Different Sensitivity of Macrophages to Phospholipidosis Induction by Amphiphilic Cationic Drugs
Abstract
:1. Introduction
2. Results
2.1. Characterization of Macrophages (Cell Area, Lysosome Area, Lysosome Number/Cell, Cytotoxicity to CADs)
2.2. Identification of Optimal Staining
2.3. Effect of CADs on Cell Area
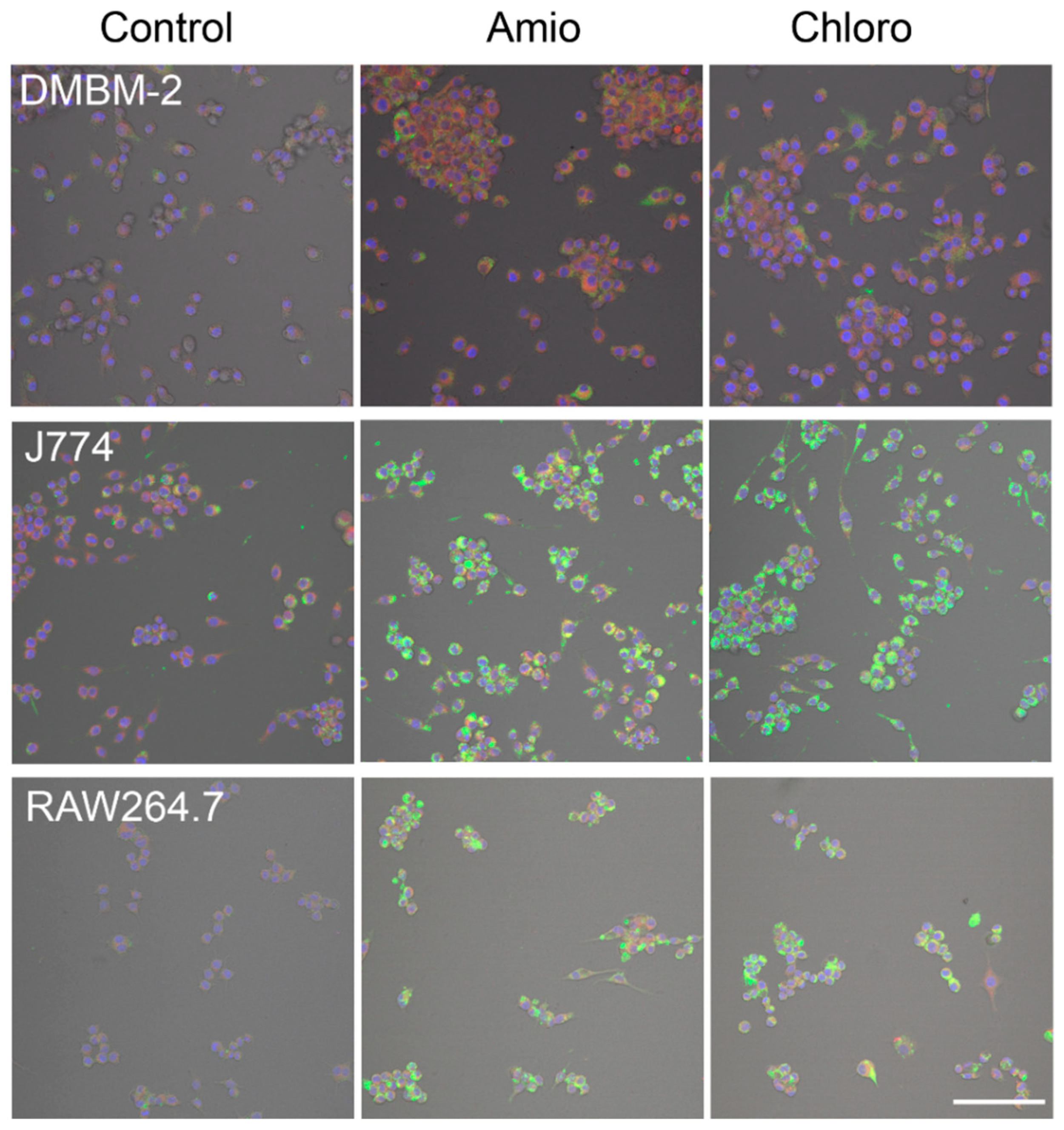
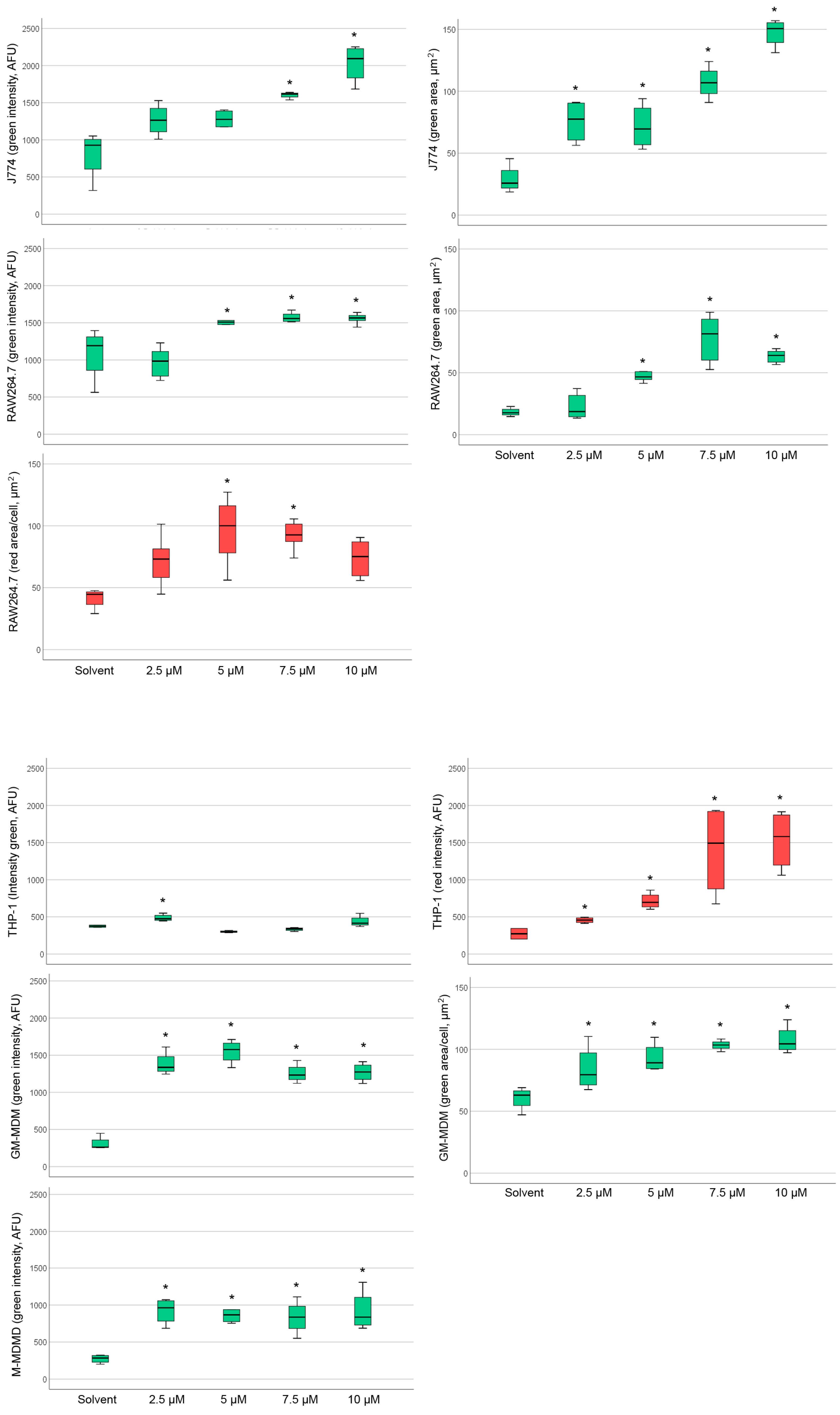
2.4. Detection of Phospholipidosis and Neutral Lipids
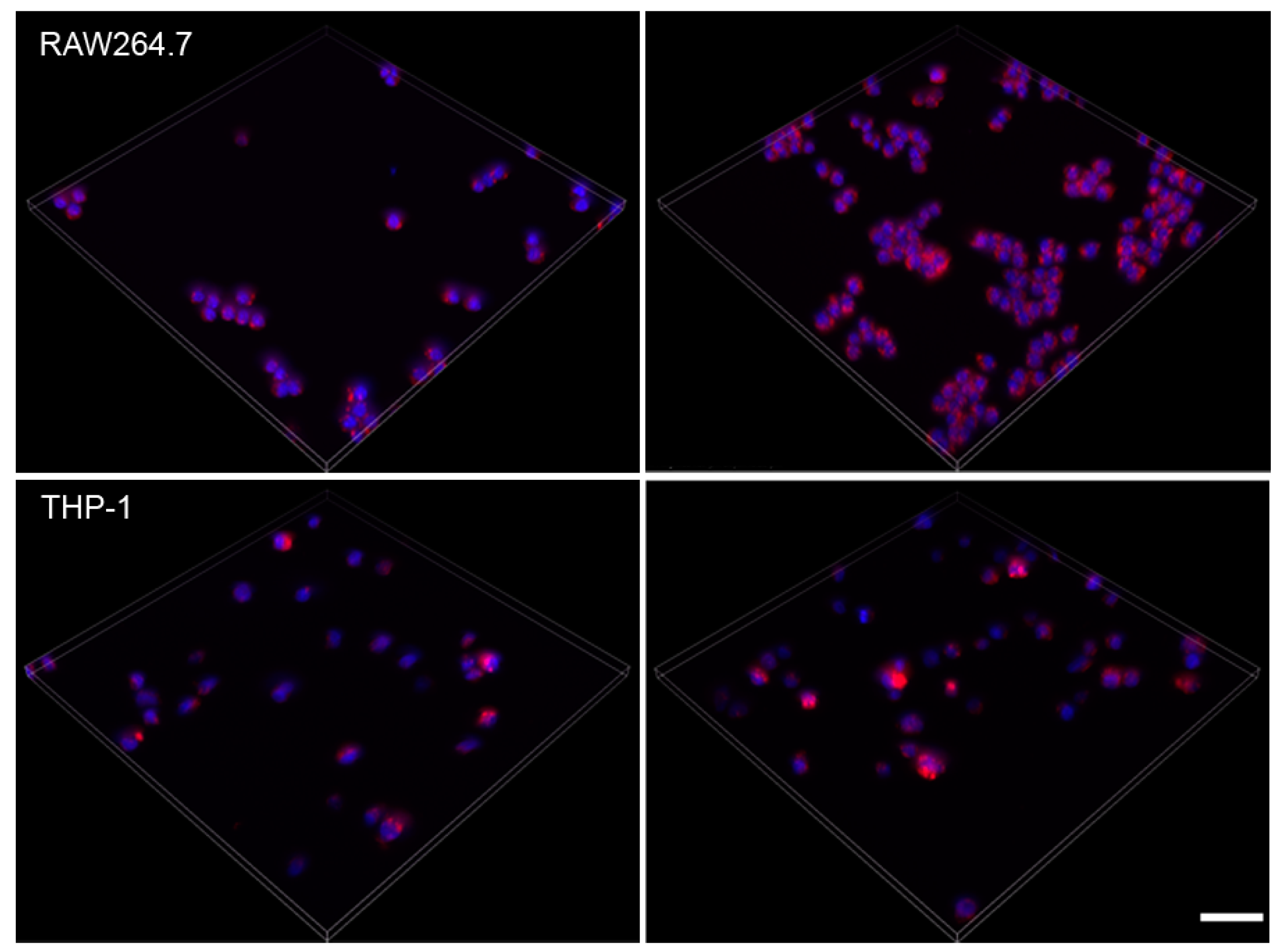
2.5. Effect of Amiodarone and Chloroquine on Number, Area and Staining Intensity of Lysosomes
3. Discussion
3.1. Establishment of Macrophage Models and Staining
3.2. Action of Amiodarone and Chloroquine on Lysosomes
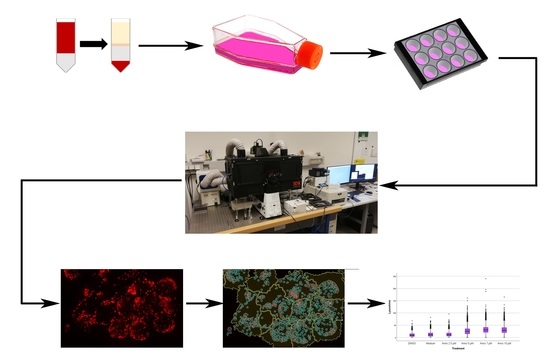
4. Materials and Methods
4.1. Cell Culture
4.2. Differentiation to THP-1 Macrophages
Characterization of THP-1 Macrophages
4.3. Generation of Monocyte-Derived Macrophages (MDMs)
4.3.1. Differentiation to M1 and M2 State
4.3.2. Cytotoxicity Screening Using Measurement of Dehydrogenase Activity
4.3.3. Identification of Lysosomes by LysoSensor Staining
4.3.4. Measurement of Membrane Potential by Acridine Orange Staining
4.3.5. Lipid Accumulation by Nile Red Staining
4.3.6. Neutral Lipid and Phospholipid Accumulation by HCS LipidTOX Dyes
4.4. Lysosome Morphology by LysoID Staining
4.5. Statistics
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Amio | Amiodarone |
| AD | Distilled water |
| CAD | Cationic amphiphilic drug |
| Chloro | Chloroquine |
| EDTA | Ethylenediaminetetraacetic acid |
| DIILD | Drug induced interstitial lung disease |
| DMEM | Dulbecco’s Modified Eagle Medium |
| DMSO | Dimethylsulfoxide |
| DPBS | Dulbecco’s PBS |
| DPPC | Dipalmitoylphosphatidylcholine |
| FBS | Fetal bovine serum |
| GM-CSF | Granulocyte-macrophage colony stimulating factor |
| GM-MDM | M1 macrophages |
| HCS | High content screening |
| ILD | Interstitial lung disease |
| LDL | Low density lipoprotein |
| LPS | Lipopolysaccharide |
| LRS | Leukoreduction system |
| M-CSF | Macrophage colony stimulating factor |
| M-MDM | M2 macrophages |
| MDM | Monocyte-derived macrophage |
| NEAA | non-essential amino acids |
| PLD | Phospholipidosis |
