The Importance of Hypoxia-Inducible Factors (HIF-1 and HIF-2) for the Pathophysiology of Inflammatory Bowel Disease
Abstract
1. Introduction
2. Results
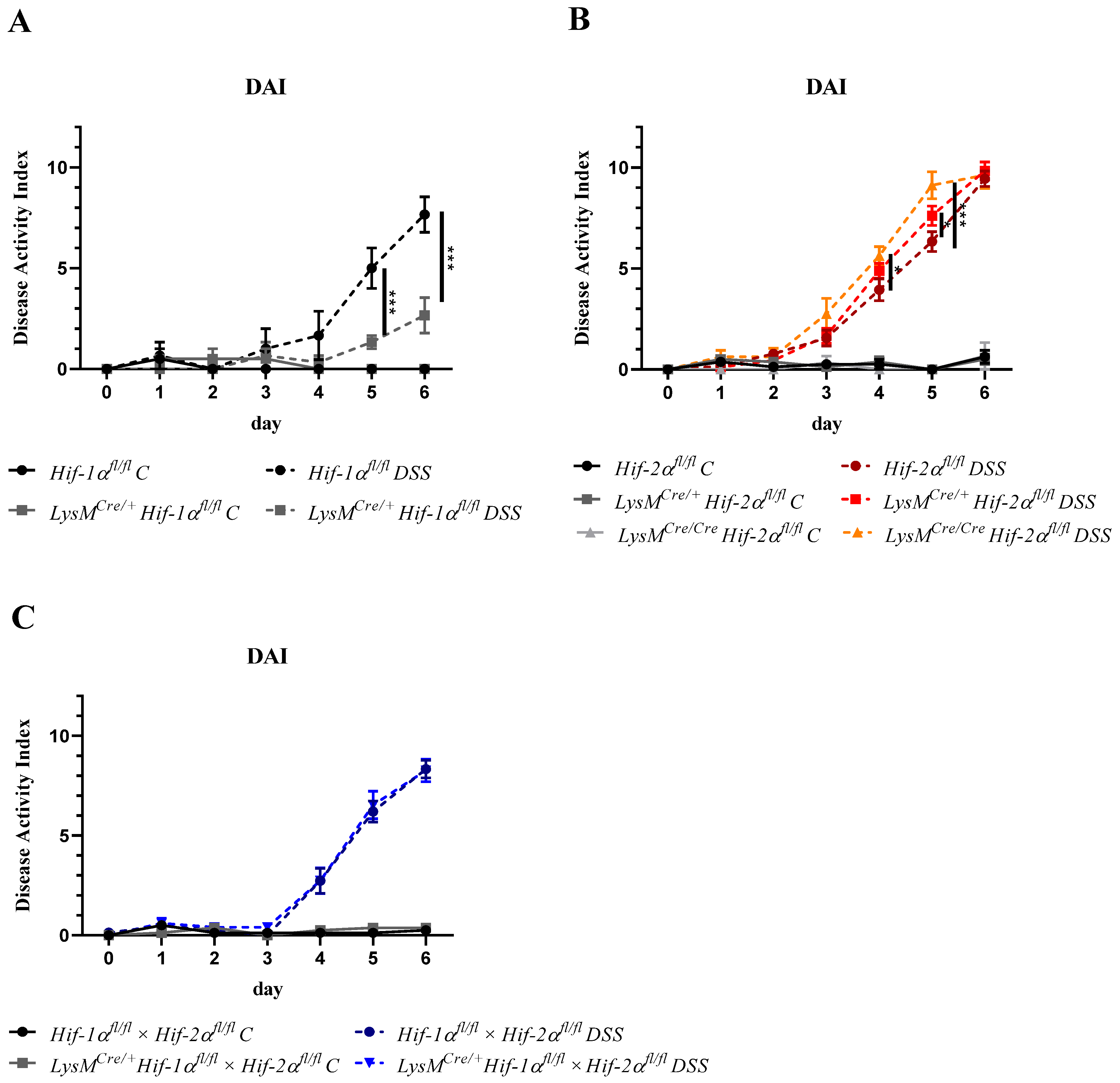
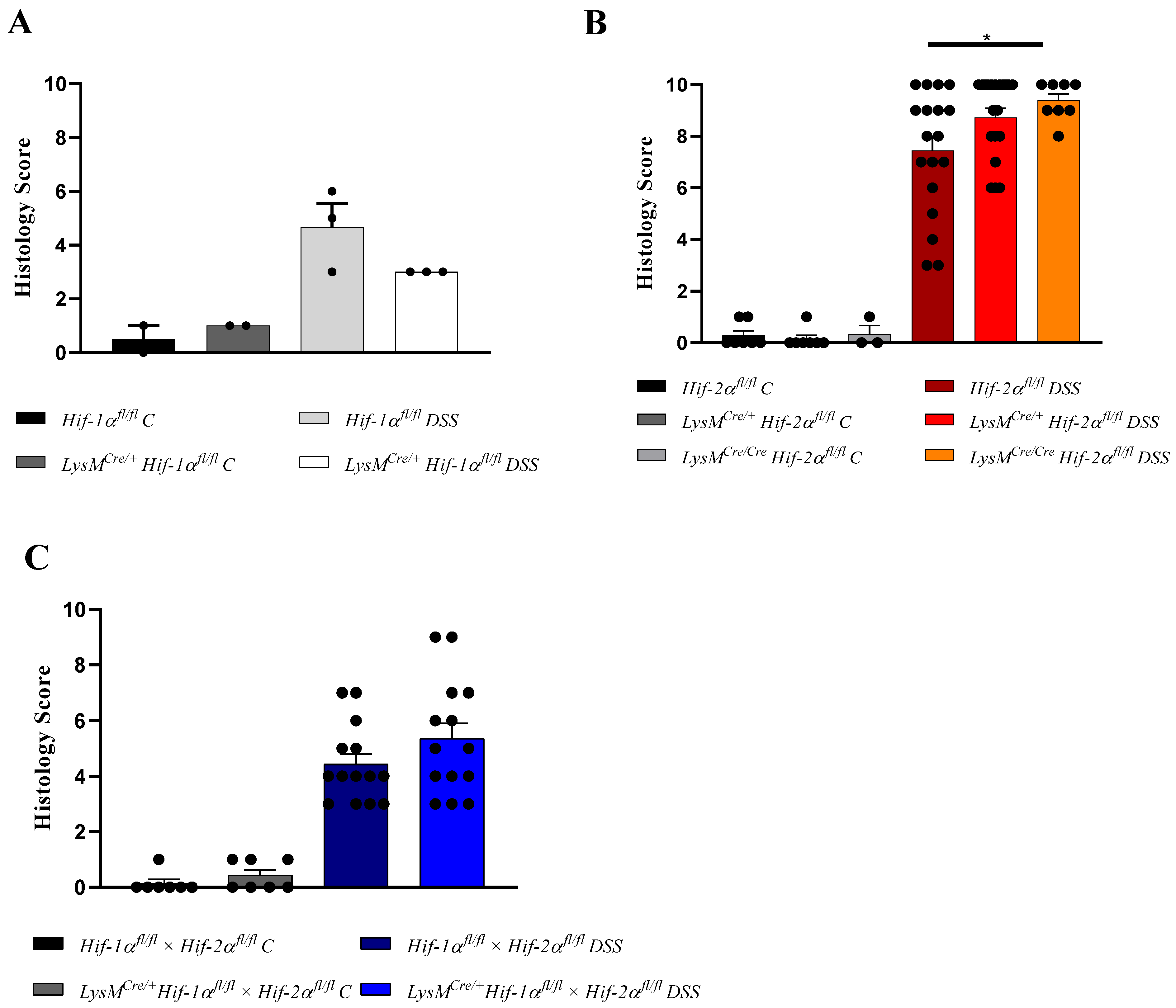
2.1. The Severity of Colon Inflammation is Reduced in Mice Lacking Functional HIF-1α, but Higher in Mice Lacking Functional HIF-2α in Myeloid Cells
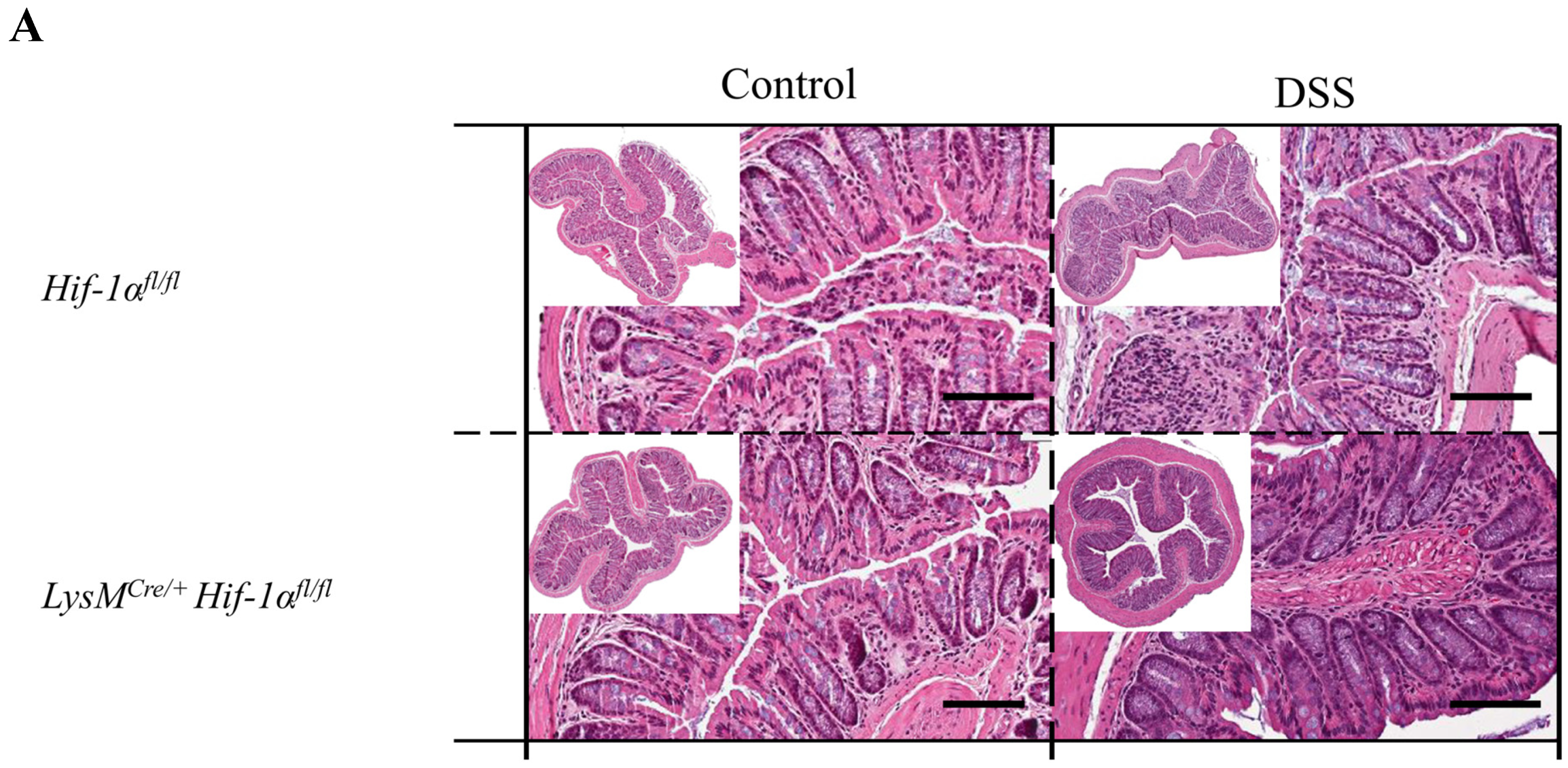
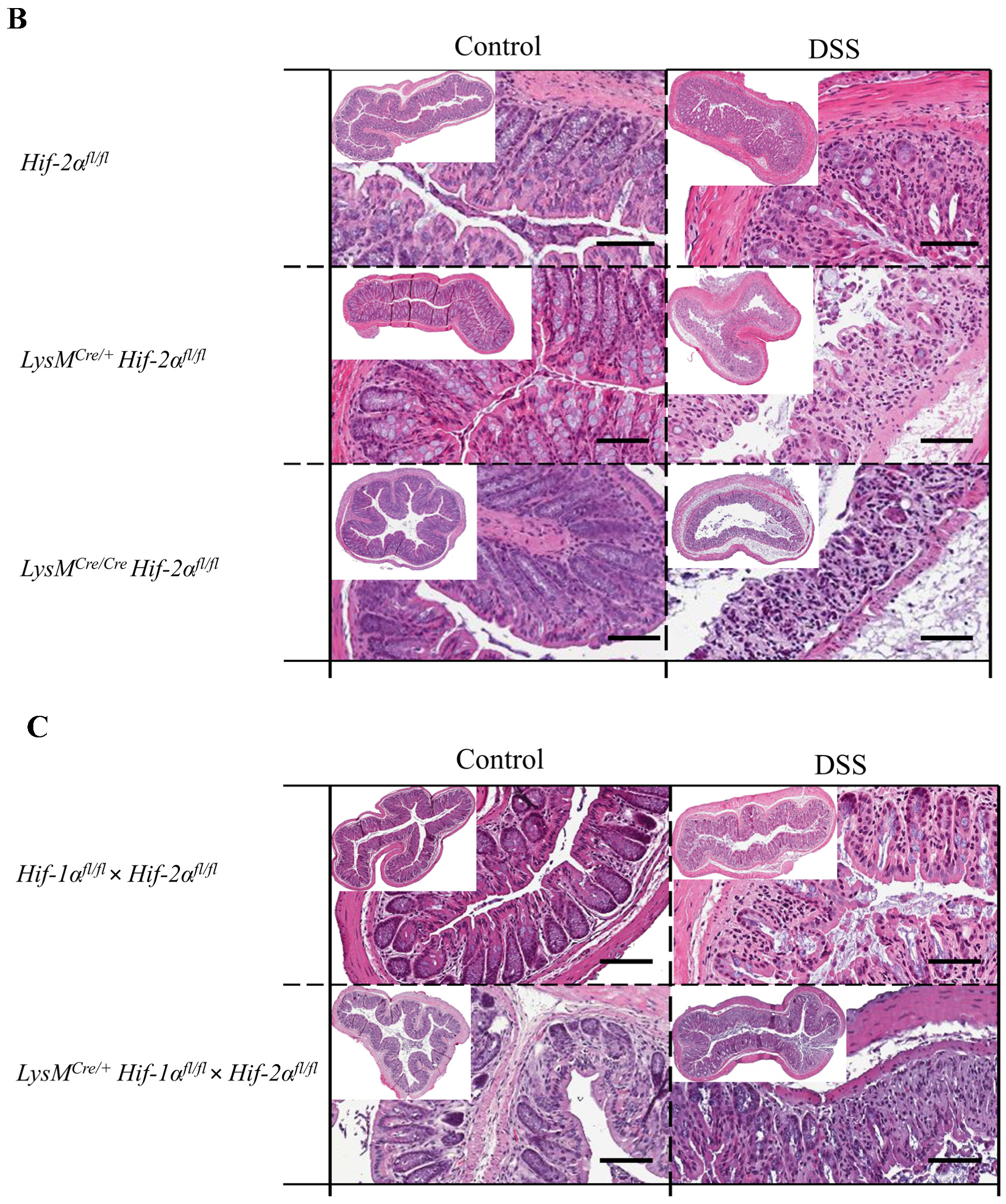
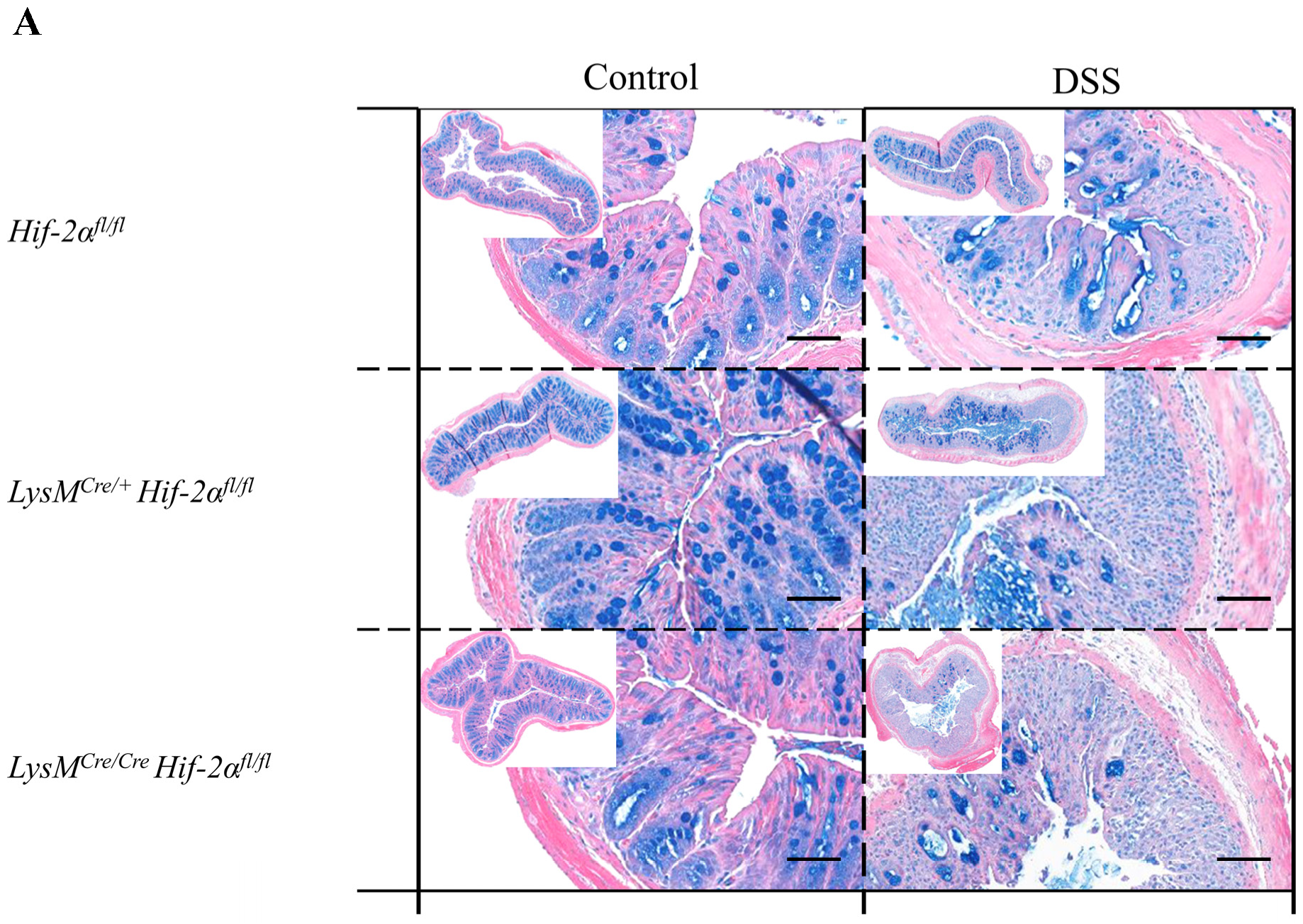
2.2. Tissue Destruction after DSS Treatment in Mice without Functional HIF-1α is Reduced, but Higher in Mice without Functional HIF-2α in Myeloid Cells
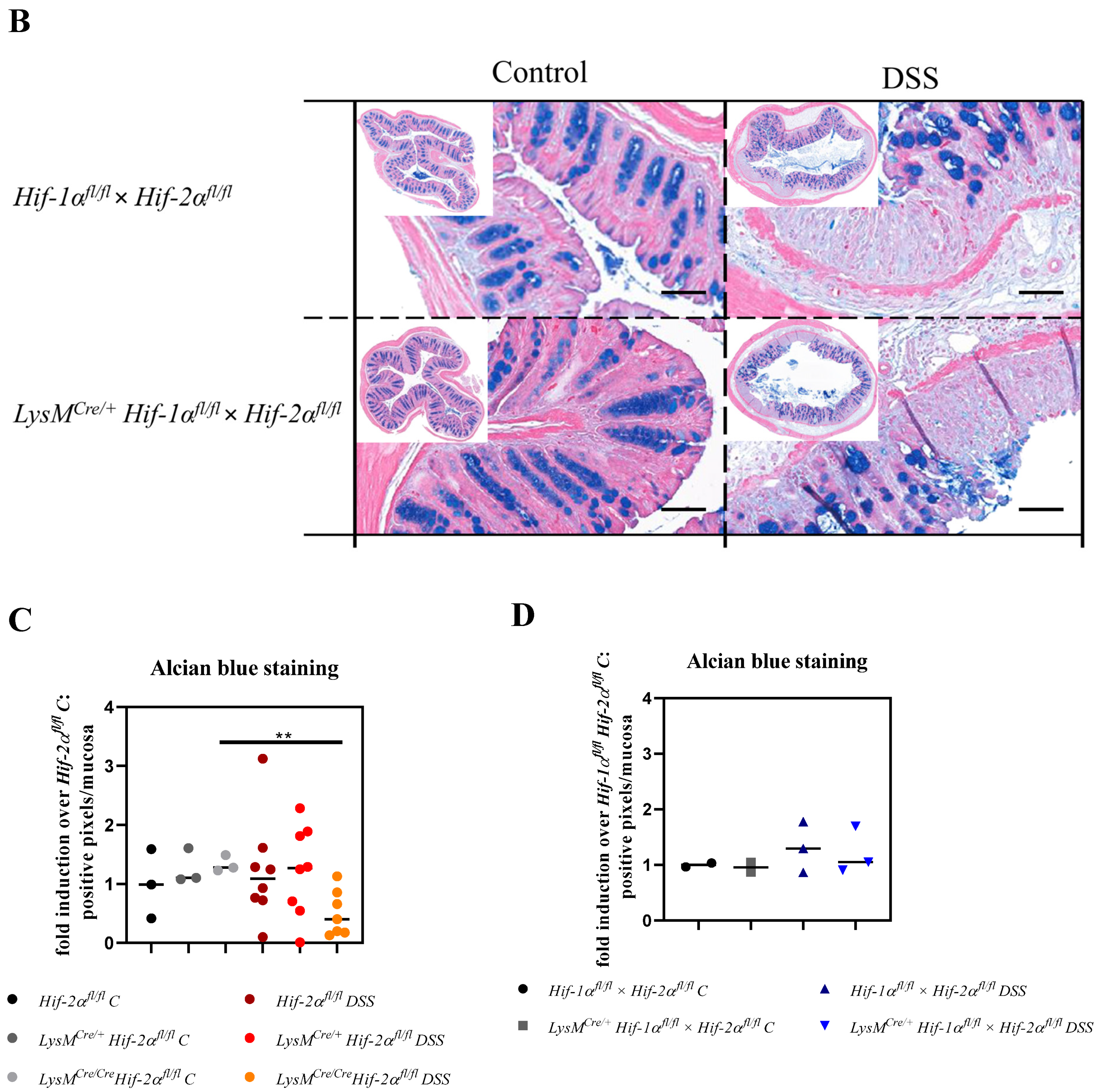
2.3. Tissue Destruction Reduced the Mucin Production in the Colon
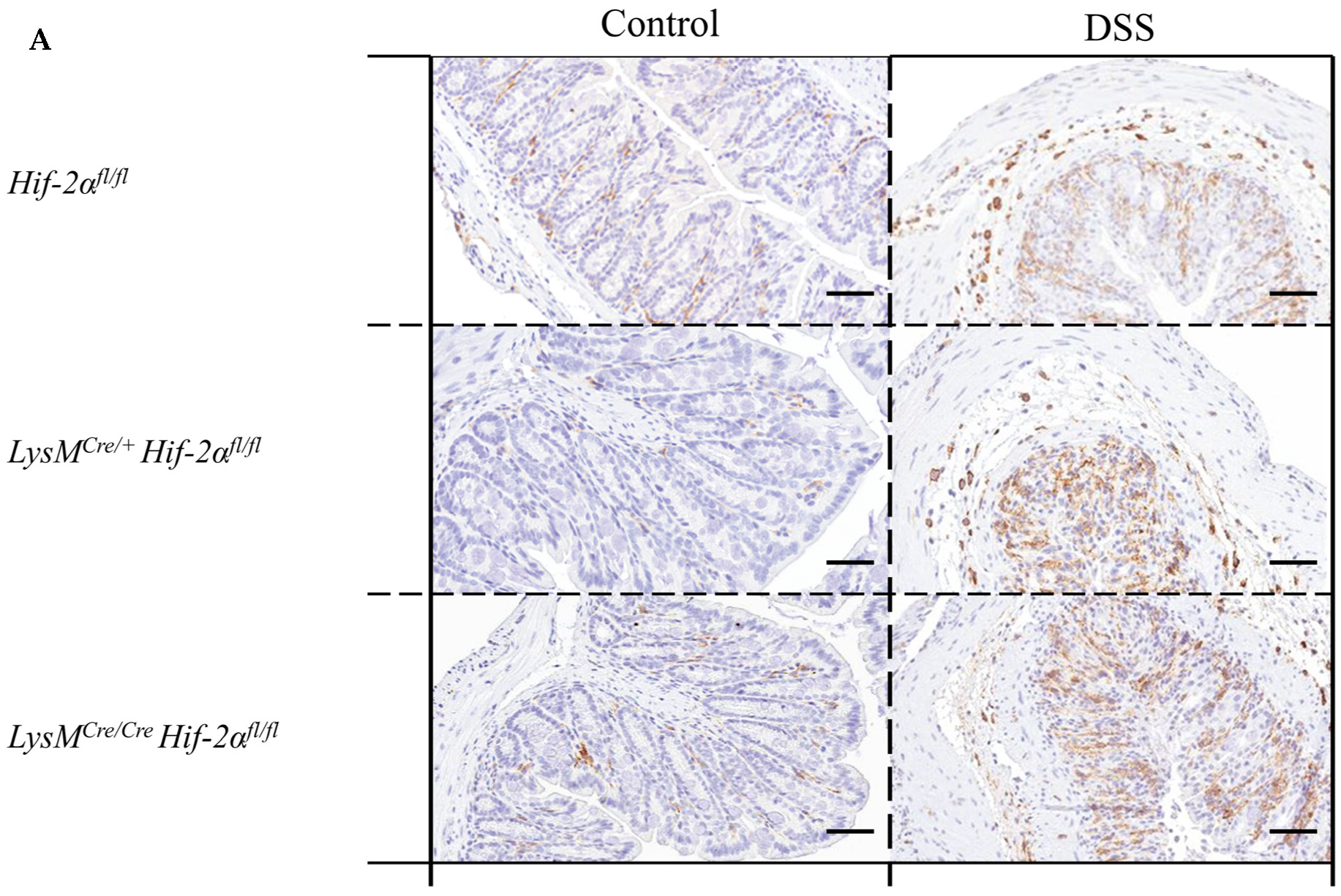
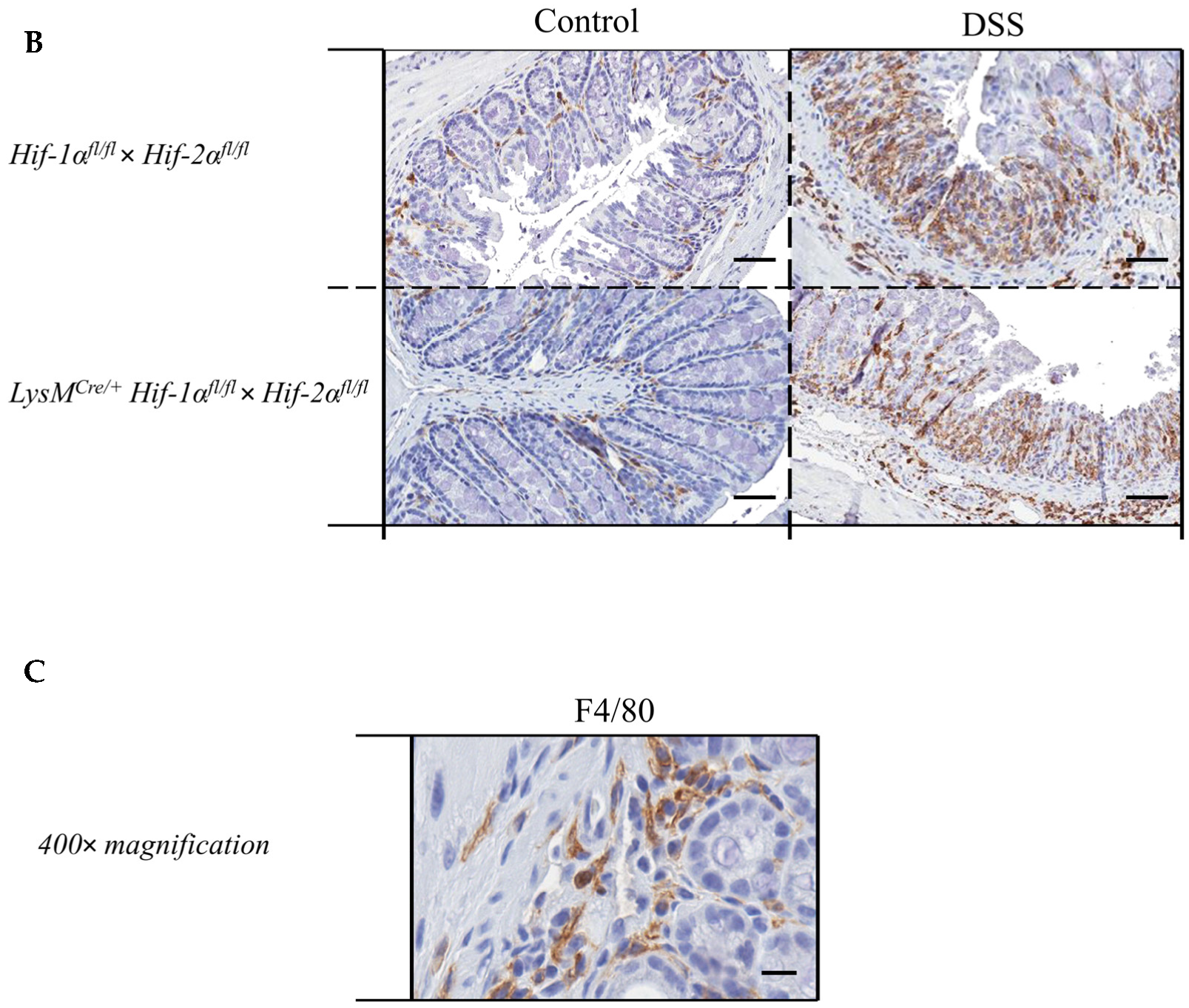
2.4. Macrophage Migration towards the Inflamed Colon is not Altered by Myeloid HIF-2α Knockout
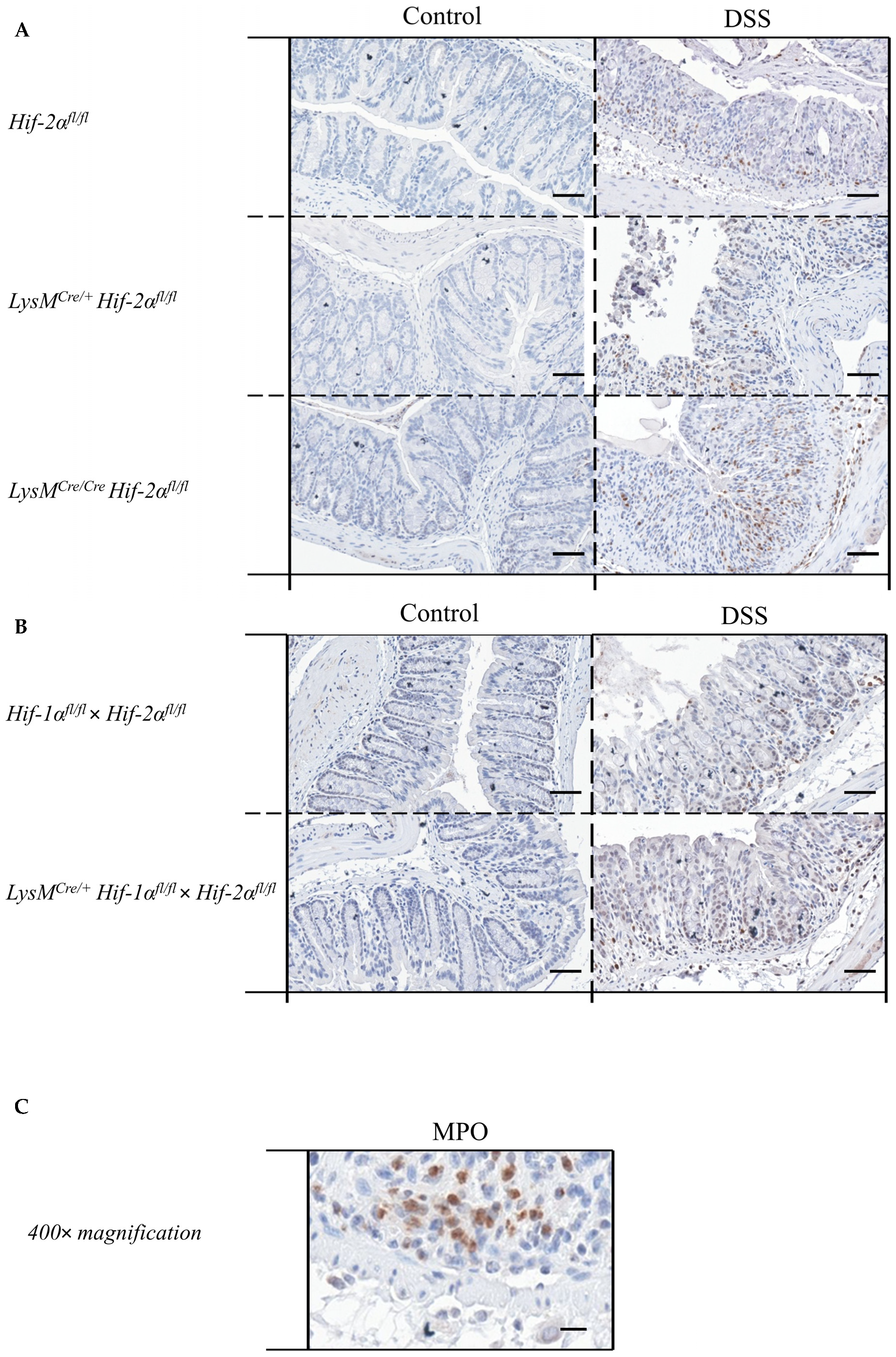
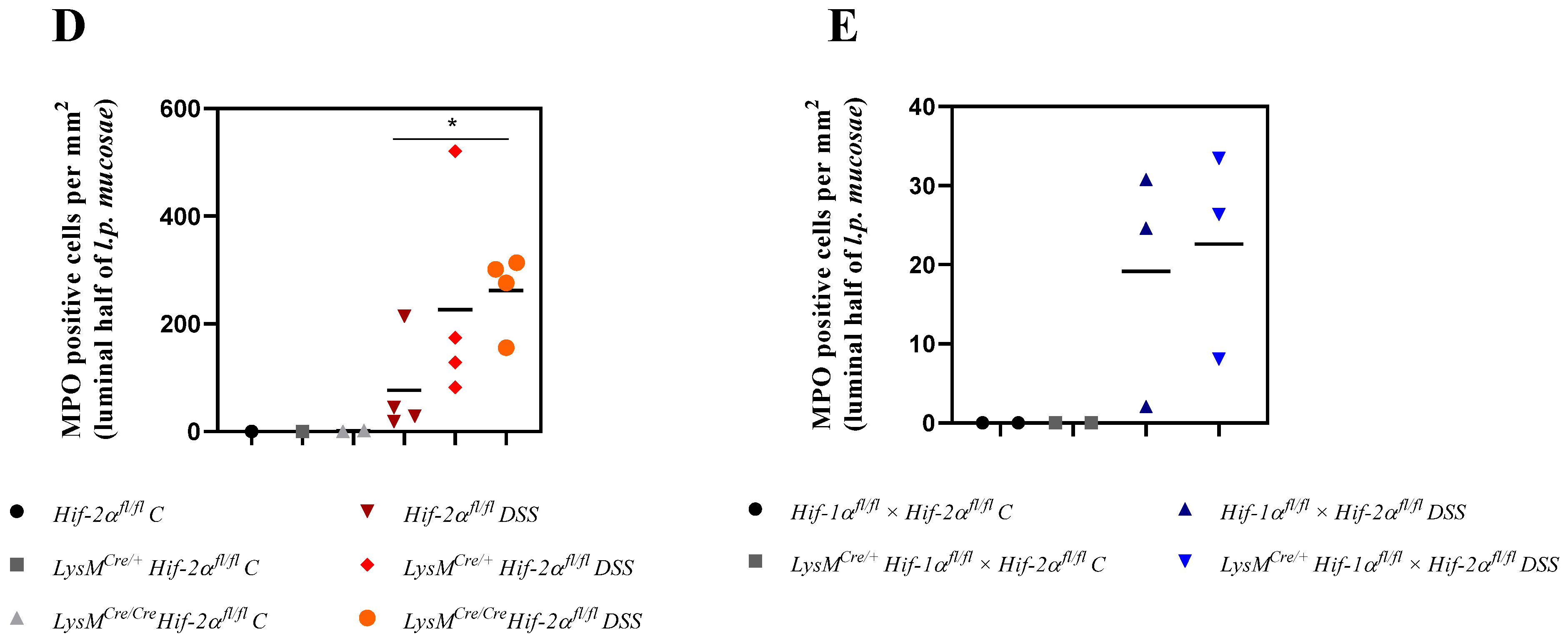
2.5. Mice without Functional HIF-2α in Myeloid Cells Recruit more Neutrophils towards the Gut Lumen
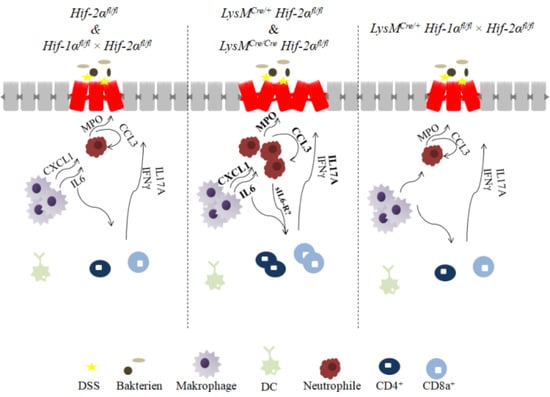
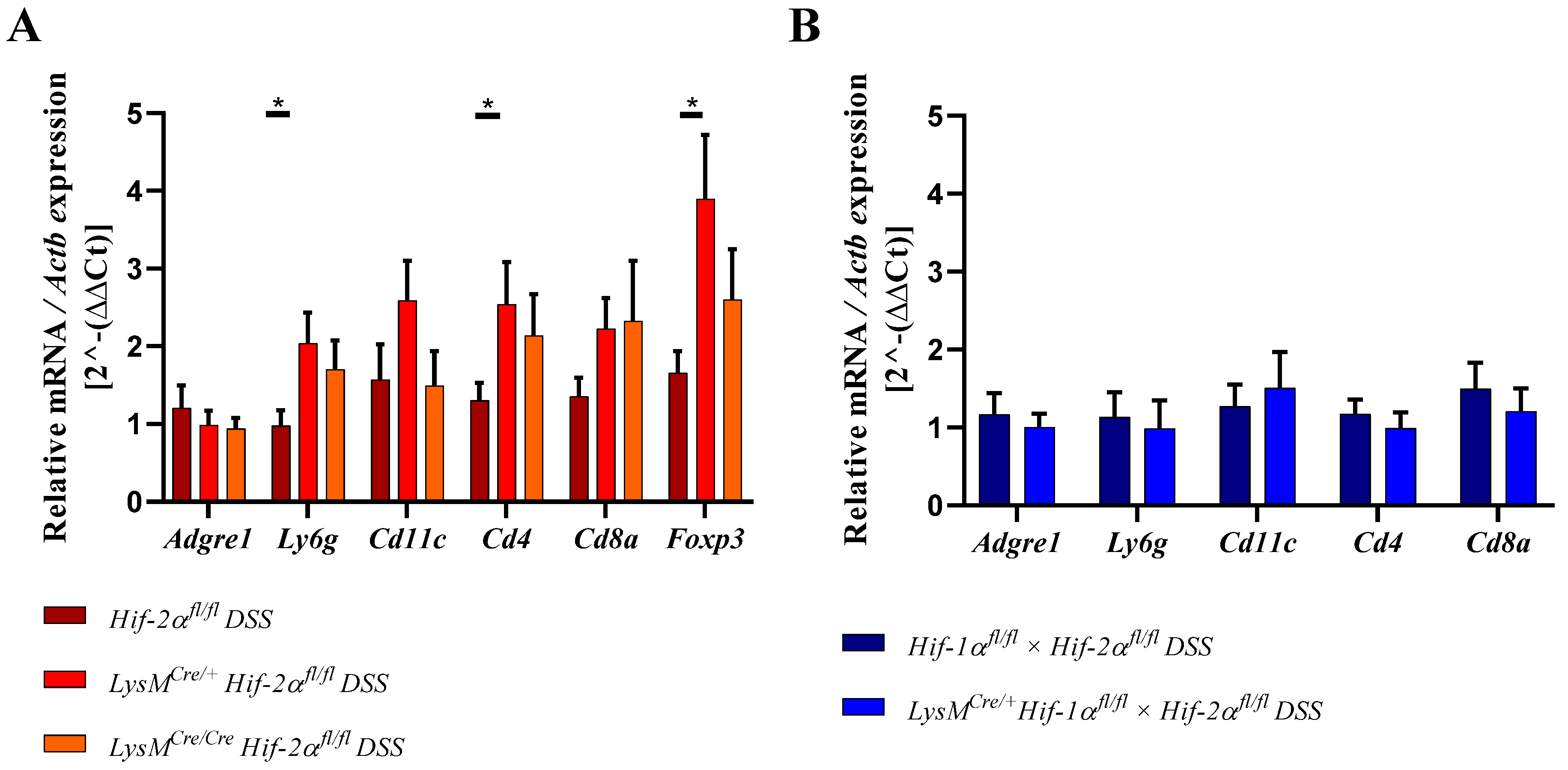
2.6. Mice without Functional HIF-2α in Myeloid Cells Recruit more Neutrophils, CD4+, and CD8a+ T-Cells and Tregs towards the Inflamed Colon
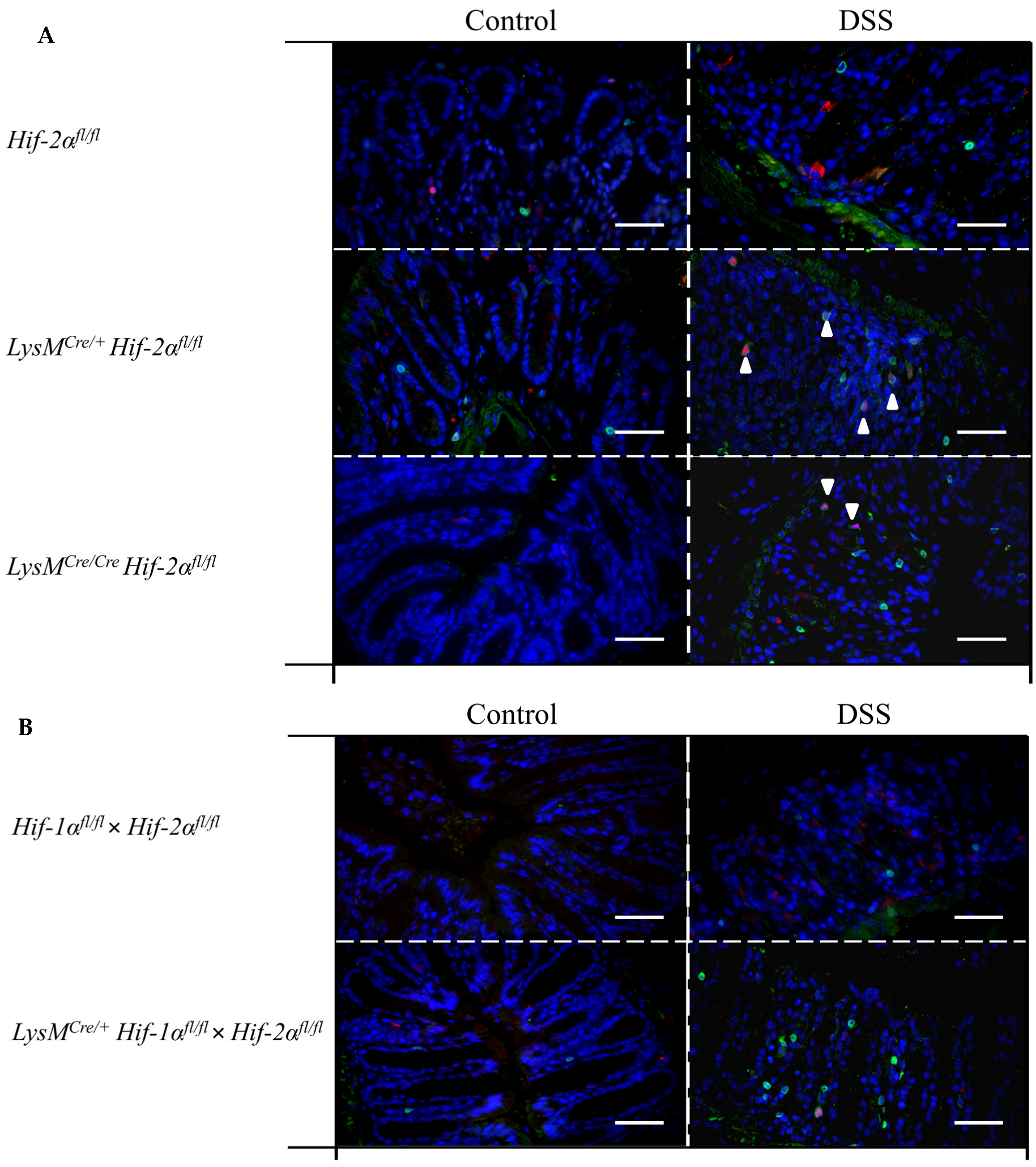
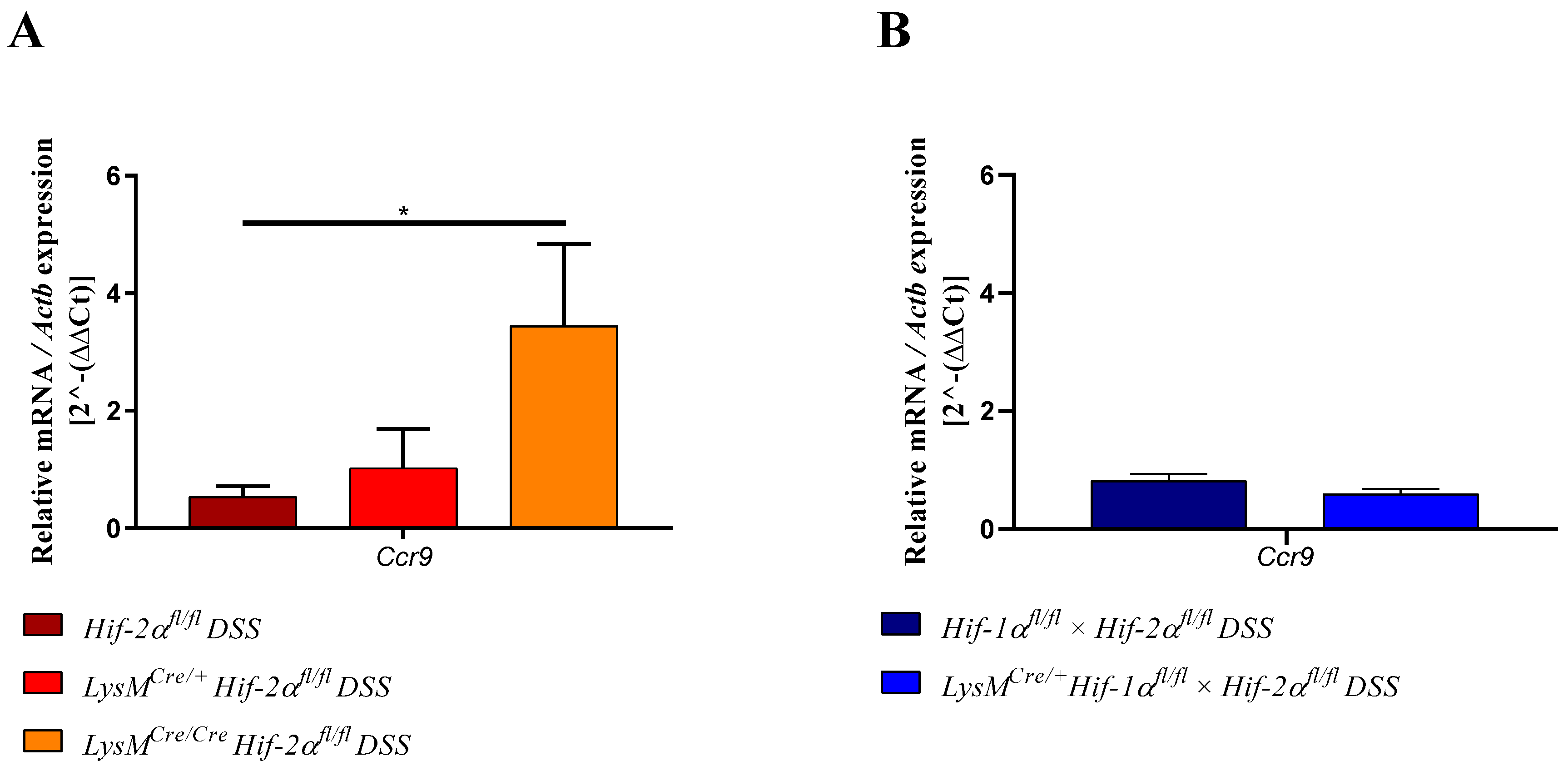
2.7. Mice without Functional HIF-2α in Myeloid Cells Induce Migration of Lymphocytes towards the Gut
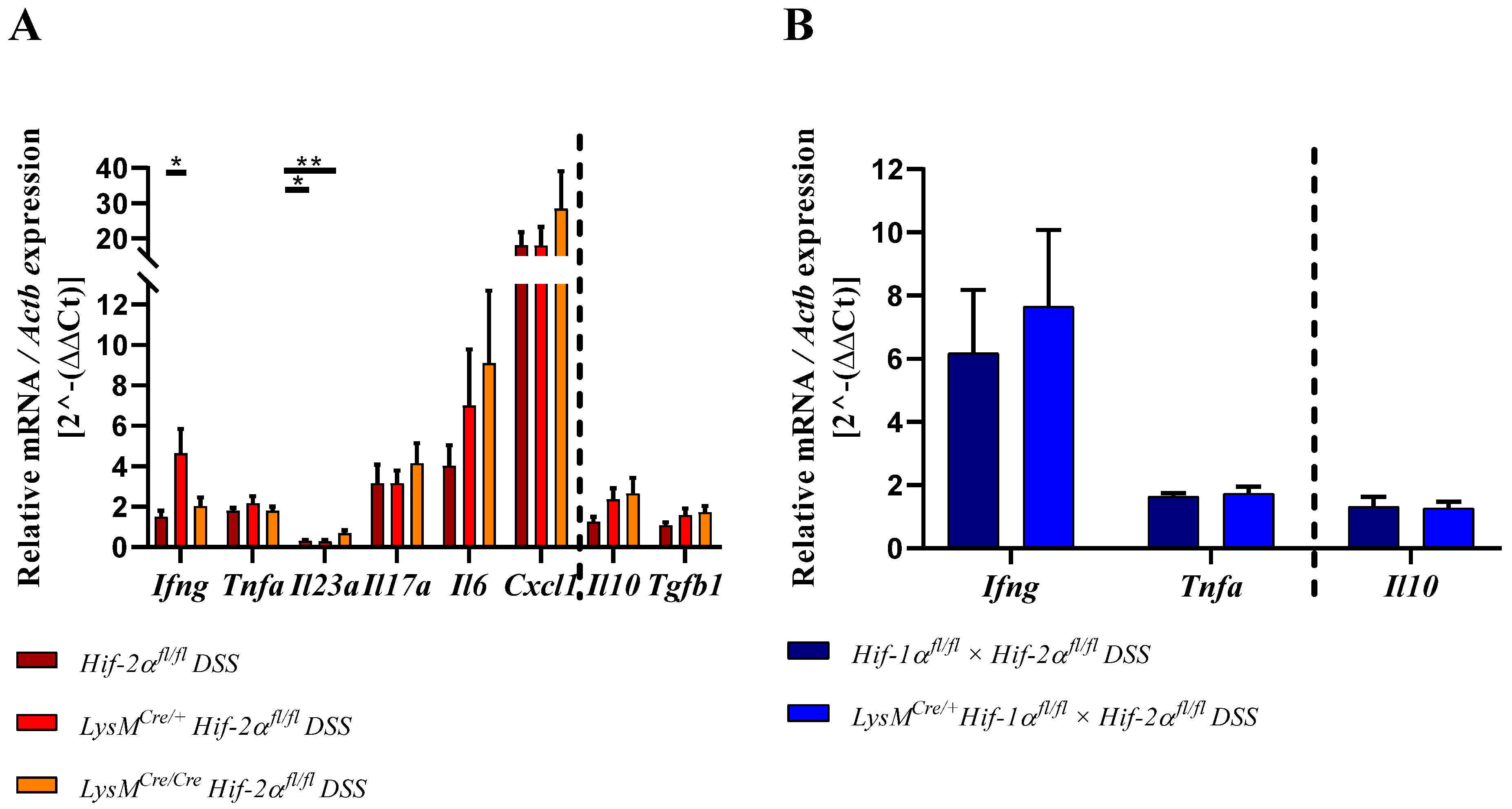
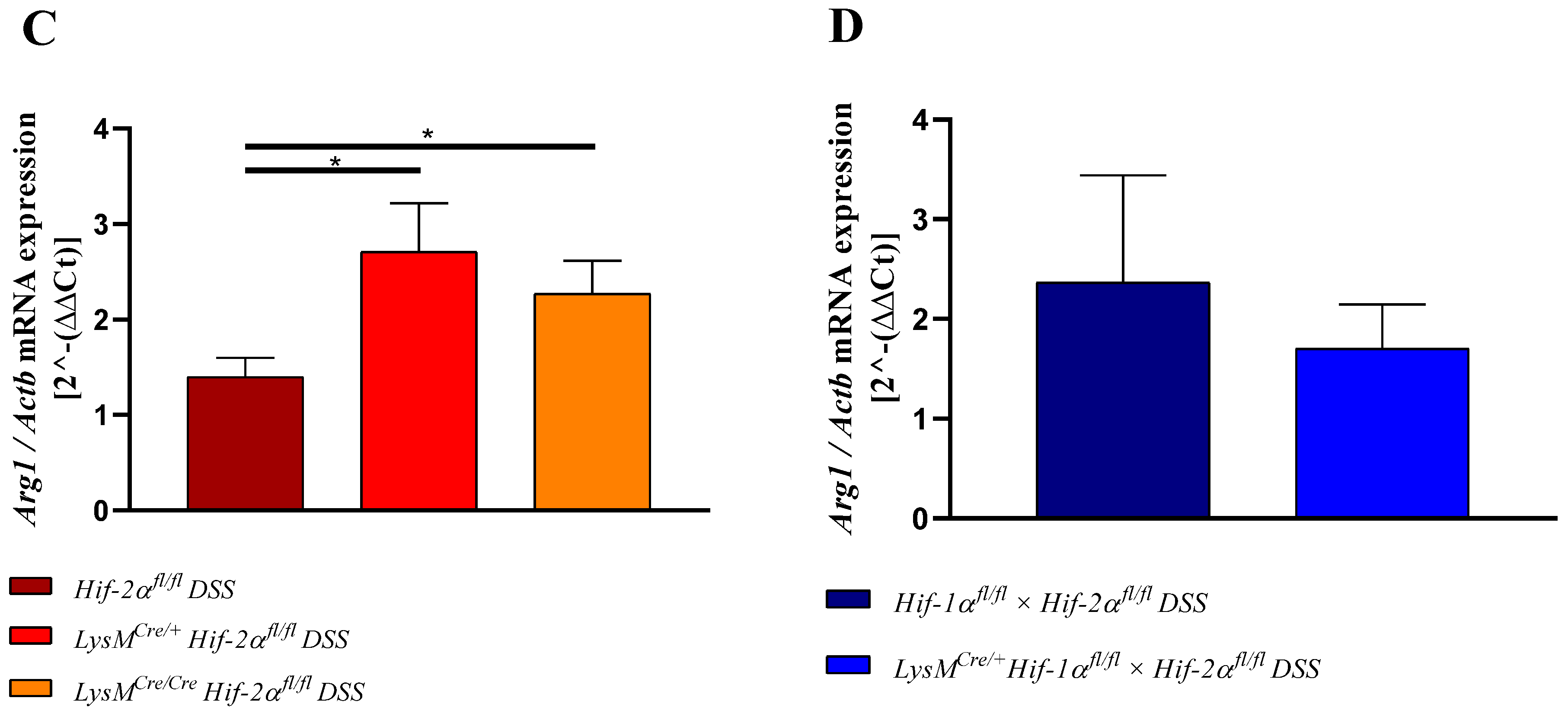
2.8. HIF-2α Loss in Myeloid Cells induced High Pro- and Anti-Inflammatory Gene Expressions in the Inflamed Gut
3. Discussion
4. Materials and Methods
4.1. Animal Model
4.2. Cell Culture
4.3. Histology
4.4. Real-Time PCR
4.5. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BMDM | Bone-marrow-derived macrophage |
| CCR9 | C-C chemokine receptor type 9 |
| C-TAD | C-terminal transactivation domain |
| CXCL1 | chemokine (C-X-C motif) ligand 1 |
| DAI | Disease Activity Index |
| DC | Dendritic cell |
| DNA | deoxyribonucleic acid |
| DSS | dextran sodium sulfate |
| FIH | Factor Inhibiting HIF |
| HIF | Hypoxia-inducible factor |
| HRE | Hypoxia-Response Element |
| IBD | Inflammatory bowel disease |
| IFNγ | Interferon-γ |
| IL | Interleukin |
| ODD | Oxygen-dependent degradation |
| O2 | Oxygen |
| PHD | Prolyl hydroxylase |
| pVHL | von-Hippel Lindau protein |
| TGFβ | Transforming growth factor beta |
| TNFα | Tumor necrosis factor alpha |
References
- Dignass, A.; Schulze, H.; Preiß, J.C.; Zeitz, M.; Aust, D.E.; Barretton, G.; Autschbach, F.; Ballauff, A.; Bokemeyer, B.; Fichtner-Feigl, S.; et al. Updated German guideline on diagnosis and treatment of ulcerative colitis, 2011. Z. Gastroenterol. 2011, 49, 1276. [Google Scholar] [CrossRef] [PubMed]
