Long Non-Coding RNA Associated with Cholesterol Homeostasis and Its Involvement in Metabolic Diseases
Abstract
:1. Introduction
2. LncRNA in Cholesterol Synthesis and Metabolism
3. LncRNAs in Cholesterol Efflux
4. Disease Associated Cholesterol Dysregulation via LncRNAs
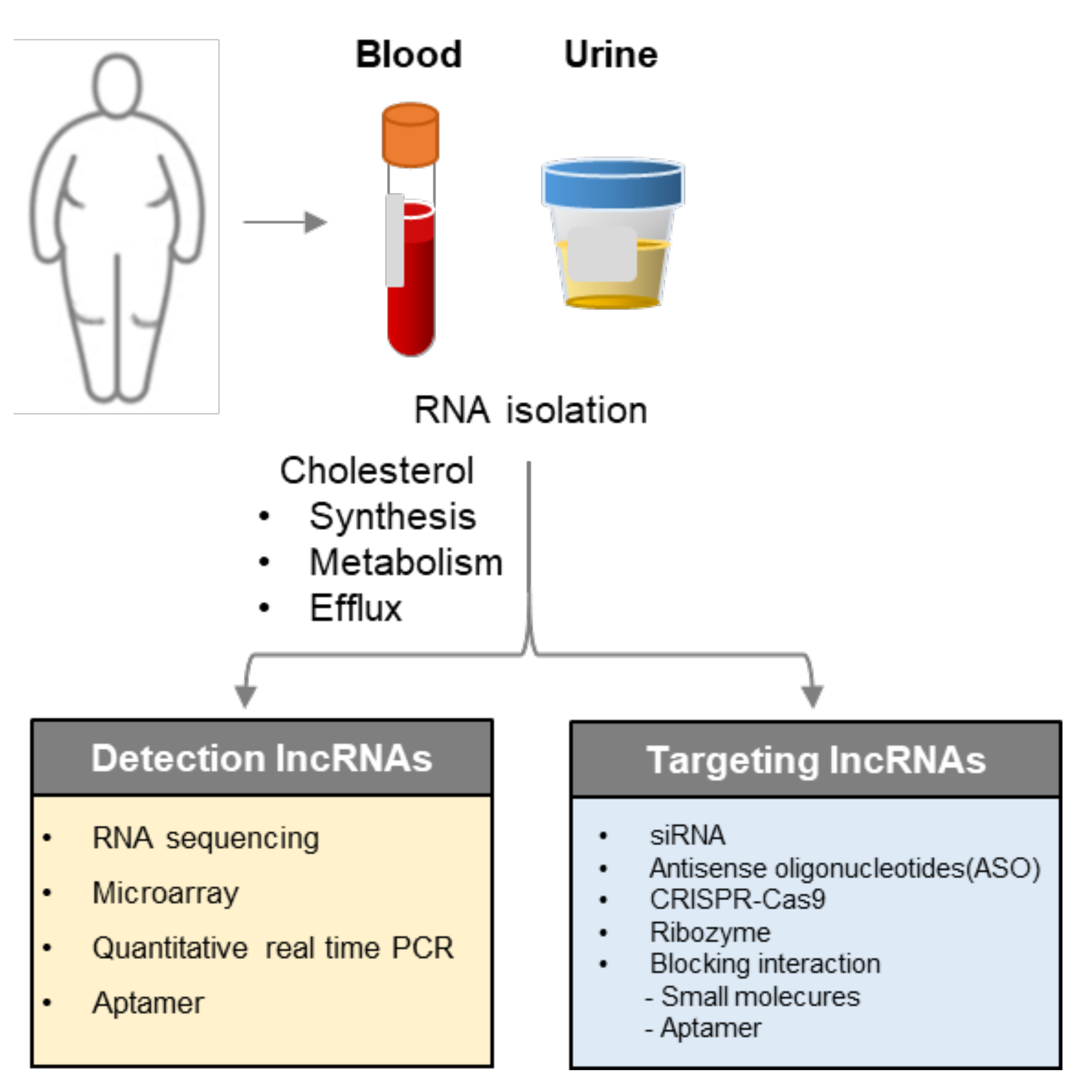
5. Implementation for Diagnosis and Therapeutic Targets
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| lncRNA | long non-coding RNA |
| VLDL | very-low-density lipoprotein |
| LDL | low density lipoprotein |
| IDL | intermediate density lipoprotein |
| HDL | high density lipoprotein |
| ENCODE | Encyclopedia of DNA Elements |
| DNMT1 | DNA methyltransferase1 |
| NF1A | nuclear factor 1A |
| LPL | lipoprotein lipase |
| HCC | hepatocellular carcinoma |
| PPAR-α | peroxisome proliferator-activated receptor alpha |
| LDLR | low-density lipoprotein receptor |
| PCSK9 | proprotein convertase subtilisin/kexin type 9 |
| HMGCR | 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase |
| ABC | ATP-binding cassette |
| ADAM10 | A Disintegrin and metalloproteinase domain-containing protein 10 |
| DNMT1 | DNA methyltransferase 1 |
| CYP27A1 | sterol 27-hydroxylase |
| HUVEC | human umbilical vein endothelial cells |
| VSMC | vascular smooth muscle cell |
| MEG3 | material representation gene 3 |
| PTBP1 | polypyrimidine track binding protein 1 |
| SHP | small heterodimer partner |
| CREB | cAMP response element-binding protein |
| TUG1 | taurine-up-regulated gene 1 |
| FGF1 | fibroblast growth factor 1 |
| ITGB3 | integrin beta-3 |
| TBXA2R | thromboxane A2 receptor |
| CAD | coronary artery disease |
| SREBP | Sterol regulatory element binding transcription factor |
| SNPs | single nucleotide polymorphisms |
| MI | myocardial infarction |
References
- Martini, C.; Pallottini, V. Cholesterol: From feeding to gene regulation. Genes Nutr. 2007, 2, 181–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, R.A.; Garcia-Palmieri, M.R. Cholesterol, Triglycerides, and Associated Lipoproteins. In Clinical Methods: The History, Physical, and Laboratory Examinations, 3rd ed.; Butterworths: Boston, MA, USA, 1990. [Google Scholar]
- Cohen, D.E. Balancing cholesterol synthesis and absorption in the gastrointestinal tract. J. Clin. Lipidol. 2008, 2, S1–S3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casares, D.; Escriba, P.V.; Rossello, C.A. Membrane Lipid Composition: Effect on Membrane and Organelle Structure, Function and Compartmentalization and Therapeutic Avenues. Int. J. Mol. Sci. 2019, 20, 2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, C.L.; Walch, L.; Verbavatz, J.M. Lipids and Their Trafficking: An Integral Part of Cellular Organization. Dev. Cell 2016, 39, 139–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soffientini, U.; Graham, A. Intracellular cholesterol transport proteins: Roles in health and disease. Clin. Sci. 2016, 130, 1843–1859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Liu, Q. Cholesterol metabolism and homeostasis in the brain. Protein Cell 2015, 6, 254–264. [Google Scholar] [CrossRef] [Green Version]