| NBD-PE | N-(7-nitrobenz-2-oxa-1,3 diazol-4-yl)1,2 dihexadecanoyl-sn-glycero-3-phosphoethanolamine |
| PBMC | Peripheral blood mononuclear cells |
| PMA | Phorbol 12-myristate 13-acetate |
| P/S | Penicillin/Streptomycin |
| RPMI | Roswell Park Memorial Institute |
References
- Schwaiblmair, M.; Behr, W.; Haeckel, T.; Markl, B.; Foerg, W.; Berghaus, T. Drug induced interstitial lung disease. Open Respir. Med. J. 2012, 6, 63–74. [Google Scholar] [CrossRef] [Green Version]
- Halliwell, W.H. Cationic amphiphilic drug-induced phospholipidosis. Toxicol. Pathol. 1997, 25, 53–60. [Google Scholar] [CrossRef]
- Barone, L.R.; Boyer, S.; Damewood, J.R., Jr.; Fikes, J.; Ciaccio, P.J. Phospholipogenic pharmaceuticals are associated with a higher incidence of histological findings than nonphospholipogenic pharmaceuticals in preclinical toxicology studies. J. Toxicol. 2012, 2012, 308594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, A.; Hoffman, E.; Ball, D.; Klapwijk, J.; Steven, R.T.; Dexter, A.; Bunch, J.; Baker, D.; Murnane, D.; Hutter, V.; et al. Comparison of Oral, Intranasal and Aerosol Administration of Amiodarone in Rats as a Model of Pulmonary Phospholipidosis. Pharmaceutics 2019, 11, 345. [Google Scholar] [CrossRef] [Green Version]
- Morelli, J.K.; Buehrle, M.; Pognan, F.; Barone, L.R.; Fieles, W.; Ciaccio, P.J. Validation of an in vitro screen for phospholipidosis using a high-content biology platform. Cell Biol. Toxicol. 2006, 22, 15–27. [Google Scholar] [CrossRef]
- Reynolds, G.; Haniffa, M. Human and Mouse Mononuclear Phagocyte Networks: A Tale of Two Species? Front. Immunol. 2015, 6, 330. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, N.; Yu, X.; Zhang, X.; Li, S.; Lei, Z.; Hu, R.; Li, H.; Mao, Y.; Wang, X.; et al. Tumacrophage: Macrophages transformed into tumor stem-like cells by virulent genetic material from tumor cells. Oncotarget 2017, 8, 82326–82343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taciak, B.; Bialasek, M.; Braniewska, A.; Sas, Z.; Sawicka, P.; Kiraga, L.; Rygiel, T.; Krol, M. Evaluation of phenotypic and functional stability of RAW 264.7 cell line through serial passages. PLoS ONE 2018, 13, e0198943. [Google Scholar] [CrossRef]
