Abstract
Pancreatic cancer remains one of the most difficult malignancies to treat. Minimal improvements in patient outcomes and persistently abysmal patient survival rates underscore the great need for new treatment strategies. Currently, there is intense interest in therapeutic strategies that target tyrosine protein kinases. Here, we employed kinome arrays and bioinformatic pipelines capable of identifying differentially active protein tyrosine kinases in different patient-derived pancreatic ductal adenocarcinoma (PDAC) cell lines and wild-type pancreatic tissue to investigate the unique kinomic networks of PDAC samples and posit novel target kinases for pancreatic cancer therapy. Consistent with previously described reports, the resultant peptide-based kinome array profiles identified increased protein tyrosine kinase activity in pancreatic cancer for the following kinases: epidermal growth factor receptor (EGFR), fms related receptor tyrosine kinase 4/vascular endothelial growth factor receptor 3 (FLT4/VEGFR-3), insulin receptor (INSR), ephrin receptor A2 (EPHA2), platelet derived growth factor receptor alpha (PDGFRA), SRC proto-oncogene kinase (SRC), and tyrosine kinase non receptor 2 (TNK2). Furthermore, this study identified increased activity for protein tyrosine kinases with limited prior evidence of differential activity in pancreatic cancer. These protein tyrosine kinases include B lymphoid kinase (BLK), Fyn-related kinase (FRK), Lck/Yes-related novel kinase (LYN), FYN proto-oncogene kinase (FYN), lymphocyte cell-specific kinase (LCK), tec protein kinase (TEC), hemopoietic cell kinase (HCK), ABL proto-oncogene 2 kinase (ABL2), discoidin domain receptor 1 kinase (DDR1), and ephrin receptor A8 kinase (EPHA8). Together, these results support the utility of peptide array kinomic analyses in the generation of potential candidate kinases for future pancreatic cancer therapeutic development.
1. Introduction
In vivo, kinases are heavily trafficked and compartmentalized to microdomains and cellular substructures where they act in concert with other kinases, receptors, and effector proteins to realize intricate signaling mechanisms that give rise to complex cellular behaviors. Despite this complexity, kinases are traditionally studied in isolation: A kinase of interest is typically purified from a sample and the enzymatic activity of the isolated kinase is subsequently interrogated. Kinase isolation is often dependent upon antibody binding and subject to the challenges and expenses of antibody-based purification techniques. Traditional assays remove each kinase of interest from its interacting partners and the physiologic conditions in which it normally operates. By breaking these networks into discrete components and examining individual kinases in isolation, researchers discover initiators of kinase phosphorylation activity and catalogue the peptide targets that are activated or deactivated by direct phosphorylation by a specific kinase. Unfortunately, these purification techniques incentivize researchers to minimize the integrity of a kinase’s normal physiologic environment in favor of a clean, easily assayable sample. However, the cleaner a purified kinase sample becomes, the less it can represent that kinase’s activity under physiological endogenous conditions. While this reductionism continues to allow researchers to meaningfully investigate discrete components of kinomic signaling networks, the knowledge gained remains incomplete. In this study, we combined PamGene multiplexed kinome activity array data with four bioinformatic pipelines to identify protein kinases responsible for the differential phosphorylation activity observed in patient-derived and commercial pancreatic ductal adenocarcinoma (PDAC) cell lines compared to patient-derived wild-type pancreatic tissue specimens (Figure 1).
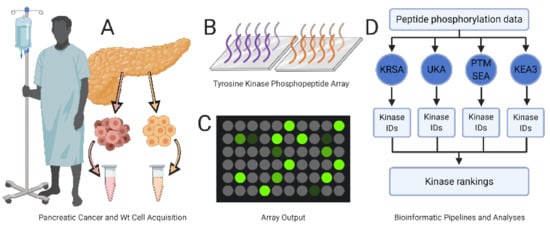
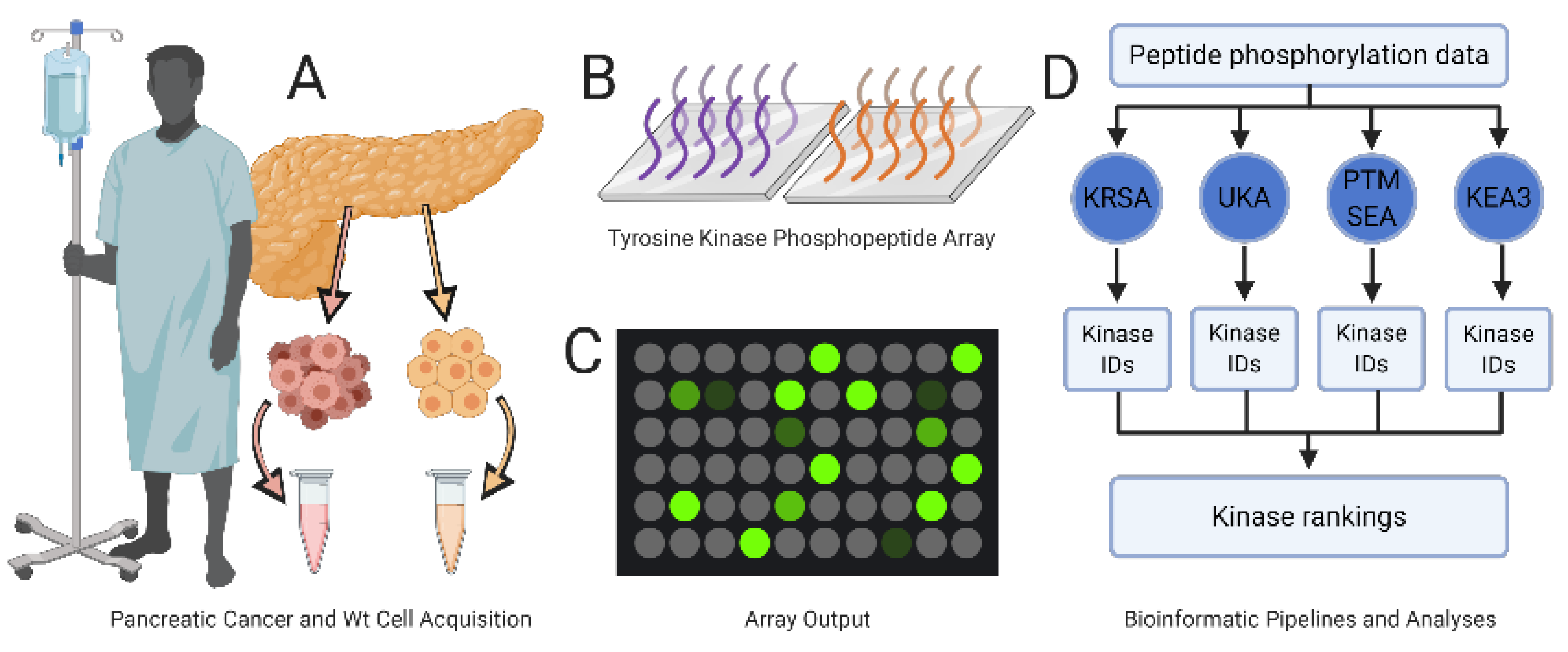
Figure 1.
Experimental design. (A) Patient-derived pancreatic cancer cells (light red) and wild-type pancreatic tissue specimens (yellow) are processed and diluted to a uniform protein concentration. (B) Samples are added to the PamChip array containing 196 consensus phosphopeptide sequences immobilized on porous ceramic membranes; two (purple and orange) such sequences are illustrated here. (C) Quantification of peptide phosphorylation levels. (D) Peptide phosphorylation data are analyzed with each of four independent bioinformatic pipelines (KRSA, UKA, PTM-SEA, KEA3) and then combined to generate a list of tyrosine protein kinases’ targets.
Protein kinases are enzymes capable of phosphorylating other proteins to regulate biochemical signaling pathways and modulate cellular behavior [1]. In cancer, many protein kinases are associated with cancer cell initiation, progression, and metastasis, as well as relapse and survival. Small-molecule protein kinase inhibitors represent an increasingly successful therapeutic strategy for a range of cancer types with over 50 FDA-approved protein kinase inhibitors in clinical use [2,3,4] and hundreds more undergoing clinical or preclinical trials. Protein kinase inhibitors improve patient outcomes significantly and protein kinases are, therefore, a popular target for difficult-to-treat malignancies such as pancreatic cancer. Pancreatic cancer patients demonstrate the lowest five-year relative survival rates of any cancer type, with decades of medical research unable to increase these rates beyond 9% [5]. Protein kinase inhibitors erlotinib (OSI-774; Tarceva), everolimus (RAD001; Afinitor), and sumitinib (SU11248; Sutent) are approved for the treatment of pancreatic cancers [4], though the majority of these approvals are for a rare (1% to 2%) subtype of pancreatic cancer known as pancreatic neuroendocrine tumors [6] rather than the far more common PDAC.
Previous, excellent genomic and proteomic studies have characterized PDAC subtypes [7,8,9,10,11]. Different and new PDAC subtypes continue to be defined, with myriad genetic or molecular factors increasing the resolution of the disease spectrum. While the present study could not comprehensively address the complexity and heterogeneity of PDAC tumor subtyping, our results inform growing understanding of the unique protein tyrosine kinase networks that play a role in one or more of the commercial and patient-derived cell lines investigated (Figure 2, Figure 3 and Figure 4).
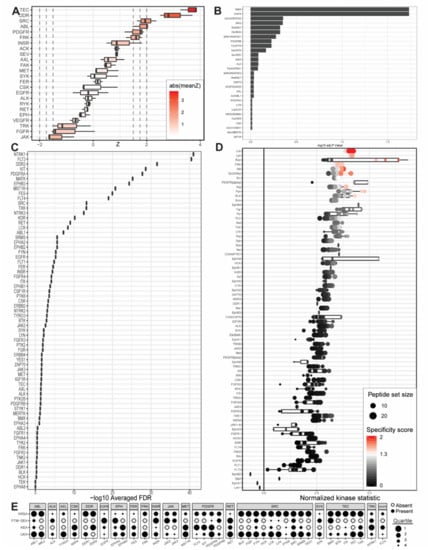
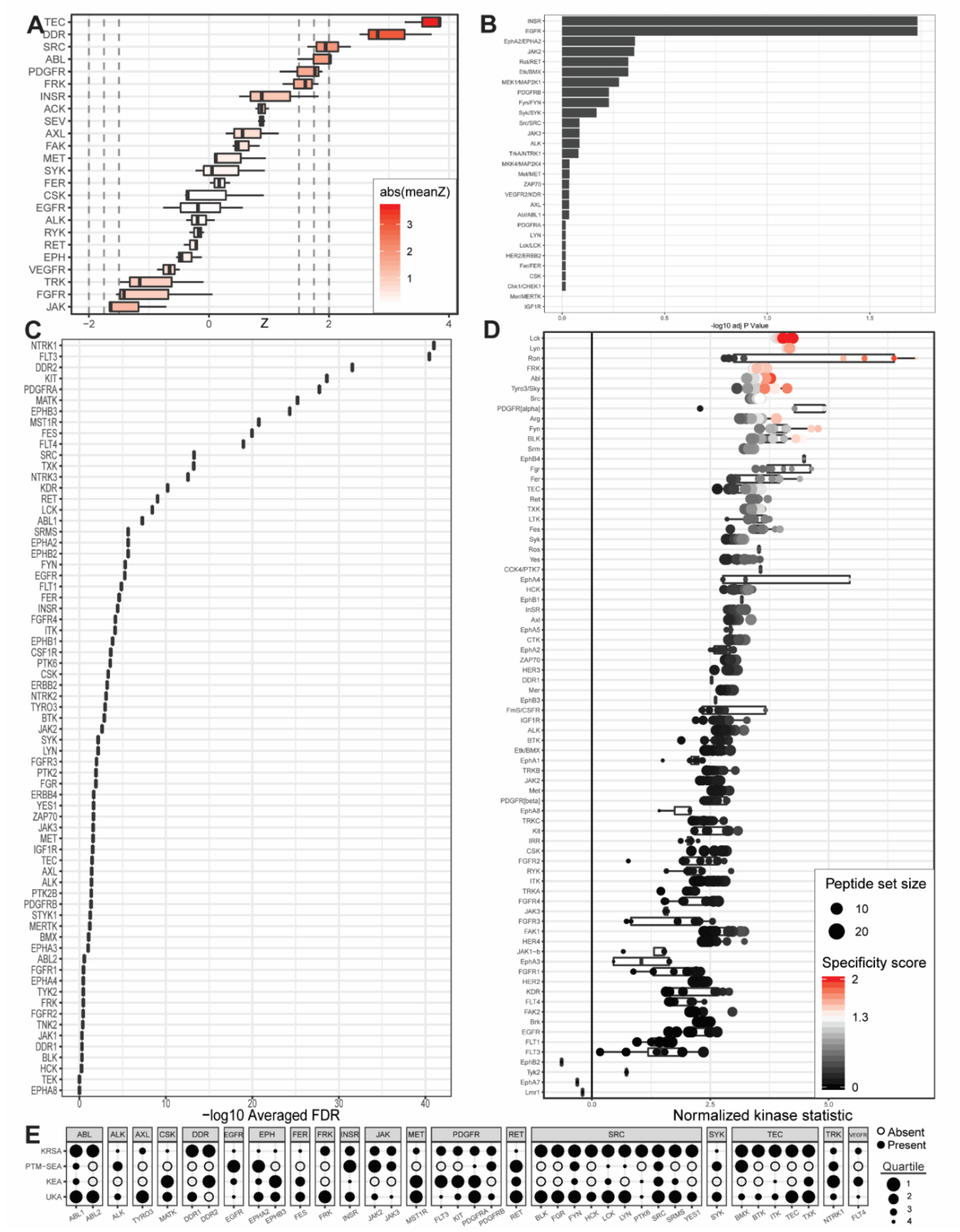
Figure 2.
Outputs from upstream kinase identification pipelines for the commercially available PANC1 PDAC cell line compared to patient-derived wild-type pancreatic tissue. (A) Kinome Random Sampling Analyzer (KRSA); (B) Post-Translational Modification Signature Enrichment Analysis (PTM-SEA); (C) Kinase Enrichment Analysis Version 3 (KEA3); (D) Upstream Kinase Analysis (UKA); (E) Quartile summary. A more detailed figure legend can be found in Appendix B.

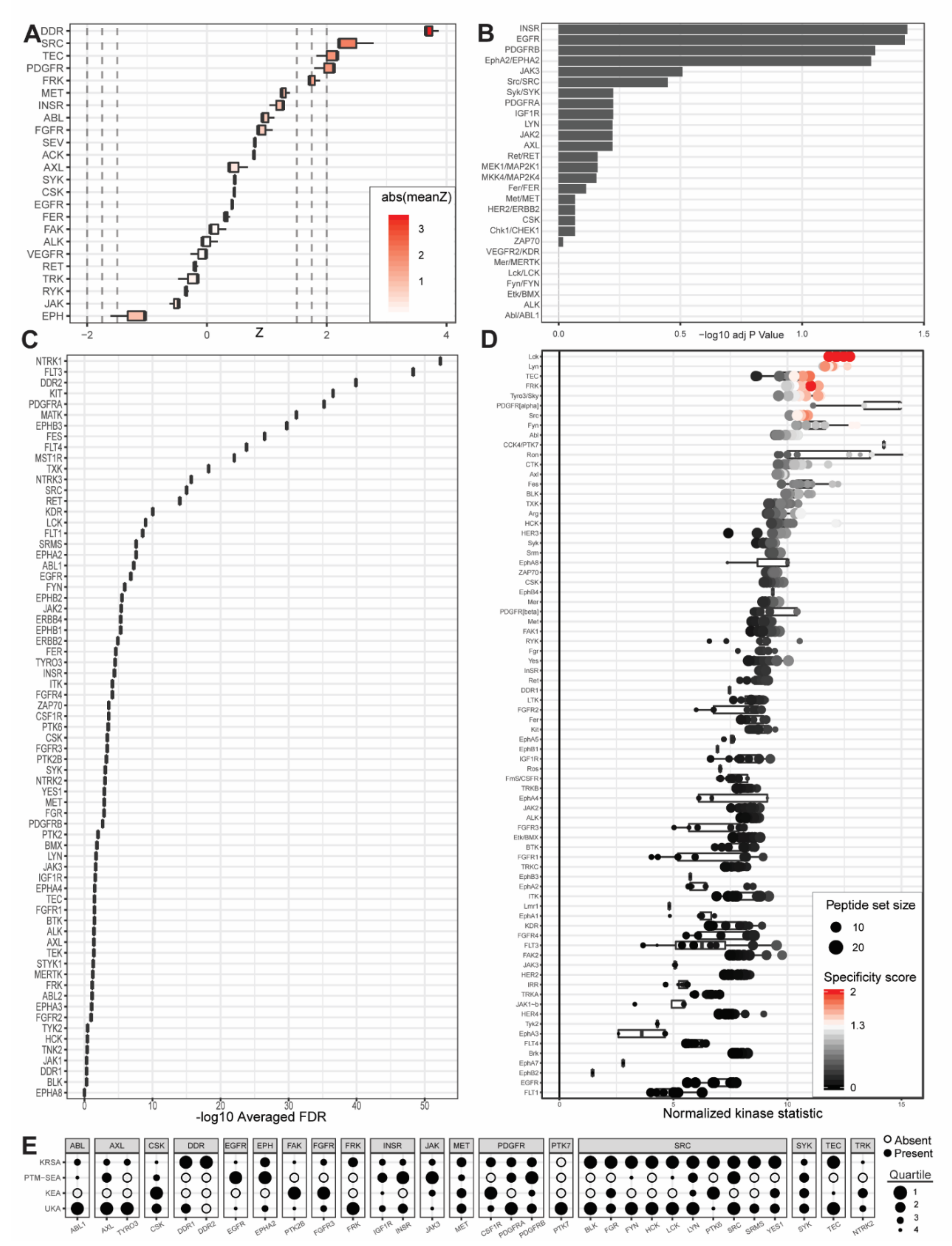
Figure 3.
Outputs from upstream kinase identification pipelines for the patient-derived PDCL15 PDAC cell line compared to patient-derived wild-type pancreatic tissue. (A) Kinome Random Sampling Analyzer (KRSA); (B) Post-Translational Modification Signature Enrichment Analysis (PTM-SEA); (C) Kinase Enrichment Analysis Version 3 (KEA3); (D) Upstream Kinase Analysis (UKA). (E) Quartile summary. A more detailed figure legend can be found in Appendix B.