- Litvinov, D.Y.; Savushkin, E.V.; Dergunov, A.D. Intracellular and Plasma Membrane Events in Cholesterol Transport and Homeostasis. J. Lipids 2018, 2018, 3965054. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Zhang, Z.; Shen, W.-J.; Azhar, S. Cellular cholesterol delivery, intracellular processing and utilization for biosynthesis of steroid hormones. Nutr. Metab. 2010, 7, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feingold, K.R.; Grunfeld, C. (Eds.) Introduction to Lipids and Lipoproteins. In Endotext; MDText.com Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Olson, R.E. Discovery of the lipoproteins, their role in fat transport and their significance as risk factors. J. Nutr. 1998, 128, 439s–443s. [Google Scholar] [CrossRef] [PubMed]
- De Isla, L.P.; Alonso, R.; Mata, N.; Saltijeral, A.; Muñiz, O.; Rubio-Marin, P.; Diaz-Diaz, J.L.; Fuentes, F.; De Andrés, R.; Zambón, D.; et al. Coronary Heart Disease, Peripheral Arterial Disease, and Stroke in Familial Hypercholesterolaemia: Insights From the SAFEHEART Registry (Spanish Familial Hypercholesterolaemia Cohort Study). Arterioscler. Thromb. Vasc. Biol. 2016, 36, 2004–2010. [Google Scholar] [CrossRef] [Green Version]
- Ormazabal, V.; Nair, S.; Elfeky, O.; Aguayo, C.; Salomon, C.; Zuñiga, F.A. Association between insulin resistance and the development of cardiovascular disease. Cardiovasc. Diabetol. 2018, 17, 122. [Google Scholar] [CrossRef]
- Daniels, T.F.; Killinger, K.M.; Michal, J.J.; Wright, R.W., Jr.; Jiang, Z. Lipoproteins, cholesterol homeostasis and cardiac health. Int. J. Biol. Sci. 2009, 5, 474–488. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.H.; Garruti, G.; Liu, M.; Portincasa, P.; Wang, D.Q.-H. Cholesterol and Lipoprotein Metabolism and Atherosclerosis: Recent Advances in Reverse Cholesterol Transport. Ann. Hepatol. 2017, 16, S27–S42. [Google Scholar] [CrossRef] [PubMed]
- Szustakowski, J.; Consor, I.H.G.S. Initial sequencing and analysis of the human genome (vol 409, pg 860, 2001). Nature 2001, 411, 720. [Google Scholar]
- Dunham, I.; Birney, E.; Lajoie, B.R.; Sanyal, A.; Dong, X.; Greven, M.; Dekker, J. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar]
- Zhang, X.Z.; Liu, H.; Chen, S.R. Mechanisms of Long Non-Coding RNAs in Cancers and Their Dynamic Regulations. Cancers 2020, 12, 1245. [Google Scholar] [CrossRef]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef] [PubMed]
- He, R.Z.; Luo, D.X.; Mo, Y.Y. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019, 6, 6–15. [Google Scholar] [CrossRef]
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long Noncoding RNAs: Past, Present, and Future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [Green Version]
- Dykes, I.M.; Emanueli, C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef]
- Gibney, E.R.; Nolan, C.M. Epigenetics and gene expression. Heredity 2010, 105, 4–13. [Google Scholar] [CrossRef] [Green Version]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Sun, H.; Wang, H.T. Long noncoding RNAs in DNA methylation: New players stepping into the old game. Cell Biosci. 2016, 6, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kar, S.; Deb, M.; Sengupta, D.; Shilpi, A.; Parbin, S.; Torrisani, J.; Pradhan, S.; Patra, S.K. An insight into the various regulatory mechanisms modulating human DNA methyltransferase 1 stability and function. Epigenetics 2012, 7, 994–1007. [Google Scholar] [CrossRef] [Green Version]
- Davidovich, C.; Cech, T.R. The recruitment of chromatin modifiers by long noncoding RNAs: Lessons from PRC2. RNA 2015, 21, 2007–2022. [Google Scholar] [CrossRef] [Green Version]
- Saxena, A.; Carninci, P. Long non-coding RNA modifies chromatin Epigenetic silencing by long non-coding RNAs. Bioessays 2011, 33, 830–839. [Google Scholar] [CrossRef] [Green Version]