- Tran, T.H.; Rastogi, R.; Shelke, J.; Amiji, M.M. Modulation of Macrophage Functional Polarity towards Anti-Inflammatory Phenotype with Plasmid DNA Delivery in CD44 Targeting Hyaluronic Acid Nanoparticles. Sci. Rep. 2015, 5, 16632. [Google Scholar] [CrossRef]
- Monner, D.A.; Denker, B. Characterization of clonally derived, spontaneously transformed bone marrow macrophage cell lines from lipopolysaccharide hyporesponsive LPS(d) and normal LPS(n) mice. J. Leukoc. Biol. 1997, 61, 469–480. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, E.; Patel, A.; Ball, D.; Klapwijk, J.; Millar, V.; Kumar, A.; Martin, A.; Mahendran, R.; Dailey, L.A.; Forbes, B.; et al. Morphometric Characterization of Rat and Human Alveolar Macrophage Cell Models and their Response to Amiodarone using High Content Image Analysis. Pharm. Res. 2017, 34, 2466–2476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, A.E.; Finney-Hayward, T.K.; Quint, J.K.; Thomas, C.M.; Tudhope, S.J.; Wedzicha, J.A.; Barnes, P.J.; Donnelly, L.E. Defective macrophage phagocytosis of bacteria in COPD. Eur. Respir. J. 2010, 35, 1039–1047. [Google Scholar] [CrossRef] [Green Version]
- Starr, T.; Bauler, T.J.; Malik-Kale, P.; Steele-Mortimer, O. The phorbol 12-myristate-13-acetate differentiation protocol is critical to the interaction of THP-1 macrophages with Salmonella Typhimurium. PLoS ONE 2018, 13, e0193601. [Google Scholar] [CrossRef]
- Park, E.K.; Jung, H.S.; Yang, H.I.; Yoo, M.C.; Kim, C.; Kim, K.S. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm. Res. 2007, 56, 45–50. [Google Scholar] [CrossRef]
- Lescoat, A.; Ballerie, A.; Augagneur, Y.; Morzadec, C.; Vernhet, L.; Fardel, O.; Jégo, P.; Jouneau, S.; Lecureur, V. Distinct Properties of Human M-CSF and GM-CSF Monocyte-Derived Macrophages to Simulate Pathological Lung Conditions In Vitro: Application to Systemic and Inflammatory Disorders with Pulmonary Involvement. Int. J. Mol. Sci. 2018, 19, 894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambrecht, B.N. Alveolar macrophage in the driver’s seat. Immunity 2006, 24, 366–368. [Google Scholar] [CrossRef] [Green Version]
- Mitsi, E.; Kamng’ona, R.; Rylance, J.; Solorzano, C.; Jesus Reine, J.; Mwandumba, H.C.; Ferreira, D.M.; Jambo, K.C. Human alveolar macrophages predominately express combined classical M1 and M2 surface markers in steady state. Respir. Res. 2018, 19, 66. [Google Scholar] [CrossRef] [Green Version]
