. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Willem, M.; Werner, A.; Raivich, G.; Emerson, M.; Neyses, L.; Mayer, U. Absence of α7 integrin in dystrophin-deficient mice causes a myopathy similar to Duchenne muscular dystrophy. Hum. Mol. Genet. 2006, 15, 989–998. [Google Scholar] [CrossRef]
- Grady, R.M.; Teng, H.; Nichol, M.C.; Cunningham, J.C.; Wilkinson, R.S.; Sanest, J.R. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: A model for Duchenne muscular dystrophy. Cell 1997, 90, 729–738. [Google Scholar] [CrossRef]
- Susman, R.D.; Quijano-Roy, S.; Yang, N.; Webster, R.; Clarke, N.F.; Dowling, J.; Kennerson, M.; Nicholson, G.; Biancalana, V.; Ilkovski, B.; et al. Expanding the clinical, pathological and MRI phenotype of DNM2-related centronuclear myopathy. Neuromuscul. Disord. 2010, 20, 229–237. [Google Scholar] [CrossRef]
- Li, D.; Long, C.; Yue, Y.; Duan, D. Sub-physiological sarcoglycan expression contributes to compensatory muscle protection in mdx mice. Hum. Mol. Genet. 2009, 18, 1209–1220. [Google Scholar] [CrossRef]
- Banks, G.B.; Combs, A.C.; Odom, G.L.; Bloch, R.J.; Chamberlain, J.S. Muscle Structure Influences Utrophin Expression in mdx Mice. PLoS Genet. 2014, 10, 1–16. [Google Scholar] [CrossRef]
- Martins, P.C.M.; Ayub-Guerrieri, D.; Martins-Bach, A.B.; Onofre-Oliveira, P.; Malheiros, J.M.; Tannus, A.; De Sousa, P.L.; Carlier, P.G.; Vainzof, M. Dmdmdx/Largemyd: A new mouse model of neuromuscular diseases useful for studying physiopathological mechanisms and testing therapies. DMM Dis. Model. Mech. 2013, 6, 1167–1174. [Google Scholar] [CrossRef]
- Nishimura, D.; Sakai, H.; Sato, T.; Sato, F.; Nishimura, S.; Toyama-Sorimachi, N.; Bartsch, J.W.; Sehara-Fujisawa, A. Roles of ADAM8 in elimination of injured muscle fibers prior to skeletal muscle regeneration. Mech. Dev. 2015, 135, 58–67. [Google Scholar] [CrossRef]
- Han, R.; Rader, E.P.; Levy, J.R.; Bansal, D.; Campbell, K.P. Dystrophin deficiency exacerbates skeletal muscle pathology in dysferlin-null mice. Skelet. Muscle 2011, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hosur, V.; Kavirayani, A.; Riefler, J.; Carney, L.M.B.; Lyons, B.; Gott, B.; Cox, G.A.; Shultz, L.D. Dystrophin and dysferlin double mutant mice: A novel model for rhabdomyosarcoma. Cancer Genet. 2012, 205, 232–241. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nitahara-Kasahara, Y.; Hayashita-Kinoh, H.; Chiyo, T.; Nishiyama, A.; Okada, H.; Takeda, S.; Okada, T. Dystrophic mdx mice develop severe cardiac and respiratory dysfunction following genetic ablation of the anti-inflammatory cytokine IL-10. Hum. Mol. Genet. 2014, 23, 3990–4000. [Google Scholar] [CrossRef] [PubMed]
- Sacco, A.; Mourkioti, F.; Tran, R.; Choi, J.; Llewellyn, M.; Kraft, P.; Shkreli, M.; Delp, S.; Pomerantz, J.H.; Artandi, S.E.; et al. Short telemeres and stem cell exhaustion model in mdx mice. Cell 2010, 143, 1059–1071. [Google Scholar] [CrossRef]
- Megeney, L.A.; Kablar, B.; Garrett, K.; Anderson, J.E.; Rudnicki, M.A. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996, 10, 1173–1183. [Google Scholar] [CrossRef]
- Deconinck, A.E.; Rafael, J.A.; Skinner, J.A.; Brown, S.C.; Potter, A.C.; Metzinger, L.; Watt, D.J.; Dickson, J.G.; Tinsley, J.M.; Davies, K.E. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell 1997, 90, 717–727. [Google Scholar] [CrossRef]
- Ardite, E.; Perdiguero, E.; Vidal, B.; Gutarra, S.; Serrano, A.L.; Muñoz-Cánoves, P. PAI-1-regulated miR-21 defines a novel age-associated fibrogenic pathway in muscular dystrophy. J. Cell Biol. 2012, 196, 163–175. [Google Scholar] [CrossRef]
- Cox, G.A.; Cole, N.M.; Matsumura, K.; Phelps, S.F.; Hauschka, S.D.; Campbell, K.P.; Faulkner, J.A.; Chamberlain, J.S. Overexpression of dystrophin in transgenic mdx mice eliminates dystrophic symptoms without toxicity. Nature 1993, 364, 725–729. [Google Scholar] [CrossRef]
- Phelps, S.F.; Hauser, M.A.; Cole, N.M.; Rafael, J.A.; Hinkle, R.T.; Faulkner, J.A.; Chamberlain, J.S. Expression of full-length and truncated dystrophin mini-genes in transgenic mdx mice. Hum. Mol. Genet. 1995, 4, 1251–1258. [Google Scholar] [CrossRef]
- Dunckley, M.G.; Wells, D.J.; Walsh, F.S.; Dickson, G. Direct retroviral-mediated transfer of a dystrophin minigene into mdx mouse muscle in vivo. Hum. Mol. Genet. 1993, 2, 717–723. [Google Scholar] [CrossRef]
- Cox, G.A.; Sunada, Y.; Campbell, K.P.; Chamberlain, J.S. Dp71 can restore the dystrophin-associated glycoprotein complex in muscle but fails to prevent dystrophy. Nat. Genet. 1994, 8, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, D.S.; Sunada, Y.; Campbell, K.P.; Yaffe, D.; Nudel, U. Exogenous Dp71 restores the levels of dystrophin associated proteins but does not alleviate muscle damage in mdx mice. Nat. Genet. 1994, 8, 340–344. [Google Scholar] [CrossRef]
- Judge, L.M.; Arnett, A.L.H.; Banks, G.B.; Chamberlain, J.S. Expression of the dystrophin isoform Dp116 preserves functional muscle mass and extends lifespan without preventing dystrophy in severely dystrophic mice. Hum. Mol. Genet. 2011, 20, 4978–4990. [Google Scholar] [CrossRef] [PubMed]
- Judge, L.M.; Haraguchi, M.; Chamberlain, J.S. Dissecting the signalling and mechanical functions of the dystrophin-glycoprotein complex. J. Cell Sci. 2006, 119, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Warner, L.E.; DelloRusso, C.T.; Crawford, R.W.; Rybakova, I.N.; Patel, J.R.; Ervasti, J.M.; Chamberlain, J.S. Expression of Dp260 in muscle tethers the actin cytoskeleton to the dystrophin-glycoprotein complex and partially prevents dystrophy. Hum. Mol. Genet. 2002, 11, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Gaedigk, R.; Law, D.J.; Fitzgerald-Gustafson, K.M.; McNulty, S.G.; Nsumu, N.N.; Modrcin, A.C.; Rinaldi, R.J.; Pinson, D.; Fowler, S.C.; Bilgen, M.; et al. Improvement in survival and muscle function in an mdx/utrn-/- double mutant mouse using a human retinal dystrophin transgene. Neuromuscul. Disord. 2006, 16, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Charlet-B, N.; Savkur, R.S.; Singh, G.; Philips, A.V.; Grice, E.A.; Cooper, T.A. Loss of the Muscle-Specific Chloride Channel in Type 1 Myotonic Dystrophy Due to Misregulated Alternative Splicing several lines of evidence indicate that a gain of function for RNA CUG)n. Mol. Cell 2002, 10, 45–53. [Google Scholar] [CrossRef]
- Hakim, C.H.; Duan, D. Truncated dystrophins reduce muscle stiffness in the extensor digitorum longus muscle of mdx mice. J. Appl. Physiol. 2013, 114, 482–489. [Google Scholar] [CrossRef][Green Version]
- Wang, B.; Li, J.; Fu, F.H.; Chen, C.; Zhu, X.; Zhou, L.; Jiang, X.; Xiao, X. Construction and analysis of compact muscle-specific promoters for AAV vectors. Gene Ther. 2008, 15, 1489–1499. [Google Scholar] [CrossRef]