- Alatab, S.; Sepanlou, S.G.; Ikuta, K.; Vahedi, H.; Bisignano, C.; Safiri, S.; Sadeghi, A.; Nixon, M.R.; Abdoli, A.; Abolhassani, H.; et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 17–30. [Google Scholar] [CrossRef]
- Ekbom, A.; Helmick, C.; Zack, M.; Adami, H.O. Ulcerative colitis and colorectal cancer: A Population-Based Study. N. Engl. J. Med. 1990, 323, 1228–1233. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, S.R. Colorectal carcinoma in patients with Crohn’s Disease. Gastroenterology 1985, 89, 398–407. [Google Scholar] [CrossRef]
- Fakhoury, M.; Negrulj, R.; Mooranian, A.; Al-Salami, H. Inflammatory bowel disease: Clinical aspects and treatments. J. Inflamm. Res. 2014, 7, 113. [Google Scholar] [CrossRef]
- Ott, A. Inflammation and transcutaneous measurement of oxygen pressure in dermatology. Adv. Exp. Med. Biol. 1987, 220, 79–82. [Google Scholar]
- Sawyer, R.G.; Spengler, M.D.; Adams, R.B.; Pruett, T.L. The peritoneal environment during infection: The effect of monomicrobial and polymicrobial bacteria on pO2 and pH. Ann. Surg. 1991, 213, 253–260. [Google Scholar] [CrossRef]
- Silver, I.A. Measurement of pH and ionic composition of pericellular sites. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1975, 271, 261–272. [Google Scholar]
- Semenza, G.L. Hypoxia-inducible factor 1: Master regulator of O2 homeostasis. Curr. Opin. Genet. Dev. 1998, 8, 588–594. [Google Scholar] [CrossRef]
- Wang, G.L.; Jiang, B.H.; Rue, E.A.; Semenza, G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 1995, 92, 5510–5514. [Google Scholar] [CrossRef]
- Appelhoffl, R.J.; Tian, Y.M.; Raval, R.R.; Turley, H.; Harris, A.L.; Pugh, C.W.; Ratcliffe, P.J.; Gleadle, J.M. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 2004, 279, 38458–38465. [Google Scholar] [CrossRef] [PubMed]
- Berra, E.; Benizri, E.; Ginouvès, A.; Volmat, V.; Roux, D.; Pouysségur, J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 2003, 22, 4082–4090. [Google Scholar] [CrossRef] [PubMed]
- Ivan, M.; Kondo, K.; Yang, H.; Kim, W.; Valiando, J.; Ohh, M.; Salic, A.; Asara, J.M.; Lane, W.S.; Kaelin, J. HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 2001, 292, 464–468. [Google Scholar] [CrossRef]
- Bishop, T.; Ratcliffe, P.J. Benefits and risks of manipulating the HIF hydroxylase pathway in ischemic heart disease. In Translational Vascular Medicine: Pathogenesis, Diagnosis, and Treatment; Springer: London, UK, 2013; pp. 17–25. ISBN 9780857299208. [Google Scholar]
- Bäcker, V.; Cheung, F.Y.; Siveke, J.T.; Fandrey, J.; Winning, S. Knockdown of myeloid cell hypoxia-inducible factor-1α ameliorates the acute pathology in DSS-induced colitis. PLoS ONE 2017, 12, e0190074. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.L.; Bruyninckx, W.J.; Kelly, C.J.; Glover, L.E.; McNamee, E.N.; Bowers, B.E.; Bayless, A.J.; Scully, M.; Saeedi, B.J.; Golden-Mason, L.; et al. Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity 2014, 40, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Cramer, T.; Yamanishi, Y.; Clausen, B.E.; Förster, I.; Pawlinski, R.; Mackman, N.; Haase, V.H.; Jaenisch, R.; Corr, M.; Nizet, V.; et al. HIF-1α is essential for myeloid cell-mediated inflammation. Cell 2003, 112, 645–657. [Google Scholar] [CrossRef]