- Froberg, J.E.; Yang, L.; Lee, J.T. Guided by RNAs: X-Inactivation as a Model for lncRNA Function. J. Mol. Biol. 2013, 425, 3698–3706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Ruscio, A.; Ebralidze, A.K.; Benoukraf, T.; Amabile, G.; Goff, L.A.; Terragni, J.; Figueroa, M.E.; Pontes, L.L.D.F.; Alberich-Jorda, M.; Zhang, P.; et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 2013, 503, 371–376. [Google Scholar] [CrossRef] [Green Version]
- Pastori, C.; Wahlestedt, C. Involvement of long noncoding RNAs in diseases affecting the central nervous system. RNA Biol. 2012, 9, 860–870. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Zhuang, Y.; Zhao, X.; Li, X. Long Non-coding RNA in Neuronal Development and Neurological Disorders. Front. Genet. 2019, 9, 744. [Google Scholar] [CrossRef] [Green Version]
- Baskaran, R.; Velmurugan, B.K. Role of lncRNAs in the cancer development and progression and their regulation by various phytochemicals. Biomed. Pharmacother. 2018, 102, 242–248. [Google Scholar]
- Sparber, P.; Filatova, A.; Khantemirova, M.; Skoblov, M. The role of long non-coding RNAs in the pathogenesis of hereditary diseases. BMC Med. Genom. 2019, 12, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Degirmenci, U.; Lei, S. Role of lncRNAs in Cellular Aging. Front. Endocrinol. 2016, 7, 151. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Tu, C.; Liu, Y. Role of lncRNAs in aging and age-related diseases. Aging Med. 2018, 1, 158–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.W.; Zhao, J.Y.; Li, S.F.; Huang, J.L.; Qiu, Y.R.; Ma, X.; Wang, Y.C. RP5-833A20.1/miR-382-5p/NFIA-Dependent Signal Transduction Pathway Contributes to the Regulation of Cholesterol Homeostasis and Inflammatory Reaction. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 87–101. [Google Scholar] [CrossRef] [Green Version]
- Zhen, Z.; Ren, S.; Ji, H.; Ding, X.; Zou, P.; Lu, J. The lncRNA DAPK-IT1 regulates cholesterol metabolism and inflammatory response in macrophages and promotes atherogenesis. Biochem. Biophys. Res. Commun. 2019, 516, 1234–1241. [Google Scholar] [CrossRef]
- Wang, L.; Xia, J.-W.; Ke, Z.-P.; Zhang, B. Blockade of NEAT1 represses inflammation response and lipid uptake via modulating miR-342-3p in human macrophages THP-1 cells. J. Cell. Physiol. 2019, 234, 5319–5326. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Wu, L.; Wu, N.; Chen, Q.; Li, Y.; Du, X.; Sun, M. Long Noncoding RNA lnc-HC Regulates PPAR gamma-Mediated Hepatic Lipid Metabolism through miR-130b-3. Mol. Ther. Nucleic Acids 2019, 18, 954–965. [Google Scholar] [CrossRef] [Green Version]
- Lan, X.; Yan, J.; Ren, J.; Zhong, B.; Li, J.; Li, Y.; Liu, L.; Yi, J.; Sun, Q.; Yang, X.; et al. A Novel Long Noncoding RNA Lnc-HC Binds hnRNPA2B1 to Regulate Expressions of Cyp7a1 and Abca1 in Hepatocytic Cholesterol Metabolism. Hepatology 2016, 64, 58–72. [Google Scholar] [CrossRef] [Green Version]
- Sallam, T.; Jones, M.C.; Gilliland, T.; Zhang, L.; Wu, X.; Eskin, A.; Sandhu, J.; Casero, D.; Vallim, T.S.X.W.T.Q.D.A.; Hong, C.; et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 2016, 534, 124–128. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.; Xiao, Z.; Wang, Y.; Zheng, M.; Song, T.; Cai, X.; Sun, B.; Ye, L.; Zhang, X.-D. Long Noncoding RNA HULC Modulates Abnormal Lipid Metabolism in Hepatoma Cells through an miR-9-Mediated RXRA Signaling Pathway. Cancer Res. 2015, 75, 846–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Go, G.W.; Mani, A. Low-density lipoprotein receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J. Biol. Med. 2012, 85, 19–28. [Google Scholar]