- Ferrer, A.; Foster, H.; Wells, K.E.; Dickson, G.; Wells, D.J. Long-term expression of full-length human dystrophin in transgenic mdx mice expressing internally deleted human dystrophins. Gene Ther. 2004, 11, 884–893. [Google Scholar] [CrossRef]
- Tinsley, J.; Deconinck, N.; Fisher, R.; Kahn, D.; Phelps, S.; Gillis, J.M.; Davies, K. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat. Med. 1998, 4, 1441–1444. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Bareja, A.; Judge, L.; Yue, Y.; Lai, Y.; Fairclough, R.; Davies, K.E.; Chamberlain, J.S.; Duan, D. Sarcolemmal nNOS anchoring reveals a qualitative difference between dystrophin and utrophin. J. Cell Sci. 2010, 123, 2008–2013. [Google Scholar] [CrossRef] [PubMed]
- Gawlik, K.I.; Oliveira, B.M.; Durbeej, M. Transgenic expression of laminin α1 chain does not prevent muscle disease in the mdx mouse model for Duchenne muscular dystrophy. Am. J. Pathol. 2011, 178, 1728–1737. [Google Scholar] [CrossRef] [PubMed]
- Moens, P.; Baatsen, P.H.W.W.; Maréchal, G. Increased susceptibility of EDL muscles from mdx mice to damage induced by contractions with stretch. J. Muscle Res. Cell Motil. 1993, 14, 446–451. [Google Scholar] [CrossRef]
- Fong, P.; Turner, P.; Denetclaw, W.; Steinhardt, R. Increased activity of calcium leak channels in myotubes of Duchenne human and mdx mouse origin. Science 1990, 250, 673–676. [Google Scholar] [CrossRef]
- Turner, P.R.; Schultz, R.; Ganguly, B.; Steinhardt, R.A. Proteolysis results in altered leak channel kinetics and elevated free calcium in mdx muscle. J. Membr. Biol. 1993, 133, 243–251. [Google Scholar] [CrossRef]
- Vandebrouck, C.; Constantin, B.; Raymond, G.; Cognard, C.; Duport, G. Normal calcium homeostasis in dystrophin-expressing facioscapulohumeral muscular dystrophy myotubes. Neuromuscul. Disord. 2002, 12, 266–272. [Google Scholar] [CrossRef]
- Avila, G.; O’Brien, J.J.; Dirksen, R.T. Excitation - Contraction uncoupling by a human central core disease mutation in the ryanodine receptor. Proc. Natl. Acad. Sci. USA 2001, 98, 4215–4220. [Google Scholar] [CrossRef]
- Bostick, B.; Yue, Y.; Long, C.; Duan, D. Prevention of dystrophin-deficient cardiomyopathy in twenty-one-month-old carrier mice by mosaic dystrophin expression or complementary dystrophin/utrophin expression. Circ. Res. 2008, 102, 121–130. [Google Scholar] [CrossRef]
- Bostick, B.; Yue, Y.; Long, C.; Marschalk, N.; Fine, D.M.; Chen, J.; Duan, D. Cardiac expression of a mini-dystrophin that normalizes skeletal muscle force only partially restores heart function in aged Mdx mice. Mol. Ther. 2009, 17, 253–261. [Google Scholar] [CrossRef]
- Hakim, C.H.; Grange, R.W.; Duan, D. The passive mechanical properties of the extensor digitorum longus muscle are compromised in 2-to 20-mo-old mdx mice. J. Appl. Physiol. 2011, 110, 1656–1663. [Google Scholar] [CrossRef] [PubMed]
- Lefaucheur, J.P.; Pastoret, C.; Sebille, A. Phenotype of dystrophinopathy in old MDX mice. Anat. Rec. 1995, 242, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Pastoret, C.; Sebille, A. Mdx Mice Show Progressive Weakness and Muscle Deterioration With Age. J. Neurol. Sci. 1995, 129, 97–105. [Google Scholar] [CrossRef]
- Im, W.B.; Phelps, S.F.; Copen, E.H.; Adams, E.G.; Slightom, J.L.; Chamberlain, J.S. Differential expression of dystrophin isoforms in strains of mdx mice with different mutations. Hum. Mol. Genet. 1996, 5, 1149–1153. [Google Scholar] [CrossRef]
- Megeney, L.A.; Kablar, B.; Perry, R.L.S.; Ying, C.; May, L.; Rudnicki, M.A. Severe cardiomyopathy in mice lacking dystrophin and MyoD. Proc. Natl. Acad. Sci. USA 1999, 96, 220–225. [Google Scholar] [CrossRef]
- Mourkioti, F.; Kustan, J.; Kraft, P.; Day, J.W.; Zhao, M.-M.; Kost-Alimova, M.; Protopopov, A.; DePinho, R.A.; Bernstein, D.; Meeker, A.K.; et al. Role of Telomere Dysfunction in Cardiac Failure in Duchenne Muscular Dystrophy. Nat. Cell Biol. 2013, 15, 895–904. [Google Scholar] [CrossRef]
- Matsumura, C.Y.; Taniguti, A.P.T.; Pertille, A.; Neto, H.S.; Marques, M.J. Stretch-activated calcium channel protein TRPC1 is correlated with the different degrees of the dystrophic phenotype in mdx mice. Am. J. Physiol.-Cell Physiol. 2011, 301, 1344–1350. [Google Scholar] [CrossRef]
- Rafael-Fortney, J.A.; Chimanji, N.S.; Schill, K.E.; Martin, C.D.; Murray, J.D.; Ganguly, R.; Stangland, J.E.; Tran, T.; Xu, Y.; Canan, B.D.; et al. Early Treatment with Lisinopril and Spironolactone Preserves Cardiac and Skeletal Muscle in Duchenne Muscular Dystrophy Mice. Circulation 2011, 124, 582–588. [Google Scholar] [CrossRef]
- van Putten, M.; Kumar, D.; Hulsker, M.; Hoogaars, W.M.H.; Plomp, J.J.; van Opstal, A.; van Iterson, M.; Admiraal, P.; van Ommen, G.J.B.; AC’t Hoen, P.; et al. Comparison of skeletal muscle pathology and motor function of dystrophin and utrophin deficient mouse strains. Neuromuscul. Disord. 2012, 22, 406–417. [Google Scholar] [CrossRef]
- Selsby, J.T.; Ross, J.W.; Nonneman, D.; Hollinger, K. Porcine models of muscular dystrophy. ILAR J. 2015, 56, 116–126. [Google Scholar] [CrossRef]
- Straub, V.; Murphy, A.; Udd, B. 229th ENMC international workshop: Limb girdle muscular dystrophies–Nomenclature and reformed classification Naarden, The Netherlands, 17–19 March 2017. Neuromuscul. Disord. 2018, 28, 702–710. [Google Scholar] [CrossRef]
- Hornsey, M.A.; Laval, S.H.; Barresi, R.; Lochmüller, H.; Bushby, K. Muscular dystrophy in dysferlin-deficient mouse models. Neuromuscul. Disord. 2013, 23, 377–387. [Google Scholar] [CrossRef]
- van Putten, M.; Lloyd, E.M.; de Greef, J.C.; Raz, V.; Willmann, R.; Grounds, M.D. Mouse models for muscular dystrophies: An overview. Dis. Model. Mech. 2020, 13. [Google Scholar] [CrossRef]
- Fu, Y.-H.; Friedman, D.L.; Richards, S.; Pearlman, J.A.; Gibbs, R.A.; Pizzuti, A.; Ashizawa, T.; Rerryman, M.B.; Scarlato, G.; Fenwick, R.G.; et al. Decreased expression of myotonin-protein kinase messenger RNA and protein in adult form of myotonic dystrophy. Science 1993, 260, 235–238. [Google Scholar] [CrossRef]
- Jansen, G.; Groenen, P.J.T.A.; Bächner, D.; Jap, P.H.K.; Coerwinkel, M.; Oerlemans, F.; Van Den Broek, W.; Gohlsch, B.; Pette, D.; Plomp, J.J.; et al. Abnormal myotonic dystrophy protein kinase levels produce only mild myopathy in mice. Nat. Genet. 1996, 13, 316–324. [Google Scholar] [CrossRef]
- Alwazzan, M.; Newman, E.; Hamshere, M.G.; Brook, J.D. Myotonic dystrophy is associated with a reduced level of RNA from the DMWD allele adjacent to the expanded repeat. Hum. Mol. Genet. 1999, 8, 1491–1497. [Google Scholar] [CrossRef]
- Junghans, R.P.; Ebralidze, A.; Tiwari, B. Does (CUG)n repeat in DMPK mRNA “paint” chromosome 19 to suppress distant genes to create the diverse phenotype of myotonic dystrophy?: A new hypothesis of long-range cis autosomal inactivation. Neurogenetics 2001, 3, 59–67. [Google Scholar] [CrossRef]