- Flück, K.; Breves, G.; Fandrey, J.; Winning, S. Hypoxia-inducible factor 1 in dendritic cells is crucial for the activation of protective regulatory T cells in murine colitis. Mucosal Immunol. 2015, 9, 379–390. [Google Scholar] [CrossRef]
- Walmsley, S.R.; Print, C.; Farahi, N.; Peyssonnaux, C.; Johnson, R.S.; Cramer, T.; Sobolewski, A.; Condliffe, A.M.; Cowburn, A.S.; Johnson, N.; et al. Hypoxia-induced neutrophil survival is mediated by HIF-1α-dependent NF-κB activity. J. Exp. Med. 2005, 201, 105–115. [Google Scholar] [CrossRef]
- Kapitsinou, P.P.; Sano, H.; Michael, M.; Kobayashi, H.; Davidoff, O.; Bian, A.; Yao, B.; Zhang, M.Z.; Harris, R.C.; Duffy, K.J.; et al. Endothelial HIF-2 mediates protection and recovery from ischemic kidney injury. J. Clin. Investig. 2014, 124, 2396–2409. [Google Scholar] [CrossRef]
- Walmsley, S.; Harris, A.; Thompson, A.A.R.; Whyte, M.K.B. HIF-mediated innate immune responses: Cell signaling and therapeutic implications. Hypoxia 2014, 2, 47. [Google Scholar] [CrossRef]
- Cooper, H.S.; Murthy, S.N.; Shah, R.S.; Sedergran, D.J. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab. Investig. 1993, 69, 238–249. [Google Scholar] [PubMed]
- Rosales, C. Neutrophil: A cell with many roles in inflammation or several cell types? Front. Physiol. 2018, 9, 113. [Google Scholar] [CrossRef] [PubMed]
- Babyatsky, M.W.; Rossiter, G.; Podolsky, D.K. Expression of transforming growth factors α and β in colonic mucosa in inflammatory bowel disease. Gastroenterology 1996, 110, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Drakes, M.L. Colon lamina propria dendritic cells induce a proinflammatory cytokine response in lamina propria T cells in the SCID mouse model of colitis. J. Leukoc. Biol. 2005, 78, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Fleming, T.J.; Fleming, M.L.; Malek, T.R. Selective expression of Ly-6G on myeloid lineage cells in mouse bone marrow. RB6-8C5 mAb to granulocyte-differentiation antigen (Gr-1) detects members of the Ly-6 family. J. Immunol. 1993, 151, 2399–2408. [Google Scholar] [PubMed]
- Contor, H.; Boyse, E.A. Functional subclasses of T lymphocytes bearing different Ly antigens. I. The generation of functionally distinct T cell subclasses is a differentiative process independent of antigen. J. Exp. Med. 1975, 141, 1376–1389. [Google Scholar] [CrossRef]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. J. Immunol. 2017, 198, 986–992. [Google Scholar] [CrossRef]
- Hori, S.; Nomura, T.; Science, S.S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef]