- Ray, R.M.; Hansen, A.H.; Slott, S.; Taskova, M.; Astakhova, I.K.; Morris, K.V. Control of LDL Uptake in Human Cells by Targeting the LDLR Regulatory Long Non-coding RNA BM450697. Mol. Ther. Nucleic Acids 2019, 17, 264–276. [Google Scholar] [CrossRef] [Green Version]
- Mitchel, K.; Theusch, E.; Cubitt, C.; Dosé, A.C.; Stevens, K.; Naidoo, D.; Medina, M.W. RP1-13D10.2 Is a Novel Modulator of Statin-Induced Changes in Cholesterol. Circ. Cardiovasc. Genet. 2016, 9, 223–230. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Hu, Z.; Zhang, W.; Yu, J.; Yang, Y.; Xu, Z.; Luo, H.; Liu, X.; Liu, Y.; Chen, C.; et al. Regulation of Cholesterol Homeostasis by a Novel Long Non-coding RNA LASER. Sci. Rep. 2019, 9, 1–12. [Google Scholar]
- Liu, G.; Zheng, X.; Xu, Y.; Yanlu, X.; Chen, J.; Huang, X. Long Non-coding RNAs Expression Profile in HepG2 Cells Reveals the Potential Role of Long Non-coding RNAs in the Cholesterol Metabolism. Chin. Med. J. 2015, 128, 91–97. [Google Scholar] [CrossRef]
- Huang, J.B.; Chen, S.; Cai, D.; Bian, D.; Wang, F. Long noncoding RNA lncARSR promotes hepatic cholesterol biosynthesis via modulating Akt/SREBP-2/HMGCR pathway. Life Sci. 2018, 203, 48–53. [Google Scholar] [CrossRef]
- Huang, C.; Hu, Y.-W.; Zhao, J.-J.; Ma, X.; Zhang, Y.; Guo, F.-X.; Kang, C.-M.; Lu, J.-B.; Xiu, J.-C.; Sha, Y.-H.; et al. Long Noncoding RNA HOXC-AS1 Suppresses Ox-LDL-Induced Cholesterol Accumulation Through Promoting HOXC6 Expression in THP-1 Macrophages. DNA Cell Biol. 2016, 35, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Molina, E.; Chew, G.S.; Myers, S.A.; Clarence, E.M.; Eales, J.M.; Tomaszewski, M.; Charchar, F.J. A Novel Y-Specific Long Non-Coding RNA Associated with Cellular Lipid Accumulation in HepG2 cells and Atherosclerosis-related Genes. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Li, Y.; Shen, S.; Ding, S.; Wang, L. LincRNA DYN-LRB2-2 upregulates cholesterol efflux by decreasing TLR2 expression in macrophages. J. Cell. Biochem. 2018, 119, 1911–1921. [Google Scholar] [CrossRef]
- Cai, C.; Zhu, H.; Ning, X.; Li, L.; Yang, B.; Chen, S.; Wang, L.; Lu, X.-F.; Gu, N. LncRNA ENST00000602558,1 regulates ABCG1 expression and cholesterol efflux from vascular smooth muscle cells through a p65-dependent pathway. Atherosclerosis 2019, 285, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Yin, R.; Shi, H.; Wang, X.; Shen, D.; Wang, X.; Pan, C. LncRNA ZFAS1 confers inflammatory responses and reduces cholesterol efflux in atherosclerosis through regulating miR-654-3p-ADAM10/ RAB22A axis. Int. J. Cardiol. 2020, 315, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Han, S.; Sun, Q.; Yao, Y.; Li, S.; Yuan, C.; Zhang, B.; Jing, B.; Wu, J.; Song, Y.; et al. Long non-coding RNA CDKN2B-AS1 reduces inflammatory response and promotes cholesterol efflux in atherosclerosis by inhibiting ADAIVA10 expression. Aging 2019, 11, 1695–1715. [Google Scholar] [CrossRef]
- Hennessy, E.J.; Van Solingen, C.; Scacalossi, K.R.; Ouimet, M.; Afonso, M.S.; Prins, J.; Koelwyn, G.J.; Sharma, M.; Ramkhelawon, B.; Carpenter, S.; et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primates. Nat. Metab. 2019, 1, 98–110. [Google Scholar] [CrossRef]
