Genome-Wide Identification of the Tify Gene Family and Their Expression Profiles in Response to Biotic and Abiotic Stresses in Tea Plants (Camellia sinensis)
Abstract
:1. Introduction
2. Results
2.1. Identification of the Members of the TIFY Gene Family in the Tea Plant
2.2. Phylogenetic Analysis of the TIFY Family
2.3. Sequence Analysis of the CsTIFY Family
2.4. Expression Profile of CsTIFY Genes in Different Organs
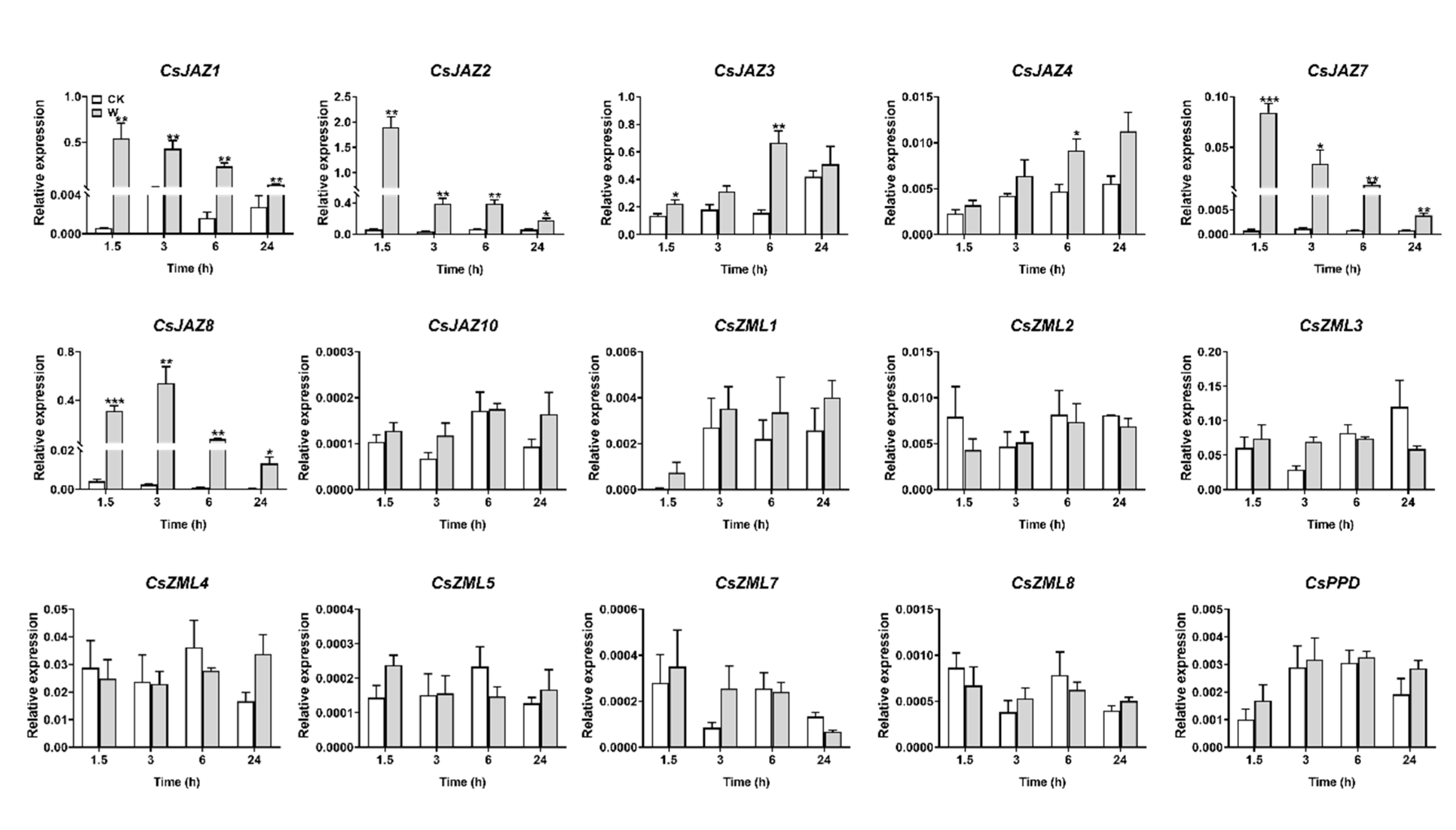
2.5. Expression Profiles of CsTIFY Genes under JA and Mechanical Wounding Treatment
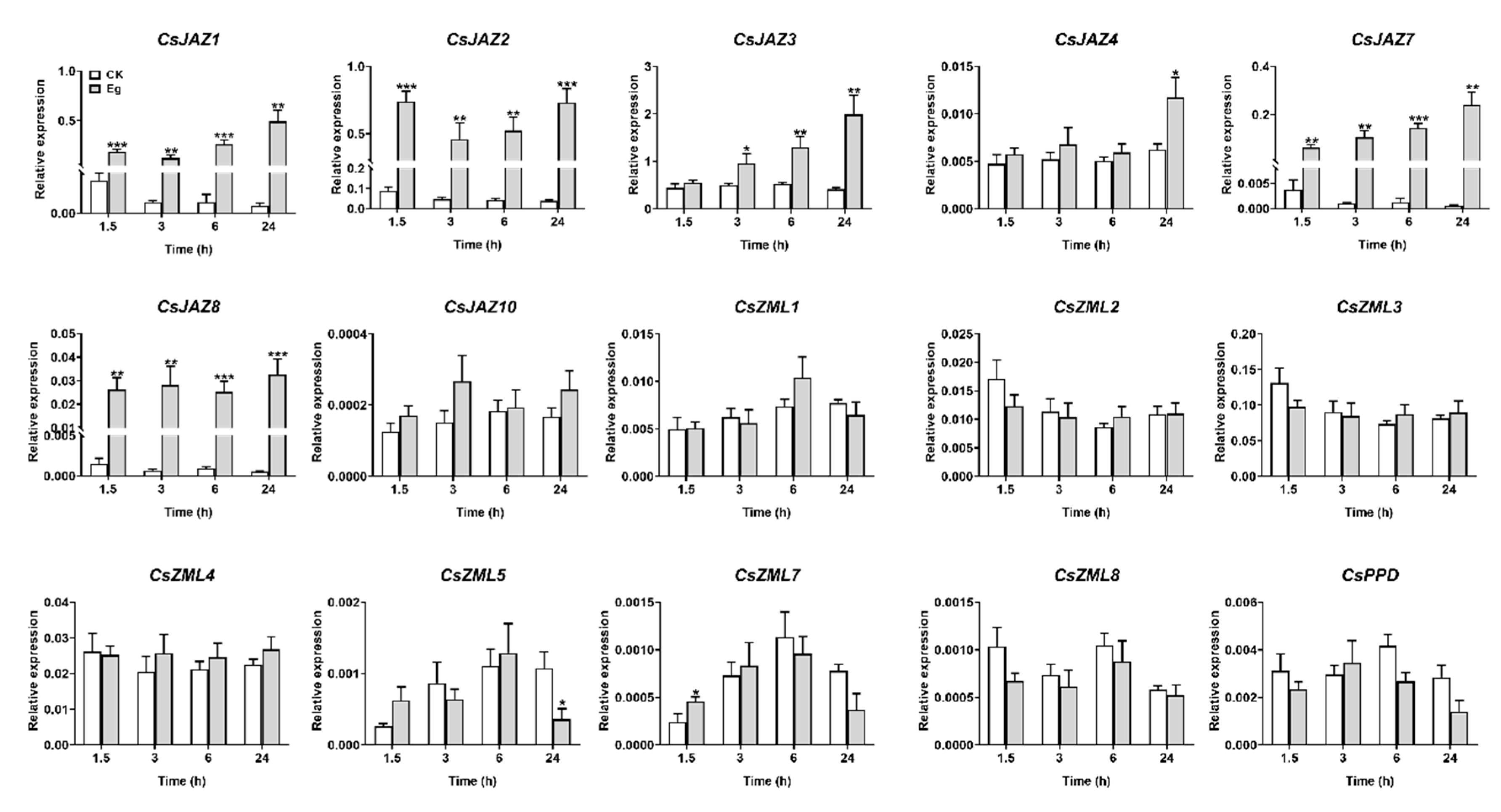
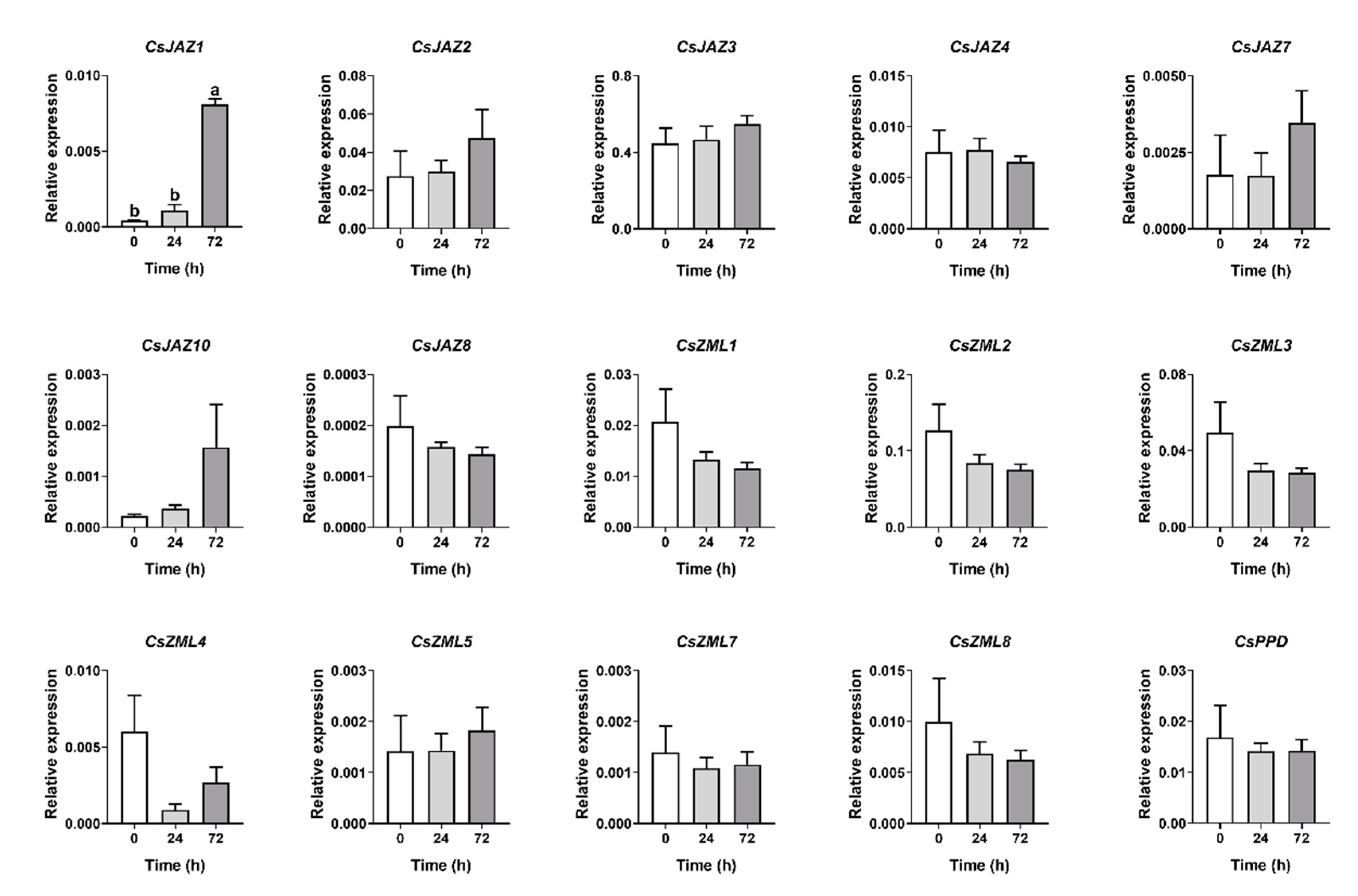
2.6. Expression Profiles of CsTIFY Genes in Response to Attack by the Tea Geometrid and Tea Anthracnose
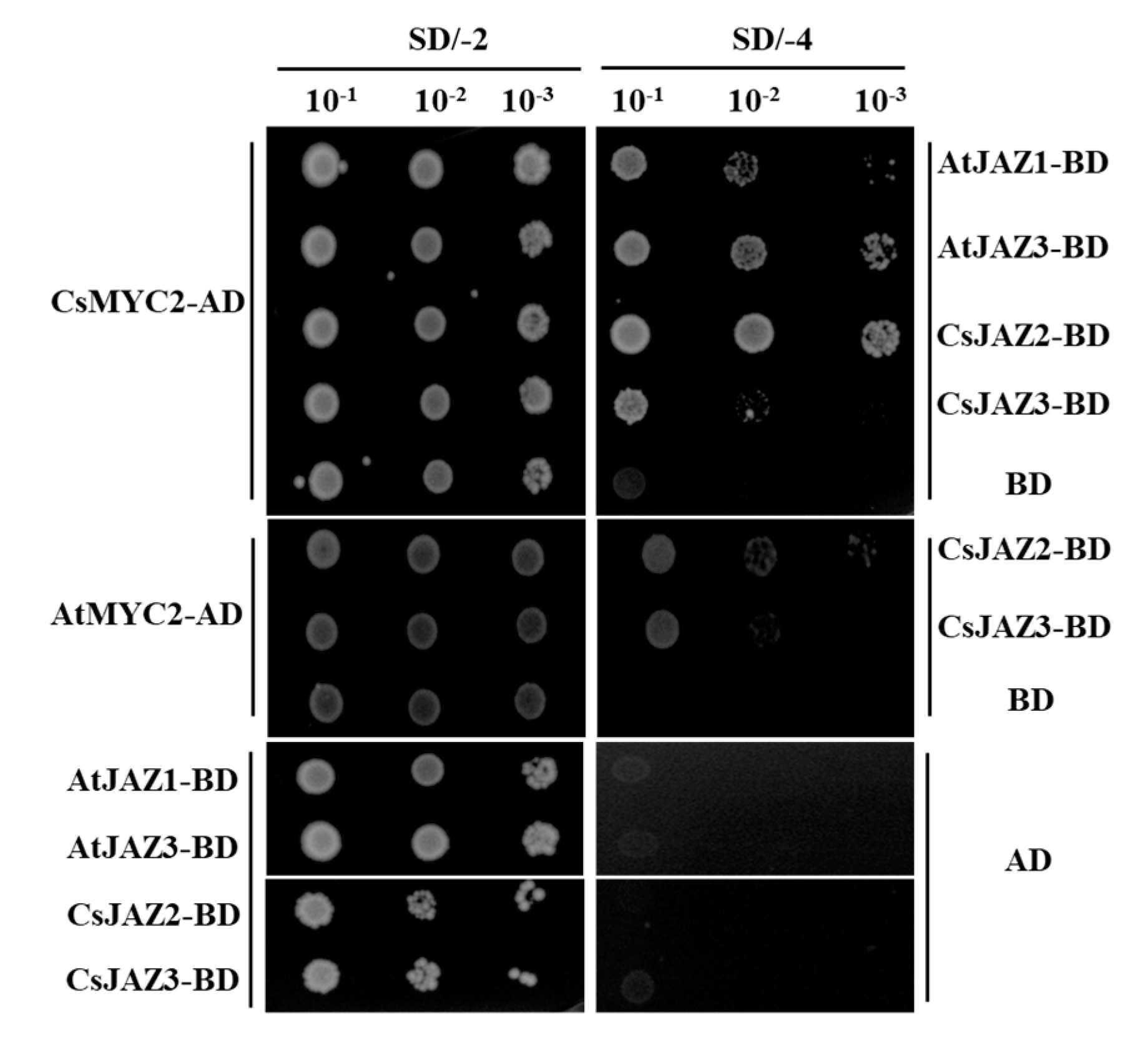
2.7. Interactions of CsJAZ with CsMYC2
3. Discussion
4. Materials and Methods
4.1. Insect and Fungi Pathogen
4.2. Plant Materials and Treatments
4.2.1. Different Organs
4.2.2. JA Treatment
4.2.3. Mechanical Wounding Treatment
4.2.4. E. obliqua Treatment
4.2.5. C. camelliae Infection
4.3. Identification of TIFY Family in Tea
4.4. Bioinformatics Analysis of TIFY Family in Tea
4.5. RNA Extraction and qRT-PCR Anlaysis
4.6. Yeast Two-Hybrid Assays
4.7. Data Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vanholme, B.; Grunewald, W.; Bateman, A.; Kohchi, T.; Gheysen, G. The tify family previously known as ZIM. Trends Plant Sci. 2007, 12, 239–244. [Google Scholar] [CrossRef]