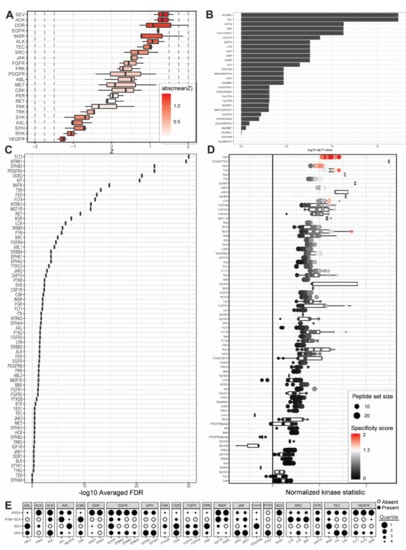
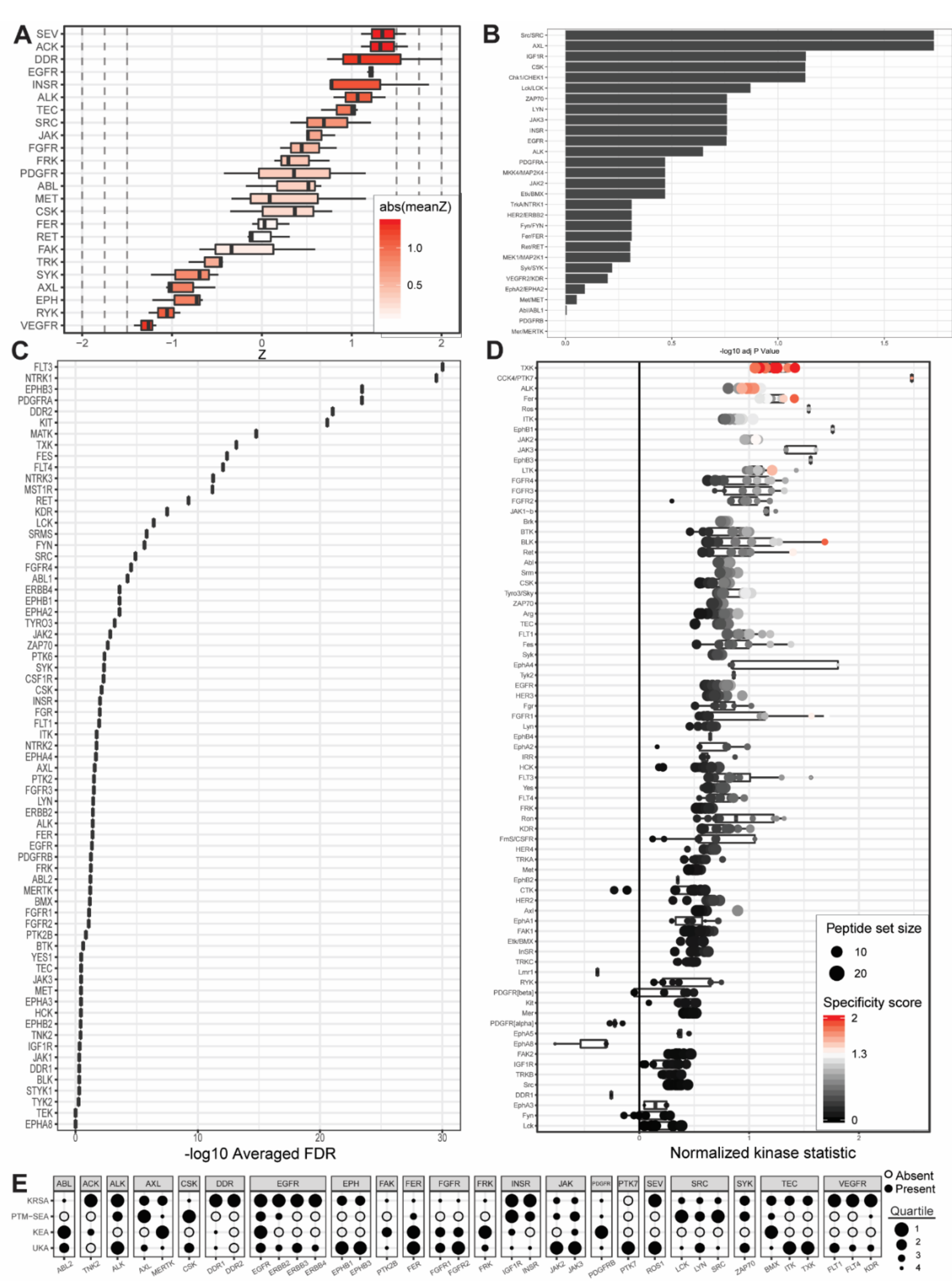
Figure 4.
Outputs from upstream kinase identification pipelines for the patient-derived PDCL5 PDAC cell line compared to patient-derived wild-type pancreatic tissue. (A) Kinome Random Sampling Analyzer (KRSA); (B) Post-Translational Modification Signature Enrichment Analysis (PTM-SEA); (C) Kinase Enrichment Analysis Version 3 (KEA3); (D) Upstream Kinase Analysis (UKA). (E) Quartile summary. A more detailed figure legend can be found in Appendix B.
To define kinases that are differentially active in one or more PDAC cell lines and identify potentially actionable drug targets, we sought to break away from linear, one-by-one investigations of individual PDAC kinases and consider instead the totality of PDAC kinase networks within each group. To accomplish this, we combined emerging laboratory technologies and contemporary bioinformatic pipelines to identify lead candidate kinases based on peptide phosphorylation signatures. These technologies include the PamStation and the Protein Tyrosine Kinase PamChip. The bioinformatic pipelines include the Kinome Random Sampling Analyzer (KRSA) pipeline, developed by our own laboratory [12,13,14,15,16,17,18,19], and the Upstream Kinase Analysis (UKA) pipeline, which is part of the BioNavigator software tool developed by collaborators at PamGene [20,21]. To passively validate and further contextualize the results of this approach, two additional bioinformatic pipelines were utilized. These additional bioinformatic pipelines include the Post-Translational Modification Signature Enrichment Analysis (PTM-SEA) pipeline developed at the Broad Institute of MIT and Harvard [22], as well as the Kinase Enrichment Analysis Version 3 (KEA3) developed by the Ma’ayan laboratory [23]. Using laboratory equipment designed by PamGene, we measured the relative phosphorylation levels of 198 representative peptide substrates for PDAC cells and wild-type pancreatic tissue before running the resultant data through each of these four self-contained analytical platforms. The analytical results from each pipeline were interpreted individually as well as in combination to maximize the strengths of each pipeline’s respective algorithm and reference database set. The results were synthesized to identify kinases likely responsible for the unique phosphorylation signatures of PDAC (Figure 2, Figure 3 and Figure 4).
2. Results
2.1. PamGene Kinome Activity Profiling Using Protein Tyrosine Kinase PamChip®
The PamChip is a kind of array. Groups of identical peptide fragments populate a single spot on the chip (Figure 1). The peptide spots contain amino acid sequences of sufficient length and complexity to represent the phosphorylation sites of specific proteins. When these peptide spots are exposed to experimental samples, they become phosphorylated by the activated kinases within that sample. Phosphorylation events correspond precisely with the enzymatic activity of the sample’s kinase networks. Fluorescently tagged phosphoantibodies produce a signal proportional to the phosphorylation activity of the kinases contained within the sample. Using specific collections of peptide fragments, the intensities reported by a PamChip provide a signature that can be used to identify the kinases responsible for the observed phosphorylation activity. UKA or KRSA pipelines process differentially phosphorylated peptide fragment identities and signal intensities. For PTM-SEA and KEA3 pipelines to evaluate PamChip signatures, the peptide fragments must first be converted to protein identities. Because some peptide spots on the PamChip represent multiple proteins and because multiple peptide spots may represent different sites on a single protein, this conversion results in a loss of information. The signal intensities on the PamChip—which quantify the degree to which a peptide fragment has been phosphorylated—are also lost when PamChip signatures are converted to the binary “differentially expressed” input categories required for PTM-SEA or KEA3 pipelines. In other words, instead of interpreting “peptide-fragment-1 with signal intensity 10.3,” PTM-SEA or KEA3 only process “protein-a.” For this reason, we preferentially used the UKA and KRSA pipelines for candidate kinase identification, using the less well-suited (but excellent in their own right) PTM-SEA and KEA3 pipelines for passive validation.
Unabridged comparison of the upstream kinases that each pipeline identified as being responsible for the observed phosphorylation patterns of each pancreatic cancer cell line compared to control wild-type patient-derived pancreatic cells are presented in Figure 2, Figure 3 and Figure 4 as well as Supplementary Table S1. The databases associated with each pipeline offer different levels of kinase coverage. This is advantageous for identification of true positives, although the absence of a kinase from a pipeline’s identification output cannot be considered a negative indication of that kinase serving as a causative factor in the phosphorylation patterns observed. This approach, therefore, minimizes type I error (i.e., erroneous rejection of a true null hypothesis) and accommodates type II error (i.e., erroneous acceptance of a false null hypothesis). UKA and KRSA bioinformatic pipelines are specifically designed to analyze PamChip kinome activity data. As such, each respective algorithm can integrate multiple PamChip data metrics into its final analytical output. The PTM-SEA and KEA3 pipelines, although capable of excellent analytic activity, are designed for slightly different applications. In their native state, significant programmatic alterations are required to generate results that meaningfully interpret PamChip experimental data. The most significant differences between UKA or KRSA bioinformatic pipelines and PTM-SEA or KEA3 bioinformatic pipelines relate to how these two groups handle multiple dimensions of data and how these two groups natively interpret phosphorylated peptides.
2.2. UKA and KRSA Combinatory Analysis
In Table 1, we average the percentile rankings without any concern for false negatives. The goal was to increase positive identification of kinases whose activity is truly a causative factor in the differential phosphorylation patterns observed between tumor and wild-type cells. In Table 2, we attempt to decrease the presentation of false positives by dividing these averages by the number of pipelines employed—in this case, two. In Table 1 and Table 2, we also combine the results from the two patient-derived pancreatic cancer cell lines (PDCL15, PDCL5; Patient-Derived) and we combine the results of all pancreatic cancer cell lines (PDCL15, PDCL5, PANC1; All), in order to generate average percentile rankings that inform the general kinase activity of pancreatic cancer. These tables contain only the highest scoring kinases according to UKA and KRSA bioinformatic pipelines. If two or more kinases received identical average percentile ranks or identical weighted average percentile ranks, then rank order was determined arbitrarily. While our complete output data may be found in Table S1, the present tables include only the top 10 highest ranked kinases. Kinases with identical scores were arbitrarily assigned sequential ranks.

Table 1.
UKA and KRSA average percentile rankings.

Table 2.
UKA and KRSA weighted average percentile rankings.
2.3. Expanded PTM-SEA and KEA3 Combinatory Analysis
Table 3 and Table 4 expand our analyses to include results obtained through PTM-SEA and KEA3 bioinformatic pipelines. Table 3 provides unweighted average percentile rankings. Table 4 attempts to decrease false positives by weighting these averages according to the number of pipelines which identify a given kinase as being responsible for the observed phosphorylation differences between a pancreatic cancer cell line and patient-derived wild-type pancreas. As above, Table 3 and Table 4 also combine results from our two patient-derived pancreatic cancer cell lines (PDCL15, PDCL5; Patient-Derived) as well as from all pancreatic cancer cell lines (PDCL15, PDCL5, PANC1; All).

Table 3.
All pipelines’ (KRSA, UKA, PTM-SEA, and KEA3) average percentile rankings.

Table 4.
All pipelines’ (KRSA, UKA, PTM-SEA, and KEA3) weighted average percentile rankings.
3. Discussion
3.1. Identification of Lead Candidate Kinases
Our results confirm the activity of known protein tyrosine kinase-related pathways previously reported as perturbed in pancreatic cancer [24,25,26,27,28]. These results also identify protein tyrosine kinases as yet understudied or unreported in pancreatic cancer. Because our experimental model allows us to maintain the integrity of kinase networks, our data suggest involvement of signaling pathways and regulatory cascades in pancreatic cancer pathophysiology. This study presents evidence in support of the continued development of previously established inhibitory therapeutics that target select protein kinases, such as epidermal growth factor receptor (EGFR) [25], ephrin receptor A2 (EPHA2) [29], and SRC proto-oncogene kinase (SRC) [27], and our experimental results identify new PDAC targets, such as B lymphoid kinase (BLK), lymphocyte cell-specific kinase (LCK), and ABL proto-oncogene 2 kinase (ABL2), which may play a critical role in cancer cell biochemistry or desmoplastic inflammatory cell behavior.
While data gained from reductionist kinase investigations have previously been used to support other complex biochemical studies such as mass spectrometry-based proteomic studies [30], peptide-based kinome array profiling offers unique advantages. Traditionally, peptides identified as kinase targets are probed in vitro and in vivo to examine the behavior of a given kinase. Following that, genetic techniques that knockdown or constitutively express a given kinase further probe the activity of that kinase within normal or experimental biological milieus. But many of these strategies are unable to measure multiple kinases simultaneously while also maintaining kinomic network integrity in complex biological samples. Peptide-based kinome array profiling can accomplish this by overcoming kinase isolation requirements in order to evaluate hundreds of kinases simultaneously. This study was designed as a series of hypotheses generating experiments. Although our experimental designs did not allow us to draw definitive conclusions, our experimental results fall into one of two major categories, which we have termed “reference kinases” and “neoteric kinases.” The first category, “reference kinases,” represents kinases with well-established roles in human cancer pathophysiology. The second category, “neoteric kinases,” represents candidate kinases potentially contributing to PDAC pathology in new or previously understudied ways.
3.2. Reference Kinases
Reference kinases include protein tyrosine kinases identified by our study and subsequent bioinformatic analyses that recapitulate previously reported findings in the field of human cancer biology. Kinases in this category provide reference data that passively validate our experimental observations and contextualize our results within the scope of verified kinase discoveries.
One notable reference kinase identified as differentially active in our study of PDAC cells is the EGFR tyrosine kinase. Previously, EGFR has been linked to pancreatic tumor size, advanced clinical staging, and poor survival [28]. The expression frequency of EGFR in human pancreatic carcinomas is reported as 43% [28] and 68.4% in primary invasive ductal carcinoma of the pancreas [31] with elevated expression of EGFR activating ligands also reported. Consistent with these reports, our results show differential EGFR activity in weighted analyses of PDCL5 (Figure 4, Table 2 and Table 4) and aggregated patient-derived cell lines (Table 4). Directionality (increased kinase phosphorylation activity or decreased kinase phosphorylation activity) within each cell line can be gleaned from KRSA’s report of the log2-fold change of phosphorylated peptide substrates attributed to each kinase family (Figure A1, Figure A2, or Figure A3) or by UKA’s report of an individual kinase’s mean kinase statistic (Table A1). In our study, EGFR demonstrated increased phosphorylation activity in pancreatic cancer compared to control. Inhibition of the EGFR tyrosine kinase improves survival in PDAC animal models [25]. As such, targeted inhibition of tyrosine kinases, including EGFR, is a popular goal of many emerging therapeutic strategies [2]. Erlotinib (OSI-774) is an FDA-approved small-molecule EGFR tyrosine kinase inhibitor for use in pancreatic cancer [3]. Many additional small-molecule tyrosine kinase inhibitors are currently under study or in various stages of clinical trial for their putative role in pancreatic cancer pathophysiology.
The vascular endothelial growth factor receptor (VEGFR) tyrosine kinase family is also heavily implicated in the development of pancreatic cancer [32]. VEGFR-3, also known as fms related receptor tyrosine kinase 4 (FLT4), has been explored as a target for pancreatic cancer therapy [24,33] with significantly upregulated FLT4 expression documented in pancreatic cancer specimens [34,35]. Beyond PDAC, single nucleotide polymorphisms (SNPs) of FLT4 correlate with decreased progression-free survival of patients with gastroenteropancreatic neuroendocrine neoplasms [36]. Our results demonstrate increased FLT4 kinase activity in PDAC cells. In the present study, we identified FLT4 as one of the most differentially active kinases in PDCL5 patient-derived PDAC samples according to average percentile rankings across all pipelines (Table 3). KRSA and UKA directionality metrics (Figure A3, Table A1) demonstrate increased FLT4 activity in pancreatic cancer compared to wild-type controls.
Several kinases consistently identified as differentially active across multiple pipelines, cell lines, or final combinatorial analyses recapitulate kinases previously identified as playing well-established roles in a variety of human cancer pathologies. These kinases include insulin receptor (INSR) kinase [37,38] (Figure 2 and Figure 3, Table 3 and Table 4), EPHA2 kinase [29,39,40,41,42,43,44,45,46] (Figure 2 and Figure 3, Table 4), platelet-derived growth factor receptor alpha (PDGFRA) kinase [47,48,49,50,51] (Figure 2 and Figure 3, Table 1, Table 2, Table 3 and Table 4), SRC kinase [26,27,52,53,54,55,56,57] (Figure 2 and Figure 3, Table 1, Table 2, Table 3 and Table 4), and tyrosine kinase nonreceptor 2 (TNK2) kinase [58,59,60,61,62,63,64] (Figure 4, Table 1). Of these, EPHA2 is particularly well characterized in PDAC with other groups recently presenting evidence of EPHA2-mediated drug resistance in pancreatic cancer cells [29] and proposing EPHA2 as a potential biomarker or therapeutic target in pancreatic cancer [29,40]. SRC, too, has a well-established evidence base supporting its role in PDAC [27]. Furthermore, many of these kinases are known to constitute important signaling axes in pancreatic cancer. PDGFR/SRC signaling is a therapeutic target in pancreatic cancer [26] with reports of SRC also potentiating PDGFRA activity in other cancer models. TNK2 (also known as ACK1) associates with EGFR in cancer cells to maintain EGFR on the cell surface and enhance human breast cancer cell migration and invasion [65]. Again, this relationship seems to have some degree of bidirectionality, with EGFR influencing TNK2 activation [66,67]. Identification of multiple kinase pairs constituting previously reported signaling axes is encouraging and supports the validity of our experimental design in maintaining the integrity of kinomic signaling networks.
These results lend strength not only to our experimental and bioinformatic identification of differentially active kinases in pancreatic cancer, but also suggest that kinases identified as among the most strongly differential (e.g., in the top 10) in unweighted average percentile rankings (Table 1 or Table 3) and in the corresponding weighted average percentile rankings (Table 2 or Table 4) may represent kinases highly likely to contribute to the pathophysiologic processes of PDAC.
3.3. Neoteric Kinases
Neoteric kinases represent a second, smaller category of experimental findings that include kinases whose identification in our study and bioinformatic analyses suggest new, hitherto unidentified, or otherwise understudied kinase functionalities in PDAC. Because these kinases are not strictly “novel,” we instead call this group “neoteric” in reference to the emerging roles these kinases may play in pancreatic tumor desmoplasia, immune response, and oncometabolism. Beyond the passive validation that our “reference kinase” group provides, this group of “neoteric kinases” provides potentially novel insights. It became evident, after identifying lead candidate kinases, that our data highlight several potential players in unique aspects of PDAC tumor development. Kinases that appear both in our final unweighted average percentile rankings and in our final weighted average percentile rankings are defined as lead candidate kinases and include BLK (Figure 2 and Figure 3, Table 1 and Table 2), Fyn-related kinase (FRK) (Figure 3, Table 1 and Table 2), Lck/Yes-related novel kinase (LYN) (Figure 2 and Figure 3, Table 1, Table 2, and Table 4), FYN proto-oncogene kinase (FYN) (Figure 2 and Figure 3, Table 1, Table 2, Table 3 and Table 4), LCK (Figure 2 and Figure 3, Table 1, Table 2, Table 3 and Table 4), and tec protein kinase (TEC) (Figure 2 and Figure 3, Table 1, Table 2 and Table 3). Additional kinases identified by one or more bioinformatic pipelines define candidate kinases and include hemopoietic cell kinase (HCK) (Figure 3, Table 2), ABL2 (Figure 2, Table 2), discoidin domain receptor 1 kinase (DDR1) (Table S1), and ephrin receptor A8 kinase (EPHA8) (Table S1). While some of these kinases have been previously identified in kinome or phosphorylome studies of pancreatic cancer (e.g., DDR1 [9], FYN [68]), we classified them as neoteric for the purposes of this discussion because sufficient questions remain as to how these kinases relate to PDAC pathology, treatment, or molecular signaling.