- Sallam, T.; Jones, M.; Thomas, B.J.; Wu, X.; Gilliland, T.; Qian, K.; Eskin, A.; Casero, D.; Zhang, Z.; Sandhu, J.; et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat. Med. 2018, 24, 304–312. [Google Scholar] [CrossRef]
- Xu, B.; Xiao, L.; Kang, C.; Ding, L.; Guo, F.; Li, P.; Lu, Z.; Wu, Q.; Xu, Y.; Bai, H. LncRNA AC096664,3/PPAR-gamma/ABCG1-dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis. J. Cell. Biochem. 2019, 120, 13775–13782. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Tan, L.; Yao, J.; Yang, L. Long non-coding RNA MALAT1 regulates cholesterol accumulation in ox-LDL-induced macrophages via the microRNA-17-5p/ABCA1 axis. Mol. Med. Rep. 2020, 21, 1761–1770. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Li, T. LncRNA TUG1 regulates ApoM to promote atherosclerosis progression through miR-92a/FXR1 axis. J. Cell. Mol. Med. 2020, 24, 8836–8848. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, Z.; Trottier, J.; Barbier, O.; Wang, L. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology 2017, 65, 604–615. [Google Scholar] [CrossRef]
- Wang, X.; Guo, S.; Hu, Y.; Guo, H.; Zhang, X.; Yan, Y.; Ma, J.; Li, Y.; Wang, H.; He, J.; et al. Microarray analysis of long non-coding RNA expression profiles in low high-density lipoprotein cholesterol disease. Lipids Health Dis. 2020, 19, 175. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, X.; Gong, Y.; Li, T.; Yang, L.; Xu, W.; Dong, L. Role of the lncRNA-mRNA network in atherosclerosis using ox-low-density lipoprotein-induced macrophage-derived foam cells. Mol. Omics 2020. [Google Scholar] [CrossRef]
- Li, P.; Yan, X.; Xu, G.; Pang, Z.; Weng, J.; Yin, J.; Li, M.; Yu, L.; Chen, Q.; Sun, K. A novel plasma lncRNA ENST00000416361 is upregulated in coronary artery disease and is related to inflammation and lipid metabolism. Mol. Med. Rep. 2020, 21, 2375–2384. [Google Scholar]
- Li, Y.; Zhang, D.; Zhang, Y.; Xu, X.; Bi, L.; Zhang, M.; Yu, B.; Zhang, Y. Association of lncRNA polymorphisms with triglyceride and total cholesterol levels among myocardial infarction patients in Chinese population. Gene 2020, 724, 143684. [Google Scholar] [CrossRef]
- Zhang, L.; Cheng, H.; Yue, Y.; Li, S.; Zhang, D.; He, R. TUG1 knockdown ameliorates atherosclerosis via up-regulating the expression of miR-133a target gene FGF1. Cardiovasc. Pathol. 2018, 33, 6–15. [Google Scholar] [CrossRef]
- Phillips, M.C. Molecular Mechanisms of Cellular Cholesterol Efflux. J. Biol. Chem. 2014, 289, 24020–24029. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Zhu, H.M.; Ge, J.B. Long Noncoding RNA: Recent Updates in Atherosclerosis. Int. J. Biol. Sci. 2016, 12, 898–910. [Google Scholar] [CrossRef] [Green Version]
- Linton, M.R.F.; Yancey, P.G.; Davies, S.S.; Jerome, W.G.J.; Linton, E.F.; Vickers, K.C. The Role of Lipids and Lipoproteins in Atherosclerosis. In Endotext; Feingold, K.R., Grunfeld, C., Eds.; MDText.com Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Gotto, A.M., Jr. Jeremiah Metzger Lecture: Cholesterol, inflammation and atherosclerotic cardiovascular disease: Is it all LDL? Trans. Am. Clin. Climatol. Assoc. 2011, 122, 256–289. [Google Scholar]
- Paredes, S.; Fonseca, L.; Ribeiro, L.; Ramos, H.; Oliveira, J.C.; Palma, I. Novel and traditional lipid profiles in Metabolic Syndrome reveal a high atherogenicity. Sci. Rep. 2019, 9, 11792. [Google Scholar] [CrossRef] [Green Version]
- Shi, T.; Gao, G.; Cao, Y.L. Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics. Dis. Markers 2016, 2016, 9085195. [Google Scholar] [CrossRef] [Green Version]
