Silencing of the Ortholog of DEFECTIVE IN ANTHER DEHISCENCE 1 Gene in the Woody Perennial Jatropha curcas Alters Flower and Fruit Development
Abstract
1. Introduction
2. Results
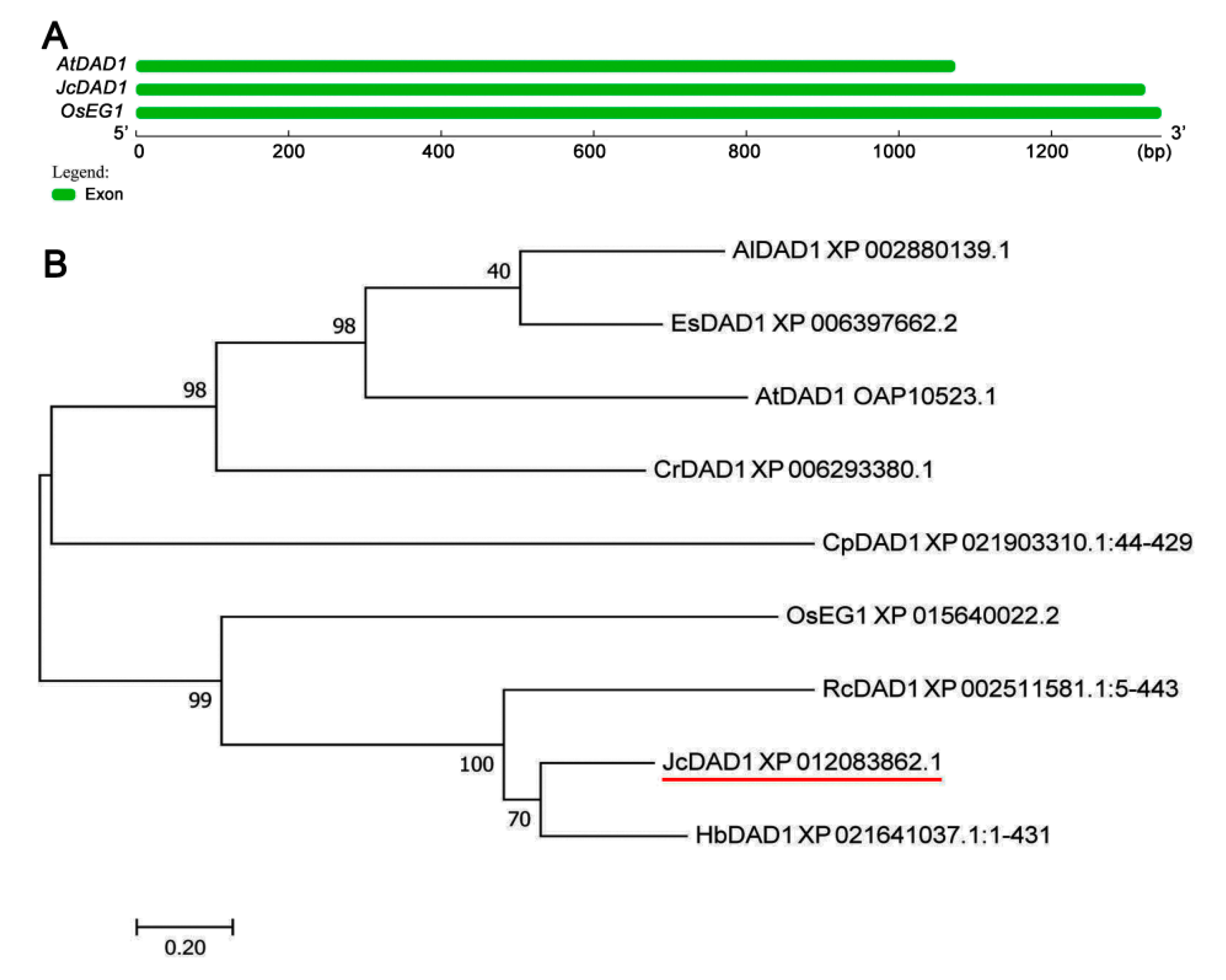
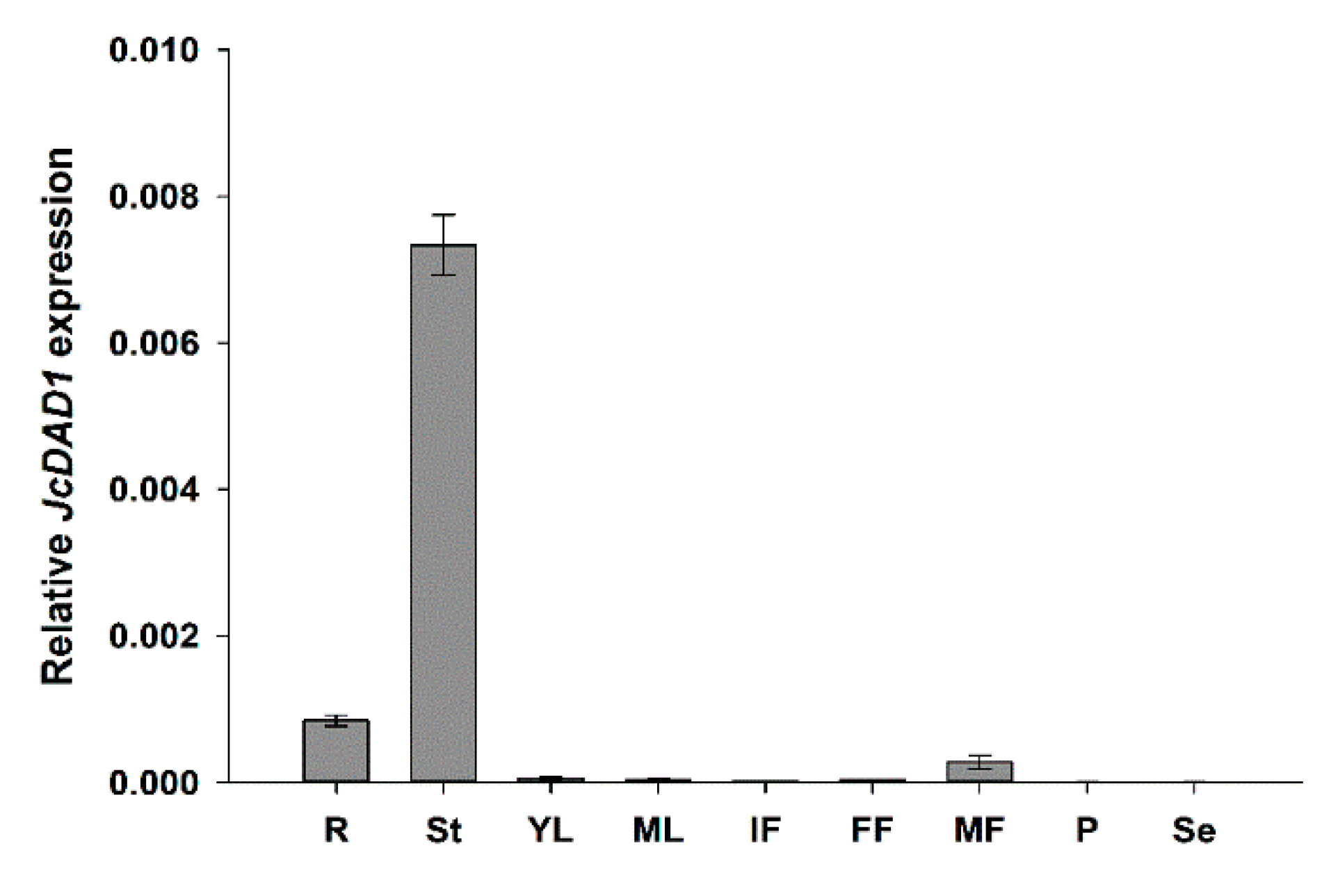
2.1. Characterization of the JcDAD1 Gene
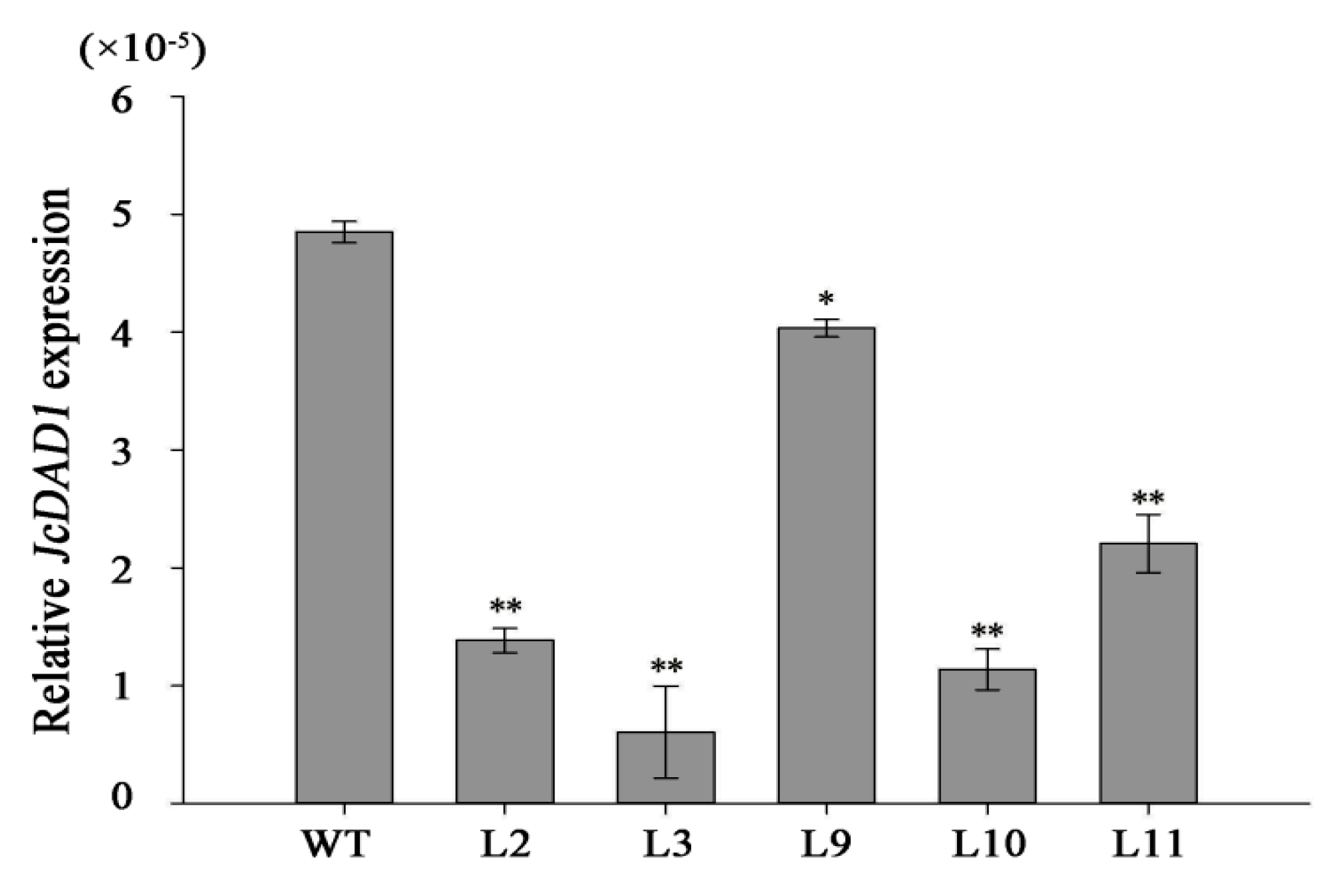
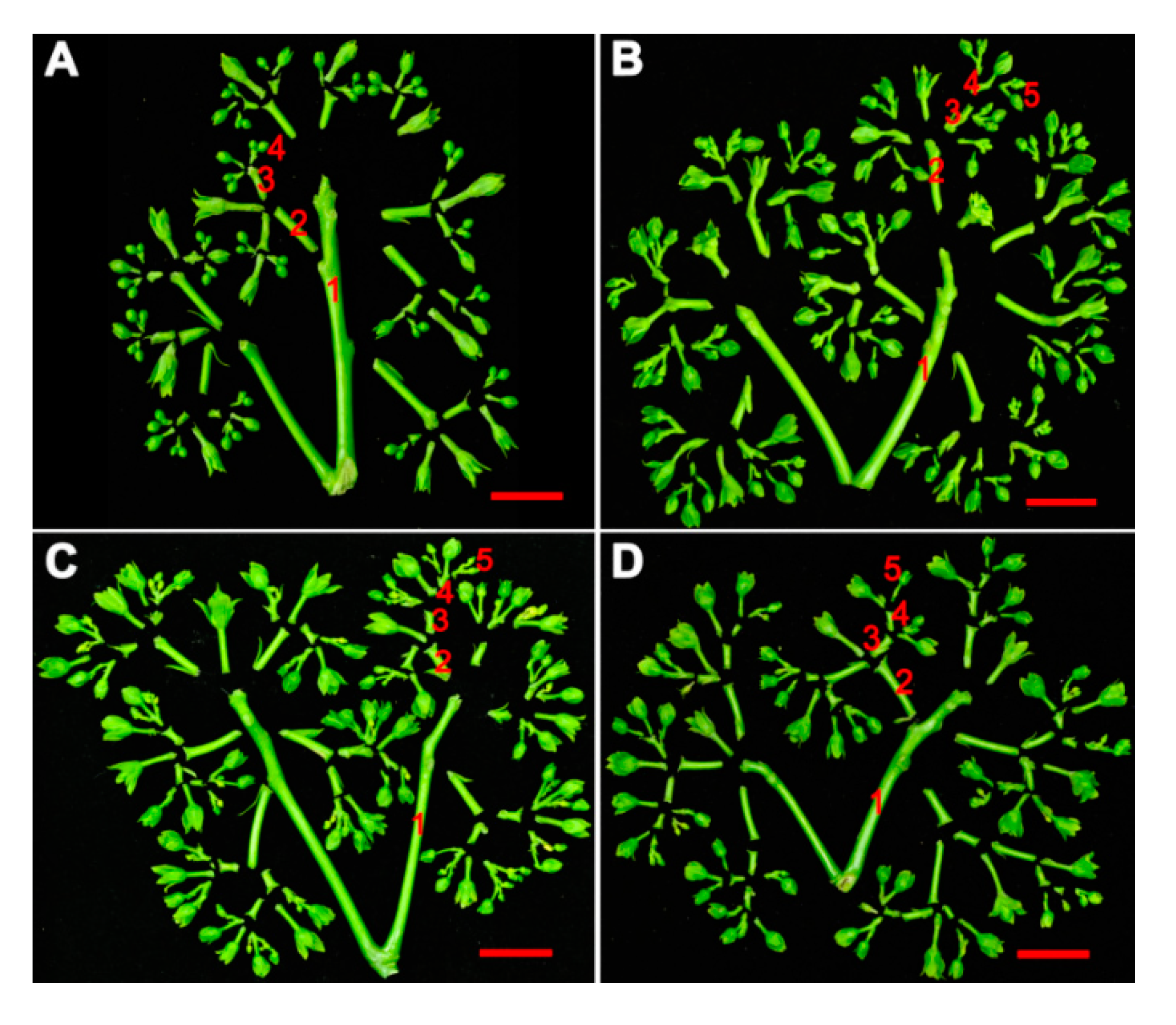
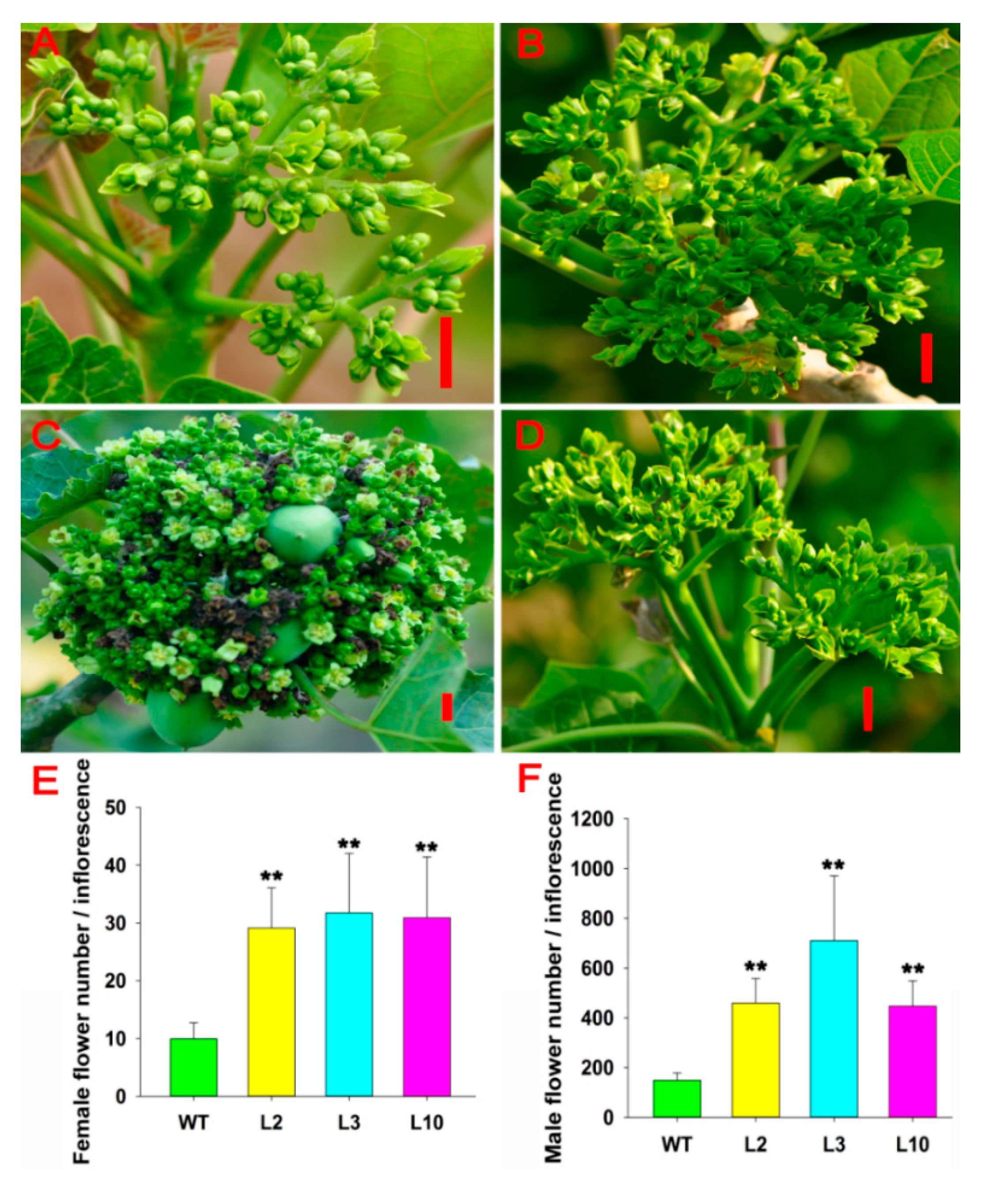
2.2. JcDAD1 Gene Silencing Increased Inflorescence Branching, Flower Number, and Flower Size
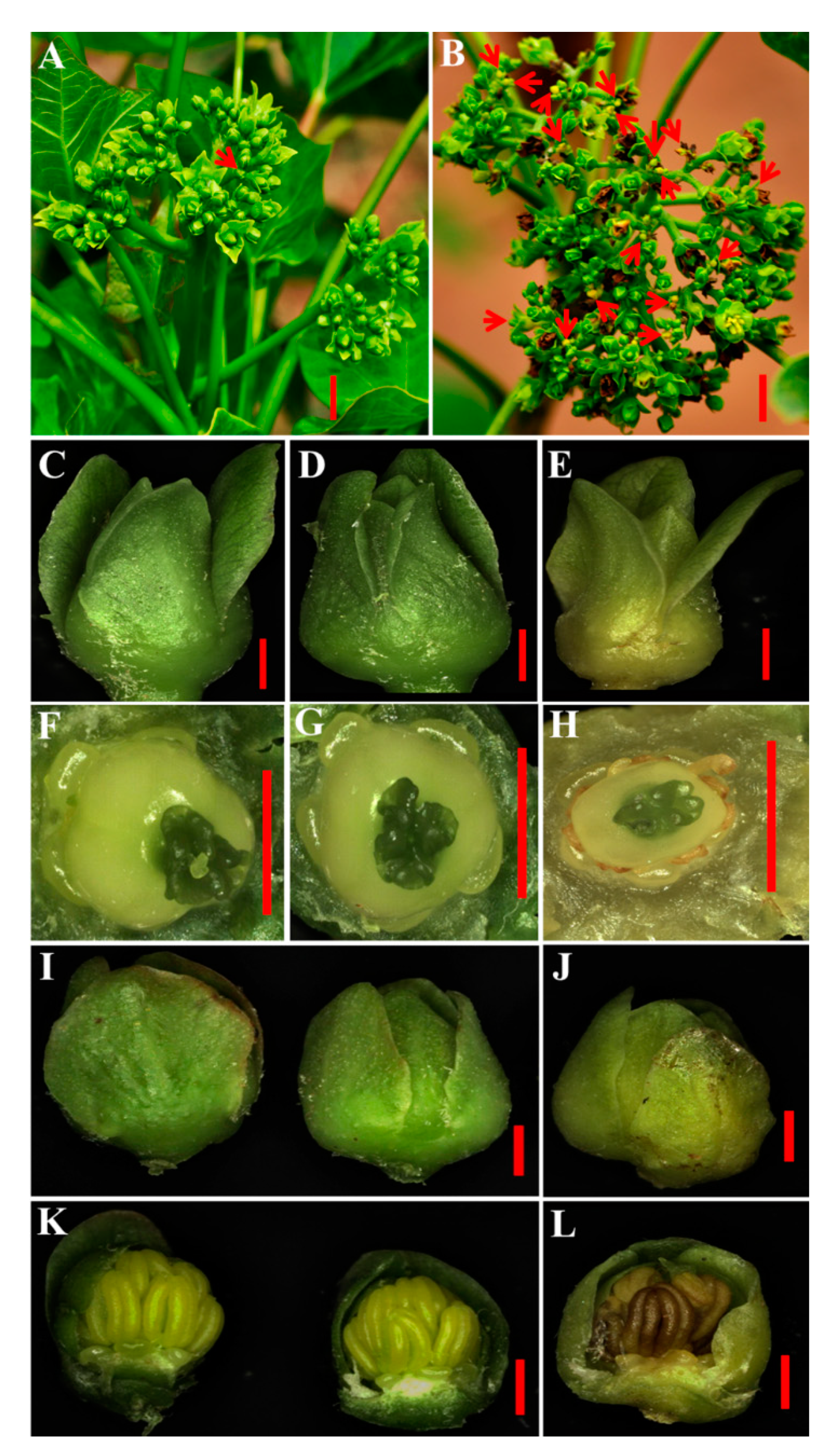
2.3. JcDAD1 Gene Silencing Caused the Abortion of Some Female Flowers but Did Not Affect Anther Dehiscence
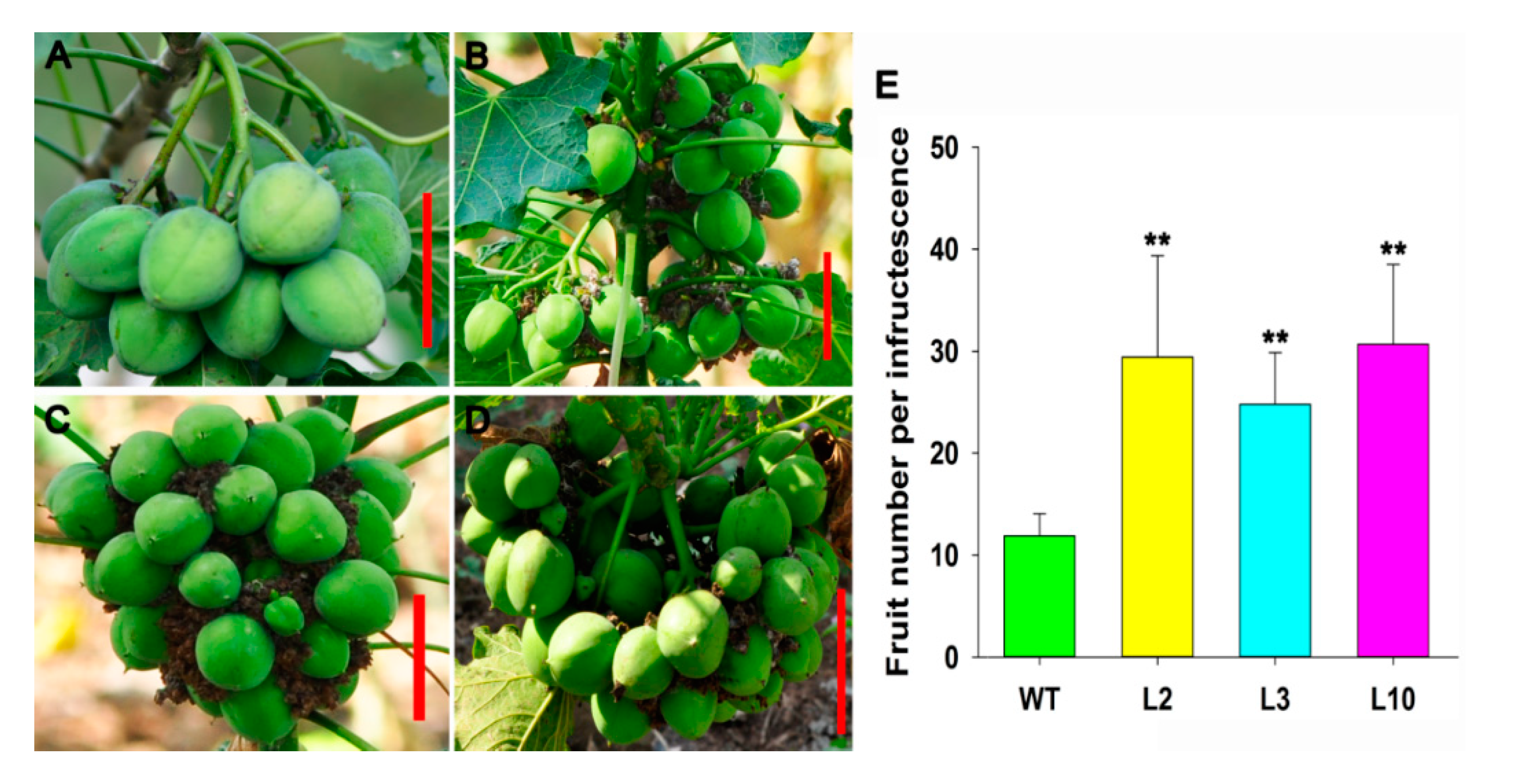
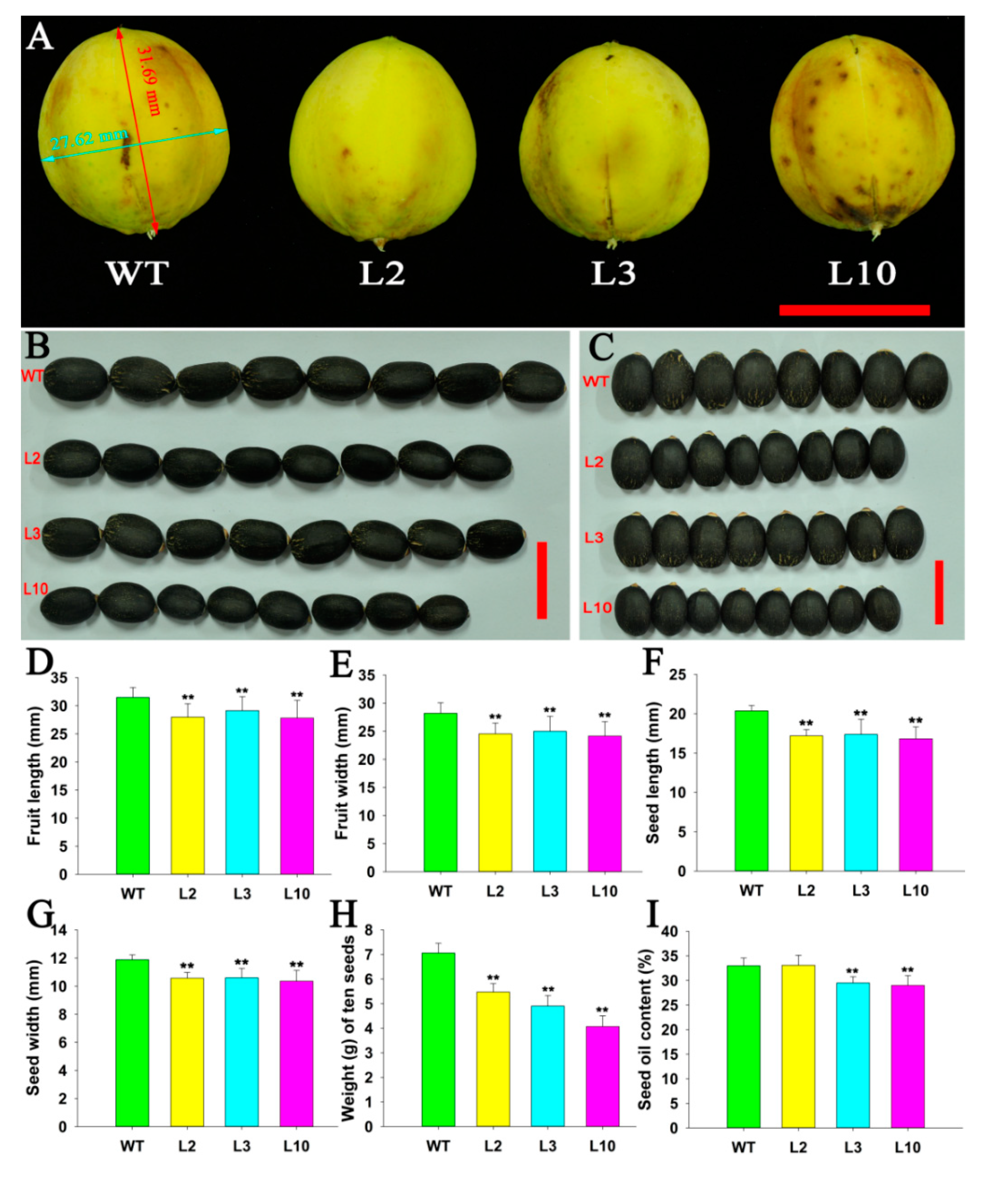
2.4. JcDAD1 Gene Silencing Affected Jatropha Yield Traits
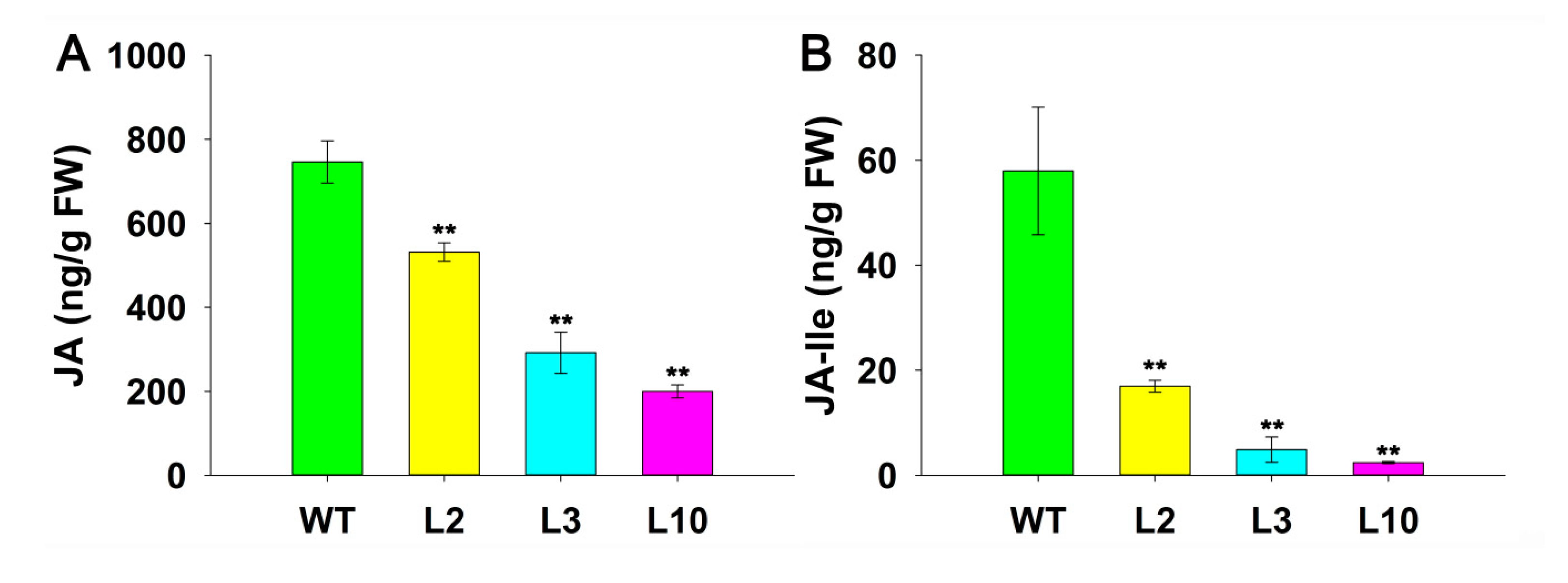
2.5. JcDAD1 Gene Silencing Reduced Endogenous JA and JA-Ile Contents in Jatropha Inflorescences
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Sequence Alignment and Phylogenetic Analysis
4.3. Isolation of JcDAD1 cDNA
4.4. RNAi Silencing Vector Construction and Transformation
4.5. Quantitative RT-PCR (qRT-PCR)
4.6. Phenotypic Analysis of Flowers
4.7. Quantitation of the JA and JA-Ile Contents
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| aos | allene oxide synthase |
| DAD1 | DEFECTIVE IN ANTHER DEHISCENCE 1 |
| EG1 | EXTRA GLUME 1 |
| fad | fatty acid desaturase |
| JA | jasmonic acid |
| JA-Ile | jasmonic acid-isoleucine |
| JcDAD1-RNAi | Jatropha curcas DEFECTIVE IN ANTHER DEHISCENCE 1-RNA interference |
| lox | lipoxygenase |
| qRT-PCR | quantitative reverse transcriptase-polymerase chain reaction |
References
- Šimura, J.; Antoniadi, I.; Široká, J.; Tarkowská, D.; Strnad, M.; Ljung, K.; Novák, O. Plant Hormonomics: Multiple Phytohormone Profiling by Targeted Metabolomics. Plant Physiol. 2018, 177, 476–489. [Google Scholar] [CrossRef]