Pronounced deposition of extracellular matrix constituents and the aberrant propagation of fibroblasts characteristic of desmoplasia are common in PDAC tumor microenvironments. Desmoplasia acts as a biophysical barrier contributing to pancreatic cancer therapeutic resistance. Desmoplastic stroma and pancreatic tumor cells interact with one another to elicit complex cellular behaviors with seemingly contradictory roles in PDAC progression [69]. At times pro-tumorigenic [70] and at times anti-tumoral [69], the role of desmoplasia in PDAC remains an active area of study. Our identification of increased HCK, ABL2, DDR1, FYN, and LYN suggests a role for these kinases in the desmoplastic reactions that contribute to the poor survival rates of PDAC patients. HCK overexpression activates fibrotic pathways [71]. ABL2 signaling regulates fibroblast proliferation [72]. DDR1 inhibitors reduce fibrosis in other fibrotic diseases [73,74,75,76]. FYN regulates downstream serine-threonine kinase activities involved in the modulation of fibroblast–epithelial cell interactions and the promotion of organ fibrosis [77,78]. Serotonin promotes fibroblast activation and collagen deposition [79]. LYN mediates pro-tumor serotonin signaling [80].
Desmoplasia serves as the primary source of the cytokines and chemokines that facilitate tumor progression in PDAC [81]. While immunotherapeutic strategies have significantly impacted clinical success in many other human malignancies, pancreatic cancer remains resistant. LCK is an important regulator of immune cell functionality [82]. TEC is a key player in the inflammatory response of pancreatitis [83]. The LCK and TEC kinases identified in the present study may also play a role in the anti-cancer immune response elicited and frequently evaded by PDAC.
The final effector proteins and terminal nodes for many kinase cascades are transcription factors. Our identification of BLK as a differentially active tyrosine kinase in PDAC cells compared to wild-type pancreatic cells presents new insight into the role that the pancreatic and duodenal homeobox 1 (PDX1) transcription factor plays in tumor progression. Overexpression of BLK induces an increase in the PDX1 transcription factor in the cytoplasm and nucleus [84]. The biological functionalities of the PDX1 transcription factor are multitudinous and context dependent. Our group has published extensively on the pro-tumorigenic role of the PDX1 transcription factor in pancreatic cancer progression [85,86,87,88,89,90,91], while other groups have demonstrated antimetastatic [92] and tumor-suppressive effects [93]. Recent evidence shows PDX1 functionality and its multiple—often antagonistic—effects on pancreatic cancer are stage-specific [93,94]. Our data suggest potential mechanistic relationships by which BLK kinase signaling cascades may contribute to PDX1′s multifaceted role in PDAC.
PDX1, originally known as insulin promoter factor 1 (IPF1), also serves as a transcriptional activator for metabolic genes such as insulin and glucose transporter type 2. Intersection between the PDX1 transcription factor, the BLK, and INSR kinases, as well as the role these kinases play in oncometabolic processes, will be examined in future studies. PDAC cells experience extreme deprivation of nutrient and oxygen delivery. Our data also implicate LYN, EPHA8, and FYN kinases as potential actors in oncometabolic PDAC signaling pathways and suggest mechanisms by which these kinases may facilitate oncogenic behavior.
Identification of these kinases in the present study contributes to growing understanding of abnormal fibrotic processes prominent in PDAC, dysregulated transcription factor activity, anti-cancer immune response, and the complex kinomic signaling networks responsible for pathometabolic tumor regulation and nutrient delivery. As novel PDAC subtypes continue to be defined [7,8,10,11,95,96,97], it is clear that therapeutic strategies for PDAC must take different genetic backgrounds into account. The mutational profiles for the patient-derived cell lines used in this study (Table A2) provide useful insights into which protein tyrosine kinases may serve as effective targets of personalized/precision therapeutic intervention (Figure A4).
4. Materials and Methods
4.1. Experimental Design
The experimental design is illustrated in Figure 1. In brief, PDAC epithelial cells and normal pancreatic ductal epithelial cells derived from patients were subjected to kinome array analysis using the PamStation 12 platform. All samples were prepared and assayed sequentially using Tyrosine Kinase PamChips consisting of 196 peptides with known phosphorylation sequences representing over 100 different proteins associated with the activity of upstream kinases. Each sample was assayed in triplicate with the results averaged across three identical kinome array runs.
4.2. Cell Lines and Patient-Derived Tissue
We used three different pancreatic cancer cell lines: one commercial cell line and two patient-derived cell lines (PDCL). We used commercial (ATCC CRL-1469) PANC1 cells originating from human pancreatic ductal cells carrying a TP53_R273H mutation and a KRAS_G12D mutation [98]. Two patient-derived cell lines (PDCL5, original name TKCC-05; PDCL-15, original name TKCC-15-Lo) were kindly provided by Andrew Biankin from Wolfson Wohl Cancer Research Centre, UK, with authentication by STR [90]. PDCL5 carries a TP53_G245S mutation and a KRAS_G12V mutation, while PDCL15 carries only a KRAS_G12D mutation (Table A2). These mutational profiles were kindly provided by Andrew Biankin and confirmed by F. Charles Brunicardi and Shi-He Liu. Normal patient-derived pancreatic ductal tissue was harvested from a healthy donor and kindly provided by Camillo Ricordi at the Diabetes Research Institute, University of Miami Miller School of Medicine, under the material transfer agreement. Because traditional two-dimensional cell culture models fail to accurately represent cancer microenvironments, we endeavored to obtain control tissue that more accurately represents physiological conditions. The pancreatic tissue contains ductal cells, acinar cells, and other elements included in the pancreatic microenvironment. While our decision to compare cell lines with wild-type pancreatic tissue may introduce a degree of bias into the study (cell lines and tissue samples contain different cellular contexts and environments), we used identical control wild-type tissue for each cell line comparison. All experiments and procedures were performed in strict compliance with all relevant laws and institutional guidelines.
Cell lines were cultured and lysed 72 h after plating, and tissue samples were processed as previously described [90]. All procedures were performed on ice. Tissue homogenization was performed using a D2400 Homogenizer and 1.5-mm Triple-Pure Zirconium Beads, with five rounds of homogenization and liquid nitrogen cooling to maintain low temperatures and minimize protein degradation. Each round of homogenization consisted of three cycles, with each cycle consisting of 30 s of active homogenization at 7 m/s and 30-second intervals between cycles. Tissue and cell lysate protein extractions were performed using M-PER (mammalian protein extraction reagent) (ThermoFisher, Waltham, MA, USA) and Halt Protease and Phosphatase Inhibitor Cocktails (ThermoFisher). Samples were centrifuged (14,000 RPM, 10 min, 4 °C) before supernatant collection. Total protein concentrations were assayed (Pierce BCA Protein Assay Kit, ThermoFisher) and samples were diluted to 1 µg/µL. All samples were prepared and measured simultaneously. Because freeze–thaw cycles decrease kinase activity [99], multiple aliquots were stored at −80 °C to minimize freeze–thaw cycles, with frozen aliquots used only once for kinome array assays.
4.3. Tyrosine Kinase Array
Tyrosine kinase activity was measured with the PamStation 12 instrument (PamGene International, ’s-Hertogenbosch, The Netherlands) and PTK (4-well) array PamChips using fluorescently labeled antibodies to detect differential phosphorylation of 196 reporter peptides (including three internal controls) per well. These 196 consensus phosphopeptide sequences were immobilized on porous ceramic membranes. The PamChip wells were blocked with 2% bovine serum albumin (BSA) prior to addition of 1 µg of protein suspended in manufacturer’s kinase buffer (PamGene). Next, we added 157 µM adenosine triphosphate (ATP) and FITC-labeled anti-phospho tyrosine antibodies (PamGene) to each well. Homogenized lysates containing active kinases and assay solution were pumped back and forth through PamChip wells in order to facilitate interactions between the active kinases and the 196 immobilized consensus phosphopeptide sequences. Evolve (PamGene) software captured FITC-labeled anti-phospho-antibodies bound to the phosphorylated consensus sequences. Image capture occurred every six seconds for 60 min. After washing, peptide signal intensity was recorded across several exposure times (10, 20, 50, 100, 200 milliseconds). The linear regression slope was calculated in order to provide the peptide phosphorylation intensity signal used in downstream comparative analyses. Signal ratios between pairs of samples were used to calculate fold change (FC) for each peptide. Differential peptide signals greater than or equal to 30% (FC ≥ 1.30 or FC ≤ 0.70) were considered demonstrative of minimum threshold changes in the degree of phosphorylation. This threshold value derived from conservative interpretation of previous literature suggesting even smaller orders of magnitude are sufficiently correlated with biologically relevant signaling changes [16,100,101]. Nonlinear (R2 values less than 0.90) or undetectable peptides in the post-wash phase were not selected for further analysis. Kinome assays were performed in triplicate with the calculated FC per peptide averaged across three replicates.
4.4. Upstream Kinase Identification
Kinase activity corresponded to the degree of consensus peptide phosphorylation as measured using real-time Evolve kinetic image capture software. The raw data generated by the PamStation platform were minimally processed (using threshold changes described above) to generate a list of differentially phosphorylated peptide sequences, which served as input for subsequent bioinformatic analyses. To expand coverage of peptide sequences and maximize identification of candidate upstream kinases, we used four distinct bioinformatic pipelines, each with a semi-overlapping set of databases by which their respective algorithms queried our list of differentially phosphorylated peptide sequences. Because each pipeline also relied upon a unique pipeline-specific algorithm, evaluating the results of one pipeline within the context of the results obtained through the other three pipelines allowed for enhanced perspective. Shared identification of upstream kinases responsible for the observed peptide phosphorylation patterns between pipelines could, therefore, be weighted and integrated into the final analysis. These pipelines include (1) Upstream Kinase Analysis (UKA) from PamGene, (2) Post-Translational Modification Signature Enrichment Analysis (PTM-SEA) from the Broad Institute of MIT and Harvard, (3) Kinase Enrichment Analysis Version 3 (KEA3) from the Ma’ayan laboratory, and (4) Kinome Random Sampling Analyzer (KRSA) developed by our own laboratory [12,13,14,15,16,17]. Meaningful differences between pipelines were compared in the Discussion section, while the methods by which we deployed each pipeline are presented below.
4.4.1. Upstream Kinase Analysis (UKA) Pipeline
Using PamGene’s BioNavigator software, we ran our data sets through the Protein Tyrosine Kinase (PTK) Upstream Kinase Analysis (UKA) Knowledge Integration PamApp. These data sets included (1) mean phosphorylated peptide sequences of PDCL15 vs. wild-type, (2) mean phosphorylated peptide sequences of PDCL5 vs. wild-type, and (3) mean phosphorylated peptide sequences of PANC1 vs. wild-type. Sample names served as the factor uniquely defining each observation. The “treatment off chip” factor served as the factor defining the experimental groupings. The default scan rank (4 to 12) and permutation (500) parameters were applied to the analysis, in addition to the default in vitro/in vivo (1), in silico (PhosphoNET) (1), minimal sequence homology (0.9), minimal PhosphoNET prediction score (300), and minimal peptide set (3) parameters. Inclusive percentile ranks were calculated according to the absolute value of UKA’s Median Final Score output.
4.4.2. Post-Translational Modification Signature Enrichment Analysis (PTM-SEA) Pipeline
We used the Broad Institute’s Single sample Gene Set Enrichment Analysis (ssGSEA) and Post-Translational Modification Signature Enrichment Analysis (PTM-SEA) publicly available repository (https://github.com/broadinstitute/ssGSEA2.0), RStudio Desktop 1.2.5042 (https://rstudio.com), and underlaying R 3.3.1 software environment (https://cran.rstudio.com). We ran our data sets through the PTM-SEA pipeline after modifying the peptide database to include only the peptide sequences present on the PamChip plus all peptide sequences with minimal sequence homologies of 0.9. These data sets include (1) mean phosphorylated peptide sequences of PDCL15 vs. wild-type, (2) mean phosphorylated peptide sequences of PDCL5 vs. wild-type, and (3) mean phosphorylated peptide sequences of PANC1 vs. wild-type. Results were filtered to include only protein tyrosine kinases. PTM-SEA inclusive percentile ranks were determined according to each kinase’s respective 1/fdr.pvalue.totalGeoMeanLFC output value.
4.4.3. Kinase Enrichment Analysis Version 3 (KEA3) Pipeline
We used the Ma’ayan laboratory’s Kinase Enrichment Analysis Version 3 (KEA3) (https://amp.pharm.mssm.edu/kea3/#) to process our data sets. These data sets include (1) mean phosphorylated peptide sequences of PDCL15 vs. wild-type, (2) mean phosphorylated peptide sequences of PDCL5 vs. wild-type, and (3) mean phosphorylated peptide sequences of PANC1 vs. wild-type. To accommodate the input parameters of KEA3, these peptide sequences were converted to HGNC-approved gene symbols before the data sets were entered into the KEA3 pipeline. Results were filtered to include only protein tyrosine kinases. Average FDR p-values from 0.2, 0.3, and 0.4 LFC cutoff input lists were averaged according to ChengKSIN, PTMsigDB, or PhosDAll database outputs. The resultant ChengKSIN, PTMsigDB, and PhosDAll average values were themselves averaged and −log10 transformations of these averages were used for inclusive percentile ranking calculations.
4.4.4. Kinome Random Sampling Analyzer (KRSA) Pipeline
Our laboratory developed Kinome Random Sampling Analyzer (KRSA) (version 2.0, Toledo, OH, USA) to associate differentially phosphorylated peptide sequences with specific kinases [12,13,14,15,16,17]. To accomplish this, we mapped phosphorylation sites within the reporter peptides to individual protein kinases that phosphorylate these sites. To this end, multiple databases were queried including GPS 5.0 (http://gps.biocuckoo.cn), Kinexus Phosphonet (http://www.kinexus.ca), PhosphoELM (http://phospho.elm.eu.org), and PhosphoSite Plus (https://www.phosphosite.org). In this way, peptide sequences and kinases were matched such that ranked predictions were generated to identify tyrosine kinases most likely to have produced the observed phosphorylation results. Kinases with scores greater than twice the prediction threshold for each phosphorylation site in the GPS 5.0 database were carried forward for downstream analysis. The top five kinase predictions in the Kinexus database were also carried forward for downstream analysis. Additional kinases reported to act on specific phosphorylation sites were identified using PhosphoELM and Phosphosite Plus public databases and carried forward for downstream analysis. Downstream analysis consists of 3000 random sampling iterations in which an equal number of differentially phosphorylated peptide sequences are randomly selected from the total list of 196 phosphopeptide sites on the tyrosine kinase PamChip. Predicted kinases were then mapped to each iteration in order to generate comparative controls to which the experimentally generated kinase lists could be compared. This allowed meaningful approximations of the direction of activity (increased or decreased) and significance for each experimentally identified kinase. Inclusive percentile ranks were calculated according to mean LFC output. KRSA is publicly available at https://github.com/kalganem/KRSA. Because KRSA identifies kinase families rather than individual kinases, all kinases within a family were given identical scores.
4.5. Combinatory Analyses
To resolve different output metrics, the results of each respective pipeline were converted to inclusive percentile rankings. These inclusive percentile rankings were aggregated according to cell line and protein tyrosine kinase and averaged per cell line (PANC1, PDCL15, PDCL5) or per cell line group (Patient-derived cell line group: PDCL15 and PDCL5; All group: PANC1, PDCL15, PDCL5). Weighted averages were calculated by dividing average percentile rankings by the number of pipelines under consideration.
4.6. Peptide Identities, Gene Synonyms, Family Designations, and Other Mapped Data
To resolve different output terms and provide additional contextual information, several sources were consulted for peptide identities, gene synonyms, kinase family designations, and other categorical or descriptive terms. These sources include UniProt’s Human and mouse protein kinases: classification and index (https://www.uniprot.org) [102,103,104], as well as kinase.com (http://kinase.com/) [103], GPS 5.0 (http://gps.biocuckoo.cn) [105,106], The GeneCards’ human gene database (https://www.genecards.org/), and HUGO Gene Nomenclature Committee (HGNC) (https://www.genenames.org/). Full listing of approved human gene nomenclature can be found in Appendix B, Table A3. Nomenclature mapping can be found in Appendix B, Table A4.
4.7. Figure Generation
Figures created with BioRender.com (Toronto, Ontario, Canada), Adobe Creative Suite (San Jose, CA, USA), and R (version 3.6.3). Additional figure panels created with KRSA, UKA/BioNavigator, PTM-SEA, or KEA3.