- Shikata, M.; Matsuda, Y.; Ando, K.; Nishii, A.; Takemura, M.; Yokota, A.; Kohchi, T. Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 2004, 55, 631–639. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garcia-Casado, G.; Lopez Vidriero, I.; Lozano, F.M.; Ponce, M.R. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Yan, Y.; Stolz, S.; Chetelat, A.; Reymond, P.; Pagni, M.; Dubugnon, L.; Farmer, E. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 2007, 19, 2470–2483. [Google Scholar] [CrossRef] [Green Version]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Bai, Y.; Meng, Y.; Huang, D.; Qi, Y.; Chen, M. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 2011, 98, 128–136. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Gao, M.; Singer, S.D.; Fei, Z.; Wang, H.; Wang, X. Genome-wide identification and analysis of the TIFY gene family in grape. PLoS ONE 2012, 7, e44465. [Google Scholar] [CrossRef] [Green Version]
- Ebel, C.; BenFeki, A.; Hanin, M.; Solano, R.; Chini, A. Characterization of wheat (Triticum aestivum) TIFY family and role of Triticum Durum TdTIFY11a in salt stress tolerance. PLoS ONE 2018, 13, e0200566. [Google Scholar] [CrossRef] [Green Version]
- Chung, H.S.; Niu, Y.; Browse, J.; Howe, G. Top hits in contemporary JAZ: An update on jasmonate signaling. Phytochemistry 2009, 70, 1547–1559. [Google Scholar] [CrossRef] [Green Version]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef]
- Sun, J.Q.; Jiang, H.L.; Li, C.Y. Systemin/Jasmonate-mediated systemic defense signaling in tomato. Mol. Plant 2011, 4, 607–615. [Google Scholar] [CrossRef]