- Roelandt, M.; Demedts, M.; Callebaut, W.; Coolen, D.; Slabbynck, H.; Bockaert, J.; Kips, J.; Brie, J.; Ulburghs, M.; De Boeck, K.; et al. Epidemiology of interstitial lung disease (ILD) in flanders: Registration by pneumologists in 1992–1994. Working group on ILD, VRGT. Vereniging voor Respiratoire Gezondheidszorg en Tuberculosebestrijding. Acta Clin. Belg. 1995, 50, 260–268. [Google Scholar] [CrossRef]
- Casartelli, A.; Bonato, M.; Cristofori, P.; Crivellente, F.; Dal Negro, G.; Masotto, I.; Mutinelli, C.; Valko, K.; Bonfante, V. A cell-based approach for the early assessment of the phospholipidogenic potential in pharmaceutical research and drug development. Cell Biol. Toxicol. 2003, 19, 161–176. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.J.; Herman, P.; Kang, J.S.; Lakowicz, J.R. Fluorescence lifetime characterization of novel low-pH probes. Anal. Biochem. 2001, 294, 118–125. [Google Scholar] [CrossRef]
- Zelenin, A. Biological Techniques Series. Fluorescent and Luminescent Probes for Biological Activity. A Practical Guide to Technology for Quantitative Real-Time Analysis; Zelenin, A.V., Ed.; Academic Press: San Diego, CA, USA, 1999; pp. 117–135. [Google Scholar]
- Thomé, M.P.; Filippi-Chiela, E.C.; Villodre, E.S.; Migliavaca, C.B.; Onzi, G.R.; Felipe, K.B.; Lenz, G. Ratiometric analysis of Acridine Orange staining in the study of acidic organelles and autophagy. J. Cell Sci. 2016, 129, 4622–4632. [Google Scholar] [CrossRef] [Green Version]
- Grandl, M.; Schmitz, G. Fluorescent high-content imaging allows the discrimination and quantitation of E-LDL-induced lipid droplets and Ox-LDL-generated phospholipidosis in human macrophages. Cytom. A 2010, 77, 231–242. [Google Scholar] [CrossRef]
- Coleman, J.; Xiang, Y.; Pande, P.; Shen, D.; Gatica, D.; Patton, W.F. A live-cell fluorescence microplate assay suitable for monitoring vacuolation arising from drug or toxic agent treatment. J. Biomol. Screen. 2010, 15, 398–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodell, C.B.; Koch, P.D.; Weissleder, R. Screening for new macrophage therapeutics. Theranostics 2019, 9, 7714–7729. [Google Scholar] [CrossRef]