5. Conclusions
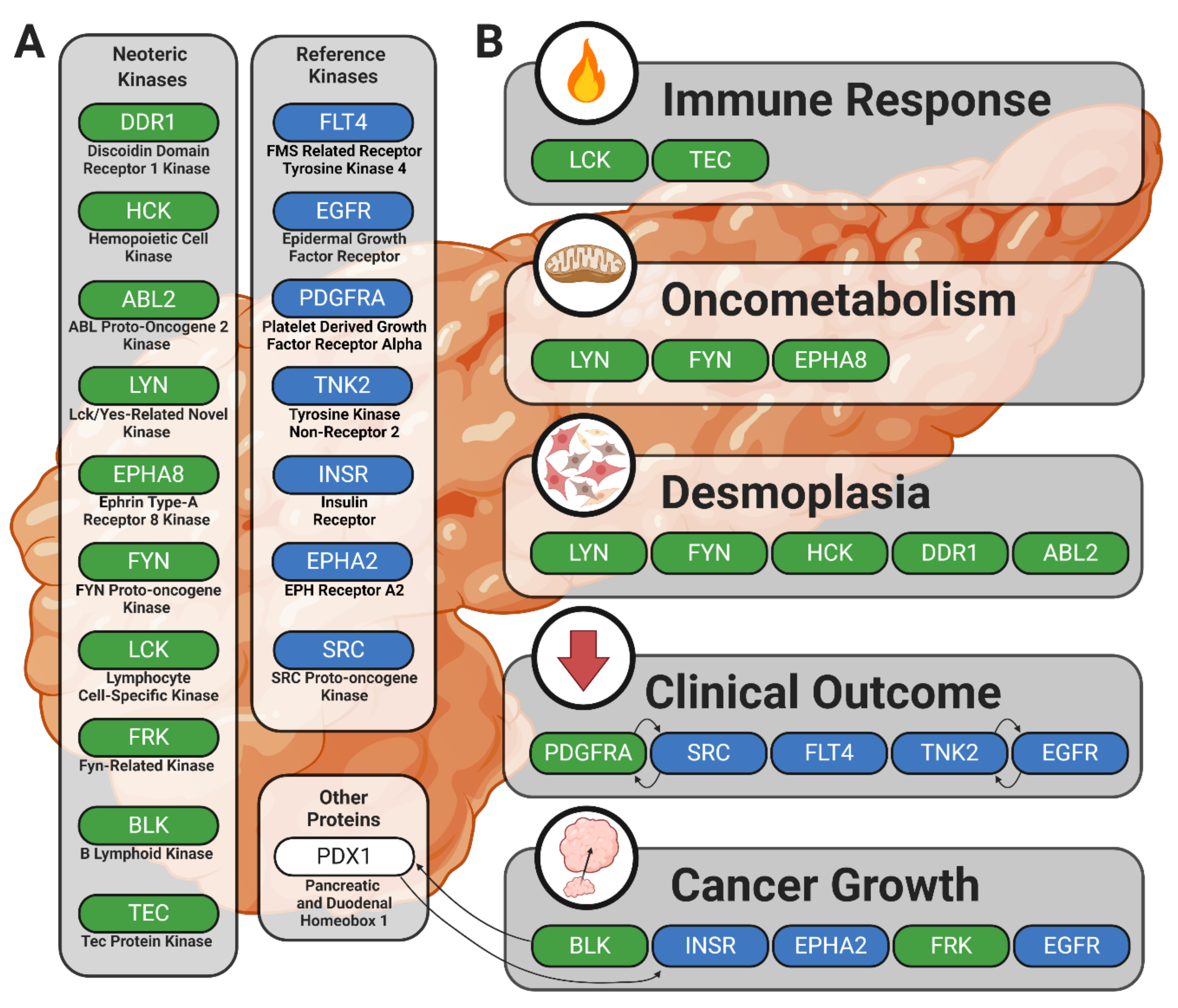
This study provides evidence in support of previously reported kinases in human cancer with an emphasis on PDAC. This passive validation supports the strength of ongoing drug development strategies that target protein tyrosine kinases and propounds the utility and accuracy of peptide-based kinomic analytical platforms. Furthermore, our identification and contextualization of candidate or lead candidate kinases responsible for the differential phosphorylation signatures observed between PDAC commercial or patient-derived cell lines compared to wild-type pancreatic patient samples provides evidence of unique kinomic relationships between pancreatic tumor cells and the desmoplastic stromal environments that support tumor progression and cause significant obstacles in pancreatic cancer therapy. Identification of the BLK, HCK, FRK, ABL2, DDR1, LYN, EPHA8, FYN, LCK, and TEC kinases as potentially significant mediators of pancreatic cancer progression and fibrotic/desmoplastic development fits well into established knowledge while also advancing new avenues of drug development and discovery. Additionally, our data provide increased understanding of the relationship between BLK protein tyrosine kinase, PDX1 transcription factor, and pancreatic disease. This study also outlines additional mechanisms by which HCK, ABL2, and DDR1 may play a role in pancreatic cancer and fibrosis. These results also support the role of LYN in oncometabolic processes and posit pathways by which LYN, EPHA8, and FYN may facilitate oncogenic cellular behavior. Lastly, we provide a rationale for continued investigation of the complex interplay between anti-cancer immune response and the activity of LCK and TEC kinases. These findings are summarized in Figure 5, and our companion review piece provides additional information on the role of BLK, HCK, FRK, ABL2, DDR1, LYN, EPHA8, FYN, LCK, and TEC kinases in PDAC and pancreatic cancer desmoplasia [107].
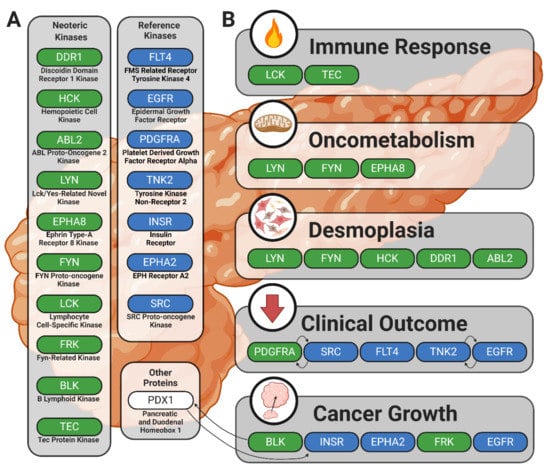
Figure 5.
Summary figure illustrating kinases showing increased enzymatic phosphorylation activity in PDAC and their potential roles in the disease process. Solid black arrows indicate relationships between kinases or other proteins. (A) The neoteric kinase group includes candidate kinases potentially contributing to PDAC pathology in new or previously understudied ways; the reference kinase group includes kinases with well-established roles in human cancer pathophysiology. (B) Kinases are clustered by linkage to the processes that might underlie their involvement in PDAC.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/22/8679/s1, Table S1: Output data table.
Author Contributions
Conceptualization, R.E.M., K.A., J.F.C, T.T., and F.N.; methodology, K.A., R.E.M., J.F.C., A.S.I., T.T., and F.N.; software, R.E.M., K.A., J.F.C., A.S.I., T.T., and F.N.; validation, K.A., A.S.I., and J.F.C.; formal analysis, K.A., A.S.I., and J.F.C.; investigation, J.F.C., K.A., and A.S.I.; resources, R.E.M., J.F.C., K.A., A.S.I., T.T., F.N., F.C.B., and S.-H.L.; data curation, K.A., J.F.C., and A.S.I.; writing—original draft preparation, J.F.C.; writing—review and editing, R.E.M, K.A., A.S.I, T.T., and F.N.; visualization, J.F.C.; supervision, R.E.M. and R.S.; project administration, R.E.M.; funding acquisition, R.E.M. and F.C.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by NIMH R01 MH107487, NIH R01 AG057598, and NIMH R01 MH121102, as well as the University of Toledo Foundation
Acknowledgments
The authors thank the Broad Institute and the Ma’ayan laboratory for their independent development of the PTM-SEA and KEA3 pipelines, respectively. The authors thank Andrew Biankin at the Wolfson Wohl Cancer Research Centre, University of Glasgow, Glasgow, for gifting PDCL5 and PDCL15 cell lines.
Conflicts of Interest
T.T. and F.N. are employed by PamGene International B.V. The remaining authors have declared that no conflicts of interests exist.
Abbreviations
| PDAC | Pancreatic ductal adenocarcinoma |
| KRSA | Kinome Random Sampling Analyzer |
| UKA | Upstream Kinase Analysis |
| PTM-SEA | Post-Translational Modification Signature Enrichment Analysis |
| KEA3 | Kinase Enrichment Analysis Version 3 |
| Z | Standard score |
| FC | Fold change |
| LFC | Log fold change |
| R2 | R-squared statistical measure |
| HGNC | HUGO Gene Nomenclature Committee |
| ssGSEA | Single sample Gene Set Enrichment Analysis |
| PDCL5 | Patient-derived pancreatic ductal adenocarcinoma cell line 5 |
| PDCL15 | Patient-derived pancreatic ductal adenocarcinoma cell line 15 |
| SNP | Single nucleotide polymorphism |
| PDX1, IPF1 | Pancreatic and duodenal homeobox 1 transcription factor |
| BSA | Bovine serum albumin |
| LCK | LCK proto-oncogene, Src family tyrosine kinase |
| DDR2 | Discoidin domain receptor tyrosine kinase 2 |
| LYN | LYN proto-oncogene, Src family tyrosine kinase |
| SRC | SRC proto-oncogene, non-receptor tyrosine kinase |
| ABL1 | ABL proto-oncogene 1, non-receptor tyrosine kinase |
| TEC | Tec protein tyrosine kinase |
| FYN | FYN proto-oncogene, Src family tyrosine kinase |
| BLK | BLK proto-oncogene, Src family tyrosine kinase |
| TXK | TXK tyrosine kinase |
| SRMS | Src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites |
| PDGFRA | Platelet-derived growth factor receptor alpha |
| FRK | Fyn-related Src family tyrosine kinase |
| PTK7 | Protein tyrosine kinase 7 (inactive) |
| ROS1 | ROS proto-oncogene 1, receptor tyrosine kinase |
| TNK2 | Tyrosine kinase non receptor 2 |
| ALK | ALK receptor tyrosine kinase |
| LTK | Leukocyte receptor tyrosine kinase |
| ITK | IL2 inducible T cell kinase |
| FLT1 | Fms-related receptor tyrosine kinase 1 |
| EPHB1 | EPH receptor B1 |
| ABL2 | ABL proto-oncogene 2, non-receptor tyrosine kinase |
| HCK | HCK proto-oncogene, Src family tyrosine kinase |
| EPHB3 | EPH receptor B3 |
| BTK | Bruton tyrosine kinase |
| EGFR | Epidermal growth factor receptor |
| MST1R | Macrophage stimulating 1 receptor |
| INSR | Insulin receptor |
| FGR | FGR proto-oncogene, Src family tyrosine kinase |
| KIT | KIT proto-oncogene, receptor tyrosine kinase |
| FLT4 | Fms-related receptor tyrosine kinase 4 |
| FLT3 | Fms-related receptor tyrosine kinase 3 |
| RET | Ret proto-oncogene |
| EPHA2 | EPH receptor A2 |
| PDGFRB | Platelet-derived growth factor receptor beta |
| ZAP70 | Zeta chain of T cell receptor-associated protein kinase 70 |
| JAK2 | Janus kinase 2 |
| KDR | Kinase insert domain receptor |
| AXL | AXL receptor tyrosine kinase |
| CSK | C-terminal Src kinase |
| MET | MET proto-oncogene, receptor tyrosine kinase |
| SEV | Sevenless |
| SYK | Spleen-associated tyrosine kinase |
| VEGFR | Vascular endothelial growth factor receptor |
| TYRO3 | TYRO3 protein tyrosine kinase |
| EPHB4 | EPH receptor B4 |
| PTK6 | Protein tyrosine kinase 6 |
| YES1 | YES proto-oncogene 1, Src family tyrosine kinase |
| CSF1R | Colony stimulating factor 1 receptor |
| FES | FES proto-oncogene, tyrosine kinase |
| INSRR | Insulin receptor-related receptor |
| FGFR4 | Fibroblast growth factor receptor 4 |
| JAK3 | Janus kinase 3 |
| MATK | Megakaryocyte-associated tyrosine kinase |
| FGFR3 | Fibroblast growth factor receptor 3 |
| ERBB3 | Erb-b2 receptor tyrosine kinase 3 |
| BMX | BMX nonreceptor tyrosine kinase |
| IGF1R | Insulin-like growth factor 1 receptor |
| NTRK1 | Neurotrophic receptor tyrosine kinase 1 |
| EPHA4 | EPH receptor A4 |
| EPHB2 | EPH receptor B2 |
| NTRK3 | Neurotrophic receptor tyrosine kinase 3 |
| FER | FER tyrosine kinase |
| FGFR2 | Fibroblast growth factor receptor 2 |
| EPHA1 | EPH receptor A1 |
| ERBB4 | Erb-b2 receptor tyrosine kinase 4 |
| FGFR1 | Fibroblast growth factor receptor 1 |
| DDR1 | Discoidin domain receptor tyrosine kinase 1 |
| EPHA5 | EPH receptor A5 |
| JAK1 | Janus kinase 1 |
| EPHA7 | EPH receptor A7 |
| ERBB2 | Erb-b2 receptor tyrosine kinase 2 |
| NTRK2 | Neurotrophic receptor tyrosine kinase 2 |
| TYK2 | Tyrosine kinase 2 |
| PTK2 | Protein tyrosine kinase 2 |
| SLTM | SAFB-like transcription modulator |
| EPHA8 | EPH receptor A8 |
| EPHA3 | EPH receptor A3 |
| MERTK | MER proto-oncogene, tyrosine kinase |
| RYK | Receptor-like tyrosine kinase |
| PTK2B | Protein tyrosine kinase 2 beta |
| STYK1 | Serine/threonine/tyrosine kinase 1 |
| TEK | TEK receptor tyrosine kinase |
| AATK | Apoptosis-associated tyrosine kinase |
| MTTP | Microsomal triglyceride transfer protein |
| TPM3 | Tropomyosin 3 |
Appendix A
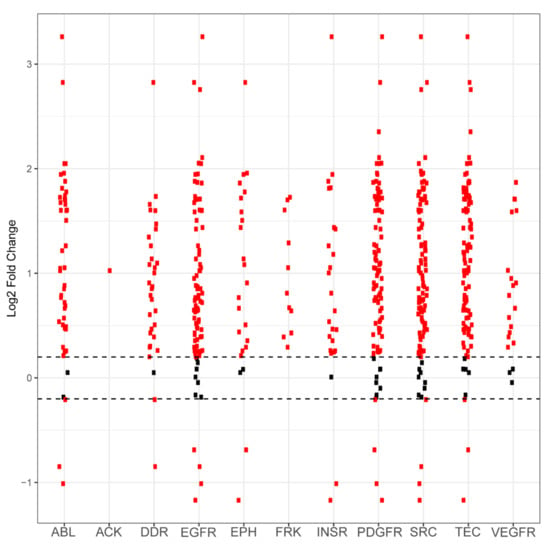
Figure A1.
Differential phosphorylation levels of peptide sequences attributed to kinase family activity in PANC1 vs. wild-type control. Each red dot in a column represents one peptide sequence whose phosphorylation is performed by that column’s kinase family. The y axis reports log2-fold change and the x axis identifies the kinase family. Black dots represent peptides that did not demonstrate differential phosphorylation in PANC1 samples compared to wild-type samples, with the dashed horizontal line representing a positive or negative 0.2 log2-fold change cutoff. Red dots above these lines represent peptides that are more phosphorylated in PANC1 compared to control. Red dots that are below these lines represent peptides that are less phosphorylated in PANC1 compared to control.
Figure A1.
Differential phosphorylation levels of peptide sequences attributed to kinase family activity in PANC1 vs. wild-type control. Each red dot in a column represents one peptide sequence whose phosphorylation is performed by that column’s kinase family. The y axis reports log2-fold change and the x axis identifies the kinase family. Black dots represent peptides that did not demonstrate differential phosphorylation in PANC1 samples compared to wild-type samples, with the dashed horizontal line representing a positive or negative 0.2 log2-fold change cutoff. Red dots above these lines represent peptides that are more phosphorylated in PANC1 compared to control. Red dots that are below these lines represent peptides that are less phosphorylated in PANC1 compared to control.
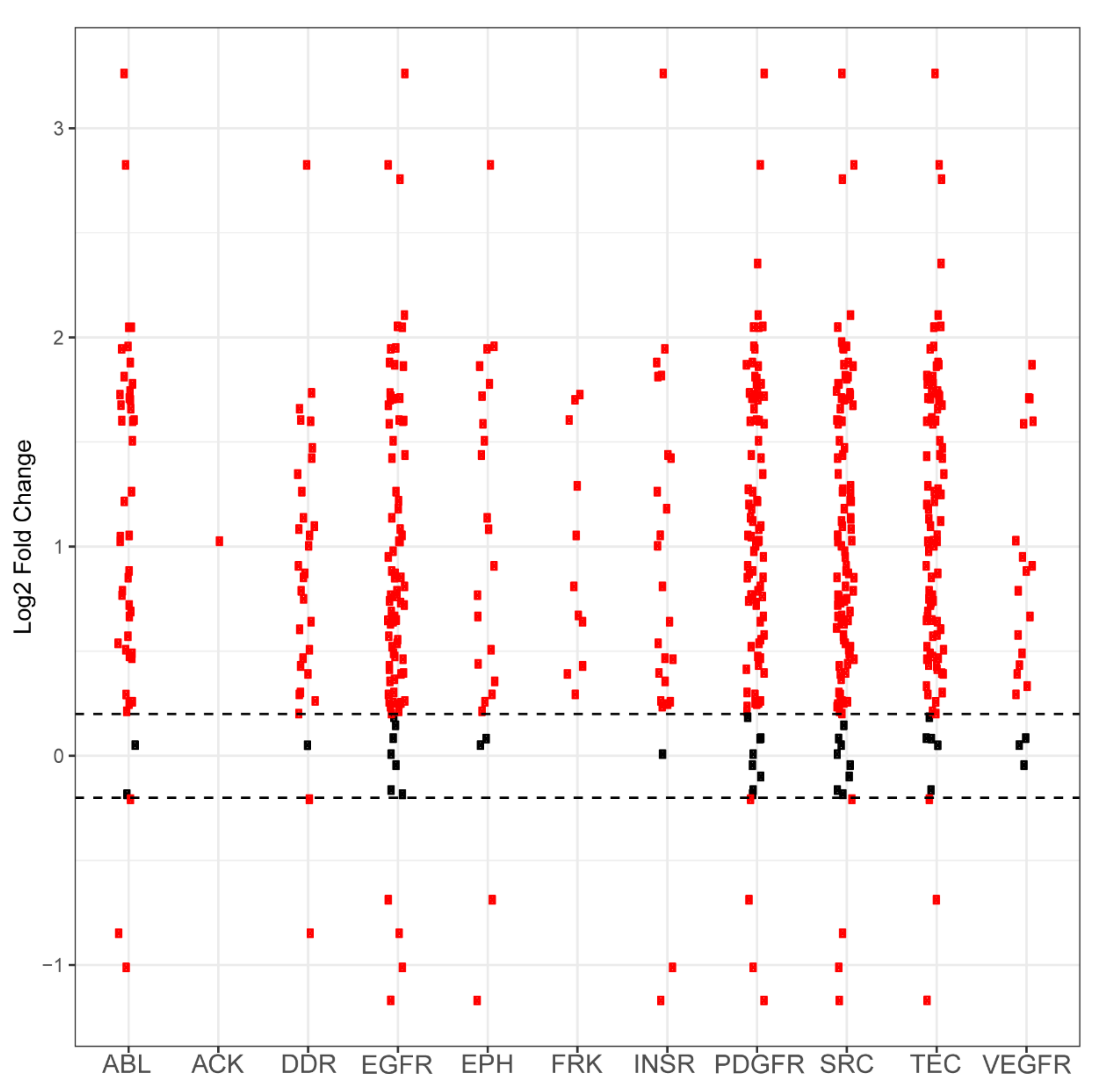
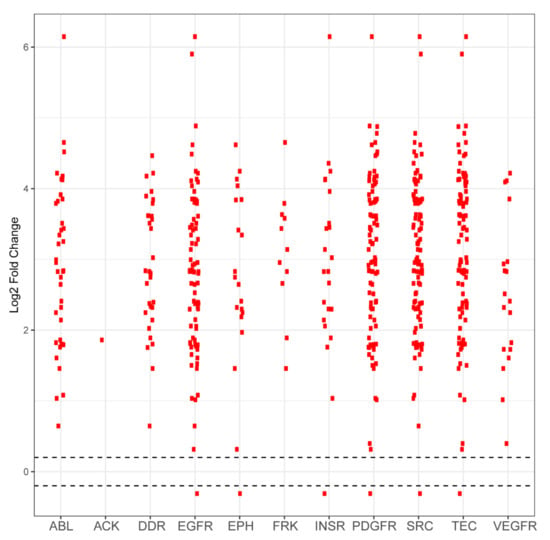
Figure A2.
Differential phosphorylation levels of peptide sequences attributed to kinase family activity in PDCL15 vs. wild-type control. Each red dot in a column represents one peptide sequence whose phosphorylation is performed by that column’s kinase family. The y axis reports log2-fold change and the x axis identifies the kinase family. Black dots represent peptides that did not demonstrate differential phosphorylation in PDCL15 samples compared to wild-type samples, with the dashed horizontal line representing a positive or negative 0.2 log2-fold change cutoff. Red dots above these lines represent peptides that are more phosphorylated in PDCL15 compared to control. Red dots that are below these lines represent peptides that are less phosphorylated in PDCL15 compared to control.