- Balfagón, D.; Sengupta, S.; Gómez-Cadenas, A.; Fritschi, F.B.; Azad, R.K.; Mittler, R.; Zandalinas, S.I. Jasmonic Acid Is Required for Plant Acclimation to a Combination of High Light and Heat Stress. Plant Physiol. 2019, 181, 1668–1682. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; Li, C.; Li, J. Hormone function in plants. In Hormone Metabolism and Signaling in Plants, 1st ed.; Li, J., Li, C., Smith, S.M., Eds.; Academic Press: London, UK, 2017; pp. 1–38. [Google Scholar]
- Creelman, R.A.; Mullet, J.E. Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. USA 1995, 92, 4114–4119. [Google Scholar] [CrossRef] [PubMed]
- Vick, B.A.; Zimmerman, D.C. Biosynthesis of Jasmonic Acid by Several Plant Species. Plant Physiol. 1984, 75, 458–461. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.K.; Tamogami, S.; Han, O.; Iwahashi, H.; Rakwal, R. Rice octadecanoid pathway. Biochem. Biophys. Res. Commun. 2004, 317, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, D.; Stingl, N.; Kubigsteltig, I.I.; Bals, T.; Juenger, M.; Pollmann, S.; Berger, S.; Schuenemann, D.; Mueller, M.J. DONGLE and DEFECTIVE IN ANTHER DEHISCENCE1 lipases are not essential for wound-and pathogen-induced jasmonate biosynthesis: Redundant lipases contribute to jasmonate formation. Plant Physiol. 2010, 153, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Bannenberg, G.; Martínez, M.; Hamberg, M.; Castresana, C. Diversity of the Enzymatic Activity in the Lipoxygenase Gene Family of Arabidopsis thaliana. Lipids 2008, 44, 85. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.A.; Schilmiller, A.L. Oxylipin metabolism in response to stress. Curr. Opin. Plant Biol. 2002, 5, 230–236. [Google Scholar] [CrossRef]
- Laudert, D.; Pfannschmidt, U.; Lottspeich, F.; Holländer-Czytko, H.; Weiler, E.W. Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol. Biol. 1996, 31, 323–335. [Google Scholar] [CrossRef]
- Brash, A.R.; Baertschi, S.W.; Ingram, C.D.; Harris, T.M. Isolation and characterization of natural allene oxides: Unstable intermediates in the metabolism of lipid hydroperoxides. Proc. Natl. Acad. Sci. USA 1988, 85, 3382–3386. [Google Scholar] [CrossRef]