- Zhu, D.; Bai, X.; Luo, X.; Chen, Q.; Cai, H.; Ji, W.; Zhu, Y. Identification of wild soybean (Glycine soja) TIFY family genes and their expression profiling analysis under bicarbonate stress. Plant Cell Rep. 2013, 32, 263–272. [Google Scholar] [CrossRef]
- Zhang, L.; You, J.; Chan, Z. Identification and characterization of TIFY family genes in Brachypodium distachyon. J. Plant Res. 2015, 128, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- White, D.W.R. PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 13238–13243. [Google Scholar] [CrossRef] [Green Version]
- Velez-Bermudez, I.C.; Salazar-Henao, J.E.; Fornale, S.; Lopez-Vidriero, I.; Franco-Zorrilla, J.M.; Grotewold, E.; Gray, J.; Solano, R.; Schmidt, W.; Pages, M. A MYB/ZML complex regulates wound-induced lignin genes in maize. Plant Cell 2015, 27, 3245–3259. [Google Scholar] [CrossRef] [Green Version]
- Ge, L.; Yu, J.; Wang, H.; Luth, D.; Bai, G.; Wang, K.; Chen, R. Increasing seed size and quality by manipulating BIG SEEDS1 in legume species. Proc. Natl. Acad. Sci. USA 2016, 113, 12414–12419. [Google Scholar] [CrossRef] [Green Version]
- Cuellar Perez, A.; Nagels Durand, A.; Vanden Bossche, R.; De Clercq, R.; Persiau, G.; Van Wees, S.C.; Pieterse, C.M.; Gevaert, K.; Jaeger, G.D.; Goossens, A. The non-JAZ TIFY protein TIFY8 from Arabidopsis thaliana is a transcriptional repressor. PLoS ONE 2014, 9, e84891. [Google Scholar] [CrossRef] [Green Version]