- Klesert, T.R.; Cho, D.H.; Clark, J.I.; Maylie, J.; Adelman, J.; Snider, L.; Yuen, E.C.; Soriano, P.; Tapscott, S.J. Mice deficient in Six5 develop cataracts: Implications for myotonic dystrophy. Nat. Genet. 2000, 25, 105–109. [Google Scholar] [CrossRef]
- Sarkar, P.S.; Appukuttan, B.; Han, J.; Ito, Y.; Ai, C.; Tsai, W.; Chai, Y.; Stout, J.T.; Reddy, S. Heterozygous loss of Six5 in mice is sufficient to cause ocular cataracts. Nat. Genet. 2000, 25, 110–114. [Google Scholar] [CrossRef]
- Mankodi, A.; Logigian, E.; Callahan, L.; McClain, C.; White, R.; Henderson, D.; Krym, M.; Thornton, C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science 2000, 289, 1769–1772. [Google Scholar] [CrossRef]
- Ranum, L.P.W.; Rasmussen, P.F.; Benzow, K.A.; Koob, M.D.; Day, J.W. Genetic mapping of a second myotonic dystrophy locus. Nat. Genet. 1998, 19, 196–198. [Google Scholar] [CrossRef]
- Liquori, C.L.; Ricker, K.; Moseley, M.L.; Jacobsen, J.F.; Kress, W.; Naylor, S.L.; Day, J.W.; Ranum, L.P.W. Myotonic dystrophy type 2 caused by a CCTG expansion in intron I of ZNF9. Science 2001, 293, 864–867. [Google Scholar] [CrossRef]
- Timchenko, L.T.; Miller, J.W.; Timchenko, N.A.; Devore, D.R.; Datar, K.V.; Lin, L.; Roberts, R.; Thomas Caskey, C.; Swanson, M.S. Identification of a (CUG)(n) triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996, 24, 4407–4414. [Google Scholar] [CrossRef]
- Philips, A.V.; Timchenko, L.T.; Cooper, T.A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 1998, 280, 737–741. [Google Scholar] [CrossRef]
- Miller, J.W.; Urbinati, C.R.; Teng-Umnuay, P.; Stenberg, M.G.; Byrne, B.J.; Thornton, C.A.; Swanson, M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000, 19, 4439–4448. [Google Scholar] [CrossRef]
- Kanadia, R.N.; Shin, J.; Yuan, Y.; Beattie, S.G.; Wheeler, T.M.; Thornton, C.A.; Swanson, M.S. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl. Acad. Sci. USA 2006, 103, 11748–11753. [Google Scholar] [CrossRef]
- Ward, A.J.; Rimer, M.; Killian, J.M.; Dowling, J.J.; Cooper, T.A. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum. Mol. Genet. 2010, 19, 3614–3622. [Google Scholar] [CrossRef]
- Koshelev, M.; Sarma, S.; Price, R.E.; Wehrens, X.H.T.; Cooper, T.A. Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum. Mol. Genet. 2010, 19, 1066–1075. [Google Scholar] [CrossRef]
- Mankodi, A.; Takahashi, M.P.; Jiang, H.; Beck, C.L.; Bowers, W.J.; Moxley, R.T.; Cannon, S.C.; Thornton, C.A. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell 2002, 10, 35–44. [Google Scholar] [CrossRef]
- Fugier, C.; Klein, A.F.; Hammer, C.; Vassilopoulos, S.; Ivarsson, Y.; Toussaint, A.; Tosch, V.; Vignaud, A.; Ferry, A.; Messaddeq, N.; et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 2011, 17, 720–725. [Google Scholar] [CrossRef]
- Kimura, T.; Nakamori, M.; Lueck, J.D.; Pouliquin, P.; Aoike, F.; Fujimura, H.; Dirksen, R.T.; Takahashi, M.P.; Dulhunty, A.F.; Sakoda, S. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum. Mol. Genet. 2005, 14, 2189–2200. [Google Scholar] [CrossRef] [PubMed]
- López-Martínez, A.; Soblechero-Martín, P.; De-la-Puente-Ovejero, L.; Nogales-Gadea, G.; Arechavala-Gomeza, V. An Overview of Alternative Splicing Defects Implicated in Myotonic Dystrophy Type I. Genes (Basel) 2020, 11, 1109. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.; Smith, D.B.J.; Rich, M.M.; Leferovich, J.M.; Reilly, P.; Davis, B.M.; Tran, K.; Rayburn, H.; Bronson, R.; Cros, D.; et al. Mice lacking the myotonic dystrophy protein kinase develop a late onset progressive myopathy. Nat. Genet. 1996, 13, 325–335. [Google Scholar] [CrossRef]
- Sarkar, P.S.; Paul, S.; Han, J.; Reddy, S. Six5 is required for spermatogenic cell survival and spermiogenesis. Hum. Mol. Genet. 2004, 13, 1421–1431. [Google Scholar] [CrossRef]
- Wakimoto, H.; Maguire, C.T.; Sherwood, M.C.; Vargas, M.M.; Sarkar, P.S.; Han, J.; Reddy, S.; Berul, C.I. Characterization of cardiac conduction system abnormalities in mice with targeted disruption of Six5 gene. J. Interv. Card. Electrophysiol. 2002, 7, 127–135. [Google Scholar] [CrossRef]
- O’Cochlain, D.F.; Perez-Terzic, C.; Reyes, S.; Kane, G.C.; Behfar, A.; Hodgson, D.M.; Strommen, J.A.; Liu, X.K.; van den Broek, W.; Wansink, D.G.; et al. Transgenic overexpression of human DMPK accumulates into hypertrophic cardiomyopathy, myotonic myophaty and hypotension traits of myotonic dystrophy. Hum. Mol. Genet. 2004, 13, 2505–2518. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.M.; Krym, M.C.; Thornton, C.A. Ribonuclear foci at the neuromuscular junction in myotonic dystrophy type 1. Neuromuscul. Disord. 2007, 17, 242–247. [Google Scholar] [CrossRef]
- Gourdon, G.; Radvanyi, F.; Lia, A.S.; Duros, C.; Blanche, M.; Abitbol, M.; Junien, C.; Hofmann-Radvanyi, H. Moderate intergenerational and somatic instability of a 55-CTG repeat in transgenic mice. Nat. Genet. 1997, 15, 190–192. [Google Scholar] [CrossRef]
- Seznec, H.; Lia-Baldini, A.S.; Duros, C.; Fouquet, C.; Lacroix, C.; Hofmann-Radvanyi, H.; Junien, C.; Gourdon, G. Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum. Mol. Genet. 2000, 9, 1185–1194. [Google Scholar] [CrossRef]
- Gomes-Pereira, M.; Foiry, L.; Nicole, A.; Huguet, A.; Junien, C.; Munnich, A.; Gourdon, G. CTG trinucleotide repeat “big jumps”: Large expansions, small mice. PLoS Genet. 2007, 3, 0488–0491. [Google Scholar] [CrossRef]
- Mahadevan, M.S.; Yadava, R.S.; Yu, Q.; Balijepalli, S.; Frenzel-McCardell, C.D.; Bourne, T.D.; Phillips, L.H. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat. Genet. 2006, 38, 1066–1070. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-S.; Kearney, D.L.; Biasi, M.; De Taffet, G.; Cooper, T.A. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J. Clin. Invest. 2007, 117, 2802–2811. [Google Scholar] [CrossRef] [PubMed]
- Orengo, J.P.; Chambon, P.; Metzger, D.; Mosier, D.R.; Snipes, G.J.; Cooper, T.A. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc. Natl. Acad. Sci. USA 2008, 105, 2646–2651. [Google Scholar] [CrossRef] [PubMed]
- Yadava, R.S.; Frenzel-McCardell, C.D.; Yu, Q.; Srinivasan, V.; Tucker, A.L.; Puymirat, J.; Thornton, C.A.; Prall, O.W.; Harvey, R.P.; Mahadevan, M.S. RNA toxicity in myotonic muscular dystrophy induces NKX2-5 expression. Nat. Genet. 2008, 40, 61–68. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kanadia, R.N.; Johnstone, K.A.; Mankodi, A.; Lungu, C.; Thornton, C.A.; Esson, D.; Timmers, A.M.; Hauswirth, W.W.; Swanson, M.S. A Muscleblind Knockout Model for Myotonic Dystrophy. Science 2003, 302, 1978–1980. [Google Scholar] [CrossRef]
- Lueck, J.D.; Mankodi, A.; Swanson, M.S.; Thornton, C.A.; Dirksen, R.T. Muscle chloride channel dysfunction in two mouse models of myotonic dystrophy. J. Gen. Physiol. 2007, 129, 79–94. [Google Scholar] [CrossRef]