- Schaller, A.; Stintzi, A. Enzymes in jasmonate biosynthesis–Structure, function, regulation. Phytochemistry 2009, 70, 1532–1538. [Google Scholar] [CrossRef] [PubMed]
- Stenzel, I.; Hause, B.; Miersch, O.; Kurz, T.; Maucher, H.; Weichert, H.; Ziegler, J.; Feussner, I.; Wasternack, C. Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol. Biol. 2003, 51, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.; Carrier, D.J.; Schaedler, T.; Waterham, H.R.; van Roermund, C.W.; Theodoulou, F.L. Peroxisomal ABC transporters: Functions and mechanism. Biochem. Soc. Trans. 2015, 43, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Browse, J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Koo, A.J.K.; Howe, G.A. Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Front. Plant Sci. 2012, 3, 19. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi Pirbalouti, A.; Sajjadi, S.E.; Parang, K. A review (research and patents) on jasmonic acid and its derivatives. Arch. der Pharm. 2014, 347, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.; Yan, C.; Li, L.; Xie, D.; Li, C. Jasmonates. In Hormone Metabolism and Signaling in Plants, 1st ed.; Li, J., Li, C., Smith, S.M., Eds.; Academic Press: London, UK, 2017; pp. 243–272. [Google Scholar]
- Ghelli, R.; Brunetti, P.; Napoli, N.; De Paolis, A.; Cecchetti, V.; Tsuge, T.; Serino, G.; Matsui, M.; Mele, G.; Rinaldi, G. A newly identified flower-specific splice variant of AUXIN RESPONSE FACTOR8 regulates stamen elongation and endothecium lignification in Arabidopsis. Plant Cell 2018, 30, 620–637. [Google Scholar] [CrossRef]
- Zhang, C.; Lei, Y.; Lu, C.; Wang, L.; Wu, J. MYC2, MYC3, and MYC4 function additively in wounding-induced jasmonic acid biosynthesis and catabolism. J. Integr. Plant Biol. 2020, 62, 1159–1175. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, Perception, Signal Transduction and Action in Plant Stress Response, Growth and Development. An Update to the 2007 Review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef]