Figure A2.
Differential phosphorylation levels of peptide sequences attributed to kinase family activity in PDCL15 vs. wild-type control. Each red dot in a column represents one peptide sequence whose phosphorylation is performed by that column’s kinase family. The y axis reports log2-fold change and the x axis identifies the kinase family. Black dots represent peptides that did not demonstrate differential phosphorylation in PDCL15 samples compared to wild-type samples, with the dashed horizontal line representing a positive or negative 0.2 log2-fold change cutoff. Red dots above these lines represent peptides that are more phosphorylated in PDCL15 compared to control. Red dots that are below these lines represent peptides that are less phosphorylated in PDCL15 compared to control.
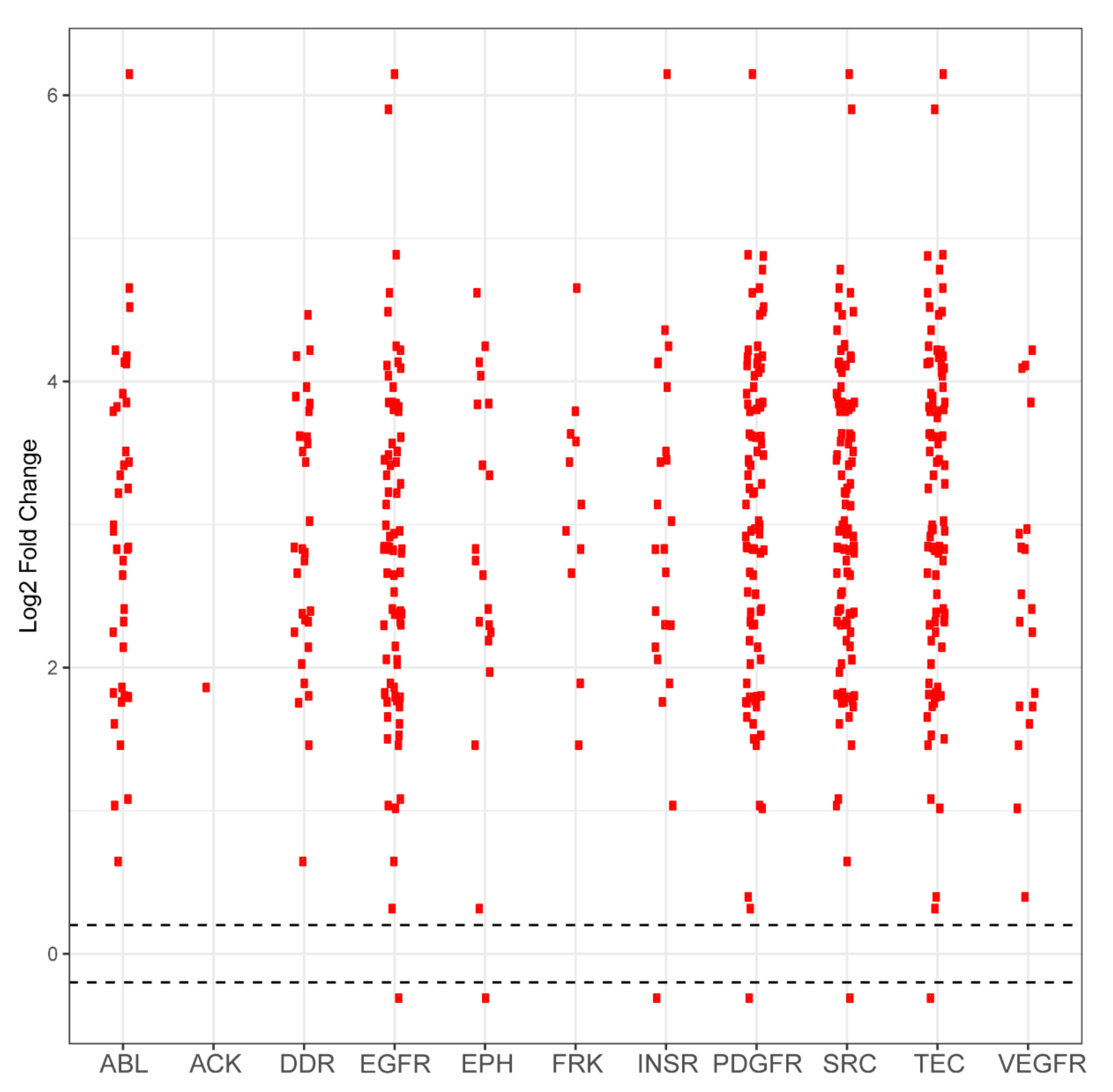
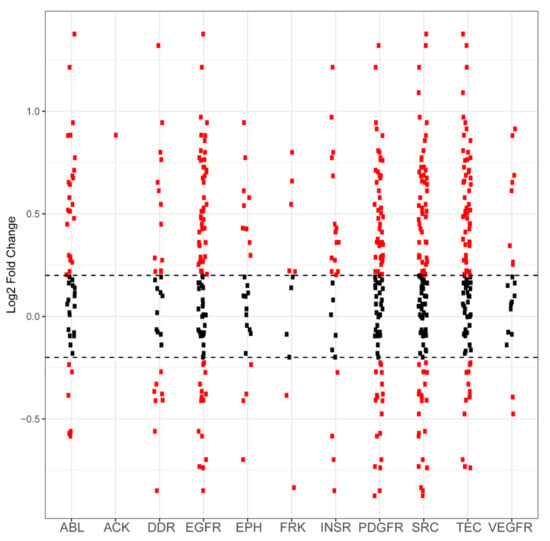
Figure A3.
Differential phosphorylation levels of peptide sequences attributed to kinase family activity in PDCL5 vs. wild-type control. Each red dot in a column represents one peptide sequence whose phosphorylation is performed by that column’s kinase family. The y axis reports log2-fold change and the x axis identifies the kinase family. Black dots represent peptides that did not demonstrate differential phosphorylation in PDCL5 samples compared to wild-type samples, with the dashed horizontal line representing a positive or negative 0.2 log2-fold change cutoff. Red dots above these lines represent peptides that are more phosphorylated in PDCL5 compared to control. Red dots that are below these lines represent peptides that are less phosphorylated in PDCL5 compared to control.
Figure A3.
Differential phosphorylation levels of peptide sequences attributed to kinase family activity in PDCL5 vs. wild-type control. Each red dot in a column represents one peptide sequence whose phosphorylation is performed by that column’s kinase family. The y axis reports log2-fold change and the x axis identifies the kinase family. Black dots represent peptides that did not demonstrate differential phosphorylation in PDCL5 samples compared to wild-type samples, with the dashed horizontal line representing a positive or negative 0.2 log2-fold change cutoff. Red dots above these lines represent peptides that are more phosphorylated in PDCL5 compared to control. Red dots that are below these lines represent peptides that are less phosphorylated in PDCL5 compared to control.
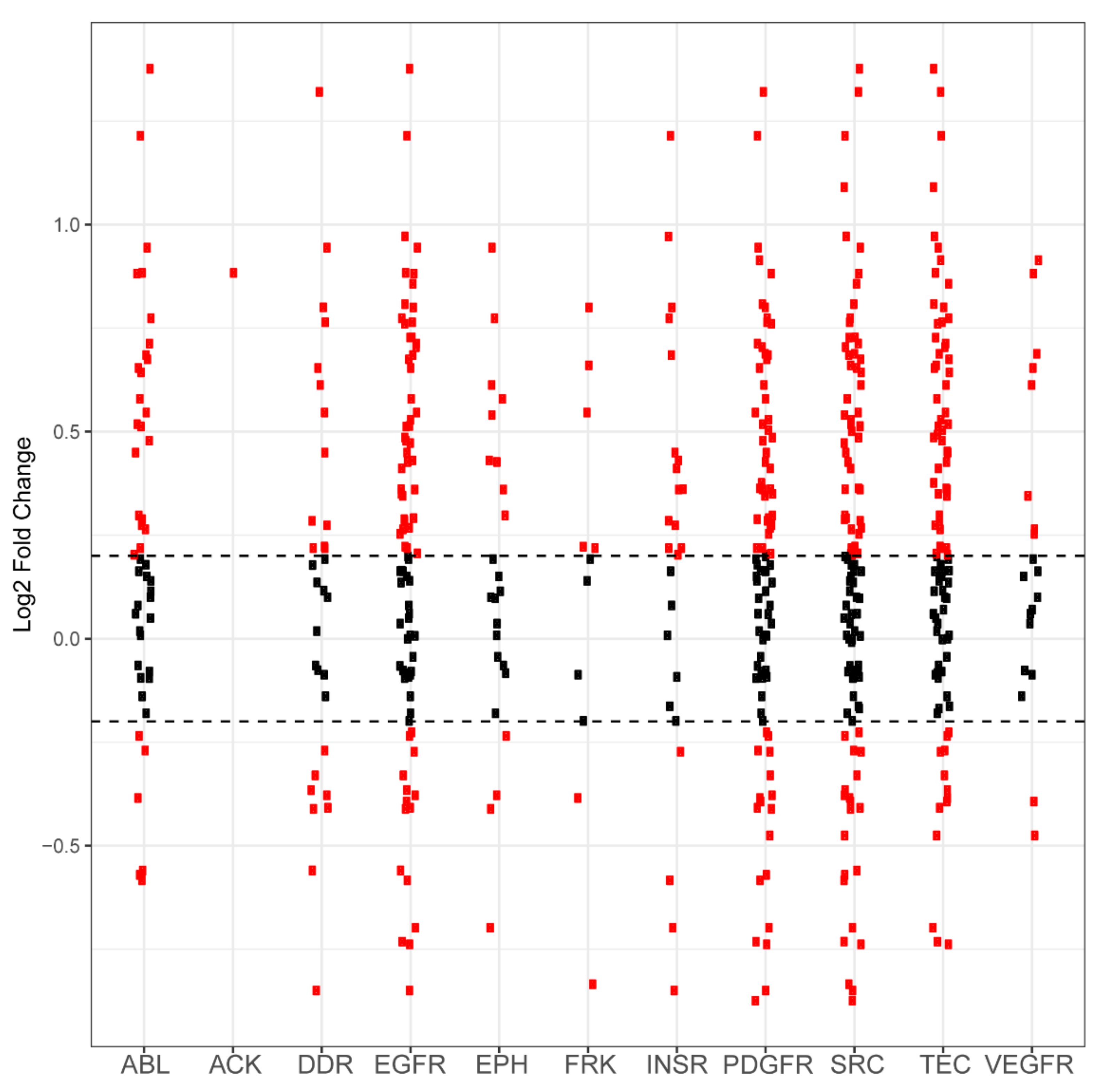
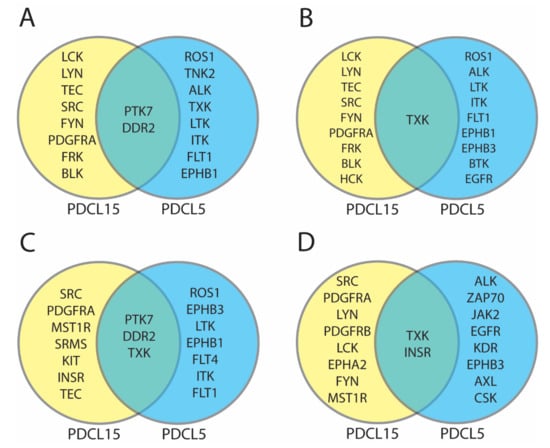
Figure A4.
Comparison of protein tyrosine kinases identified in patient-derived cell lines. For each comparison yellow circles represent differentially active protein tyrosine kinases in PDCL15, blue circles represent differentially active protein tyrosine kinases in PDCL5, and green overlapping area represents differentially active protein tyrosine kinases in PDCL15 and PDCL5. (A) Comparison of top 10 differentially active protein tyrosine kinases according to UKA and KRSA average percentile rankings. (B) Comparison of top 10 differentially active protein tyrosine kinases according to UKA and KRSA weighted average percentile rankings. (C) Comparison of top 10 differentially active protein tyrosine kinases according to all pipelines (KRSA, UKA, PTM-SEA, and KEA3) average percentile rankings. (D) Comparison of top 10 differentially active protein tyrosine kinases according to all pipelines (KRSA, UKA, PTM-SEA, and KEA3) weighted average percentile rankings.
Figure A4.
Comparison of protein tyrosine kinases identified in patient-derived cell lines. For each comparison yellow circles represent differentially active protein tyrosine kinases in PDCL15, blue circles represent differentially active protein tyrosine kinases in PDCL5, and green overlapping area represents differentially active protein tyrosine kinases in PDCL15 and PDCL5. (A) Comparison of top 10 differentially active protein tyrosine kinases according to UKA and KRSA average percentile rankings. (B) Comparison of top 10 differentially active protein tyrosine kinases according to UKA and KRSA weighted average percentile rankings. (C) Comparison of top 10 differentially active protein tyrosine kinases according to all pipelines (KRSA, UKA, PTM-SEA, and KEA3) average percentile rankings. (D) Comparison of top 10 differentially active protein tyrosine kinases according to all pipelines (KRSA, UKA, PTM-SEA, and KEA3) weighted average percentile rankings.
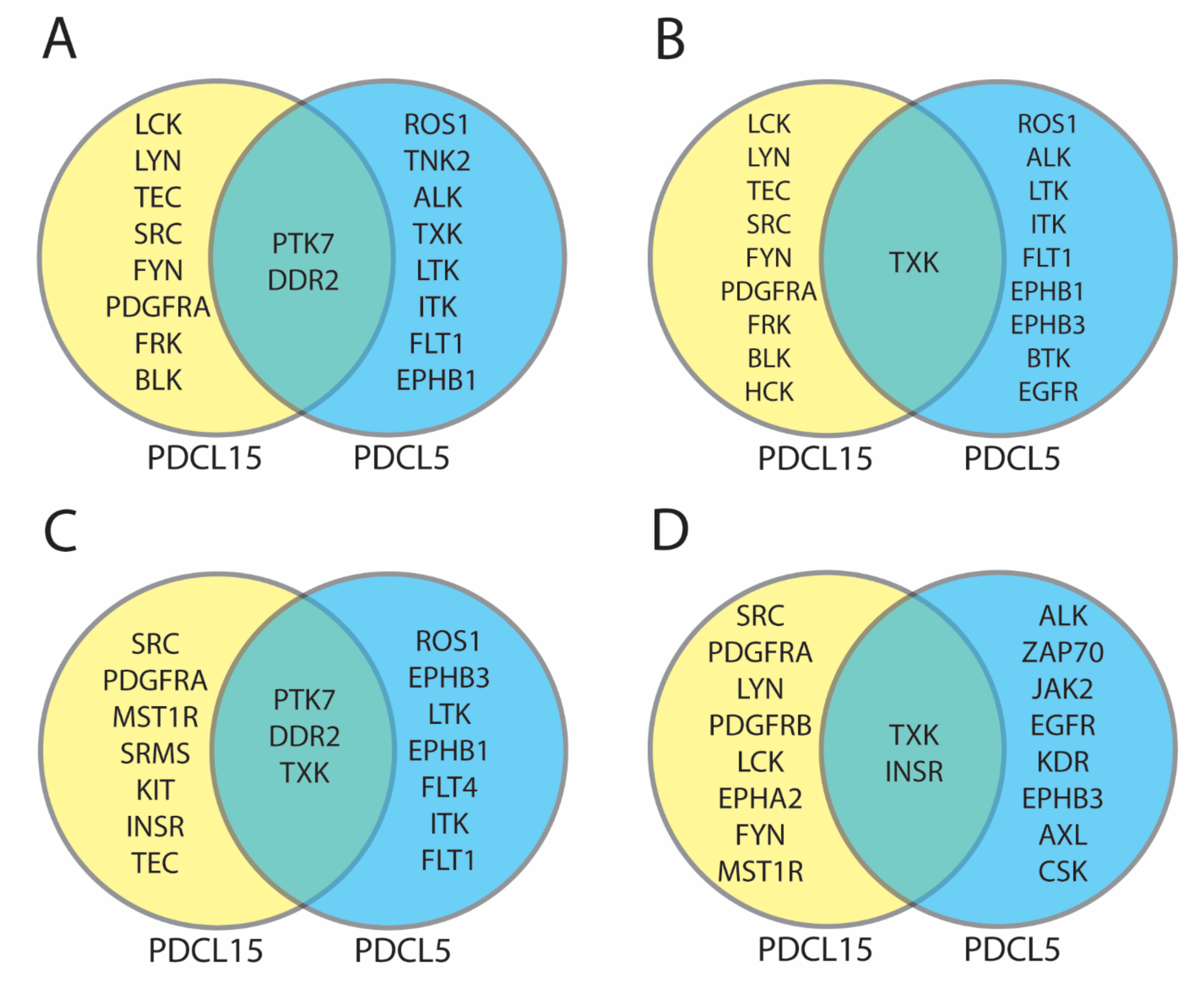

Table A1.
UKA mean kinase statistic describes direction of differential kinase activity.
Table A1.
UKA mean kinase statistic describes direction of differential kinase activity.
| Cell Line | Kinase | Mean Kinase Statistic | Direction |
|---|---|---|---|
| PDCL15 | BLK | 10.30884201 | Increased |
| PANC1 | BLK | 3.809077325 | Increased |
| PDCL5 | BLK | 0.963873271 | Increased |
| PDCL15 | EGFR | 6.92042103 | Increased |
| PANC1 | EGFR | 2.196020549 | Increased |
| PDCL5 | EGFR | 0.723998698 | Increased |
| PDCL15 | EphA2 | 6.538112723 | Increased |
| PANC1 | EphA2 | 2.767994333 | Increased |
| PDCL5 | EphA2 | 0.663688934 | Increased |
| PDCL15 | FLT4 | 5.969120078 | Increased |
| PANC1 | FLT4 | 1.888408842 | Increased |
| PDCL5 | FLT4 | 0.754741786 | Increased |
| PDCL15 | FRK | 10.54460498 | Increased |
| PANC1 | FRK | 3.517514717 | Increased |
| PDCL5 | FRK | 0.581814545 | Increased |
| PDCL15 | Fyn | 11.52215741 | Increased |
| PANC1 | Fyn | 4.046633984 | Increased |
| PDCL5 | Fyn | 0.080983021 | Increased |
| PDCL15 | InSR | 8.923873567 | Increased |
| PANC1 | InSR | 3.024794143 | Increased |
| PDCL5 | InSR | 0.508668865 | Increased |
| PDCL15 | Lck | 12.35321414 | Increased |
| PANC1 | Lck | 4.06073018 | Increased |
| PDCL5 | Lck | 0.177602219 | Increased |
| PDCL15 | Lyn | 11.88473844 | Increased |
| PANC1 | Lyn | 4.172287974 | Increased |
| PDCL5 | Lyn | 0.620188468 | Increased |
| PDCL15 | PDGFR[alpha] | 14.1657858 | Increased |
| PANC1 | PDGFR[alpha] | 4.47805184 | Increased |
| PDCL5 | PDGFR[alpha] | −0.21206998 | Decreased |
| PDCL15 | Src | 10.51520452 | Increased |
| PANC1 | Src | 3.487086518 | Increased |
| PDCL5 | Src | 0.34051653 | Increased |
| PDCL15 | TEC | 10.09732985 | Increased |
| PANC1 | TEC | 3.229557798 | Increased |
| PDCL5 | TEC | 0.732187188 | Increased |
| PDCL15 | HCK | 10.2005583 | Increased |
| PANC1 | HCK | 3.13041308 | Increased |
| PDCL5 | HCK | 0.537561913 | Increased |
| PDCL15 | Arg | 9.644954083 | Increased |
| PANC1 | Arg | 3.436290625 | Increased |
| PDCL5 | Arg | 0.713079917 | Increased |
| PDCL15 | DDR1 | 7.462484096 | Increased |
| PANC1 | DDR1 | 2.528789422 | Increased |
| PDCL5 | DDR1 | −0.255600871 | Decreased |
| PDCL15 | EphA8 | 9.12751897 | Increased |
| PANC1 | EphA8 | 1.855262187 | Increased |
| PDCL5 | EphA8 | −0.456838117 | Decreased |

Table A2.