- Carter, P.B.; Collins, F.M. The route of enteric infection in normal mice. J. Exp. Med. 1974, 139, 1189–1203. [Google Scholar] [CrossRef]
- Van den Broeck, W.; Derore, A.; Simoens, P. Anatomy and nomenclature of murine lymph nodes: Descriptive study and nomenclatory standardization in BALB/cAnNCrl mice. J. Immunol. Methods 2006, 312, 12–19. [Google Scholar] [CrossRef]
- Wendland, M.; Czeloth, N.; Mach, N.; Malissen, B.; Kremmer, E.; Pabst, O.; Förster, R. CCR9 is a homing receptor for plasmacytoid dendritic cells to the small intestine. Proc. Natl. Acad. Sci. USA 2007, 104, 6347–6352. [Google Scholar] [CrossRef] [PubMed]
- Wurbel, M.A.; Philippe, J.M.; Nguyen, C.; Victorero, G.; Freeman, T.; Wooding, P.; Malissen, B. The chemokine TECK is expressed by thymic and intestinal epithelial cells and attracts double-and single-positive thymocytes expressing the TECK receptor CCR9. Eur. J. Immunol. 2000, 30, 262–271. [Google Scholar] [CrossRef]
- Duvallet, E.; Semerano, L.; Assier, E.; Falgarone, G.; Boissier, M.C. Interleukin-23: A key cytokine in inflammatory diseases. Ann. Med. 2011, 43, 503–511. [Google Scholar] [CrossRef]
- Sawant, K.V.; Poluri, K.M.; Dutta, A.K.; Sepuru, K.M.; Troshkina, A.; Garofalo, R.P.; Rajarathnam, K. Chemokine CXCL1 mediated neutrophil recruitment: Role of glycosaminoglycan interactions. Sci. Rep. 2016, 6, 33123. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.D.; Nedjai, B.; Hurst, T.; Pennington, D.J. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 2563–2582. [Google Scholar] [CrossRef] [PubMed]
- Shouval, D.S.; Ouahed, J.; Biswas, A.; Goettel, J.A.; Horwitz, B.H.; Klein, C.; Muise, A.M.; Snapper, S.B. Interleukin 10 receptor signaling: Master regulator of intestinal mucosal homeostasis in mice and humans. In Advances in Immunology; Academic Press Inc.: Cambridge, MA, USA, 2014; Volume 122, pp. 177–210. [Google Scholar]
- Higashiyama, M.; Hokari, R.; Hozumi, H.; Kurihara, C.; Ueda, T.; Watanabe, C.; Tomita, K.; Nakamura, M.; Komoto, S.; Okada, Y.; et al. HIF-1 in T cells ameliorated dextran sodium sulfate-induced murine colitis. J. Leukoc. Biol. 2012, 91, 901–909. [Google Scholar] [CrossRef]
- Karhausen, J.; Furuta, G.T.; Tomaszewski, J.E.; Johnson, R.S.; Colgan, S.P.; Haase, V.H. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J. Clin. Investig. 2004, 114, 1098–1106. [Google Scholar] [CrossRef]
- Lin, N.; Shay, J.E.S.; Xie, H.; Lee, D.S.M.; Skuli, N.; Tang, Q.; Zhou, Z.; Azzam, A.; Meng, H.; Wang, H.; et al. Myeloid Cell Hypoxia-Inducible Factors Promote Resolution of Inflammation in Experimental Colitis. Front. Immunol. 2018, 9, 2565. [Google Scholar] [CrossRef]
- Kim, Y.-E.; Lee, M.; Gu, H.; Kim, J.; Jeong, S.; Yeo, S.; Lee, Y.J.; Im, S.-H.; Sung, Y.-C.; Kim, H.J.; et al. Hypoxia-inducible factor-1 (HIF-1) activation in myeloid cells accelerates DSS-induced colitis progression in mice. Dis. Model. Mech. 2018, 1, dmm.033241. [Google Scholar]