- McConn, M.; Browse, J. The Critical Requirement for Linolenic Acid Is Pollen Development, Not Photosynthesis, in an Arabidopsis Mutant. Plant Cell 1996, 8, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Halitschke, R.; Kim, H.B.; Baldwin, I.T.; Feldmann, K.A.; Feyereisen, R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002, 31, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Caldelari, D.; Wang, G.; Farmer, E.E.; Dong, X. Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 2011, 75, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Browse, J. The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry 2009, 70, 1539–1546. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, K.; Ishiguro, S.; Okada, K.; Takasaki, T.; Hinata, K. Antisense inhibition of a nuclear gene, BrDAD1, in Brassica causes male sterility that is restorable with jasmonic acid treatment. Mol. Breed. 2003, 11, 325–336. [Google Scholar] [CrossRef]
- Cai, Q.; Yuan, Z.; Chen, M.; Yin, C.; Luo, Z.; Zhao, X.; Liang, W.; Hu, J.; Zhang, D. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lunde, C.; Kimberlin, A.; Leiboff, S.; Koo, A.J.; Hake, S. Tasselseed5 overexpresses a wound-inducible enzyme, ZmCYP94B1, that affects jasmonate catabolism, sex determination, and plant architecture in maize. Commun. Biol. 2019, 2, 1–11. [Google Scholar] [CrossRef]
- Acosta, I.F.; Laparra, H.; Romero, S.P.; Schmelz, E.; Hamberg, M.; Mottinger, J.P.; Moreno, M.A.; Dellaporta, S.L. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 2009, 323, 262–265. [Google Scholar] [CrossRef]
- Yan, Y.; Christensen, S.; Isakeit, T.; Engelberth, J.; Meeley, R.; Hayward, A.; Emery, R.N.; Kolomiets, M.V. Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell 2012, 24, 1420–1436. [Google Scholar] [CrossRef]