Cell line profiles.
Table A2.
Cell line profiles.
| Cell Line | Category | Clinicopathological Data of the Patient of Origin | Standard of Care | Mutational Profile | Ref. |
|---|---|---|---|---|---|
| PANC1 | Commercial | Age: 56; Gender: Male; Ethnicity: Caucasian; Disease: Epithelioid Carcinoma of Ductal Cell Origin | Surgical resection with or without post-surgical adjuvant therapy | KRAS_G12D; TP53_R273H | [98] |
| PDCL15 | Patient Derived | Age: 66; Gender: Male; Ethnicity: Caucasian; Disease: Pancreatic Ductal Adenocarcinoma | Surgical resection with or without post-surgical adjuvant therapy | KRAS_G12D; TP53_WT; | [7,9,11,108] Data Repo |
| PDCL5 | Patient Derived | Age: 56; Gender: Male; Ethnicity: Caucasian; Disease: Pancreatic Ductal Adenocarcinoma | Surgical resection with or without post-surgical adjuvant therapy | KRAS_G12V; TP53_G245S | [7,9,11,108] Data Repo |
Appendix B

Table A3.
Approved human gene nomenclature.
Table A3.
Approved human gene nomenclature.
| Cell Line | Kinase | Mean Kinase Statistic | Direction |
|---|---|---|---|
| AATK | apoptosis associated tyrosine kinase | HGNC:21 | 17q25.3 |
| ABL1 | ABL proto-oncogene 1, non-receptor tyrosine kinase | HGNC:76 | 9q34.12 |
| ABL2 | ABL proto-oncogene 2, non-receptor tyrosine kinase | HGNC:77 | 1q25.2 |
| ALK | ALK receptor tyrosine kinase | HGNC:427 | 2p23.2-p23.1 |
| AXL | AXL receptor tyrosine kinase | HGNC:905 | 19q13.2 |
| BLK | BLK proto-oncogene, Src family tyrosine kinase | HGNC:1057 | 8p23.1 |
| BMX | BMX non-receptor tyrosine kinase | HGNC:1079 | Xp22.2 |
| BTK | Bruton tyrosine kinase | HGNC:1133 | Xq22.1 |
| CSF1R | colony stimulating factor 1 receptor | HGNC:2433 | 5q32 |
| CSK | C-terminal Src kinase | HGNC:2444 | 15q24.1 |
| DDR1 | discoidin domain receptor tyrosine kinase 1 | HGNC:2730 | 6p21.33 |
| DDR2 | discoidin domain receptor tyrosine kinase 2 | HGNC:2731 | 1q23.3 |
| EGFR | epidermal growth factor receptor | HGNC:3236 | 7p11.2 |
| EPHA1 | EPH receptor A1 | HGNC:3385 | 7q34-q35 |
| EPHA2 | EPH receptor A2 | HGNC:3386 | 1p36.13 |
| EPHA3 | EPH receptor A3 | HGNC:3387 | 3p11.1 |
| EPHA4 | EPH receptor A4 | HGNC:3388 | 2q36.1 |
| EPHA5 | EPH receptor A5 | HGNC:3389 | 4q13.1-q13.2 |
| EPHA7 | EPH receptor A7 | HGNC:3390 | 6q16.1 |
| EPHA8 | EPH receptor A8 | HGNC:3391 | 1p36.12 |
| EPHB1 | EPH receptor B1 | HGNC:3392 | 3q22.2 |
| EPHB2 | EPH receptor B2 | HGNC:3393 | 1p36.12 |
| EPHB3 | EPH receptor B3 | HGNC:3394 | 3q27.1 |
| EPHB4 | EPH receptor B4 | HGNC:3395 | 7q22.1 |
| ERBB2 | erb-b2 receptor tyrosine kinase 2 | HGNC:3430 | 17q12 |
| ERBB3 | erb-b2 receptor tyrosine kinase 3 | HGNC:3431 | 12q13.2 |
| ERBB4 | erb-b2 receptor tyrosine kinase 4 | HGNC:3432 | 2q34 |
| FER | FER tyrosine kinase | HGNC:3655 | 5q21.3 |
| FES | FES proto-oncogene, tyrosine kinase | HGNC:3657 | 15q26.1 |
| FGFR1 | fibroblast growth factor receptor 1 | HGNC:3688 | 8p11.23 |
| FGFR2 | fibroblast growth factor receptor 2 | HGNC:3689 | 10q26.13 |
| FGFR3 | fibroblast growth factor receptor 3 | HGNC:3690 | 4p16.3 |
| FGFR4 | fibroblast growth factor receptor 4 | HGNC:3691 | 5q35.2 |
| FGR | FGR proto-oncogene, Src family tyrosine kinase | HGNC:3697 | 1p35.3 |
| FLT1 | fms related receptor tyrosine kinase 1 | HGNC:3763 | 13q12.3 |
| FLT3 | fms related receptor tyrosine kinase 3 | HGNC:3765 | 13q12.2 |
| FLT4 | fms related receptor tyrosine kinase 4 | HGNC:3767 | 5q35.3 |
| FRK | fyn related Src family tyrosine kinase | HGNC:3955 | 6q22.1 |
| FYN | FYN proto-oncogene, Src family tyrosine kinase | HGNC:4037 | 6q21 |
| HCK | HCK proto-oncogene, Src family tyrosine kinase | HGNC:4840 | 20q11.21 |
| IGF1R | insulin like growth factor 1 receptor | HGNC:5465 | 15q26.3 |
| INSR | insulin receptor | HGNC:6091 | 19p13.2 |
| INSRR | insulin receptor related receptor | HGNC:6093 | 1q23.1 |
| ITK | IL2 inducible T cell kinase | HGNC:6171 | 5q33.3 |
| JAK1 | Janus kinase 1 | HGNC:6190 | 1p31.3 |
| JAK2 | Janus kinase 2 | HGNC:6192 | 9p24.1 |
| JAK3 | Janus kinase 3 | HGNC:6193 | 19p13.11 |
| KDR | kinase insert domain receptor | HGNC:6307 | 4q12 |
| KIT | KIT proto-oncogene, receptor tyrosine kinase | HGNC:6342 | 4q12 |
| LCK | LCK proto-oncogene, Src family tyrosine kinase | HGNC:6524 | 1p35.2 |
| LTK | leukocyte receptor tyrosine kinase | HGNC:6721 | 15q15.1 |
| LYN | LYN proto-oncogene, Src family tyrosine kinase | HGNC:6735 | 8q12.1 |
| MATK | megakaryocyte-associated tyrosine kinase | HGNC:6906 | 19p13.3 |
| MERTK | MER proto-oncogene, tyrosine kinase | HGNC:7027 | 2q13 |
| MET | MET proto-oncogene, receptor tyrosine kinase | HGNC:7029 | 7q31 |
| MST1R | macrophage stimulating 1 receptor | HGNC:7381 | 3p21.31 |
| NTRK1 | neurotrophic receptor tyrosine kinase 1 | HGNC:8031 | 1q23.1 |
| NTRK2 | neurotrophic receptor tyrosine kinase 2 | HGNC:8032 | 9q21.33 |
| NTRK3 | neurotrophic receptor tyrosine kinase 3 | HGNC:8033 | 15q25.3 |
| PDGFRA | platelet derived growth factor receptor alpha | HGNC:8803 | 4q12 |
| PDGFRB | platelet derived growth factor receptor beta | HGNC:8804 | 5q32 |
| PTK2 | protein tyrosine kinase 2 | HGNC:9611 | 8q24.3 |
| PTK2B | protein tyrosine kinase 2 beta | HGNC:9612 | 8p21.2 |
| PTK6 | protein tyrosine kinase 6 | HGNC:9617 | 20q13.33 |
| PTK7 | protein tyrosine kinase 7 (inactive) | HGNC:9618 | 6p21.1 |
| RET | ret proto-oncogene | HGNC:9967 | 10q11.21 |
| ROS1 | ROS proto-oncogene 1, receptor tyrosine kinase | HGNC:10261 | 6q22.1 |
| RYK | receptor like tyrosine kinase | HGNC:10481 | 3q22.2 |
| SLTM | SAFB like transcription modulator | HGNC:20709 | 15q22.1 |
| SRC | SRC proto-oncogene, non-receptor tyrosine kinase | HGNC:11283 | 20q11.23 |
| SRMS | src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites | HGNC:11298 | 20q13.33 |
| STYK1 | serine/threonine/tyrosine kinase 1 | HGNC:18889 | 12p13.2 |
| SYK | spleen associated tyrosine kinase | HGNC:11491 | 9q22.2 |
| TEC | tec protein tyrosine kinase | HGNC:11719 | 4p12-p11 |
| TEK | TEK receptor tyrosine kinase | HGNC:11724 | 9p21.2 |
| TNK2 | tyrosine kinase non receptor 2 | HGNC:19297 | 3q29 |
| TXK | TXK tyrosine kinase | HGNC:12434 | 4p12 |
| TYK2 | tyrosine kinase 2 | HGNC:12440 | 19p13.2 |
| TYRO3 | TYRO3 protein tyrosine kinase | HGNC:12446 | 15q15.1 |
| YES1 | YES proto-oncogene 1, Src family tyrosine kinase | HGNC:12841 | 18p11.32 |
| ZAP70 | zeta chain of T cell receptor associated protein kinase 70 | HGNC:12858 | 2q11.2 |

Table A4.
Nomenclature mapping.
Table A4.
Nomenclature mapping.
| Pipeline | Default | Standardized |
|---|---|---|
| KRSA | TEC | TEC |
| KRSA | DDR | DDR |
| KRSA | SRC | SRC |
| KRSA | ABL | ABL |
| KRSA | PDGFR | PDGFR |
| KRSA | FRK | FRK |
| KRSA | JAK | JAK |
| KRSA | INSR | INSR |
| KRSA | FGFR | FGFR |
| KRSA | TRK | TRK |
| KRSA | ACK | ACK |
| KRSA | SEV | SEV |
| KRSA | VEGFR | VEGFR |
| KRSA | AXL | AXL |
| KRSA | FAK | FAK |
| KRSA | MET | MET |
| KRSA | EPH | EPH |
| KRSA | RET | RET |
| KRSA | SYK | SYK |
| KRSA | RYK | RYK |
| KRSA | FER | FER |
| KRSA | ALK | ALK |
| KRSA | EGFR | EGFR |
| KRSA | CSK | CSK |
| UKA | Lck | Lck |
| UKA | Lyn | Lyn |
| UKA | TEC | TEC |
| UKA | FRK | FRK |
| UKA | Tyro3/Sky | TYRO3 |
| UKA | PDGFR[alpha] | PDGFRa |
| UKA | Src | Src |
| UKA | Fyn | Fyn |
| UKA | Abl | ABL1 |
| UKA | CCK4/PTK7 | PTK7 |
| UKA | Ron | MST1R |
| UKA | CTK | MATK |
| UKA | Axl | Axl |
| UKA | Fes | Fes |
| UKA | BLK | BLK |
| UKA | TXK | TXK |
| UKA | Arg | ABL2 |
| UKA | HCK | HCK |
| UKA | HER3 | ERBB3 |
| UKA | Syk | Syk |
| UKA | Srm | SRMS |
| UKA | EphA8 | EphA8 |
| UKA | ZAP70 | ZAP70 |
| UKA | CSK | CSK |
| UKA | EphB4 | EphB4 |
| UKA | Mer | MERTK |
| UKA | PDGFR[beta] | PDGFRb |
| UKA | Met | Met |
| UKA | FAK1 | PTK2 |
| UKA | RYK | RYK |
| UKA | Fgr | Fgr |
| UKA | Yes | YES1 |
| UKA | InSR | InSR |
| UKA | Ret | Ret |
| UKA | DDR1 | DDR1 |
| UKA | LTK | LTK |
| UKA | FGFR2 | FGFR2 |
| UKA | Fer | Fer |
| UKA | Kit | Kit |
| UKA | EphA5 | EphA5 |
| UKA | EphB1 | EphB1 |
| UKA | IGF1R | IGF1R |
| UKA | Ros | ROS1 |
| UKA | FmS/CSFR | CSF1R |
| UKA | TRKB | NTRK2 |
| UKA | EphA4 | EphA4 |
| UKA | JAK2 | JAK2 |
| UKA | ALK | ALK |
| UKA | FGFR3 | FGFR3 |
| UKA | Etk/BMX | BMX |
| UKA | BTK | BTK |
| UKA | FGFR1 | FGFR1 |
| UKA | TRKC | NTRK3 |
| UKA | EphB3 | EphB3 |
| UKA | EphA2 | EphA2 |
| UKA | ITK | ITK |
| UKA | Lmr1 | AATK |
| UKA | EphA1 | EphA1 |
| UKA | KDR | KDR |
| UKA | FGFR4 | FGFR4 |
| UKA | FLT3 | FLT3 |
| UKA | FAK2 | PTK2B |
| UKA | JAK3 | JAK3 |
| UKA | HER2 | ERBB2 |
| UKA | IRR | INSRR |
| UKA | TRKA | NTRK1 |
| UKA | JAK1~b | JAK1 |
| UKA | HER4 | ERBB4 |
| UKA | Tyk2 | Tyk2 |
| UKA | EphA3 | EphA3 |
| UKA | FLT4 | FLT4 |
| UKA | Brk | PTK6 |
| UKA | EphA7 | EphA7 |
| UKA | EphB2 | EphB2 |
| UKA | EGFR | EGFR |
| UKA | FLT1 | FLT1 |
| PTM-SEA | ZAP70 | ZAP70 |
| PTM-SEA | VEGFR2/KDR | KDR |
| PTM-SEA | TrkA/NTRK1 | NTRK1 |
| PTM-SEA | Syk/SYK | SYK |
| PTM-SEA | Src/SRC | SRC |
| PTM-SEA | Ret/RET | RET |
| PTM-SEA | PDGFRB | PDGFRB |
| PTM-SEA | PDGFRA | PDGFRA |
| PTM-SEA | MKK4/MAP2K4 | MAP2K4 |
| PTM-SEA | Met/MET | MET |
| PTM-SEA | Mer/MERTK | MERTK |
| PTM-SEA | MEK1/MAP2K1 | MAP2K1 |
| PTM-SEA | LYN | LYN |
| PTM-SEA | Lck/LCK | LCK |
| PTM-SEA | JAK3 | JAK3 |
| PTM-SEA | JAK2 | JAK2 |
| PTM-SEA | INSR | INSR |
| PTM-SEA | IGF1R | IGF1R |
| PTM-SEA | HER2/ERBB2 | ERBB2 |
| PTM-SEA | Fyn/FYN | FYN |
| PTM-SEA | Fer/FER | FER |
| PTM-SEA | Etk/BMX | BMX |
| PTM-SEA | EphA2/EPHA2 | EPHA2 |
| PTM-SEA | EGFR | EGFR |
| PTM-SEA | CSK | CSK |
| PTM-SEA | Chk1/CHEK1 | CHEK1 |
| PTM-SEA | AXL | AXL |
| PTM-SEA | ALK | ALK |
| PTM-SEA | Abl/ABL1 | ABL1 |
| KEA3 | NTRK1 | NTRK1 |
| KEA3 | FLT3 | FLT3 |
| KEA3 | DDR2 | DDR2 |
| KEA3 | KIT | KIT |
| KEA3 | PDGFRA | PDGFRA |
| KEA3 | MATK | MATK |
| KEA3 | EPHB3 | EPHB3 |
| KEA3 | MST1R | MST1R |
| KEA3 | FES | FES |
| KEA3 | FLT4 | FLT4 |
| KEA3 | SRC | SRC |
| KEA3 | TXK | TXK |
| KEA3 | NTRK3 | NTRK3 |
| KEA3 | KDR | KDR |
| KEA3 | RET | RET |
| KEA3 | LCK | LCK |
| KEA3 | ABL1 | ABL1 |
| KEA3 | EPHA2 | EPHA2 |
| KEA3 | SRMS | SRMS |
| KEA3 | EPHB2 | EPHB2 |
| KEA3 | FYN | FYN |
| KEA3 | EGFR | EGFR |
| KEA3 | FLT1 | FLT1 |
| KEA3 | FER | FER |
| KEA3 | INSR | INSR |
| KEA3 | FGFR4 | FGFR4 |
| KEA3 | ITK | ITK |
| KEA3 | EPHB1 | EPHB1 |
| KEA3 | CSF1R | CSF1R |
| KEA3 | PTK6 | PTK6 |
| KEA3 | CSK | CSK |
| KEA3 | ERBB2 | ERBB2 |
| KEA3 | NTRK2 | NTRK2 |
| KEA3 | TYRO3 | TYRO3 |
| KEA3 | BTK | BTK |
| KEA3 | JAK2 | JAK2 |
| KEA3 | SYK | SYK |
| KEA3 | LYN | LYN |
| KEA3 | FGFR3 | FGFR3 |
| KEA3 | PTK2 | PTK2 |
| KEA3 | FGR | FGR |
| KEA3 | ERBB4 | ERBB4 |
| KEA3 | YES1 | YES1 |
| KEA3 | ZAP70 | ZAP70 |
| KEA3 | JAK3 | JAK3 |
| KEA3 | MET | MET |
| KEA3 | IGF1R | IGF1R |
| KEA3 | TEC | TEC |
| KEA3 | AXL | AXL |
| KEA3 | ALK | ALK |
| KEA3 | PTK2B | PTK2B |
| KEA3 | PDGFRB | PDGFRB |
| KEA3 | STYK1 | STYK1 |
| KEA3 | MERTK | MERTK |
| KEA3 | BMX | BMX |
| KEA3 | EPHA3 | EPHA3 |
| KEA3 | ABL2 | ABL2 |
| KEA3 | FGFR1 | FGFR1 |
| KEA3 | EPHA4 | EPHA4 |
| KEA3 | TYK2 | TYK2 |
| KEA3 | FRK | FRK |
| KEA3 | FGFR2 | FGFR2 |
| KEA3 | TNK2 | TNK2 |
| KEA3 | JAK1 | JAK1 |
| KEA3 | DDR1 | DDR1 |
| KEA3 | BLK | BLK |
| KEA3 | HCK | HCK |
| KEA3 | EPHA8 | EPHA8 |
| KEA3 | TEK | TEK |
Figure 2, Figure 3 and Figure 4, expanded figure legend. Default outputs from upstream kinase identification pipelines for PDAC cell lines compared to patient-derived wild-type pancreatic tissue. (A) The Kinome Random Sampling Analyzer (KRSA) pipeline identifies differentially active upstream kinases according to mean standard score (Z) values on the x axis, with kinase family on the y axis, and color gradation representing absolute mean standard score. Full red represents an absolute mean standard score above 3 and full white represents an absolute mean standard score below 1. Absolute mean standard scores for each kinase family are calculated by averaging the absolute standard score values of nine standard scores, each derived from input lists consisting of peptide sequences with log-fold change (LFC) in phosphorylation above 0.2, 0.3, or 0.4 in one of our three biological replicates. Each plotted box represents values within and including the first and third percentiles (the 25th and 75th percentiles). Upper or lower whiskers extend from the box to the largest or smallest kinase family standard score that is less than or equal to 150% of the distance between the first and third percentiles, respectively. Outliers defined as greater than 150% of the distance between the first and third percentiles are plotted individually. Dashed lines delineate positive values above 1.5, 1.7, and 2.0 or negative values below −1.5, −1.7, and −2.0. Kinase families appear in descending order of standard score with the most significant differential kinase family activity appearing at the top and bottom of the list. Any zero-value standard score represents a kinase family having identical mean log-fold change values between the phosphorylation levels of representative peptide sequences measured in the PDAC cell line group and the phosphorylation levels of those same representative peptide sequences when measured in the control wild-type patient sample group. (B) The Post-Translational Modification Signature Enrichment Analysis (PTM-SEA) pipeline identifies differentially active upstream kinases according to negative decadic logarithms of the adjusted probability values. These values allow the most significant differentially active kinases to be listed from top to bottom in order of descending significance. Adjusted probability values less than 0.05 are, therefore, represented by x axis values above 1.30. Adjusted probability values correcting for multiple comparisons use the Bonferroni correction method on comparisons between the phosphorylated peptide sequences of the experimental PDAC cell line group and the phosphorylated peptide sequences of the control wild-type patient group in order to determine the extent to which these differences may be attributed to each individual kinase listed on the left. (C) The Kinase Enrichment Analysis Version 3 (KEA3) pipeline also identifies differentially activated upstream kinases according to negative decadic logarithms of the average adjusted probability values and allows the most significant differentially active kinases to be listed from top to bottom in order of descending significance. Adjusted probability values less than 0.05 are, therefore, represented by x axis values above 1.30. Adjusted probability values less than 0.01 are represented by x axis values above 2. Adjusted probability values correcting for multiple comparisons use the false discovery rate (FDR) correction method to compare the genes containing phosphorylated peptide sequences of the experimental PDAC cell line group with the genes containing phosphorylated peptide sequences of the control wild-type patient group in order to determine the extent to which these differences may be attributed to each individual kinase listed on the left. (D) The Upstream Kinase Analysis (UKA) pipeline identifies differentially activated upstream kinases according to the normalized kinase statistic for change in phosphorylation between the peptide sequences of the experimental PDAC cell line group and the control wild-type patient group. This normalized kinase statistic is calculated by subtracting the mean phosphorylation levels of the peptide sequences associated with a given kinase in the control group from the mean phosphorylation levels of the peptide sequences associated with that same kinase in the experimental group and dividing the resulting value by the standard deviation. A quotient above 0 indicates increased activity of a kinase from the experimental group. A quotient below 0 indicates decreased activity of a kinase from the experimental group. Each graphed point represents the normalized kinase statistic when the UKA algorithm uses only one of the pipeline’s available databases. The diameter of a circle increases with the size of the peptide sequences associated with the corresponding kinases in the corresponding database. Color gradation represents specificity scores with full red representing values of two and full black representing specificity scores of zero. Specificity scores are informed by the quotient produced with a dividend corresponding to the number of times that a permuted normalized kinase statistic greatly exceeded the nonpermuted normalized kinase statistic and a divisor corresponding to the number of random permutations, or the quotient produced by the number 1 as a dividend and the number of random permutations as the divisor. Specificity scores represent the negative decadic logarithm of whichever quotient is larger. Individual kinases are listed along the y axis from top to bottom in descending order of median “final score.” UKA calculates the “final score” as the summation of the significance score and specificity score of each individual kinase. (E) Comparison of upstream kinase activity as determined by KRSA, PTM-SEA, KEA3, and UKA, each listed on the y axis. Kinase family names appear along the top of the x axis. Individual kinase family members appear along the bottom of the x axis. The comparison includes only kinases identified as one of the top 10 most differentially active upstream kinases by any one of the pipelines. Kinases with identical scores were arbitrarily assigned sequential ranks. However, if two or more kinases tied for the last (10th) position in this list, then the list was extended so as not to arbitrarily exclude an equivalently ranked kinase. The diameter of each black circle corresponds with the percentile in which the labeled kinase appears in the labeled pipeline. The kinase family scores of the KRSA pipeline are repeated for each individual kinase. A white circle demonstrates not only absence from a given pipeline’s top 10 list, but also demonstrates absence from a given pipeline’s unfiltered exhaustive output.