- El-Mohandes, E.M.; Moustafa, A.M.; Khalaf, H.A.; Hassan, Y.F. The role of mast cells and macrophages in amiodarone induced pulmonary fibrosis and the possible attenuating role of atorvastatin. Biotech. Histochem. 2017, 92, 467–480. [Google Scholar] [CrossRef]
- Hoffman, E.; Martin, A.; Chau, D.; Murnane, D.; Hutter, V. Investigating the impact of polarization on foamy macrophage induction by amiodarone in vitro. In Proceedings of the Drug Delivery to the Lungs Conference, Edinburgh, Scotland, UK, 6–8 December 2017. [Google Scholar]
- Lea, S.; Dungwa, J.; Ravi, A.; Singh, D. Alveolar macrophage size is increased in COPD patients compared to controls. Eur. Respir. J. 2017, 50, PA1011. [Google Scholar] [CrossRef]
- Fathi, M.; Johansson, A.; Lundborg, M.; Orre, L.; Sköld, C.M.; Camner, P. Functional and morphological differences between human alveolar and interstitial macrophages. Exp. Mol. Pathol. 2001, 70, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Wolkove, N.; Baltzan, M. Amiodarone pulmonary toxicity. Can. Respir. J. 2009, 16, 43–48. [Google Scholar] [CrossRef]
- Spano, A.; Barni, S.; Sciola, L. PMA withdrawal in PMA-treated monocytic THP-1 cells and subsequent retinoic acid stimulation, modulate induction of apoptosis and appearance of dendritic cells. Cell Prolif. 2013, 46, 328–347. [Google Scholar] [CrossRef]
- Chanput, W.; Mes, J.J.; Savelkoul, H.F.; Wichers, H.J. Characterization of polarized THP-1 macrophages and polarizing ability of LPS and food compounds. Food Funct. 2013, 4, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, T.; Inagawa, H.; Kohchi, C.; Hirashima, M.; Soma, G. Functional characterization of lipopolysaccharide derived from symbiotic bacteria in rice as a macrophage-activating substance. Anticancer Res. 2011, 31, 2467–2476. [Google Scholar]
- Hayman, Y.A.; Sadofsky, L.R.; Williamson, J.D.; Hart, S.P.; Morice, A.H. The effects of exogenous lipid on THP-1 cells: An in vitro model of airway aspiration? ERJ Open Res. 2017, 3. [Google Scholar] [CrossRef]