- Browse, J. Jasmonate: Preventing the maize tassel from getting in touch with his feminine side. Sci. Signal. 2009, 2, pe9. [Google Scholar] [CrossRef]
- Ito, T.; Ng, K.-H.; Lim, T.-S.; Yu, H.; Meyerowitz, E.M. The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell 2007, 19, 3516–3529. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-G.; Lee, S.; Kim, Y.-S.; Yun, D.-J.; Woo, J.-C.; Park, C.-M. Activation tagging of an Arabidopsis SHI-RELATED SEQUENCE gene produces abnormal anther dehiscence and floral development. Plant Mol. Biol. 2010, 74, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Cecchetti, V.; Altamura, M.M.; Brunetti, P.; Petrocelli, V.; Falasca, G.; Ljung, K.; Costantino, P.; Cardarelli, M. Auxin controls Arabidopsis anther dehiscence by regulating endothecium lignification and jasmonic acid biosynthesis. Plant J. 2013, 74, 411–422. [Google Scholar] [CrossRef]
- Ruduś, I.; Terai, H.; Shimizu, T.; Kojima, H.; Hattori, K.; Nishimori, Y.; Tsukagoshi, H.; Kamiya, Y.; Seo, M.; Nakamura, K.; et al. Wound-induced expression of DEFECTIVE IN ANTHER DEHISCENCE1 and DAD1-like lipase genes is mediated by both CORONATINE INSENSITIVE1-dependent and independent pathways in Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.J.; Shih, C.F.; Yang, J.Y.; Tan, C.M.; Hsu, W.H.; Huang, Y.P.; Liao, P.C.; Yang, C.H. A RING-type E3 ligase controls anther dehiscence by activating the jasmonate biosynthetic pathway gene DEFECTIVE IN ANTHER DEHISCENCE1 in Arabidopsis. Plant J. 2013, 74, 310–327. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.Z.; Xu, Z.F. Benzyladenine Treatment Significantly Increases the Seed Yield of the Biofuel Plant Jatropha curcas. J. Plant Growth Regul. 2011, 30, 166–174. [Google Scholar] [CrossRef]
- Zhang, X.; Pan, B.-Z.; Chen, M.; Chen, W.; Li, J.; Xu, Z.-F.; Liu, C. JCDB: A comprehensive knowledge base for Jatropha curcas, an emerging model for woody energy plants. BMC Genom. 2019, 20, 958. [Google Scholar] [CrossRef] [PubMed]