References
- Jagade, J.; Amrutkar, M.; Katariya, D.; Wankhade, A.; Kale, A.; Undale, V. Role of protein kinases in signal transduction and their inhibitors. Pharmacologyonline 2010. Available online: https://www.researchgate.net/profile/Manoj_Amrutkar/publication/230558661_Role_of_protein_kinases_in_signal_transduction_and_their_inhibitors/links/0912f5016fc0b4e975000000.pdf (accessed on 9 January 2020).
- Bhullar, K.S.; Lagaron, N.O.; McGowan, E.M.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-targeted cancer therapies: Progress, challenges and future directions. Mol. Cancer 2018, 17, 48. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2020 update. Pharmacol. Res. 2020, 152, 104609. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors. Pharmacol. Res. 2019, 144, 19–50. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.W.; Bennett, J.J. Clinical Presentation and Diagnosis of Pancreatic Neuroendocrine Tumors. Surg. Oncol. Clin. 2016, 25, 363–374. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef]
- Humphrey, E.S.; Su, S.P.; Nagrial, A.M.; Hochgrafe, F.; Pajic, M.; Lehrbach, G.M.; Parton, R.G.; Yap, A.S.; Horvath, L.G.; Chang, D.K.; et al. Resolution of Novel Pancreatic Ductal Adenocarcinoma Subtypes by Global Phosphotyrosine Profiling. Mol. Cell. Proteom. MCP 2016, 15, 2671–2685. [Google Scholar] [CrossRef]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef]
- Waddell, N.; Pajic, M.; Patch, A.M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; Quek, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Bentea, E.; Depasquale, E.A.K.; O’Donovan, S.M.; Sullivan, C.R.; Simmons, M.; Meador-Woodruff, J.H.; Zhou, Y.; Xu, C.; Bai, B.; Peng, J.; et al. Kinase network dysregulation in a human induced pluripotent stem cell model of DISC1 schizophrenia. Mol. Omics 2019, 15, 173–188. [Google Scholar] [CrossRef] [PubMed]
- Bentea, E.; Villers, A.; Moore, C.; Funk, A.J.; O’Donovan, S.M.; Verbruggen, L.; Lara, O.; Janssen, P.; De Pauw, L.; Declerck, N.B.; et al. Corticostriatal dysfunction and social interaction deficits in mice lacking the cystine/glutamate antiporter. Mol. Psychiatry 2020. [Google Scholar] [CrossRef] [PubMed]
- DePasquale, E.A.K.; Alganem, K.; Bentea, E.; Nawreen, N.; McGuire, J.L.; Naji, F.; Hilhorst, R.; Meller, J.; McCullumsmith, R.E. KRSA: Network-based Prediction of Differential Kinase Activity from Kinome Array Data. bioRxiv 2020. [Google Scholar] [CrossRef]
- Flaherty, E.; Zhu, S.; Barretto, N.; Cheng, E.; Deans, P.J.M.; Fernando, M.B.; Schrode, N.; Francoeur, N.; Antoine, A.; Alganem, K.; et al. Neuronal impact of patient-specific aberrant NRXN1alpha splicing. Nat. Genet. 2019, 51, 1679–1690. [Google Scholar] [CrossRef]
- McGuire, J.L.; Depasquale, E.A.; Funk, A.J.; O’Donnovan, S.M.; Hasselfeld, K.; Marwaha, S.; Hammond, J.H.; Hartounian, V.; Meador-Woodruff, J.H.; Meller, J.; et al. Abnormalities of signal transduction networks in chronic schizophrenia. NPJ Schizophr. 2017, 3, 30. [Google Scholar] [CrossRef]
- Schrode, N.; Ho, S.M.; Yamamuro, K.; Dobbyn, A.; Huckins, L.; Matos, M.R.; Cheng, E.; Deans, P.J.M.; Flaherty, E.; Barretto, N.; et al. Synergistic effects of common schizophrenia risk variants. Nat. Genet. 2019, 51, 1475–1485. [Google Scholar] [CrossRef]
- Alganem, K.; Shukla, R.; Eby, H.; Abel, M.; Zhang, X.; McIntyre, W.B.; Lee, J.; Au-Yeung, C.; Asgariroozbehani, R.; Panda, R.; et al. Kaleidoscope: A New Bioinformatics Pipeline Web Application for In Silico Hypothesis Exploration of Omics Signatures. bioRxiv 2020. [Google Scholar] [CrossRef]
- McGuire, J.L.; Hammond, J.H.; Yates, S.D.; Chen, D.; Haroutunian, V.; Meador-Woodruff, J.H.; McCullumsmith, R.E. Altered serine/threonine kinase activity in schizophrenia. Brain Res. 2014, 1568, 42–54. [Google Scholar] [CrossRef]
- Alack, K.; Weiss, A.; Kruger, K.; Horet, M.; Schermuly, R.; Frech, T.; Eggert, M.; Mooren, F.C. Profiling of human lymphocytes reveals a specific network of protein kinases modulated by endurance training status. Sci. Rep. 2020, 10, 888. [Google Scholar] [CrossRef]
- Chirumamilla, C.S.; Fazil, M.; Perez-Novo, C.; Rangarajan, S.; de Wijn, R.; Ramireddy, P.; Verma, N.K.; Vanden Berghe, W. Profiling Activity of Cellular Kinases in Migrating T-Cells. Methods Mol. Biol. 2019, 1930, 99–113. [Google Scholar] [PubMed]
- Krug, K.; Mertins, P.; Zhang, B.; Hornbeck, P.; Raju, R.; Ahmad, R.; Szucs, M.; Mundt, F.; Forestier, D.; Jane-Valbuena, J.; et al. A Curated Resource for Phosphosite-specific Signature Analysis. Mol. Cell. Proteom. MCP 2019, 18, 576–593. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, A.; Ma’ayan, A. KEA: Kinase enrichment analysis. Bioinformatics 2009, 25, 684–686. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.M.; Tsai, C.H.; Hung, W.C. Foretinib inhibits angiogenesis, lymphangiogenesis and tumor growth of pancreatic cancer in vivo by decreasing VEGFR-2/3 and TIE-2 signaling. Oncotarget 2015, 6, 14940–14952. [Google Scholar] [CrossRef]
- Durkin, A.J.; Osborne, D.A.; Yeatman, T.J.; Rosemurgy, A.S.; Armstrong, C.; Zervos, E.E. EGF receptor antagonism improves survival in a murine model of pancreatic adenocarcinoma. J. Surg. Res. 2006, 135, 195–201. [Google Scholar] [CrossRef]
- Kuo, T.L.; Cheng, K.H.; Shan, Y.S.; Chen, L.T.; Hung, W.C. beta-catenin-activated autocrine PDGF/Src signaling is a therapeutic target in pancreatic cancer. Theranostics 2019, 9, 324–336. [Google Scholar] [CrossRef]
- Parkin, A.; Man, J.; Timpson, P.; Pajic, M. Targeting the complexity of Src signalling in the tumour microenvironment of pancreatic cancer: From mechanism to therapy. FEBS J. 2019, 286, 3510–3539. [Google Scholar] [CrossRef]
- Yamanaka, Y.; Friess, H.; Kobrin, M.S.; Buchler, M.; Beger, H.G.; Korc, M. Coexpression of epidermal growth factor receptor and ligands in human pancreatic cancer is associated with enhanced tumor aggressiveness. Anticancer Res. 1993, 13, 565–569. [Google Scholar]
- Du, J.; He, Y.; Wu, W.; Li, P.; Chen, Y.; Hu, Z.; Han, Y. Targeting EphA2 with miR-124 mediates Erlotinib resistance in K-RAS mutated pancreatic cancer. J. Pharm. Pharmacol. 2019, 71, 196–205. [Google Scholar] [CrossRef]
- Sugiyama, N. Mass Spectrometry-Based Discovery of in vitro Kinome Substrates. Mass Spectrom. 2020, 9, A0082. [Google Scholar] [CrossRef]
- Dong, M.; Nio, Y.; Guo, K.J.; Tamura, K.; Tian, Y.L.; Dong, Y.T. Epidermal growth factor and its receptor as prognostic indicators in Chinese patients with pancreatic cancer. Anticancer Res. 1998, 18, 4613–4619. [Google Scholar] [PubMed]
- Costache, M.I.; Ioana, M.; Iordache, S.; Ene, D.; Costache, C.A.; Saftoiu, A. VEGF Expression in Pancreatic Cancer and Other Malignancies: A Review of the Literature. Rom. J. Intern. Med. 2015, 53, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Kurenova, E.; Liao, J.; He, D.H.; Hunt, D.; Yemma, M.; Bshara, W.; Seshadri, M.; Cance, W.G. The FAK scaffold inhibitor C4 disrupts FAK-VEGFR-3 signaling and inhibits pancreatic cancer growth. Oncotarget 2013, 4, 1632–1646. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Buchler, P.; Giese, N.; Giese, T.; Wilting, J.; Buchler, M.W.; Friess, H. Role of lymphangiogenesis and lymphangiogenic factors during pancreatic cancer progression and lymphatic spread. Int. J. Oncol. 2006, 28, 883–890. [Google Scholar] [CrossRef]
- Von Marschall, Z.; Scholz, A.; Stacker, S.A.; Achen, M.G.; Jackson, D.G.; Alves, F.; Schirner, M.; Haberey, M.; Thierauch, K.H.; Wiedenmann, B.; et al. Vascular endothelial growth factor-D induces lymphangiogenesis and lymphatic metastasis in models of ductal pancreatic cancer. Int. J. Oncol. 2005, 27, 669–679. [Google Scholar]
- Berardi, R.; Torniai, M.; Partelli, S.; Rubini, C.; Pagliaretta, S.; Savini, A.; Polenta, V.; Santoni, M.; Giampieri, R.; Onorati, S.; et al. Impact of vascular endothelial growth factor (VEGF) and vascular endothelial growth factor receptor (VEGFR) single nucleotide polymorphisms on outcome in gastroenteropancreatic neuroendocrine neoplasms. PLoS ONE 2018, 13, e0197035. [Google Scholar] [CrossRef]
- Payankaulam, S.; Raicu, A.M.; Arnosti, D.N. Transcriptional Regulation of INSR, the Insulin Receptor Gene. Genes 2019, 10, 984. [Google Scholar] [CrossRef]
- Ofer, P.; Heidegger, I.; Eder, I.E.; Schöpf, B.; Neuwirt, H.; Geley, S.; Klocker, H.; Massoner, P. Both IGF1R and INSR Knockdown Exert Antitumorigenic Effects in Prostate Cancer In Vitro and In Vivo. Mol. Endocrinol. 2015, 29, 1694–1707. [Google Scholar] [CrossRef]
- Quinn, B.A.; Wang, S.; Barile, E.; Das, S.K.; Emdad, L.; Sarkar, D.; De, S.K.; Morvaridi, S.K.; Stebbins, J.L.; Pandol, S.J.; et al. Therapy of pancreatic cancer via an EphA2 receptor-targeted delivery of gemcitabine. Oncotarget 2016, 7, 17103–17110. [Google Scholar] [CrossRef]
- Koshikawa, N.; Minegishi, T.; Kiyokawa, H.; Seiki, M. Specific detection of soluble EphA2 fragments in blood as a new biomarker for pancreatic cancer. Cell Death Dis. 2017, 8, e3134. [Google Scholar] [CrossRef]
- Fan, J.; Wei, Q.; Koay, E.J.; Liu, Y.; Ning, B.; Bernard, P.W.; Zhang, N.; Han, H.; Katz, M.H.; Zhao, Z.; et al. Chemoresistance Transmission via Exosome-Mediated EphA2 Transfer in Pancreatic Cancer. Theranostics 2018, 8, 5986–5994. [Google Scholar] [CrossRef] [PubMed]
- Markosyan, N.; Li, J.; Sun, Y.H.; Richman, L.P.; Lin, J.H.; Yan, F.; Quinones, L.; Sela, Y.; Yamazoe, T.; Gordon, N.; et al. Tumor cell-intrinsic EPHA2 suppresses anti-tumor immunity by regulating PTGS2 (COX-2). J. Clin. Investig. 2019, 129, 3594–3609. [Google Scholar] [CrossRef] [PubMed]
- Van den Broeck, A.; Vankelecom, H.; Van Eijsden, R.; Govaere, O.; Topal, B. Molecular markers associated with outcome and metastasis in human pancreatic cancer. J. Exp. Clin. Cancer Res. CR 2012, 31, 68. [Google Scholar] [CrossRef] [PubMed]
- Chee, C.E.; Krishnamurthi, S.; Nock, C.J.; Meropol, N.J.; Gibbons, J.; Fu, P.; Bokar, J.; Teston, L.; O’Brien, T.; Gudena, V.; et al. Phase II study of dasatinib (BMS-354825) in patients with metastatic adenocarcinoma of the pancreas. Oncologist 2013, 18, 1091–1092. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Yuan, W.; Lai, C.; Zhong, S.; Yang, C.; Wang, R.; Mao, L.; Chen, Z.; Chen, Z. EphA2-to-YAP pathway drives gastric cancer growth and therapy resistance. Int. J. Cancer 2020, 146, 1937–1949. [Google Scholar] [CrossRef] [PubMed]
- Hamaoka, Y.; Negishi, M.; Katoh, H. Tyrosine kinase activity of EphA2 promotes its S897 phosphorylation and glioblastoma cell proliferation. Biochem. Biophys. Res. Commun. 2018, 499, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Pettazzoni, P.; Viale, A.; Shah, P.; Carugo, A.; Ying, H.; Wang, H.; Genovese, G.; Seth, S.; Minelli, R.; Green, T.; et al. Genetic events that limit the efficacy of MEK and RTK inhibitor therapies in a mouse model of KRAS-driven pancreatic cancer. Cancer Res. 2015, 75, 1091–1101. [Google Scholar] [CrossRef]
- Xie, L.; Kassner, M.; Munoz, R.M.; Que, Q.Q.; Kiefer, J.; Zhao, Y.; Mousses, S.; Yin, H.H.; Von Hoff, D.D.; Han, H. Kinome-wide siRNA screening identifies molecular targets mediating the sensitivity of pancreatic cancer cells to Aurora kinase inhibitors. Biochem. Pharmacol. 2012, 83, 452–461. [Google Scholar] [CrossRef]
- Jouenne, F.; Chauvot de Beauchene, I.; Bollaert, E.; Avril, M.F.; Caron, O.; Ingster, O.; Lecesne, A.; Benusiglio, P.; Terrier, P.; Caumette, V.; et al. Germline CDKN2A/P16INK4A mutations contribute to genetic determinism of sarcoma. J. Med. Genet. 2017, 54, 607–612. [Google Scholar] [CrossRef]
- Knosel, T.; Chen, Y.; Altendorf-Hofmann, A.; Danielczok, C.; Freesmeyer, M.; Settmacher, U.; Wurst, C.; Schulz, S.; Yang, L.L.; Petersen, I. High KIT and PDGFRA are associated with shorter patients survival in gastroenteropancreatic neuroendocrine tumors, but mutations are a rare event. J. Cancer Res. Clin. Oncol. 2012, 138, 397–403. [Google Scholar] [CrossRef]
- Wei, T.; Zhang, L.N.; Lv, Y.; Ma, X.Y.; Zhi, L.; Liu, C.; Ma, F.; Zhang, X.F. Overexpression of platelet-derived growth factor receptor alpha promotes tumor progression and indicates poor prognosis in hepatocellular carcinoma. Oncotarget 2014, 5, 10307–10317. [Google Scholar] [CrossRef] [PubMed]
- Ahn, K.; Ji, Y.G.; Cho, H.J.; Lee, D.H. Synergistic Anti-Cancer Effects of AKT and SRC Inhibition in Human Pancreatic Cancer Cells. Yonsei Med. J. 2018, 59, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hu, B.; Wang, T.; Huang, W.; Ma, C.; Zhao, Q.; Zhuo, L.; Zhang, T.; Jiang, Y. C-Src confers resistance to mitotic stress through inhibition DMAP1/Bub3 complex formation in pancreatic cancer. Mol. Cancer 2018, 17, 174. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.T.; Pham, H.; Pandol, S.J.; Ptasznik, A. Src as the link between inflammation and cancer. Front. Physiol. 2013, 4, 416. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wei, J.; Su, G.H.; Lin, J. Dasatinib can enhance paclitaxel and gemcitabine inhibitory activity in human pancreatic cancer cells. Cancer Biol. Ther. 2019, 20, 855–865. [Google Scholar] [CrossRef]
- Ogawa, K.; Lin, Q.; Li, L.; Bai, X.; Chen, X.; Chen, H.; Kong, R.; Wang, Y.; Zhu, H.; He, F.; et al. Aspartate beta-hydroxylase promotes pancreatic ductal adenocarcinoma metastasis through activation of SRC signaling pathway. J. Hematol. Oncol. 2019, 12, 144. [Google Scholar] [CrossRef]