- Huebinger, J. Modification of cellular membranes conveys cryoprotection to cells during rapid, non-equilibrium cryopreservation. PLoS ONE 2018, 13, e0205520. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, K.; Sato, A.; Nishimura, K.; Chida, K.; Hayakawa, H. Phospholipid analysis of alveolar macrophages and bronchoalveolar lavage fluid following bleomycin administration to rabbits. Lung 1994, 172, 91–102. [Google Scholar] [CrossRef]
- Hruban, Z. Pulmonary changes induced by amphophilic drugs. Environ. Health Perspect. 1976, 16, 111–118. [Google Scholar] [CrossRef]
- Chen, D.; Xie, J.; Fiskesund, R.; Dong, W.; Liang, X.; Lv, J.; Jin, X.; Liu, J.; Mo, S.; Zhang, T.; et al. Chloroquine modulates antitumor immune response by resetting tumor-associated macrophages toward M1 phenotype. Nat. Commun. 2018, 9, 873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallagher, L.E.; Radhi, O.A.; Abdullah, M.O.; McCluskey, A.G.; Boyd, M.; Chan, E.Y.W. Lysosomotropism depends on glucose: A chloroquine resistance mechanism. Cell Death Dis. 2017, 8, e3014. [Google Scholar] [CrossRef]
- Anderson, N.; Borlak, J. Drug-induced phospholipidosis. FEBS Lett. 2006, 580, 5533–5540. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, N.A.; Downar, E.; Butany, J. Amiodarone—An inhibitor of phospholipase activity: A comparative study of the inhibitory effects of amiodarone, chloroquine and chlorpromazine. Mol. Cell. Biochem. 1987, 76, 163–172. [Google Scholar] [CrossRef]
- Kodavanti, U.P.; Mehendale, H.M. Cationic amphiphilic drugs and phospholipid storage disorder. Pharmacol. Rev. 1990, 42, 327–354. [Google Scholar]
- Hoffman, E.; Murnane, D.; Hutter, V. Investigating the Suitability of High Content Image Analysis as a Tool to Assess the Reversibility of Foamy Alveolar Macrophage Phenotypes In Vitro. Pharmaceutics 2020, 12, 262. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Kim, Y.J.; Mantel, C.; Broxmeyer, H.E. P-selectin enhances generation of CD14+CD16+ dendritic-like cells and inhibits macrophage maturation from human peripheral blood monocytes. J. Immunol. 2003, 171, 669–677. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Kruth, H.S. Culture of Macrophage Colony-stimulating Factor Differentiated Human Monocyte-derived Macrophages. J. Vis. Exp. 2016. [Google Scholar] [CrossRef]