- Pardini, B.; Sabo, A.A.; Birolo, G.; Calin, G.A. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers 2019, 11, 1170. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.-T.; Sun, Y.-M.; Huang, W.; He, B.; Zhao, Y.-N.; Chen, Y.-Q. Genome-wide Long Non-coding RNA Analysis Identified Circulating LncRNAs as Novel Non-invasive Diagnostic Biomarkers for Gynecological Disease. Sci. Rep. 2016, 6, 23343. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhong, L.; Xu, W.; Sun, Y.; Zhang, Z.; Zhao, H.; Yang, L.; Sun, J. Discovery of potential prognostic long non-coding RNA biomarkers for predicting the risk of tumor recurrence of breast cancer patients. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Shi, H.; Wang, X.; Wang, B.; Qu, Q.; Geng, H.; Sun, H. Identification of diagnostic long non-coding RNA biomarkers in patients with hepatocellular carcinoma. Mol. Med. Rep. 2019, 20, 1121–1130. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.H.; Qi, P.; Du, X. Long non-coding RNAs in cancer invasion and metastasis. Mod. Pathol. 2015, 28, 4–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bin, X.; Hongjian, Y.; Zhang, X.; Bo, C.; Shifeng, Y.; Binbin, T. Research progresses in roles of LncRNA and its relationships with breast cancer. Cancer Cell Int. 2018, 18, 1–12. [Google Scholar] [CrossRef]
- Chi, H.-C.; Tsai, C.-Y.; Tsai, M.-M.; Yeh, C.-T.; Lin, K.-H. Roles of Long Noncoding RNAs in Recurrence and Metastasis of Radiotherapy-Resistant Cancer Stem Cells. Int. J. Mol. Sci. 2017, 18, 1903. [Google Scholar] [CrossRef]
- Xie, H.; Ma, H.W.; Zhou, D.Q. Plasma HULC as a Promising Novel Biomarker for the Detection of Hepatocellular Carcinoma. Biomed Res. Int. 2013, 2013, 136106. [Google Scholar] [CrossRef]
- Lin, L.; Li, H.; Zhu, Y.; He, S.; Ge, H. Expression of metastasis-associated lung adenocarcinoma transcript 1 long non-coding RNA in vitro and in patients with non-small cell lung cancer. Oncol. Lett. 2018, 15, 9443–9449. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, L.H.; Spieker, T.; Koschmieder, S.; Humberg, J.; Jungen, D.; Bulk, E.; Hascher, A.; Wittmer, D.; Marra, A.; Hillejan, L.; et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J. Thorac. Oncol. 2011, 6, 1984–1992. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Li, Z.; Zheng, H.; Chan, M.T.V.; Wu, W.K.K. NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif. 2017, 50, e12329. [Google Scholar] [CrossRef] [Green Version]
- Lu, T.; Wang, Y.; Chen, D.; Liu, J.; Jiao, W. Potential clinical application of lncRNAs in non-small cell lung cancer. Onco Targets Ther. 2018, 11, 8045–8052. [Google Scholar] [CrossRef] [Green Version]
- Osielska, M.A.; Jagodzinski, P. Long non-coding RNA as potential biomarkers in non-small-cell lung cancer: What do we know so far? Biomed. Pharmacother. 2018, 101, 322–333. [Google Scholar] [CrossRef]
- Esfandi, F.; Taheri, M.; Omrani, M.D.; Shadmehr, M.B.; Arsang-Jang, S.; Shams, R.; Ghafouri-Fard, S. Expression of long non-coding RNAs (lncRNAs) has been dysregulated in non-small cell lung cancer tissues. BMC Cancer 2019, 19, 222. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Sun, L.N.; Wan, F.S. Molecular mechanisms of TUG1 in the proliferation, apoptosis, migration and invasion of cancer cells. Oncol. Lett. 2019, 18, 4393–4402. [Google Scholar] [CrossRef] [Green Version]