- Vaknin, Y.; Yermiyahu, U.; Bar-Tal, A.; Samocha, Y. Global maximization of Jatropha oil production under semi-arid conditions by balancing vegetative growth with reproductive capacity. GCB Bioenergy 2018, 10, 382–392. [Google Scholar] [CrossRef]
- Mazumdar, P.; Singh, P.; Babu, S.; Siva, R.; Harikrishna, J.A. An update on biological advancement of Jatropha curcas L.: New insight and challenges. Renew. Sustain. Energy Rev. 2018, 91, 903–917. [Google Scholar] [CrossRef]
- Triadiati, T.; Kurniati, K.; Widyastuti, U.; Dasumiati, D. Androgynomonoecious Jatropha curcas: Chromosomes, Isozymes, and Flowers Gender. HAYATI J. Biosci. 2019, 26, 139–146. [Google Scholar]
- Dasumiata, D.; Miftahudin, M.; Triadiati, T.; Hartana, A. Short Communication: Sex types in flowering of Jatropha curcas. Biodiversitas 2017, 18, 275–279. [Google Scholar]
- Chen, M.-S.; Pan, B.-Z.; Fu, Q.; Tao, Y.-B.; Martínez-Herrera, J.; Niu, L.; Ni, J.; Dong, Y.; Zhao, M.-L.; Xu, Z.-F. Comparative transcriptome analysis between gynoecious and monoecious plants identifies regulatory networks controlling sex determination in Jatropha curcas. Front. Plant Sci. 2017, 7, 1953. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-S.; Niu, L.; Zhao, M.-L.; Xu, C.; Pan, B.-Z.; Fu, Q.; Tao, Y.-B.; He, H.; Hou, C.; Xu, Z.-F. De novo genome assembly and Hi-C analysis reveal an association between chromatin architecture alterations and sex differentiation in the woody plant Jatropha curcas. GigaScience 2020, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zapata, L.; Ding, J.; Willing, E.-M.; Hartwig, B.; Bezdan, D.; Jiao, W.-B.; Patel, V.; Velikkakam James, G.; Koornneef, M.; Ossowski, S.; et al. Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc. Natl. Acad. Sci. USA 2016, 113, E4052–E4060. [Google Scholar] [CrossRef]
- Li, H.; Xue, D.; Gao, Z.; Yan, M.; Xu, W.; Xing, Z.; Huang, D.; Qian, Q.; Xue, Y. A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. Plant J. 2009, 57, 593–605. [Google Scholar] [CrossRef]