- Chu, S.; Zhu, X.; You, N.; Zhang, W.; Zheng, F.; Cai, B.; Zhou, T.; Wang, Y.; Sun, Q.; Yang, Z.; et al. The Fab Fragment of a Human Anti-Siglec-9 Monoclonal Antibody Suppresses LPS-Induced Inflammatory Responses in Human Macrophages. Front. Immunol. 2016, 7, 649. [Google Scholar] [CrossRef] [Green Version]
- Boyette, L.B.; Macedo, C.; Hadi, K.; Elinoff, B.D.; Walters, J.T.; Ramaswami, B.; Chalasani, G.; Taboas, J.M.; Lakkis, F.G.; Metes, D.M. Phenotype, function, and differentiation potential of human monocyte subsets. PLoS ONE 2017, 12, e0176460. [Google Scholar] [CrossRef]
- Akoglu, H. User’s guide to correlation coefficients. Turk. J. Emerg. Med. 2018, 18, 91–93. [Google Scholar] [CrossRef]


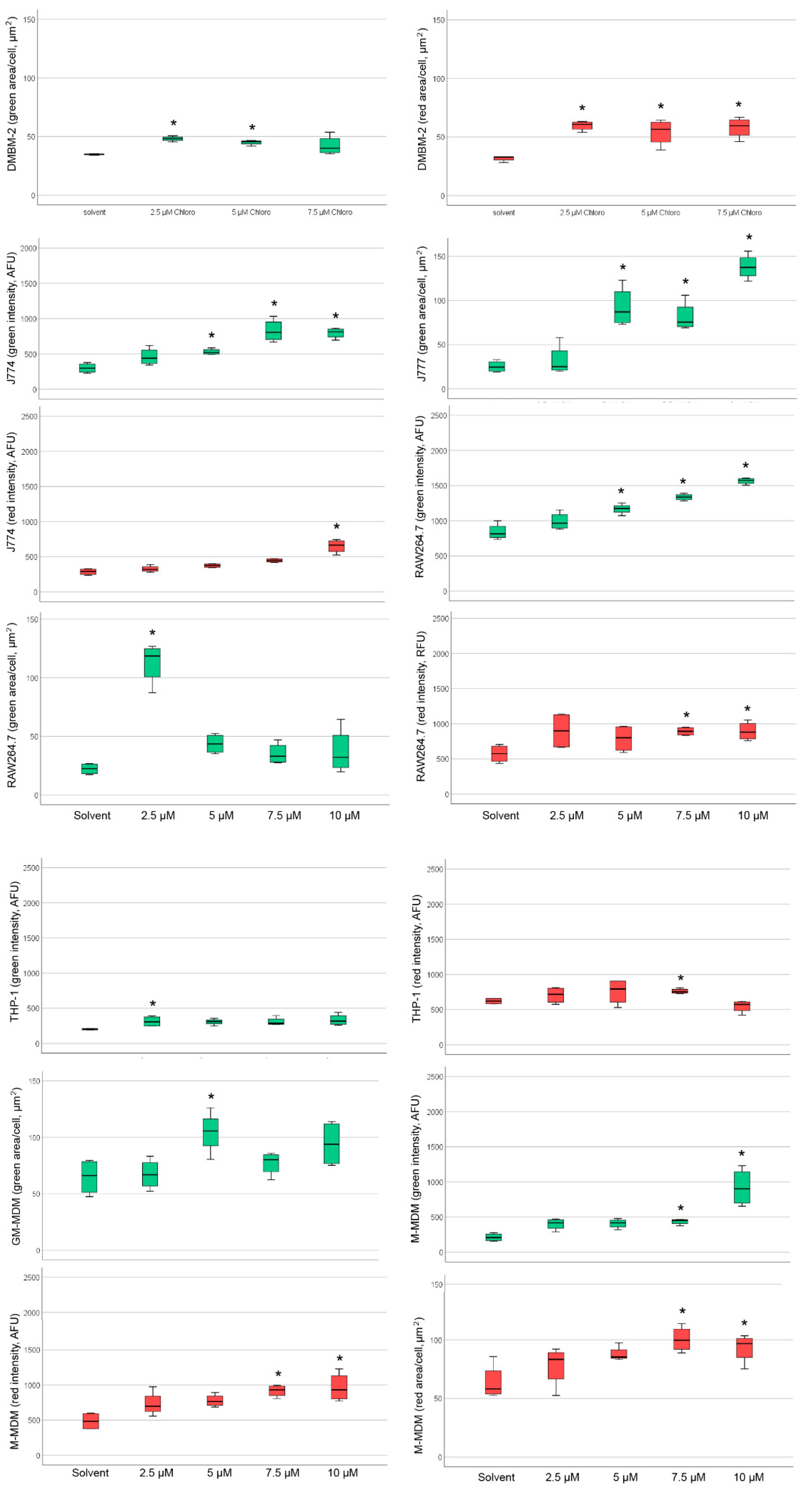
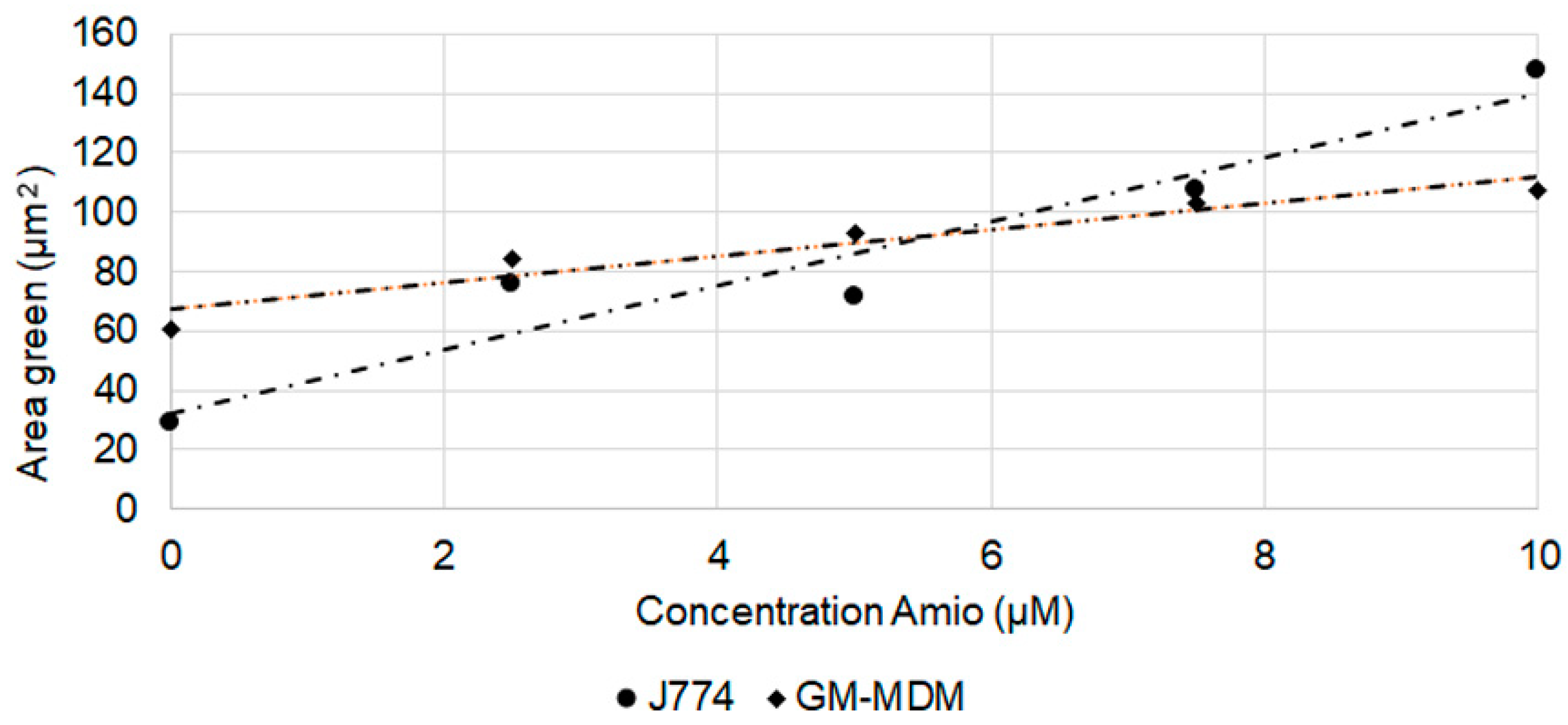
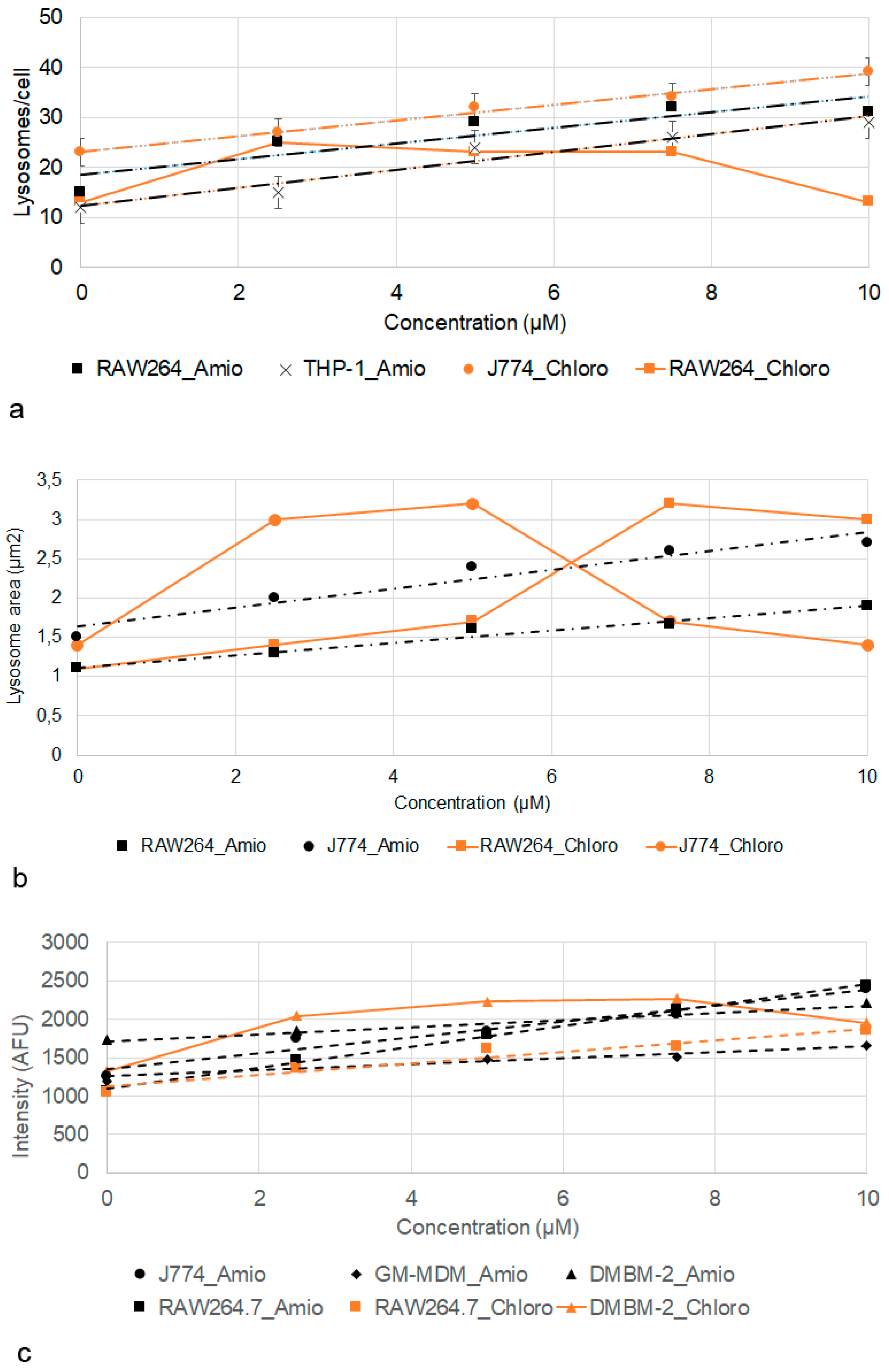
| Macrophages | Cell Area (DMSO, µm2) | Cell Area (AD, µm2) | Lysosome Number (DMSO) | Lysosome Number (AD) | Lysosome Area (DMSO, µm2) | Lysosome Area (AD, µm2) |
|---|---|---|---|---|---|---|
| DMBM-2 | 253 ± 6 * | 229 ± 2.4 | 30 ± 0.5 | 30 ± 0.4 | 3.5 ± 0.014 | 3.3 ± 0.009 |
| J774A.1 | 183 ± 1.8 * | 172 ± 1.6 | 24 ± 0.4 | 23 ± 0.4 | 1.5 ± 0.008 | 1.4 ± 0.006 |
| RAW264.7 | 131 ± 1.6 * | 120 ± 1.7 | 13 ± 0.2 | 14 ± 0.2 | 1.1 ± 0.003 | 1.1 ± 0.004 |
| THP-1 | 208 ± 3.8 | 215 ± 3.3 | 13 ± 0.4 | 14 ± 0.5 | 1.5 ± 0.01 | 1.5 ± 0.01 |
| GM-MDM | 241 ± 4.5 | 238 ± 4.0 | 11 ± 0.2 | 10 ± 0.2 | 2.1 ± 0.01 | 2.2 ± 0.01 |
| Treatment | DMBM-2 | J774 | RAW264.7 | THP-1 | GM-MDM | M-MDM |
|---|---|---|---|---|---|---|
| DMSO | 100 ± 5 | 100 ± 2 | 100 ± 3 | 100 ± 5 | 100 ± 9 | 100 ± 2 |
| Amio2.5 | 98 ± 3 | 100 ± 1 | 103 ± 4 | 86 ± 3 | 92 ± 7 | 101 ± 9 |
| Amio5.0 | 107 ± 3 | 105 ± 3 | 111 ± 2 | 89 ± 3 | 89 ± 7 | 123 ± 11 |
| Amio7.5 | 104 ± 2 | 108 ± 2 | 107 ± 3 | 93 ± 6 | 97 ± 8 | 137 ± 2 |
| Amio10.0 | 90 ± 4 | 111 ± 2 | 115 ± 2 | 105 ± 3 | 96 ± 9 | 150 ± 3 |
| AD | 100 ± 5 | 100 ± 1 | 100 ± 2 | 100 ± 4 | 100 ± 5 | 100 ± 12 |
| Chloro2.5 | 97 ± 6 | 99 ± 1 | 102 ± 2 | 97 ± 10 | 97 ± 8 | 99 ± 9 |
| Chloro5.0 | 99 ± 3 | 107 ± 2 | 104 ± 5 | 92 ± 5 | 96 ± 7 | 95 ± 11 |
| Chloro7.5 | 99 ± 7 | 108 ± 3 | 102 ± 4 | 91 ± 5 | 97 ± 7 | 108 ± 6 |
| Chloro10.0 | 98 ± 5 | 111 ± 4 | 107 ± 4 | 97 ± 6 | 99 ± 4 | 131 ± 5 |
| Amiodarone | Chloroquine | |||
|---|---|---|---|---|
| Macrophage | Phospholipid Staining | Neutral Lipid Staining | Phospholipid Staining | Neutral Lipid Staining |