- Mannu, G.S.; Zaman, M.J.S.; Gupta, A.; Rehman, H.U.; Myint, P.K. Evidence of lifestyle modification in the management of hypercholesterolemia. Curr. Cardiol. Rev. 2013, 9, 2–14. [Google Scholar]
- Enkhmaa, B.; Surampudi, P.; Anuurad, E.; Berglund, L. Lifestyle Changes: Effect of Diet, Exercise, Functional Food, and Obesity Treatment on Lipids and Lipoproteins. In Endotext; Feingold, K.R., Grunfeld, C., Eds.; MDText.com Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Agency for Healthcare Research and Quality (US). Treating High Cholesterol: A Guide for Adults. In Comparative Effectiveness Review Summary Guides for Consumers; Agency for Healthcare Research and Quality (US): Rockville, MD, USA, 2005. [Google Scholar]
- Feingold, K.R. Cholesterol Lowering Drugs. In Endotext; Feingold, K.R., Grunfeld, C., Eds.; MDText.com Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Chery, J. RNA therapeutics: RNAi and antisense mechanisms and clinical applications. Postdoc. J. 2016, 4, 35–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amodio, N.; Raimondi, L.; Juli, G.; Stamato, M.A.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 2018, 11, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Shum, K.-T.; Burnett, J.C.; Rossi, J.J. Nanoparticle-Based Delivery of RNAi Therapeutics: Progress and Challenges. Pharmaceuticals 2013, 6, 85–107. [Google Scholar] [CrossRef] [Green Version]
- Amodio, N.; Stamato, M.A.; Juli, G.; Morelli, E.; Fulciniti, M.; Manzoni, M.; Taiana, E.; Agnelli, L.; Cantafio, M.E.G.; Romeo, E.; et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia 2018, 32, 1948–1957. [Google Scholar] [CrossRef] [Green Version]
- Gutschner, T.; Hämmerle, M.; Eissmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Gross, M.; et al. The Noncoding RNA MALAT1 Is a Critical Regulator of the Metastasis Phenotype of Lung Cancer Cells. Cancer Res. 2013, 73, 1180–1189. [Google Scholar] [CrossRef] [Green Version]
- Bhat, S.A.; Ahmad, S.M.; Mumtaz, P.T.; Malik, A.A.; Dar, M.A.; Urwat, U.; Shah, R.A.; Ganai, N.A. Long non-coding RNAs: Mechanism of action and functional utility. Noncoding RNA Res. 2016, 1, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Jiang, Y.; Zhang, Y.; Xu, Y.; Zhang, C.; Han, J.; Su, F.; Liu, X.; Mi, K.; Liu, B.; et al. System level characterization of small molecule drugs and their affected long noncoding RNAs. Aging 2019, 11, 12428–12451. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, R.P.; Salah-Uddin, S.; Modarresi, F.; Khoury, N.; Wahlestedt, C.; Faghihi, M.A. Screening for Small-Molecule Modulators of Long Noncoding RNA-Protein Interactions Using AlphaScreen. J. Biomol. Screen. 2015, 20, 1132–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Murat, P.; Matak-Vinkovic, D.; Murrell, A.; Balasubramanian, S. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry 2013, 52, 9519–9527. [Google Scholar] [CrossRef]

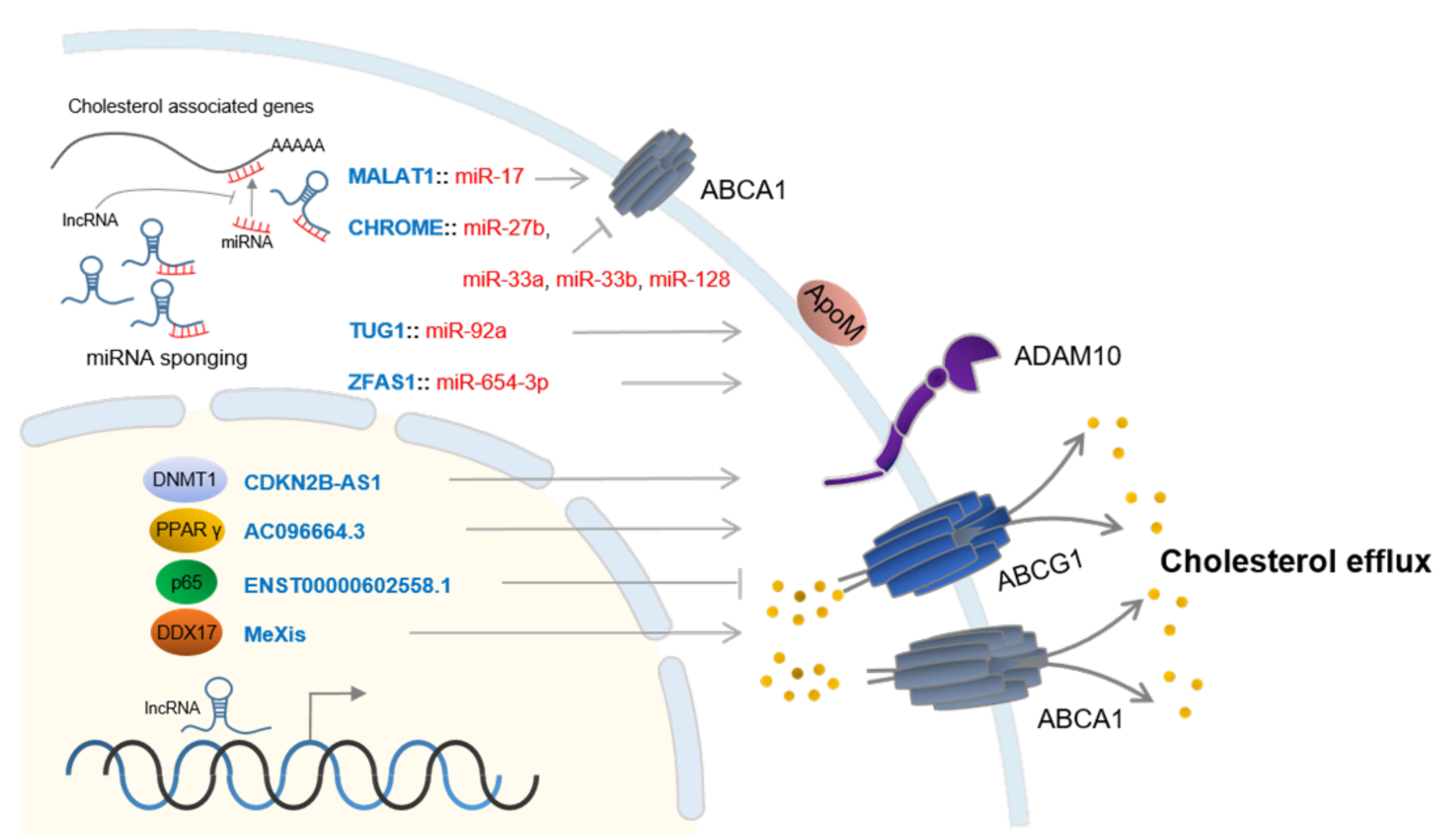


| LncRNA | Target | Findings | Ref |
|---|---|---|---|
| Cholesterol metabolism | |||
| Lnc-HC | hnRNPA2B1, Cyp7a1, Abca1 | Lnc-HC-hnRNPA2B1 complex decreases Cyp7a1, Abca1 | [41] |
| miR-130b-3p, PPARγ | Negatively regulates PPARγ expression via miR-130b-3p | [40] | |
| BM450697 | LDLR | Regulates and local scaffold for LDLR transcription | [45] |
| HOXC-AS1 | HOXC6 | Promotes HOXC6 | [50] |
| HULC | ASCL1, PPARA | miR-9/PPARA/ACSL1/cholesterol/RXRA/HULC signaling. | [43] |
| AT102202 | HMGCR | Regulates HMGCR expression | [48] |
| LASER | PCSK9 | Feedback HNF-1α/PCSK9 and LXR dependent pathway | [47] |