- Matynia, A.; Ng, C.H.; Dansithong, W.; Chiang, A.; Silva, A.J.; Reddy, S. Muscleblind1, but not Dmpk or Six5, contributes to a complex phenotype of muscular and motivational deficits in mouse models of Myotonic Dystrophy. PLoS ONE 2010, 5, 1–9. [Google Scholar] [CrossRef]
- Hao, M.; Akrami, K.; Wei, K.; De Diego, C.; Che, N.; Ku, J.H.; Tidball, J.; Graves, M.C.; Shieh, P.B.; Chen, F. Muscleblind-like 2 (Mbnl2) -deficient mice as a model for myotonic dystrophy. Dev. Dyn. 2008, 237, 403–410. [Google Scholar] [CrossRef]
- Lei, Y.; Finnell, R.H. New myotonic dystrophy type 1 mouse model. Cell Res. 2020, 30, 99–100. [Google Scholar] [CrossRef]
- Flucher, B.E. Skeletal muscle Ca V 1. 1 channelopathies. Eur. J. Physiol. 2020, 472, 739–754. [Google Scholar] [CrossRef]
- Bachinski, L.L.; Udd, B.; Meola, G.; Sansone, V.; Bassez, G.; Eymard, B.; Thornton, C.A.; Moxley, R.T.; Harper, P.S.; Rogers, M.T.; et al. Confirmation of the Type 2 Myotonic Dystrophy (CCTG)n Expansion Mutation in Patients with Proximal Myotonic Myopathy/Proximal Myotonic Dystrophy of Different European Origins: A Single Shared Haplotype Indicates an Ancestral Founder Effect. Am. J. Hum. Genet. 2003, 73, 835–848. [Google Scholar] [CrossRef] [PubMed]
- Prokic, I.; Cowling, B.S.; Kutchukian, C.; Kretz, C.; Tasfaout, H.; Gache, V.; Hergueux, J.; Wendling, O.; Ferry, A.; Toussaint, A.; et al. Differential physiological role of BIN1 isoforms in skeletal muscle development, function and regeneration. Dis. Model. Mech. 2020, 1–54. [Google Scholar] [CrossRef] [PubMed]
- Deenen, J.C.W.; Arnts, H.; Van Der Maarel, S.M.; Padberg, G.W.; Verschuuren, J.J.G.M.; Bakker, E.; Weinreich, S.S.; Verbeek, A.L.M.; Van Engelen, B.G.M. Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology 2014, 83, 1056–1059. [Google Scholar] [CrossRef]
- Gabriëls, J.; Beckers, M.C.; Ding, H.; De Vriese, A.; Plaisance, S.; Van Der Maarel, S.M.; Padberg, G.W.; Frants, R.R.; Hewitt, J.E.; Collen, D.; et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 1999, 236, 25–32. [Google Scholar] [CrossRef]
- Lek, A.; Rahimov, F.; Jones, P.L.; Kunkel, L.M. Emerging preclinical animal models for FSHD. Trends Mol. Med. 2015, 21, 295–306. [Google Scholar] [CrossRef]
- Wallace, L.M.; Garwick, S.E.; Mei, W.; Belayew, A.; Coppee, F.; Ladner, K.J.; Guttridge, D.; Yang, J.; Harper, S.Q. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol. 2011, 69, 540–552. [Google Scholar] [CrossRef]
- Krom, Y.D.; Thijssen, P.E.; Young, J.M.; den Hamer, B.; Balog, J.; Yao, Z.; Maves, L.; Snider, L.; Knopp, P.; Zammit, P.S.; et al. Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef]
- Dandapat, A.; Bosnakovski, D.; Hartweck, L.M.; Arpke, R.W.; Baltgalvis, K.A.; Vang, D.; Baik, J.; Darabi, R.; Perlingeiro, R.C.R.; Hamra, F.K.; et al. Dominant Lethal Pathologies in Male Mice Engineered to Contain an X-Linked DUX4 Transgene. Cell Rep. 2014, 8, 1484–1496. [Google Scholar] [CrossRef]
- Bosnakovski, D.; Chan, S.S.K.; Recht, O.O.; Hartweck, L.M.; Gustafson, C.J.; Athman, L.L.; Lowe, D.A.; Kyba, M. Muscle pathology from stochastic low level DUX4 expression in an FSHD mouse model. Nat. Commun. 2017, 8, 1–8. [Google Scholar] [CrossRef]
- Zhang, Y.; King, O.D.; Rahimov, F.; Jones, T.I.; Ward, C.W.; Kerr, J.P.; Liu, N.; Emerson, C.P.; Kunkel, L.M.; Partridge, T.A.; et al. Human skeletal muscle xenograft as a new preclinical model for muscle disorders. Hum. Mol. Genet. 2014, 23, 3180–3188. [Google Scholar] [CrossRef]
- Chen, J.C.J.; King, O.D.; Zhang, Y.; Clayton, N.P.; Spencer, C.; Wentworth, B.M.; Emerson, C.P.; Wagner, K.R. Morpholino-mediated Knockdown of DUX4 Toward Facioscapulohumeral Muscular Dystrophy Therapeutics. Mol. Ther. 2016, 24, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Mueller, A.L.; O’Neill, A.; Jones, T.I.; Llach, A.; Rojas, L.A.; Sakellariou, P.; Stadler, G.; Wright, W.E.; Eyerman, D.; Jones, P.L.; et al. Muscle xenografts reproduce key molecular features of facioscapulohumeral muscular dystrophy. Exp. Neurol. 2019, 320. [Google Scholar] [CrossRef] [PubMed]
- Gabellini, D.; D’Antona, G.; Moggio, M.; Prelle, A.; Zecca, C.; Adami, R.; Angeletti, B.; Ciscato, P.; Pellegrino, M.A.; Bottinelli, R.; et al. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature 2006, 439, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Caruso, N.; Herberth, B.; Bartoli, M.; Puppo, F.; Dumonceaux, J.; Zimmermann, A.; Denadai, S.; Lebossé, M.; Roche, S.; Geng, L.; et al. Deregulation of the Protocadherin Gene FAT1 Alters Muscle Shapes: Implications for the Pathogenesis of Facioscapulohumeral Dystrophy. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Van der Maarel, S.M.; Tawil, R.; Tapscott, S.J. Facioscapulohumeral muscular dystrophy and DUX4: Breaking the silence. Trends Mol. Med. 2011, 17, 252–258. [Google Scholar] [CrossRef]
- Tawil, R.; van der Maarel, S.M.; Tapscott, S.J. Facioscapulohumeral dystrophy: The path to consensus on pathophysiology. Skelet. Muscle 2014, 4, 1–15. [Google Scholar] [CrossRef]
- Richards, M.; Coppée, F.; Thomas, N.; Belayew, A.; Upadhyaya, M. Facioscapulohumeral muscular dystrophy (FSHD): An enigma unravelled? Hum. Genet. 2012, 131, 325–340. [Google Scholar] [CrossRef]
- Giesige, C.R.; Wallace, L.M.; Heller, K.N.; Eidahl, J.O.; Saad, N.Y.; Fowler, A.M.; Pyne, N.K.; Al-Kharsan, M.; Rashnonejad, A.; Chermahini, G.A.; et al. AAV-mediated follistatin gene therapy improves functional outcomes in the TIC-DUX4 mouse model of FSHD. JCI insight 2018, 3, 1–19. [Google Scholar] [CrossRef]
- Jones, T.; Jones, P.L. A cre-inducible DUX4 transgenic mouse model for investigating facioscapulohumeral muscular dystrophy. PLoS ONE 2018, 13, 1–31. [Google Scholar] [CrossRef]
- Engel, A.G.; Gomez, M.R.; Groover, R.V. Multicore disease. A recently recognized congenital myopathy associated with multifocal degeneration of muscle fibers. Mayo Clin. Proc. 1971, 46, 666–681. [Google Scholar]
- Jungbluth, H. Central core disease. Orphanet J. Rare Dis. 2007, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jungbluth, H.; Zhou, H.; Hartley, L.; Halliger-Keller, B.; Messina, S.; Longman, C.; Brockington, M.; Robb, S.A.; Straub, V.; Voit, T.; et al. Minicore myopathy with ophthalmoplegia caused by mutations in the ryanodine receptor type 1 gene. Neurology 2005, 65, 1930–1935. [Google Scholar] [CrossRef] [PubMed]
- Ferreiro, A.; Estournet, B.; Chateau, D.; Romero, N.B.; Laroche, C.; Odent, S.; Toutain, A.; Cabello, A.; Fontan, D.; dos Santos, H.G.; et al. Multi-Minicore Disease – Searching for Boundaries: Phenotype Analysis of 38 Cases. Am. Neurol. Assoc. 2000, 48, 745–757. [Google Scholar] [CrossRef]
- Shy, G.M.; Magee, K.R. A new congenital non-progressive myopathy. Brain 1956, 79, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Lawal, T.A.; Todd, J.J.; Meilleur, K.G. Ryanodine Receptor 1-Related Myopathies: Diagnostic and Therapeutic Approaches. Neurotherapeutics 2018, 15, 885–899. [Google Scholar] [CrossRef]