- Santner, A.; Estelle, M. The JAZ proteins link jasmonate perception with transcriptional changes. Plant Cell 2007, 19, 3839–3842. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.; Major, I.T.; Howe, G.A. Resolution of growth-defense conflict: Mechanistic insights from jasmonate signaling. Curr. Opin. Plant Biol. 2018, 44, 72–81. [Google Scholar] [CrossRef]
- Howe, G.A.; Yoshida, Y. Evolutionary origin of JAZ proteins and jasmonate signaling. Mol. Plant 2019, 12, 153–155. [Google Scholar] [CrossRef] [Green Version]
- Monte, I.; Franco-Zorrilla, J.M.; Garcia-Casado, G.; Zamarreno, A.M.; Garcia-Mina, J.M.; Nishihama, R.; Kohchi, T.; Solano, R. A Single JAZ repressor controls the jasmonate pathway in Marchantia polymorpha. Mol. Plant 2019, 12, 185–198. [Google Scholar] [CrossRef] [Green Version]
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant Biol. 2018, 69, 387–415. [Google Scholar] [CrossRef] [Green Version]
- Ray, R.; Li, D.P.; Rayko, H.; Ian, T.B. Using natural variation to achieve a whole-plant functional understanding of the responses mediated by jasmonate signaling. Plant J. 2019, 99, 414–425. [Google Scholar] [CrossRef]
- Guo, Q.; Yoshida, Y.; Major, I.T.; Wang, K.; Sugimoto, K.; Kapali, G.; Havko, N.E.; Benninga, C.; Howe, G.A. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 10768–10777. [Google Scholar] [CrossRef] [Green Version]