| RP1-13D10.2 | LDLR | Regulates LDLR and contributes to LDLC’s response to statin | [46] |
| RP5-833A20.1 | miR-382-5p, NFIA | RP5-833A20.1/miR-382-5p/NFIA pathway | [37] |
| DAPK-IT1 | miR-590-3p, LPL | DAPK1-IT1/hsa-miR-590-3p/LPL axis | [38] |
| Lnc-KDM5D-4 | Key processes related to fatty liver a | [51] | |
| NEAT1 | miR-342-3p | Regulates lipid uptake through modulating miR-342-3p | [39] |
| LeXis | RALY | Binds to RALY to express cholesterol synthetic genes | [42] |
| LncARSR | SREBP-2, HMGCR | Hepatic cholesterol biosynthesis via Akt/SREBP-2/HMGCR | [49] |
| Cholesterol efflux | |||
| DYN-LRB2-2 | TLR2, ABCA1 | Upregulates cholesterol efflux by ABCA1 expression | [52] |
| ENST00000602558.1 | ABCG1 | Regulates ABCG1 expression through binding to p65 | [53] |
| ZFAS1 | miR-654-3p, ADAM10, RAB22A | Elevates ADAM10/RAB22A expression to reduce cholesterol efflux in a miR-654-3p-dependent way | [54] |
| CDKN2B-AS1 | ADAM10 | Promotes cholesterol efflux by inhibiting ADAM10 | [55] |
| CHROME | miR-27b, 33a, 33b, 128 | Promotes cholesterol efflux and hepatic HDL biogenesis | [56] |
| MeXis | Abca1 | LXR-dependent transcription of Abca1 | [57] |
| AC096664.3 | ABCG1, PPAR-γ | Regulates ABCG1 expression via inhibiting PPAR-γ | [58] |
| MALAT1 | miR-17-5p, ABCA1 | MALAT1/miR-17-5p/ABCA1 axis | [59] |
| Disease | |||
| TUG1 | miR-92a, ApoM, ABCA1, ABCG1 | Regulates ApoM, ABCA1 and ABCG1 expression, and attenuates cholesterol efflux via the miR-92a/FXR1 axis | [60] |
| MEG3 | PTBP1 | Cholestasis by recruiting PTBP1 to destabilize Shp mRNA | [61] |
| LncAP001033.3-201 | ITGB3, TBXA2R | Associated with low HDL-C disease and could play a role in platelet activation in cardiovascular disease | [62] |
| LncAC068234.2-202 | |||
| Brip1os, Gm16586, AU020206,9430034N14Rik,2510016D11Rik, LNC000709, Gm15472, Gm20703, Dubr | Cdk4, Eif2ak2, Ccna2, Ccne1, Ccnd3, Mdm2, Nfkbia, Bax, Pkm, Pik3cb, Jak1, Lyn, Hdac9 | Potential therapeutic targets for atherosclerosis (AS) via an ox-low-density lipoprotein induced macrophage-derived foam cell model | [63] |
| ENST00000416361 | SREBP-1, SREBP-2 | Associated with CAD-induced lipid metabolism | [64] |
| ANRIL, MALAT1 | rs9632884 rs1537373, rs619586 rs3200401 | Genetic variation of the ANRIL rs9632884 and MALAT1 rs3200401 mediates lipid levels in MI patients | [65] |
| TUG1 | miR-133a, FGF1 | TUG1 modulates FGF1 via miR-133a | [66] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.-H.; Hwang, H.-J.; Cho, J.-Y. Long Non-Coding RNA Associated with Cholesterol Homeostasis and Its Involvement in Metabolic Diseases. Int. J. Mol. Sci. 2020, 21, 8337. https://doi.org/10.3390/ijms21218337
Lee K-H, Hwang H-J, Cho J-Y. Long Non-Coding RNA Associated with Cholesterol Homeostasis and Its Involvement in Metabolic Diseases. International Journal of Molecular Sciences. 2020; 21(21):8337. https://doi.org/10.3390/ijms21218337
Chicago/Turabian StyleLee, Kang-Hoon, Hyeon-Ji Hwang, and Je-Yoel Cho. 2020. "Long Non-Coding RNA Associated with Cholesterol Homeostasis and Its Involvement in Metabolic Diseases" International Journal of Molecular Sciences 21, no. 21: 8337. https://doi.org/10.3390/ijms21218337
APA StyleLee, K.-H., Hwang, H.-J., & Cho, J.-Y. (2020). Long Non-Coding RNA Associated with Cholesterol Homeostasis and Its Involvement in Metabolic Diseases. International Journal of Molecular Sciences, 21(21), 8337. https://doi.org/10.3390/ijms21218337