- Ferreiro, A.; Quijano-Roy, S.; Pichereau, C.; Moghadaszadeh, B.; Goemans, N.; Bönnemann, G.; Jungbluth, H.; Straub, V.; Villanova, M.; Leroy, J.P.; et al. Mutations of the selenoprotein N gene, which is implicated in rigid spine muscular dystrophy, cause the classical phenotype of multiminicore disease: Reassessing the nosology of early-onset myopathies. Am. J. Hum. Genet. 2002, 71, 739–749. [Google Scholar] [CrossRef]
- Kazamel, M.; Milone, M. Congenital myopathy with a novel SELN missense mutation and the challenge to differentiate it from congenital muscular dystrophy. J. Clin. Neurosci. 2019, 62, 238–239. [Google Scholar] [CrossRef]
- Cullup, T.; Lamont, P.J.; Cirak, S.; Damian, M.S.; Wallefeld, W.; Gooding, R.; Tan, S.V.; Sheehan, J.; Muntoni, F.; Abbs, S.; et al. Mutations in MYH7 cause Multi-minicore Disease (MmD) with variable cardiac involvement. Neuromuscul. Disord. 2012, 22, 1096–1104. [Google Scholar] [CrossRef]
- Boyden, S.E.; Mahoney, L.J.; Kawahara, G.; Myers, J.A.; Mitsuhashi, S.; Estrella, E.A.; Duncan, A.R.; Dey, F.; DeChene, E.T.; Blasko-Goehringer, J.M.; et al. Mutations in the satellite cell gene MEGF10 cause a recessive congenital myopathy with minicores. Neurogenetics 2012, 13, 115–124. [Google Scholar] [CrossRef]
- Zhou, H.; Lillis, S.; Loy, R.E.; Ghassemi, F.; Rose, M.R.; Norwood, F.; Mills, K.; Al-Sarraj, S.; Lane, R.J.M.; Feng, L.; et al. Multi-minicore disease and atypical periodic paralysis associated with novel mutations in the skeletal muscle ryanodine receptor (RYR1) gene. Neuromuscul. Disord. 2010, 20, 166–173. [Google Scholar] [CrossRef]
- Ge, L.; Fu, X.; Zhang, W.; Wang, D.; Wang, Z.; Yuan, Y.; Nonaka, I.; Xiong, H. Recessive mutations in proximal I-band of TTN gene cause severe congenital multi-minicore disease without cardiac involvement. Neuromuscul. Disord. 2019, 29, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Majczenko, K.; Davidson, A.E.; Camelo-Piragua, S.; Agrawal, P.B.; Manfready, R.A.; Li, X.; Joshi, S.; Xu, J.; Peng, W.; Beggs, A.H.; et al. Dominant mutation of CCDC78 in a unique congenital myopathy with prominent internal nuclei and atypical cores. Am. J. Hum. Genet. 2012, 91, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Kaindl, A.M.; Rüschendorf, F.; Krause, S.; Goebel, H.H.; Koehler, K.; Becker, C.; Pongratz, D.; Müller-Höcker, J.; Nürnberg, P.; Stoltenburg-Didinger, G.; et al. Missense mutations of ACTA1 cause dominant congenital myopathy with cores. J. Med. Genet. 2004, 41, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Lornage, X.; Romero, N.B.; Grosgogeat, C.A.; Malfatti, E.; Donkervoort, S.; Marchetti, M.M.; Neuhaus, S.B.; Foley, A.R.; Labasse, C.; Schneider, R.; et al. ACTN2 mutations cause “Multiple structured Core Disease” (MsCD). Acta Neuropathol 2020, 137, 501–519. [Google Scholar] [CrossRef]
- Estañ, M.C.; Fernández-Núñez, E.; Zaki, M.S.; Esteban, M.I.; Donkervoort, S.; Hawkins, C.; Caparros-Martin, J.A.; Saade, D.; Hu, Y.; Bolduc, V.; et al. Recessive mutations in muscle-specific isoforms of FXR1 cause congenital multi-minicore myopathy. Nat. Commun. 2019, 10, 1–19. [Google Scholar] [CrossRef]
- Takeshima, H.; Lino, M.; Takekura, H.; Nishi, M.; Kuno, J.; Minowa, O.; Takano, H.; Noda, T. Excitation-contraction uncoupling and muscular degeneration in mice lacking functional skeletal muscle ryanodine-receptor gene. Nature 1994, 369, 556–559. [Google Scholar] [CrossRef]
- Takekura, H.; Nishi, M.; Noda, T.; Takeshima, H.; Franzini-Armstrong, C. Abnormal junctions between surface membrane and sarcoplasmic reticulum in skeletal muscle with a mutation targeted to the ryanodine receptor. Proc. Natl. Acad. Sci. USA 1995, 92, 3381–3385. [Google Scholar] [CrossRef]
- Elbaz, M.; Ruiz, A.; Nicolay, S.; Tupini, C.; Bachmann, C.; Eckhardt, J.; Benucci, S.; Pelczar, P.; Treves, S.; Zorzato, F. Bi-allelic expression of the RyR1 p.A4329D mutation decreases muscle strength in slow-twitch muscles in mice. J. Biol. Chem. 2020, 295, 10331–10339. [Google Scholar] [CrossRef]
- Marino, M.; Stoilova, T.; Giorgi, C.; Bachi, A.; Cattaneo, A.; Auricchio, A.; Pinton, P.; Zito, E. SEPN1, an endoplasmic reticulum-localized selenoprotein linked to skeletal muscle pathology, counteracts hyperoxidation by means of redox-regulating SERCA2 pump activity. Hum. Mol. Genet. 2014, 24, 1843–1855. [Google Scholar] [CrossRef]
- Zvaritch, E.; Kraeva, N.; Bombardier, E.; McCloy, R.A.; Depreux, F.; Holmyard, D.; Kraev, A.; Seidman, C.E.; Seidman, J.G.; Russell Tupling, A.; et al. Ca2+ dysregulation in Ryr1I4895T/wt mice causes congenital myopathy with progressive formation of minicores, cores, and nemaline rods. Proc. Natl. Acad. Sci. USA 2009, 106, 21813–21818. [Google Scholar] [CrossRef]
- Fusto, A.; Moyle, L.A.; Gilbert, P.M.; Pegoraro, E. Cored in the act: The use of models to understand core myopathies. DMM Dis. Model. Mech. 2019, 12. [Google Scholar] [CrossRef] [PubMed]
- Elbaz, M.; Ruiz, A.; Bachmann, C.; Eckhardt, J.; Pelczar, P.; Venturi, E.; Lindsay, C.; Wilson, A.D.; Alhussni, A.; Humberstone, T.; et al. Quantitative RyR1 reduction and loss of calcium sensitivity of RyR1Q1970fsX16+A4329D cause cores and loss of muscle strength. Hum. Mol. Genet. 2019, 28, 2987–2999. [Google Scholar] [CrossRef] [PubMed]
- Castets, P.; Bertrand, A.T.; Beuvin, M.; Ferry, A.; Le Grand, F.; Castets, M.; Chazot, G.; Rederstorff, M.; Krol, A.; Lescure, A.; et al. Satellite cell loss and impaired muscle regeneration in selenoprotein N deficiency. Hum. Mol. Genet. 2011, 20, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Chauveau, C.; Bonnemann, C.G.; Julien, C.; Kho, A.L.; Marks, H.; Talim, B.; Maury, P.; Arne-Bes, M.C.; Uro-Coste, E.; Alexandrovich, A.; et al. Recessive TTN truncating mutations define novel forms of core myopathy with heart disease. Hum. Mol. Genet. 2014, 23, 980–991. [Google Scholar] [CrossRef]
- Maggi, L.; Scoto, M.; Cirak, S.; Robb, S.A.; Klein, A.; Lillis, S.; Cullup, T.; Feng, L.; Manzur, A.Y.; Sewry, C.A.; et al. Congenital myopathies--clinical features and frequency of individual subtypes diagnosed over a 5-year period in the United Kingdom. Neuromuscul. Disord. 2013, 23, 195–205. [Google Scholar] [CrossRef]
- Amburgey, K.; McNamara, N.; Bennett, L.R.; McCormick, M.E.; Acsadi, G.; Dowling, J.J. Prevalence of congenital myopathies in a representative pediatric united states population. Ann. Neurol. 2011, 70, 662–665. [Google Scholar] [CrossRef]
- Robinson, R.; Carpenter, D.; Shaw, M.A.; Halsall, J.; Hopkins, P. Mutations in RYR1 in Malignant Hyperthermia and Central Core Disease. Hum. Mutat. 2006, 27, 977–989. [Google Scholar] [CrossRef]