- Aligayer, H.; Boyd, D.D.; Heiss, M.M.; Abdalla, E.K.; Curley, S.A.; Gallick, G.E. Activation of Src kinase in primary colorectal carcinoma: An indicator of poor clinical prognosis. Cancer 2002, 94, 344–351. [Google Scholar] [CrossRef]
- Mahajan, K.; Mahajan, N.P. ACK1/TNK2 tyrosine kinase: Molecular signaling and evolving role in cancers. Oncogene 2015, 34, 4162–4167. [Google Scholar] [CrossRef]
- Mahajan, K.; Malla, P.; Lawrence, H.R.; Chen, Z.; Kumar-Sinha, C.; Malik, R.; Shukla, S.; Kim, J.; Coppola, D.; Lawrence, N.J.; et al. ACK1/TNK2 Regulates Histone H4 Tyr88-phosphorylation and AR Gene Expression in Castration-Resistant Prostate Cancer. Cancer Cell 2017, 31, 790–803.e8. [Google Scholar] [CrossRef]
- Mahajan, N.P.; Coppola, D.; Kim, J.; Lawrence, H.R.; Lawrence, N.J.; Mahajan, K. Blockade of ACK1/TNK2 To Squelch the Survival of Prostate Cancer Stem-like Cells. Sci. Rep. 2018, 8, 1954. [Google Scholar] [CrossRef]
- Qi, L.; Ding, Y. TNK2 as a key drug target for the treatment of metastatic colorectal cancer. Int. J. Biol. Macromol. 2018, 119, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Shinmura, K.; Kiyose, S.; Nagura, K.; Igarashi, H.; Inoue, Y.; Nakamura, S.; Maeda, M.; Baba, M.; Konno, H.; Sugimura, H. TNK2 gene amplification is a novel predictor of a poor prognosis in patients with gastric cancer. J. Surg. Oncol. 2014, 109, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, D.; Pan, L.; Sun, J. The positive feedback between lncRNA TNK2-AS1 and STAT3 enhances angiogenesis in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2018, 507, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zahari, M.S.; Renuse, S.; Kelkar, D.S.; Barbhuiya, M.A.; Rojas, P.L.; Stearns, V.; Gabrielson, E.; Malla, P.; Sukumar, S.; et al. The non-receptor tyrosine kinase TNK2/ACK1 is a novel therapeutic target in triple negative breast cancer. Oncotarget 2017, 8, 2971–2983. [Google Scholar] [CrossRef] [PubMed]
- Howlin, J.; Rosenkvist, J.; Andersson, T. TNK2 preserves epidermal growth factor receptor expression on the cell surface and enhances migration and invasion of human breast cancer cells. Breast Cancer Res. BCR 2008, 10, R36. [Google Scholar] [CrossRef]
- Galisteo, M.L.; Yang, Y.; Urena, J.; Schlessinger, J. Activation of the nonreceptor protein tyrosine kinase Ack by multiple extracellular stimuli. Proc. Natl. Acad. Sci. USA 2006, 103, 9796–9801. [Google Scholar] [CrossRef]
- Lin, Q.; Wang, J.; Childress, C.; Yang, W. The activation mechanism of ACK1 (activated Cdc42-associated tyrosine kinase 1). Biochem. J. 2012, 445, 255–264. [Google Scholar] [CrossRef]
- Britton, D.; Zen, Y.; Quaglia, A.; Selzer, S.; Mitra, V.; Lobetaner, C.; Jung, S.; Bohm, G.; Schmid, P.; Prefot, P.; et al. Quantification of pancreatic cancer proteome and phosphorylome: Indicates molecular events likely contributing to cancer and activity of drug targets. PLoS ONE 2014, 9, e90948. [Google Scholar] [CrossRef]
- Cannon, A.; Thompson, C.; Hall, B.R.; Jain, M.; Kumar, S.; Batra, S.K. Desmoplasia in pancreatic ductal adenocarcinoma: Insight into pathological function and therapeutic potential. Genes Cancer 2018, 9, 78–86. [Google Scholar] [CrossRef]
- Pandol, S.; Edderkaoui, M.; Gukovsky, I.; Lugea, A.; Gukovskaya, A. Desmoplasia of pancreatic ductal adenocarcinoma. Clin. Gastroenterol. Hepatol. 2009, 7 (Suppl. 11), S44–S47. [Google Scholar] [CrossRef]
- Wei, C.; Li, L.; Menon, M.C.; Zhang, W.; Fu, J.; Kidd, B.; Keung, K.L.; Woytovich, C.; Greene, I.; Xiao, W.; et al. Genomic Analysis of Kidney Allograft Injury Identifies Hematopoietic Cell Kinase as a Key Driver of Renal Fibrosis. J. Am. Soc. Nephrol. JASN 2017, 28, 1385–1393. [Google Scholar] [CrossRef] [PubMed]
- Torsello, B.; De Marco, S.; Bombelli, S.; Chisci, E.; Cassina, V.; Corti, R.; Bernasconi, D.; Giovannoni, R.; Bianchi, C.; Perego, R.A. The 1ALCTL and 1BLCTL isoforms of Arg/Abl2 induce fibroblast activation and extra cellular matrix remodelling differently. Biol. Open 2019, 8, bio038554. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Bian, H.; Bartual, S.G.; Du, W.; Luo, J.; Zhao, H.; Zhang, S.; Mo, C.; Zhou, Y.; Xu, Y.; et al. Structure-Based Design of Tetrahydroisoquinoline-7-carboxamides as Selective Discoidin Domain Receptor 1 (DDR1) Inhibitors. J. Med. Chem. 2016, 59, 5911–5916. [Google Scholar] [CrossRef] [PubMed]
- Richter, H.; Satz, A.L.; Bedoucha, M.; Buettelmann, B.; Petersen, A.C.; Harmeier, A.; Hermosilla, R.; Hochstrasser, R.; Burger, D.; Gsell, B.; et al. DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome. ACS Chem. Biol. 2018, 14, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, Y.; Bartual, S.G.; Luo, J.; Xu, T.; Du, W.; Xun, Q.; Tu, Z.; Brekken, R.A.; Ren, X.; et al. Tetrahydroisoquinoline-7-carboxamide Derivatives as New Selective Discoidin Domain Receptor 1 (DDR1) Inhibitors. ACS Med. Chem. Lett. 2017, 8, 327–332. [Google Scholar] [CrossRef]
- Tao, J.; Zhang, M.; Wen, Z.; Wang, B.; Zhang, L.; Ou, Y.; Tang, X.; Yu, X.; Jiang, Q. Inhibition of EP300 and DDR1 synergistically alleviates pulmonary fibrosis in vitro and in vivo. Biomed Pharm. 2018, 106, 1727–1733. [Google Scholar] [CrossRef]
- Lin, L.; Shi, C.; Sun, Z.; Le, N.-T.; Abe, J.-I.; Hu, K. The Ser/Thr kinase p90RSK promotes kidney fibrosis by modulating fibroblast–epithelial crosstalk. J. Biol. Chem. 2019, 294, 9901–9910. [Google Scholar] [CrossRef]
- Abe, J.; Berk, B. Fyn-dependent activation of p90 ribosomal S6 kinase (RSK) by H2O2: New redox sensitive pathway. In Circulation; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 1998; p. 220. [Google Scholar]
- Dolivo, D.M.; Larson, S.A.; Dominko, T. Tryptophan metabolites kynurenine and serotonin regulate fibroblast activation and fibrosis. Cell. Mol. Life Sci. 2018, 75, 3663–3681. [Google Scholar] [CrossRef]
- Jiang, S.-H.; Li, J.; Dong, F.-Y.; Yang, J.-Y.; Liu, D.-J.; Yang, X.-M.; Wang, Y.-H.; Yang, M.-W.; Fu, X.-L.; Zhang, X.-X.; et al. Increased Serotonin Signaling Contributes to the Warburg Effect in Pancreatic Tumor Cells Under Metabolic Stress and Promotes Growth of Pancreatic Tumors in Mice. Gastroenterology 2017, 153, 277–291.e19. [Google Scholar] [CrossRef]
- Martinez-Bosch, N.; Vinaixa, J.; Navarro, P. Immune Evasion in Pancreatic Cancer: From Mechanisms to Therapy. Cancers 2018, 10, 6. [Google Scholar] [CrossRef]
- Philipsen, L.; Reddycherla, A.V.; Hartig, R.; Gumz, J.; Kästle, M.; Kritikos, A.; Poltorak, M.P.; Prokazov, Y.; Turbin, E.; Weber, A.; et al. De novo phosphorylation and conformational opening of the tyrosine kinase Lck act in concert to initiate T cell receptor signaling. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Sha, S.; Yang, J.; Ren, H. Effects of Tec Tyrosine Kinase Inhibition on the Inflammatory Response of Severe Acute Pancreatitis-Associated Acute Lung Injury in Mice. Dig. Dis. Sci. 2019, 64, 2167–2176. [Google Scholar] [CrossRef] [PubMed]
- Borowiec, M.; Liew, C.W.; Thompson, R.; Boonyasrisawat, W.; Hu, J.; Mlynarski, W.M.; El Khattabi, I.; Kim, S.H.; Marselli, L.; Rich, S.S.; et al. Mutations at the BLK locus linked to maturity onset diabetes of the young and -cell dysfunction. Proc. Natl. Acad. Sci. USA 2009, 106, 14460–14465. [Google Scholar] [CrossRef] [PubMed]
- Feanny, M.A.; Fagan, S.P.; Ballian, N.; Liu, S.H.; Li, Z.; Wang, X.; Fisher, W.; Brunicardi, F.C.; Belaguli, N.S. PDX-1 expression is associated with islet proliferation in vitro and in vivo. J. Surg. Res. 2008, 144, 8–16. [Google Scholar] [CrossRef]
- Liu, S.; Ballian, N.; Belaguli, N.S.; Patel, S.; Li, M.; Templeton, N.S.; Gingras, M.C.; Gibbs, R.; Fisher, W.; Brunicardi, F.C. PDX-1 acts as a potential molecular target for treatment of human pancreatic cancer. Pancreas 2008, 37, 210–220. [Google Scholar] [CrossRef]
- Liu, S.; Wang, X.P.; Brunicardi, F.C. Enhanced cytotoxicity of RIPTK gene therapy of pancreatic cancer via PDX-1 co-delivery. J. Surg. Res. 2007, 137, 1–9. [Google Scholar] [CrossRef]
- Liu, S.H.; Rao, D.D.; Nemunaitis, J.; Senzer, N.; Zhou, G.; Dawson, D.; Gingras, M.C.; Wang, Z.; Gibbs, R.; Norman, M.; et al. PDX-1 is a therapeutic target for pancreatic cancer, insulinoma and islet neoplasia using a novel RNA interference platform. PLoS ONE 2012, 7, e40452. [Google Scholar] [CrossRef]
- Liu, S.H.; Yu, J.; Sanchez, R.; Liu, X.; Heidt, D.; Willey, J.; Nemunaitis, J.; Brunicardi, F.C. A novel synthetic human insulin super promoter for targeting PDX-1-expressing pancreatic cancer. Cancer Lett. 2018, 418, 75–83. [Google Scholar] [CrossRef]
- Liu, S.H.; Hong, Y.; Markowiak, S.; Sanchez, R.; Creeden, J.; Nemunaitis, J.; Kalinoski, A.; Willey, J.; Erhardt, P.; Lee, J.; et al. BIRC5 is a target for molecular imaging and detection of human pancreatic cancer. Cancer Lett. 2019, 457, 10–19. [Google Scholar] [CrossRef]
- Liu, S.H.; Yu, J.; Creeden, J.F.; Sutton, J.M.; Markowiak, S.; Sanchez, R.; Nemunaitis, J.; Kalinoski, A.; Zhang, J.T.; Damoiseaux, R.; et al. Repurposing metformin, simvastatin and digoxin as a combination for targeted therapy for pancreatic ductal adenocarcinoma. Cancer Lett. 2020, 491, 97–107. [Google Scholar] [CrossRef]
- Kondratyeva, L.G.; Safina, D.R.; Chernov, I.P.; Kopantzev, E.P.; Kostrov, S.V.; Sverdlov, E.D. PDX1, a key factor in pancreatic embryogenesis, can exhibit antimetastatic activity in pancreatic ductal adenocarcinoma. Cancer Manag. Res. 2019, 11, 7077–7087. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.; Takeuchi, K.K.; Ruggeri, J.M.; Bailey, P.; Chang, D.; Li, J.; Leonhardt, L.; Puri, S.; Hoffman, M.T.; Gao, S.; et al. PDX1 dynamically regulates pancreatic ductal adenocarcinoma initiation and maintenance. Genes Dev. 2016, 30, 2669–2683. [Google Scholar] [CrossRef] [PubMed]
- Vinogradova, T.V.; Sverdlov, E.D. PDX1: A Unique Pancreatic Master Regulator Constantly Changes Its Functions during Embryonic Development and Progression of Pancreatic Cancer. Biochemistry 2017, 82, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008, 321, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Raphael, B.J.; Hruban, R.H.; Aguirre, A.J.; Moffitt, R.A.; Yeh, J.J.; Stewart, C.; Robertson, A.G.; Cherniack, A.D.; Gupta, M.; Getz, G.; et al. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017, 32, 185–203.e13. [Google Scholar] [CrossRef]
- Torres, C.; Grippo, P.J. Pancreatic cancer subtypes: A roadmap for precision medicine. Ann. Med. 2018, 50, 277–287. [Google Scholar] [CrossRef]
- Lieber, M.; Mazzetta, J.; Nelson-Rees, W.; Kaplan, M.; Todaro, G. Establishment of a continuous tumor-cell line (panc-1) from a human carcinoma of the exocrine pancreas. Int. J. Cancer 1975, 15, 741–747. [Google Scholar] [CrossRef]
- Hilhorst, R.; Houkes, L.; Mommersteeg, M.; Musch, J.; van den Berg, A.; Ruijtenbeek, R. Peptide microarrays for profiling of serine/threonine kinase activity of recombinant kinases and lysates of cells and tissue samples. Methods Mol. Biol. 2013, 977, 259–271. [Google Scholar]
- Appuhamy, J.A.; Nayananjalie, W.A.; England, E.M.; Gerrard, D.E.; Akers, R.M.; Hanigan, M.D. Effects of AMP-activated protein kinase (AMPK) signaling and essential amino acids on mammalian target of rapamycin (mTOR) signaling and protein synthesis rates in mammary cells. J. Dairy Sci. 2014, 97, 419–429. [Google Scholar] [CrossRef]
- Dorsett, C.R.; McGuire, J.L.; Niedzielko, T.L.; DePasquale, E.A.; Meller, J.; Floyd, C.L.; McCullumsmith, R.E. Traumatic Brain Injury Induces Alterations in Cortical Glutamate Uptake without a Reduction in Glutamate Transporter-1 Protein Expression. J. Neurotrauma 2017, 34, 220–234. [Google Scholar] [CrossRef]
- Hunter, T. Signaling—2000 and beyond. Cell 2000, 100, 113–127. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Saavedra, D.; Barton, G.J. Classification and functional annotation of eukaryotic protein kinases. Proteins 2007, 68, 893–914. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xu, H.; Lin, S.; Deng, W.; Zhou, J.; Zhang, Y.; Shi, Y.; Peng, D.; Xue, Y. GPS 5.0: An Update on the Prediction of Kinase-specific Phosphorylation Sites in Proteins. Genom. Proteom. Bioinform. 2020, 18, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Liu, Z.; Cao, J.; Ma, Q.; Gao, X.; Wang, Q.; Jin, C.; Zhou, Y.; Wen, L.; Ren, J. GPS 2.1: Enhanced prediction of kinase-specific phosphorylation sites with an algorithm of motif length selection. Protein Eng. Des. Sel. 2011, 24, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Creeden, J.F.; Alganem, K.; Imami, A.S.; Henkel, N.D.; Brunicardi, F.C.; Liu, S.-H.; Shukla, R.; Tomar, T.; Naji, F.; McCullumsmith, R.E. Emerging Kinase Therapeutic Targets in Pancreatic Ductal Adenocarcinoma and Pancreatic Cancer Desmoplasia. Int. J. Mol. Sci. 2020. In review. [Google Scholar]
- Chou, A.; Froio, D.; Nagrial, A.M.; Parkin, A.; Murphy, K.J.; Chin, V.T.; Wohl, D.; Steinmann, A.; Stark, R.; Drury, A.; et al. Tailored first-line and second-line CDK4-targeting treatment combinations in mouse models of pancreatic cancer. Gut 2018, 67, 2142–2155. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).