- Matsui, K.; Fukutomi, S.; Ishii, M.; Kajiwara, T. A tomato lipase homologous to DAD1 (LeLID1) is induced in post-germinative growing stage and encodes a triacylglycerol lipase. FEBS Lett. 2004, 569, 195–200. [Google Scholar] [CrossRef]
- Yuan, Z.; Zhang, D. Roles of jasmonate signalling in plant inflorescence and flower development. Curr. Opin. Plant Biol. 2015, 27, 44–51. [Google Scholar] [CrossRef]
- Yan, Y.; Borrego, E.; Kolomiets, M. Jasmonate biosynthesis, perception and function in plant development and stress responses. In Lipid Metabolism; InTech: London, UK, 2013; pp. 393–442. [Google Scholar]
- Krizek, B.A.; Anderson, J.T. Control of flower size. J. Exp. Bot. 2013, 64, 1427–1437. [Google Scholar] [CrossRef]
- Nishijima, T. Large Flower Size: Molecular Basis and Role of Cytokinin. J. Jpn. Soc. Hortic. Sci. 2012, 81, 129–139. [Google Scholar] [CrossRef][Green Version]
- Makwana, V.; Shukla, P.; Robin, P. GA application induces alteration in sex ratio and cell death in Jatropha curcas. Plant Growth Regul. 2010, 61, 121–125. [Google Scholar] [CrossRef]
- Hui, W.K.; Wang, Y.; Chen, X.Y.; Zayed, M.Z.; Wu, G.J. Analysis of Transcriptional Responses of the Inflorescence Meristems in Jatropha curcas Following Gibberellin Treatment. Int. J. Mol. Sci. 2018, 19, 432. [Google Scholar] [CrossRef] [PubMed]
- Li, J.L.; Pan, B.Z.; Niu, L.J.; Chen, M.S.; Tang, M.Y.; Xu, Z.F. Gibberellin Inhibits Floral Initiation in the Perennial Woody Plant Jatropha curcas. J. Plant Growth Regul. 2018, 37, 999–1006. [Google Scholar] [CrossRef]
- Ming, X.; Tao, Y.-B.; Fu, Q.; Tang, M.; He, H.; Chen, M.-S.; Pan, B.-Z.; Xu, Z.-F. Flower-Specific Overproduction of Cytokinins Altered Flower Development and Sex Expression in the Perennial Woody Plant Jatropha curcas L. Int. J. Mol. Sci. 2020, 21, 640. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics (Oxf. Engl.) 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Fu, Q.; Niu, L.; Luo, L.; Chen, J.; Xu, Z.