| DMBM-2 | - | - | Area (2.5, 5 µM) | Area (≥2.5 µM) |
| J774 | Intensity (≥7.5 µM) Area (≥2.5 µM) | - | Intensity (≥5 µM) Area (≥5 µM) | Intensity (10 µM) |
| RAW264.7 | Intensity (≥5 µM) Area (≥5 µM) | Area (5, 7.5 µM) | Intensity (≥5 µM) Area (2.5 µM) | Intensity (≥7.5 µM) |
| THP-1 | Intensity (2.5 µM) | Intensity (≥2.5 µM) | Intensity (2.5 µM) | Intensity (7.5 µM) |
| GM-MDM | Intensity (≥2.5 µM) Area (≥2.5 µM) | - | Area (5 µM) | - |
| M-MDM | Intensity (≥2.5 µM) | - | Intensity (10 µM) | Intensity (≥7.5 µM) Area (≥7.5 µM) |
| Amiodarone | Chloroquine | |||||
|---|---|---|---|---|---|---|
| Macrophage | Number | Area | Intensity | Number | Area | Intensity |
| DMBM-2 | 2.5, 5 µM | 2.5–7.5 µM | ≥2.5 µM | 10 µM | ≥2.5 µM | ≥2.5 µM |
| J774 | ≥2.5 µM | ≥2.5 µM | ≥2.5 µM | 2.5, 10 µM | 2.5–7.5 µM | 2.5–7.5 µM |
| RAW264.7 | ≥2.5 µM | ≥2.5 µM | ≥2.5 µM | ≥2.5 µM | ≥2.5 µM | ≥2.5 µM |
| THP-1 | ≥2.5 µM | ≥5 µM | ≥2.5 µM | ≥2.5 µM | 10 µM | ≥2.5 µM |
| GM-MDM | ≥2.5 µM | 2.5, 5, 10 µM | ≥2.5 µM | 10 µM | 10 µM | ≥2.5 µM |
| Medium | DMSO | AD | Amio | Chloro | |
|---|---|---|---|---|---|
| DMBM-2 | 0.42 | 0.37 | 0.41 | 0.35 | 0.25 |
| RAW264.7 | 0.48 | 0.35 | 0.48 | 0.47 | 0.49 |
| J774 | 0.63 | 0.63 | 0.63 | 0.47 | 0.44 |
| GM-MDM | 0.56 | 0.63 | 0.53 | 0.57 | 0.55 |
| THP-1 | 0.67 | 0.65 | 0.53 | 0.66 | 0.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Öhlinger, K.; Absenger-Novak, M.; Meindl, C.; Ober, J.; Fröhlich, E. Different Sensitivity of Macrophages to Phospholipidosis Induction by Amphiphilic Cationic Drugs. Int. J. Mol. Sci. 2020, 21, 8391. https://doi.org/10.3390/ijms21218391
Öhlinger K, Absenger-Novak M, Meindl C, Ober J, Fröhlich E. Different Sensitivity of Macrophages to Phospholipidosis Induction by Amphiphilic Cationic Drugs. International Journal of Molecular Sciences. 2020; 21(21):8391. https://doi.org/10.3390/ijms21218391
Chicago/Turabian StyleÖhlinger, Kristin, Markus Absenger-Novak, Claudia Meindl, Jennifer Ober, and Eleonore Fröhlich. 2020. "Different Sensitivity of Macrophages to Phospholipidosis Induction by Amphiphilic Cationic Drugs" International Journal of Molecular Sciences 21, no. 21: 8391. https://doi.org/10.3390/ijms21218391