- Xue, X.; Ramakrishnan, S.; Anderson, E.; Taylor, M.; Zimmermann, E.M.; Spence, J.R.; Huang, S.; Greenson, J.K.; Shah, Y.M. Endothelial PAS domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology 2013, 145, 831–841. [Google Scholar] [CrossRef]
- Dougherty, E.J.; Pollenz, R.S. ARNT: A Key bHLH/PAS Regulatory Protein Across Multiple Pathways. In Comprehensive Toxicology, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2010; Volume 2–14, pp. 231–252. ISBN 9780080468846. [Google Scholar]
- Iida, N.; Grotendorst, G.R. Cloning and sequencing of a new gro transcript from activated human monocytes: Expression in leukocytes and wound tissue. Mol. Cell. Biol. 1990, 10, 5596–5599. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, K.; Henderson, R.B.; Laschinger, M.; Hogg, N. Neutrophil Chemokines KC and Macrophage-Inflammatory Protein-2 Are Newly Synthesized by Tissue Macrophages Using Distinct TLR Signaling Pathways. J. Immunol. 2008, 180, 4308–4315. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, K.; Dudeck, A.; Hasenberg, M.; Nye, E.; Van Rooijen, N.; Hartmann, K.; Gunzer, M.; Roers, A.; Hogg, N. Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood 2013, 121, 4930–4937. [Google Scholar] [CrossRef] [PubMed]
- Kolaczkowska, E.; Kubes, P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 2013, 13, 159–175. [Google Scholar] [CrossRef]
- Wright, H.L.; Moots, R.J.; Bucknall, R.C.; Edwards, S.W. Neutrophil function in inflammation and inflammatory diseases. Rheumatology 2010, 49, 1618–1631. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, R.; Kamiya, N.; Adapala, N.S.; Drissi, H.; Kim, H.K.W. HIF-1-dependent IL-6 activation in articular chondrocytes initiating synovitis in femoral head ischemic osteonecrosis. J. Bone Jt. Surg. Am. Vol. 2016, 98, 1122–1131. [Google Scholar] [CrossRef]
- Fais, S.; Capobianchi, M.R.; Pallone, F.; Di Marco, P.; Boirivant, M.; Dianzani, F.; Torsoli, A. Spontaneous release of interferon γ by intestinal lamina propria lymphocytes in Crohn’s disease. Kinetics of in vitro response to interferon γ inducers. Gut 1991, 32, 403–407. [Google Scholar] [CrossRef]
- Noguchi, M.; Hiwatashi, N.; Liu, Z.; Toyota, T. Enhanced interferon-gamma production and B7-2 expression in isolated intestinal mononuclear cells from patients with Crohn’s disease. J. Gastroenterol. 1995, 30, 52. [Google Scholar]
- Sasaki, T.; Hiwatashi, N.; Yamazaki, H.; Noguchi, M.; Toyota, T. The role of interferony in the pathogenesis of Crohn’s disease. Gastroenterol. Jpn. 1992, 27, 29–36. [Google Scholar] [CrossRef]
- Ito, R.; Shin-Ya, M.; Kishida, T.; Urano, A.; Takada, R.; Sakagami, J.; Imanishi, J.; Kita, M.; Ueda, Y.; Iwakura, Y.; et al. Interferon-gamma is causatively involved in experimental inflammatory bowel disease in mice. Clin. Exp. Immunol. 2006, 146, 330–338. [Google Scholar] [CrossRef]
- Männel, D.N.; Moore, R.N.; Mergenhagen, S.E. Macrophages as a source of tumoricidal activity (tumor-necrotizing factor). Infect. Immun. 1980, 30, 523–530. [Google Scholar] [PubMed]