- Treves, S.; Anderson, A.A.; Ducreux, S.; Divet, A.; Bleunven, C.; Grasso, C.; Paesante, S.; Zorzato, F. Ryanodine receptor 1 mutations, dysregulation of calcium homeostasis and neuromuscular disorders. Neuromuscul. Disord. 2005, 15, 577–587. [Google Scholar] [CrossRef]
- Chelu, M.G.; Goonasekera, S.A.; Durham, W.J.; Tang, W.; Lueck, J.D.; Riehl, J.; Pessah, I.N.; Zhang, P.; Bhattacharjee, M.B.; Dirksen, R.T.; et al. Heat- and anesthesia-induced malignant hyperthermia in an RyR1 knock-in mouse. FASEB J. 2006, 20, 329–330. [Google Scholar] [CrossRef]
- Durham, W.J.; Aracena-Parks, P.; Long, C.; Rossi, A.E.; Goonasekera, S.A.; Boncompagni, S.; Galvan, D.L.; Gilman, C.P.; Baker, M.R.; Shirokova, N.; et al. RyR1 S-Nitrosylation Underlies Environmental Heat Stroke and Sudden Death in Y522S RyR1 Knockin Mice. Cell 2008, 133, 53–65. [Google Scholar] [CrossRef]
- Boncompagni, S.; Rossi, A.E.; Micaroni, M.; Hamilton, S.L.; Dirksen, R.T.; Franzini-Armstrong, C.; Protasi, F. Characterization and temporal development of cores in a mouse model of malignant hyperthermia. Proc. Natl. Acad. Sci. USA 2009, 106, 21996–22001. [Google Scholar] [CrossRef] [PubMed]
- Yuen, B.; Boncompagni, S.; Feng, W.; Yang, T.; Lopez, J.R.; Matthaei, K.I.; Goth, S.R.; Protasi, F.; Franzini-Armstrong, C.; Allen, P.D.; et al. Mice expressing T4826I-RYR1 are viable but exhibit sex- and genotype-dependent susceptibility to malignant hyperthermia and muscle damage. FASEB J. 2012, 26, 1311–1322. [Google Scholar] [CrossRef] [PubMed]
- Zvaritch, E.; Depreux, F.; Kraeva, N.; Loy, R.E.; Goonasekera, S.A.; Boncompagni, S.; Kraev, A.; Gramolini, A.O.; Dirksen, R.T.; Franzini-Armstrong, C.; et al. An Ryr1I4895T mutation abolishes Ca2+ release channel function and delays development in homozygous offspring of a mutant mouse line. Proc. Natl. Acad. Sci. USA 2007, 104, 18537–18542. [Google Scholar] [CrossRef] [PubMed]
- Buj-Bello, A.; Laugel, V.; Messaddeq, N.; Zahreddine, H.; Laporte, J.; Pellissier, J.F.; Mandel, J.L. The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc. Natl. Acad. Sci. USA 2002, 99, 15060–15065. [Google Scholar] [CrossRef]
- Pierson, C.R.; Dulin-Smith, A.N.; Durban, A.N.; Marshall, M.L.; Marshall, J.T.; Snyder, A.D.; Naiyer, N.; Gladman, J.T.; Chandler, D.S.; Lawlor, M.W.; et al. Modeling the human MTM1 p.R69C mutation in murine Mtm1 results in exon 4 skipping and a less severe myotubular myopathy phenotype. Hum. Mol. Genet. 2012, 21, 811–825. [Google Scholar] [CrossRef]
- Cowling, B.S.; Chevremont, T.; Prokic, I.; Kretz, C.; Ferry, A.; Coirault, C.; Koutsopoulos, O.; Laugel, V.; Romero, N.B.; Laporte, J. Reducing dynamin 2 expression rescues X-linked centronuclear myopathy. J. Clin. Invest. 2014, 124, 1350–1363. [Google Scholar] [CrossRef]
- Chen, X.; Gao, Y.Q.; Zheng, Y.Y.; Wang, W.; Wang, P.; Liang, J.; Zhao, W.; Tao, T.; Sun, J.; Wei, L.; et al. The intragenic microRNA miR199A1 in the dynamin 2 gene contributes to the pathology of X-linked centronuclear myopathy. J. Biol. Chem. 2020, 295, 8656–8667. [Google Scholar] [CrossRef]
- Durieux, A.C.; Vignaud, A.; Prudhon, B.; Viou, M.T.; Beuvin, M.; Vassilopoulos, S.; Fraysse, B.; Ferry, A.; Lainé, J.; Romero, N.B.; et al. A centronuclear myopathy-dynamin 2 mutation impairs skeletal muscle structure and function in mice. Hum. Mol. Genet. 2010, 19, 4820–4836. [Google Scholar] [CrossRef]
- Liu, N.; Bezprozvannaya, S.; Shelton, J.M.; Frisard, M.I.; Hulver, M.W.; McMillan, R.P.; Wu, Y.; Voelker, K.A.; Grange, R.W.; Richardson, J.A.; et al. Mice lacking microRNA 133a develop dynamin 2-dependent centronuclear myopathy. J. Clin. Invest. 2011, 121, 3258–3268. [Google Scholar] [CrossRef]
- Cowling, B.S.; Prokic, I.; Tasfaout, H.; Rabai, A.; Humbert, F.; Rinaldi, B.; Nicot, A.S.; Kretz, C.; Friant, S.; Roux, A.; et al. Amphiphysin (BIN1) negatively regulates dynamin 2 for normal muscle maturation. J. Clin. Invest. 2017, 127, 4477–4487. [Google Scholar] [CrossRef]
- Muñoz, X.M.; Kretz, C.; Silva-Rojas, R.; Ochala, J.; Menuet, A.; Romero, N.B.; Cowling, B.S.; Laporte, J. Physiological impact and disease reversion for the severe form of centronuclear myopathy linked to dynamin. JCI insight 2020, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Vandersmissen, I.; Biancalana, V.; Servais, L.; Dowling, J.J.; Vander Stichele, G.; Van Rooijen, S.; Thielemans, L. An integrated modelling methodology for estimating the prevalence of centronuclear myopathy. Neuromuscul. Disord. 2018, 28, 766–777. [Google Scholar] [CrossRef] [PubMed]
- Wallgren-Pettersson, C.; Clarke, A.; Samson, F.; Fardeau, M.; Dubowitz, V.; Moser, H.; Grimm, T.; Barohn, R.J.; Barth, P.G. The myotubular myopathies: Differential diagnosis of the X linked recessive, autosomal dominant, and autosomal recessive forms and present state of DNA studies. J. Med. Genet. 1995, 32, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Pierson, C.R.; Tomczak, K.; Agrawal, P.; Moghadaszadeh, B.; Beggs, A.H. X-linked myotubular and centronuclear myopathies. J. Neuropathol. Exp. Neurol. 2005, 64, 555–564. [Google Scholar] [CrossRef]
- Jungbluth, H.; Wallgren-Pettersson, C.; Laporte, J. Centronuclear (myotubular) myopathy. Orphanet J. Rare Dis. 2008, 3, 1–13. [Google Scholar] [CrossRef]
- Taylor, G.S.; Maehama, T.; Dixon, J.E. Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. USA 2000, 97, 8910–8915. [Google Scholar] [CrossRef] [PubMed]
- Dowling, J.J.; Vreede, A.P.; Low, S.E.; Gibbs, J.Y.; Bonnemann, C.G.; Feldman, E.L. Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 2009, 5, 1–13. [Google Scholar] [CrossRef]
- Hnia, K.; Vaccari, I.; Bolino, A.; Laporte, J. Myotubularin phosphoinositide phosphatases: Cellular functions and disease pathophysiology. Trends Mol. Med. 2012, 18, 317–327. [Google Scholar] [CrossRef]
- Amoasii, L.; Hnia, K.; Laporte, J. Myotubularin Phosphoinositide Phosphatases in Human Diseases. In Phosphoinositides and Disease; Springer: Dordrecht, The Netherlands, 2012; Volume 362, pp. 209–233. [Google Scholar]
- Thomas, N.S.T.; Williams, H.; Cole, G.; Roberts, K.; Clarke, A.; Liechti-Gallati, S.; Braga, S.; Gerber, A.; Meier, C.; Moser, H.; et al. X linked neonatal centronuclear/myotubular myopathy: Evidence for linkage to Xq28 DNA marker loci. J. Med. Genet. 1990, 27, 284–287. [Google Scholar] [CrossRef]
- Blondeau, F.; Laporte, J.; Bodin, S.; Superti-Furga, G.; Payrastre, B.; Mandel, J.L. Myotubularin, a phosphatase deficient in myotubular myopathy, acts on phosphatidylinositol 3-kinase and phosphatidylinositol 3-phosphate pathway. Hum. Mol. Genet. 2000, 9, 2223–2229. [Google Scholar] [CrossRef]
- Tronchère, H.; Laporte, J.; Pendaries, C.; Chaussade, C.; Liaubet, L.; Pirola, L.; Mandel, J.L.; Payrastre, B. Production of PtdIns(5)P by the phosphoinositide 3-phosphatase myotubularin in mammalian cells Hélène. J Biol Chem 2003, 1–36. [Google Scholar]