-F. Three TFL1 homologues regulate floral initiation in the biofuel plant Jatropha curcas. Sci. Rep. 2017, 7, 43090. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Li, C.; Tang, M.; Tao, Y.-B.; Pan, B.-Z.; Zhang, L.; Niu, L.; He, H.; Wang, X.; Xu, Z.-F. An efficient protocol for Agrobacterium-mediated transformation of the biofuel plant Jatropha curcas by optimizing kanamycin concentration and duration of delayed selection. Plant Biotechnol. Rep. 2015, 9, 405–416. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wu, J.; Hettenhausen, C.; Meldau, S.; Baldwin, I.T. Herbivory Rapidly Activates MAPK Signaling in Attacked and Unattacked Leaf Regions but Not between Leaves of Nicotiana attenuata. Plant Cell 2007, 19, 1096–1122. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, C.-J.; Zhao, M.-L.; Chen, M.-S.; Xu, Z.-F. Silencing of the Ortholog of DEFECTIVE IN ANTHER DEHISCENCE 1 Gene in the Woody Perennial Jatropha curcas Alters Flower and Fruit Development. Int. J. Mol. Sci. 2020, 21, 8923. https://doi.org/10.3390/ijms21238923
Xu C-J, Zhao M-L, Chen M-S, Xu Z-F. Silencing of the Ortholog of DEFECTIVE IN ANTHER DEHISCENCE 1 Gene in the Woody Perennial Jatropha curcas Alters Flower and Fruit Development. International Journal of Molecular Sciences. 2020; 21(23):8923. https://doi.org/10.3390/ijms21238923
Chicago/Turabian StyleXu, Chuan-Jia, Mei-Li Zhao, Mao-Sheng Chen, and Zeng-Fu Xu. 2020. "Silencing of the Ortholog of DEFECTIVE IN ANTHER DEHISCENCE 1 Gene in the Woody Perennial Jatropha curcas Alters Flower and Fruit Development" International Journal of Molecular Sciences 21, no. 23: 8923. https://doi.org/10.3390/ijms21238923
APA StyleXu, C.-J., Zhao, M.-L., Chen, M.-S., & Xu, Z.-F. (2020). Silencing of the Ortholog of DEFECTIVE IN ANTHER DEHISCENCE 1 Gene in the Woody Perennial Jatropha curcas Alters Flower and Fruit Development. International Journal of Molecular Sciences, 21(23), 8923. https://doi.org/10.3390/ijms21238923