- Nakayama, T.; Kurobe, H.; Sugasawa, N.; Kinoshita, H.; Higashida, M.; Matsuoka, Y.; Yoshida, Y.; Hirata, Y.; Sakata, M.; Maxfield, M.W. Role of macrophage-derived hypoxia-inducible factor (HIF)-1α as a mediator of vascular remodelling. Cardiovasc. Res. 2013, 99, 705–715. [Google Scholar] [CrossRef]
- Neurath, M.F. IL-23 in inflammatory bowel diseases and colon cancer. Cytokine Growth Factor Rev. 2019, 45, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yen, D.; Cheung, J.; Scheerens, H.; Poulet, F.; McClanahan, T.; Mckenzie, B.; Kleinschek, M.A.; Owyang, A.; Mattson, J.; Blumenschein, W.; et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J. Clin. Investig. 2006, 116, 1310–1316. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.H.; Kljavin, N.M.; Ota, N.; Leonard, J.; Roose-Girma, M.; Diehl, L.; Ouyang, W.; Ghilardi, N. Opposing consequences of IL-23 signaling mediated by innate and adaptive cells in chemically induced colitis in mice. Mucosal Immunol. 2012, 5, 99–109. [Google Scholar] [CrossRef]
- Lin, A.M.; Rubin, C.J.; Khandpur, R.; Wang, J.Y.; Riblett, M.; Yalavarthi, S.; Villanueva, E.C.; Shah, P.; Kaplan, M.J.; Bruce, A.T. Mast Cells and Neutrophils Release IL-17 through Extracellular Trap Formation in Psoriasis. J. Immunol. 2011, 187, 490–500. [Google Scholar] [CrossRef]
- Choe, S.S.; Shin, K.C.; Ka, S.; Lee, Y.K.; Chun, J.S.; Kim, J.B. Macrophage HIF-2α ameliorates adipose tissue inflammation and insulin resistance in obesity. Diabetes 2014, 63, 3359–3371. [Google Scholar] [CrossRef]
- Takeda, N.; O’Dea, E.L.; Doedens, A.; Kim, J.W.; Weidemann, A.; Stockmann, C.; Asagiri, M.; Simon, M.C.; Hoffmann, A.; Johnson, R.S. Differential activation and antagonistic function of HIF-α isoforms in macrophages are essential for NO homeostasis. Genes Dev. 2010, 24, 491–501. [Google Scholar] [CrossRef]
- Mitsuyama, K.; Sata, M.; Tanikawa, K. Significance of interleukin-6 in patients with inflammatory bowel disease. Gastroenterol. Jpn. 1991, 26, 20–28. [Google Scholar] [CrossRef]
- Atreya, R.; Mudter, J.; Finotto, S.; Müllberg, J.; Jostock, T.; Wirtz, S.; Schütz, M.; Bartsch, B.; Holtmann, M.; Becker, C.; et al. Blockade of interleukin 6 trans signaling suppresses T-cell resistance against apoptosis in chronic intestinal inflammation: Evidence in Crohn disease and experimental colitis in vivo. Nat. Med. 2000, 6, 583–588. [Google Scholar] [CrossRef]
- Hurst, S.M.; Wilkinson, T.S.; McLoughlin, R.M.; Jones, S.; Horiuchi, S.; Yamamoto, N.; Rose-John, S.; Fuller, G.M.; Topley, N.; Jones, S.A. IL-6 and its soluble receptor orchestrate a temporal switch in the pattern of leukocyte recruitment seen during acute inflammation. Immunity 2001, 14, 705–714. [Google Scholar] [CrossRef]