- Cao, C.; Backer, J.; Laporte, J.; Bedrick, E.J.; Wandinger-Ness, A. Sequential Actions of Myotubularin Lipid Phosphatases Regulate Endosomal PI(3)P and Growth Factor Receptor Traffickin. Mol. Biol. Cell 2008, 19, 3334–3346. [Google Scholar] [CrossRef] [PubMed]
- Laporte, J.; Hu, L.J.; Kretz, C.; Mandel, J.L.; Kioschis, P.; Coy, J.F.; Klauck, S.M.; Poustka, A.; Dahl, N. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat. Genet. 1996, 13, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Laporte, J.; Biancalana, V.; Tanner, S.M.; Kress, W.; Schneider, V.; Wallgren-Pettersson, C.; Herger, F.; Buj-Bello, A.; Blondeau, F.; Liechti-Gallati, S.; et al. MTM1 mutations in X-linked myotubular myopathy. Hum. Mutat. 2000, 15, 393–409. [Google Scholar] [CrossRef]
- Biancalana, V.; Caron, O.; Gallati, S.; Baas, F.; Kress, W.; Novelli, G.; D’Apice, M.R.; Lagier-Tourenne, C.; Buj-Bello, A.; Romero, N.B.; et al. Characterisation of mutations in 77 patients with X-linked myotubular myopathy, including a family with a very mild phenotype. Hum. Genet. 2003, 112, 135–142. [Google Scholar] [CrossRef]
- McEntagart, M.; Parsons, G.; Buj-Bello, A.; Biancalana, V.; Fenton, I.; Little, M.; Krawczak, M.; Thomas, N.; Herman, G.; Clarke, A.; et al. Genotype-phenotype correlations in X-linked myotubular myopathy. Neuromuscul. Disord. 2002, 12, 939–946. [Google Scholar] [CrossRef]
- Amburgey, K.; Tsuchiya, E.; De Chastonay, S.; Glueck, M.; Alverez, R.; Nguyen, C.T.; Rutkowski, A.; Hornyak, J.; Beggs, A.H.; Dowling, J.J. A natural history study of X-linked myotubular myopathy. Neurology 2017, 89, 1355–1364. [Google Scholar] [CrossRef]
- Beggs, A.H.; Byrne, B.J.; De Chastonay, S.; Haselkorn, T.; Hughes, I.; James, E.S.; Kuntz, N.L.; Simon, J.; Swanson, L.C.; Yang, M.L.; et al. A multicenter, retrospective medical record review of X-linked myotubular myopathy: The recensus study. Muscle and Nerve 2018, 57, 550–560. [Google Scholar] [CrossRef]
- Herman, G.E.; Finegold, M.; Zhao, W.; De Gouyon, B.; Metzenberg, A. Medical complications in long-term survivors with X-linked myotubular myopathy. J. Pediatr. 1999, 134, 206–214. [Google Scholar] [CrossRef]
- Messaddeq, N.; Kretz, C.; Al-qusairi, L.; Weiss, N.; Toussaint, A.; Sanoudou, D.; Beggs, A.H.; Allard, B.; Mandel, J.; Laporte, J.; et al. T-tubule disorganization and defective excitation-contraction coupling in muscle fibers lacking myotubularin lipid phosphatase. Proc. Natl. Acad. Sci. USA 2009, 106, 18763–18768. [Google Scholar]
- Kutchukian, C.; Scrudato, M.L.; Tourneur, Y.; Poulard, K.; Vignaud, A.; Berthier, C.; Allard, B.; Lawlor, M.W.; Buj-Bello, A.; Jacquemond, V. Phosphatidylinositol 3-Kinase Inhibition restores Ca2+ release defects and prolongs survival in myotubularin-deficient mice. Proc. Natl. Acad. Sci. USA 2016, 113, 14432–14437. [Google Scholar] [CrossRef] [PubMed]
- Kutchukian, C.; Szentesi, P.; Allard, B.; Buj-bello, A.; Csernoch, L.; Jacquemond, V. Ca2+-induced sarcoplasmic reticulum Ca2+ release in myotubularin-deficient muscle fibers. Cell Calcium 2019, 80, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Buj-Bello, A.; Fougerousse, F.; Schwab, Y.; Messaddeq, N.; Spehner, D.; Pierson, C.R.; Durand, M.; Kretz, C.; Danos, O.; Douar, A.M.; et al. AAV-mediated intramuscular delivery of myotubularin corrects the myotubular myopathy phenotype in targeted murine muscle and suggests a function in plasma membrane homeostasis. Hum. Mol. Genet. 2008, 17, 2132–2143. [Google Scholar] [CrossRef]
- Lawlor, M.W.; Armstrong, D.; Viola, M.G.; Widrick, J.J.; Meng, H.; Grange, R.W.; Childers, M.K.; Hsu, C.P.; O’Callaghan, M.; Pierson, C.R.; et al. Enzyme replacement therapy rescues weakness and improves muscle pathology in mice with X-linked myotubular myopathy. Hum. Mol. Genet. 2013, 22, 1525–1538. [Google Scholar] [CrossRef]
- Dowling, J.J.; Joubert, R.; Low, S.E.; Durban, A.N.; Messaddeq, N.; Li, X.; Dulin-Smith, A.N.; Snyder, A.D.; Marshall, M.L.; Marshall, J.T.; et al. Myotubular myopathy and the neuromuscular junction: A novel therapeutic approach from mouse models. DMM Dis. Model. Mech. 2012, 5, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Sabha, N.; Volpatti, J.R.; Gonorazky, H.; Reifler, A.; Davidson, A.E.; Li, X.; Eltayeb, N.M.; Dall’Armi, C.; Di Paolo, G.; Brooks, S.V.; et al. PIK3C2B inhibition improves function and prolongs survival in myotubular myopathy animal models. J. Clin. Investig. 2016, 126, 3613–3625. [Google Scholar] [CrossRef]
- Tasfaout, H.; Buono, S.; Guo, S.; Kretz, C.; Messaddeq, N.; Booten, S.; Greenlee, S.; Monia, B.P.; Cowling, B.S.; Laporte, J. Antisense oligonucleotide-mediated Dnm2 knockdown prevents and reverts myotubular myopathy in mice. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gayi, E.; Neff, L.A.; Massana Muñoz, X.; Ismail, H.M.; Sierra, M.; Mercier, T.; Décosterd, L.A.; Laporte, J.; Cowling, B.S.; Dorchies, O.M.; et al. Tamoxifen prolongs survival and alleviates symptoms in mice with fatal X-linked myotubular myopathy. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Maani, N.; Sabha, N.; Rezai, K.; Ramani, A.; Groom, L.; Eltayeb, N.; Mavandadnejad, F.; Pang, A.; Russo, G.; Brudno, M.; et al. Tamoxifen therapy in a murine model of myotubular myopathy. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Bitoun, M.; Maugenre, S.; Jeannet, P.Y.; Lacène, E.; Ferrer, X.; Laforêt, P.; Martin, J.J.; Laporte, J.; Lochmüller, H.; Beggs, A.H.; et al. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat. Genet. 2005, 37, 1207–1209. [Google Scholar] [CrossRef]
- McNiven, M.A. Dynamin in disease. Nat. Genet. 2005, 37, 215–216. [Google Scholar] [CrossRef] [PubMed]
- Bitoun, M.; Bevilacqua, J.A.; Prudhon, B.; Maugenre, S.; Taratuto, A.L.; Monges, S.; Lubieniecki, F.; Cances, C.; Uro-Coste, E.; Mayer, M.; et al. Dynamin 2 mutations cause sporadic centronuclear myopathy with neonatal onset. Ann. Neurol. 2007, 62, 666–670. [Google Scholar] [CrossRef] [PubMed]
- Böhm, J.; Biancalana, V.; Dechene, E.T.; Bitoun, M.; Christopher, R.; Schaefer, E.; Karasoy, H.; Dempsey, M.A.; Klein, F.; Kretz, C.; et al. Mutation Spectrum in the Large GTPase Dynamin 2, and Genotype–Phenotype Correlation in Autosomal Dominant Centronuclear Myopathy. Hum Mutat 2013, 33, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Durieux, A.C.; Vassilopoulos, S.; Lainé, J.; Fraysse, B.; Briñas, L.; Prudhon, B.; Castells, J.; Freyssenet, D.; Bonne, G.; Guicheney, P.; et al. A Centronuclear Myopathy - Dynamin 2 Mutation Impairs Autophagy in Mice. Traffic 2012, 13, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Fraysse, B.; Guicheney, P.; Bitoun, M. Calcium homeostasis alterations in a mouse model of the dynamin 2-related centronuclear myopathy. Biol. Open 2016, 5, 1691–1696. [Google Scholar] [CrossRef]