- Fujimoto, M.; Nakano, M.; Terabe, F.; Kawahata, H.; Ohkawara, T.; Han, Y.; Ripley, B.; Serada, S.; Nishikawa, T.; Kimura, A.; et al. The Influence of Excessive IL-6 Production In Vivo on the Development and Function of Foxp3 + Regulatory T Cells. J. Immunol. 2011, 186, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Iness, A.; Yoon, J.; Grider, J.R.; Murthy, K.S.; Kellum, J.M.; Kuemmerle, J.F. Noncanonical STAT3 Activation Regulates Excess TGF-β1 and Collagen I Expression in Muscle of Stricturing Crohn’s Disease. J. Immunol. 2015, 194, 3422–3431. [Google Scholar] [CrossRef]
- Kühn, R.; Löhler, J.; Rennick, D.; Rajewsky, K.; Müller, W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 1993, 75, 263–274. [Google Scholar] [CrossRef]
- Chomzynski, P. Single-Step Method of RNA Isolation by Acid Guanidinium Thiocyanate–Phenol–Chloroform Extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Motulsky, H.J.; Brown, R.E. Detecting outliers when fitting data with nonlinear regression - A new method based on robust nonlinear regression and the false discovery rate. BMC Bioinform. 2006, 7, 123. [Google Scholar] [CrossRef]
| Score | Weight Loss | Stool Consistency | Hema Screen Test (Occult Blood) |
|---|---|---|---|
| 0 | none | normal | negative |
| 1 | 1–5% | normal | negative |
| 2 | 5–10% | loose | positive |
| 3 | 10–20% | loose | positive |
| 4 | >20% | diarrhea | visible bleeding |
| Score | Infiltrating Immune Cells | Extent of Injury | Crypt Damage |
|---|---|---|---|
| 0 | rare | none | intact crypts |
| 1 | slightly dispersed | mucosal | basal 1/3 damaged |
| 2 | moderately increased | mucosal and submucosal | basal 2/3 damaged |
| 3 | severely large areas: loss of tissue structure | transmural | only surface epithelium intact |
| 4 | - | - | loss of entire crypt and epithelium |
| Antibody | Concentration | Catalog Number and Company |
|---|---|---|
| rat anti-F4/80 | 0.180556 | #MCA497B, Bio-Rad AbD Serotec GmbH, Puchheim, Germany |
| goat antirat secondary Ab | 0.388889 | #A11077, Invitrogen Thermo Fisher Scientific, Dreieich, Germany |
| goat antimyeloperoxidase | 0.180556 | # AF3667, R&D systems |
| rabbit antigoat secondary Ab | 0.388889 | # sc2774, Santa Cruz Biotechnology, Heidelberg, Germany |
| goat anti-CD3-ε | 0.215278 | #sc-1127, Santa Cruz Biotechnology, Heidelberg, Germany |
| rabbit antigoat Alexa Fluor 488 | 0.388889 | #A-11078, Thermo Fisher Scientific, Dreieich, Germany |
| Rat anti-FoxP3 | 0.111111 | # FJK-16s, eBioscience Thermo Fisher Scientific, Dreieich, Germany |
| goat antirat Alexa Fluor 568 | 0.388889 | #A-11077, Thermo Fisher Scientific, Dreieich, Germany |
| rabbit anti-HIF-1alpha (C-Term) | 1:10,000 | #CAY-10006421, Cayman Chemical Biomol GmbH, Hamburg, Germany |
| rabbit anti-HIF-2 alpha/EPAS1 | 1:10,000 | #NB100-122, Novus Biologicals, Wiesbaden, Germany |
| Polyclonal goat-antirabbit Immunoglobulin/HRP | 0.388889 | #P044801-2, DAKO Agilent Technologies, Waldbronn, Germany |
| Target Gene | Accession-Number | Sequence | Product Length (bp) |
|---|---|---|---|
| 5′ Adgre1 3′ Adgre1 | NM_010130 | TCTGGGGAGCTTACGATGGA GAATCCCGCAATGATGGCAC | 237 |
| 5′ Arg1 3′ Arg1 | NM_007482 | AACACGGCAGTGGCTTTAACC GGTTTTCATGTGGCGCATTC | 117 |
| 5′ Ccr9 3′ Ccr9 | NM_001166625 | CCAAGGTGCCCACAATGAAC ACTCACAAGCCTTATTCCTGGC | 179 |
| 5′ Cd4 3′ Cd4 | NM_013488 | TGAAGGAAACGCTCCCACTC AGCAGTGCTGATGTCTTGCT | 136 |
| 5′ Cd8a 3′ Cd8a | NM_001081110 | ACCCTTGGCCGGAATCTGCG CTGTCTGACTAGCGGCCTGGGA | 112 |
| 5′ Cd11c 3′ Cd11c | NM_021334 | GGACGGTGCTGAGTTCGGACACAG CCACAAGCCAACAGCCAGGAAGG | 231 |
| 5′ Cxcl1 3′ Cxcl1 | NM_008176 | CAGGGTCAAGGCAAGCCTC CTGGGATTCACCTCAAGAACATC | 117 |
| 5′ Foxp3 3′ Foxp3 | NM_001199347 | CTGGCGAAGGGCTCGGTAGTCCT CTCCCAGAGCCCATGGCAGAAGT | 250 |
| 5′Hif-1a exon 2 3′ Hif-1a exon 2 | NM_001313919 | CATCCAGAAGTTTTCTCACACG GGCGAAGCAAAGAGTCTGAA | 138 |
| 5′Hif-2a exon 2 3′ Hif-2a exon 2 | NM_010137 | AGGAGACGGAGGTCTTCTATGA ACAGGAGCTTATGTGTCCGA | 126 |
| 5′ Ifng 3′ Ifng | NM_008337 | GGTCAACAACCCACAGGTCC CAGCGACTCCTTTTCCGCTT | 105 |
| 5′ Il6 3′ Il6 | NM_031168 | TCCTACCCCAATTTCCAATGC CATAACGCACTAGGTTTGCCG | 151 |
| 5′ Il10 3′ Il10 | NM_010548 | TGCCCCAGGCAGAGAAGCAT GGGAGAAATCGATGACAGCGCC | 109 |
| 5′ Il23a 3′ Il23a | NM_031252 | ACCAGCGGGACATATGAATCT AGACCTTGGCGGATCCTTTG | 147 |
| 5′ Il17a 3′ Il17a | NM_010552 | TCATCCCTCAAAGCTCAGCG TTCATTGCGGTGGAGAGTCC | 167 |
| 5′ Ly6g 3′ Ly6g | NM_023463 | GTACCTTGGGAAGATGTGGGT GTTCAGGCCCAGCTTATGGT | 103 |
| 5′ Tgfb1 3′ Tgfb1 | NM_011577 | TGGCCAGATCCTGTCCAAAC CATAGATGGCGTTGTTGCGG | 215 |
| 5′ Tnfa 3′ Tnfa | NM_011609 | TACCTCCTCCGCTTGCAAAT GAGTAGACTTCGGGCCTCCAC | 151 |
| 5′ Actb 3′ Actb | NM_007393 | TAGGCACCAGGGTGTGATGG CTCGGTGAGCAGCACAGG | 208 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kerber, E.L.; Padberg, C.; Koll, N.; Schuetzhold, V.; Fandrey, J.; Winning, S. The Importance of Hypoxia-Inducible Factors (HIF-1 and HIF-2) for the Pathophysiology of Inflammatory Bowel Disease. Int. J. Mol. Sci. 2020, 21, 8551. https://doi.org/10.3390/ijms21228551
Kerber EL, Padberg C, Koll N, Schuetzhold V, Fandrey J, Winning S. The Importance of Hypoxia-Inducible Factors (HIF-1 and HIF-2) for the Pathophysiology of Inflammatory Bowel Disease. International Journal of Molecular Sciences. 2020; 21(22):8551. https://doi.org/10.3390/ijms21228551
Chicago/Turabian StyleKerber, Evelyn L., Claudia Padberg, Nora Koll, Vera Schuetzhold, Joachim Fandrey, and Sandra Winning. 2020. "The Importance of Hypoxia-Inducible Factors (HIF-1 and HIF-2) for the Pathophysiology of Inflammatory Bowel Disease" International Journal of Molecular Sciences 21, no. 22: 8551. https://doi.org/10.3390/ijms21228551
APA StyleKerber, E. L., Padberg, C., Koll, N., Schuetzhold, V., Fandrey, J., & Winning, S. (2020). The Importance of Hypoxia-Inducible Factors (HIF-1 and HIF-2) for the Pathophysiology of Inflammatory Bowel Disease. International Journal of Molecular Sciences, 21(22), 8551. https://doi.org/10.3390/ijms21228551