- Kutchukian, C.; Szentesi, P.; Allard, B.; Trochet, D.; Beuvin, M.; Berthier, C.; Tourneur, Y.; Guicheney, P.; Csernoch, L.; Bitoun, M.; et al. Impaired excitation–contraction coupling in muscle fibres from the dynamin2R465W mouse model of centronuclear myopathy. J. Physiol. 2017, 595, 7369–7382. [Google Scholar] [CrossRef]
- Trochet, D.; Prudhon, B.; Beuvin, M.; Peccate, C.; Lorain, S.; Julien, L.; Benkhelifa-Ziyyat, S.; Rabai, A.; Mamchaoui, K.; Ferry, A.; et al. Allele-specific silencing therapy for Dynamin 2-related dominant centronuclear myopathy. EMBO Mol. Med. 2018, 10, 239–253. [Google Scholar] [CrossRef]
- Buono, S.; Ross, J.A.; Tasfaout, H.; Levy, Y.; Kretz, C.; Tayefeh, L.; Matson, J.; Guo, S.; Kessler, P.; Monia, B.P.; et al. Reducing dynamin 2 (DNM2) rescues DNM2-related dominant centronuclear myopathy. Proc. Natl. Acad. Sci. USA 2018, 115, 11066–11071. [Google Scholar] [CrossRef]
- Rabai, A.; Reisser, L.; Reina-San-Martin, B.; Mamchaoui, K.; Cowling, B.S.; Nicot, A.S.; Laporte, J. Allele-Specific CRISPR/Cas9 Correction of a Heterozygous DNM2 Mutation Rescues Centronuclear Myopathy Cell Phenotypes. Mol. Ther. Nucleic Acids 2019, 16, 246–256. [Google Scholar] [CrossRef]
- MacLennan, D.H.; Phillips, M.S. Malignant hyperthermia. Science 1992, 256, 789–794. [Google Scholar] [CrossRef]
- Britt, B.A. Malignant Hyperthermia; Martinus Nijhoff Publishing: Boston, MA, USA, 1987; ISBN 9781461320791. [Google Scholar]
- Rosenberg, H.; Davis, M.; James, D.; Pollock, N.; Stowell, K. Malignant hyperthermia. Orphanet J. Rare Dis. 2007, 2, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kolb, M.E.; Horne, M.L.; Martz, R. Dantrolene in human malignant hyperthermia Anesthesiology. Anesthesiology 1982, 56, 254–262. [Google Scholar] [CrossRef]
- Fujii, J.; Otsu, K.; Zorzato, F.; De Leon, S.; Khanna, V.K.; Weiler, J.E.; O’Brien, P.J.; Maclennan, D.H. Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science 1991, 253, 448–451. [Google Scholar] [CrossRef] [PubMed]
- MacLennan, D.; Duff, C.; Zorzato, F.; Fujii, J.; Phillips, M.; Korneluk, R.; Frodis, W.; Britt, B.; Worton, R. Ryanodine receptor gene is a candidate for predisposition to malignant hyperthermia. Nature 1990, 343, 559–561. [Google Scholar] [CrossRef] [PubMed]
- Riazi, S.; Kraeva, N.; Hopkins, P.M. Malignant Hyperthermia in the Post-Genomics Era: New Perspectives on an Old Concept. Anesthesiology 2018, 128, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Goebel, H.H.; Sewry, C.A.; Weller, R.O. Introduction to Muscle Disease: Pathology and Genetics; Wiley: Hoboken, NJ, USA, 2013; ISBN 9781118635469. [Google Scholar]
- Kimlicka, L.; Lau, K.; Tung, C.C.; Van Petegem, F. Disease mutations in the ryanodine receptor N-terminal region couple to a mobile intersubunit interface. Nat. Commun. 2013, 4, 1506–1510. [Google Scholar] [CrossRef]
- Feng, W.; Barrientos, G.C.; Cherednichenko, G.; Yang, T.; Padilla, I.T.; Truong, K.; Allen, P.D.; Lopez, J.R.; Pessah, I.N. Functional and biochemical properties of ryanodine receptor type 1 channels from heterozygous R163C malignant hyperthermia-susceptible mice. Mol. Pharmacol. 2011, 79, 420–431. [Google Scholar] [CrossRef]
- Richter, M.; Schleithoff, L.; Deufel, T.; Lehmann-Horn, F.; Herrmann-Frank, A. Functional characterization of a distinct ryanodine receptor mutation in human malignant hyperthermia-susceptible muscle. J. Biol. Chem. 1997, 272, 5256–5260. [Google Scholar] [CrossRef][Green Version]
- Lamb, G.D. Ca2+ inactivation, Mg2+ inhibition and malignant hyperthermia. J. Muscle Res. Cell Motil. 1993, 14, 554–556. [Google Scholar] [CrossRef]
- Choi, R.H.; Koenig, X.; Launikonis, B.S. Dantrolene requires Mg2+ to arrest malignant hyperthermia. Proc. Natl. Acad. Sci. USA 2017, 114, 4811–4815. [Google Scholar] [CrossRef]
- Diszházi, G.; Magyar, Z.É.; Mótyán, J.A.; Csernoch, L.; Jóna, I.; Nánási, P.P.; Almássy, J. Dantrolene requires MG2+ and ATP to inhibit the ryanodine receptor. Mol. Pharmacol. 2019, 96, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Hainaut, K.; Desmedt, J.E. Effect of dantrolene sodium on calcium movements in single muscle fibres. Nature 1974, 252, 728–730. [Google Scholar] [CrossRef] [PubMed]
- Lucre, J.N.; Hall, G.M.; Lister, D. Porcine malignant hyperthermia. I: Metabolic and physiological changes. Br. J. Anaesth. 1976, 48, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Hall, G.M.; Bendall, J.R.; Lucre, J.N.; Lister, D. Porcine malignant hyperthermia. II: Heat production. Br. J. Anaesth. 1976, 48, 305–308. [Google Scholar] [CrossRef]
- Nelson, T.E.; Jones, E.W.; Anderson, I.L. Porcine malignant hyperthermia. Am. J. Pathol. 1976, 84, 197–199. [Google Scholar]
- Harrison, G.G. Control of the malignant hyperpyrexic syndrome in mhs swine by dantrolene sodium. Br. J. Anaesth. 1975, 47, 62–65. [Google Scholar] [CrossRef]
- Anderson, I.L.; Jones, E.W. Porcine malignant hyperthermia: Effect of dantrolene sodium on in-vitro halothane-induced contraction of susceptible muscle. Anesthesiology 1976, 44, 57–61. [Google Scholar] [CrossRef]
- Gronert, G.A.; Milde, J.H.; Theye, R.A. Dantrolene in porcine malignant hyperthermia. Anesthesiology 1976, 44, 488–495. [Google Scholar] [CrossRef]
- Lanner, J.T.; Georgiou, D.K.; Dagnino-acosta, A.; Ainbinder, A.; Cheng, Q.; Joshi, A.D.; Chen, Z.; Yarotskyy, V.; Oakes, J.M.; Lee, C.S.; et al. AICAR Prevents Heat Induced Sudden Death in RyR1 Mutant Mice Independent of AMPK Activation. Nat. Med. 2012, 18, 244–251. [Google Scholar] [CrossRef]
- Wang, H.J.; Lee, C.S.; Yee, R.S.Z.; Groom, L.; Friedman, I.; Babcock, L.; Georgiou, D.K.; Hong, J.; Hanna, A.D.; Recio, J.; et al. Adaptive thermogenesis enhances the life-threatening response to heat in mice with an Ryr1 mutation. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Yang, T.; Riehl, J.; Esteve, E.; Matthaei, K.I.; Goth, S.; Allen, P.D.; Pessah, I.N.; Lopez, J.R. Pharmacologic and functional characterization of malignant hyperthermia in the R163C RyR1 knock-in mouse. Anesthesiology 2006, 105, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.R.; Kaura, V.; Diggle, C.P.; Hopkins, P.M.; Allen, P.D. Malignant hyperthermia, environmental heat stress, and intracellular calcium dysregulation in a mouse model expressing the p.G2435R variant of RYR1. Br. J. Anaesth. 2018, 121, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Truong, K.M.; Pessah, I.N. Comparison of Chlorantraniliprole and Flubendiamide Activity Toward Wild-Type and Malignant Hyperthermia-Susceptible Ryanodine Receptors and Heat Stress Intolerance. Toxicol. Sci. 2019, 167, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Pessah, I.N.; Truong, K.M. Additional Safety Assessments Needed for Diamide Insecticides. Toxicol. Sci. 2019, 171, 282. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).