- Oh, Y.; Baldwin, I.T.; Galis, I. NaJAZh regulates a subset of defense responses against herbivores and spontaneous leaf necrosis in Nicotiana attenuata plants. Plant Physiol. 2012, 159, 769–788. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Ye, H.; Yao, R.; Zhang, T.; Xiong, L. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015, 232, 1–12. [Google Scholar] [CrossRef]
- Lu, Q.; Wang, Y.; Li, N.; Ni, D.; Yang, Y.; Wang, X. Differences in the characteristics and pathogenicity of Colletotrichum camelliae and C. fructicola isolated from the tea plant [Camellia sinensis (L.) O. Kuntze]. Front. Microbiol. 2018, 9, 3060. [Google Scholar] [CrossRef]
- Lin, S.; Dong, Y.; Li, X.; Xing, Y.; Liu, M.; Sun, X. JA-Ile-macrolactone 5b induces tea plant (Camellia sinensis) resistance to both herbivore Ectropis obliqua and pathogen Colletotrichum camelliae. Int. J. Mol. Sci. 2020, 21, 1828. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhang, X.; Ye, M.; Li, X.W.; Lin, S.B.; Sun, X.L. The jasmonic acid pathway positively regulates the polyphenol oxidase-based defense against tea geometrid caterpillars in the tea plant (Camellia sinensis). J. Chem. Ecol. 2020, 46, 308–316. [Google Scholar] [CrossRef]
- Wang, Y.N.; Tang, L.; Hou, Y.; Wang, P.; Yang, H.; Wei, C.L. Differential transcriptome analysis of leaves of tea plant (Camellia sinensis) provides comprehensive insights into the defense responses to Ectropis oblique attack using RNA-Seq. Funct. Integr. Genom. 2016, 16, 383–398. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, X.; Wang, P.; Sun, Y.; Yue, C.; Ye, N. Genome-wide and expression pattern analysis of JAZ family involved in stress responses and postharvest processing treatments in Camellia sinensis. Sci. Rep. 2020, 10, 2792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, E.; Tong, W.; Hou, Y.; An, Y.; Chen, L.; Wu, Q.; Wan, X. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol. Plant 2020, 13, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Song, Y.; Wang, C.; Butt, H.I.; Wang, Q.; Zhang, C.; Yang, Z.; Liu, Z.; Chen, E.; Zhang, X. Genome-wide identification and functional analysis of the TIFY gene family in response to drought in cotton. Mol. Genet. Genom. 2016, 291, 2173–2187. [Google Scholar] [CrossRef] [Green Version]
- Chini, A.; Boter, M.; Solano, R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009, 276, 4682–4692. [Google Scholar] [CrossRef]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008, 55, 979–988. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Calvo, P.; Chini, A.; Fernandez-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Solano, R. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef] [Green Version]
- Chung, H.S.; Koo, A.J.; Gao, X.; Jayanty, S.; Thines, B.; Jones, A.D.; Howe, G.A. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008, 146, 952–964. [Google Scholar] [CrossRef] [Green Version]
- Moffat, C.S.; Caroline, S.M.; Robert, A.I.; Deepthi, L.W.; Nigel, J.S.; Heather, K.; Marc, R.K. ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 2012, 7, e35995. [Google Scholar] [CrossRef] [Green Version]
- Demianski, A.J.; Chung, K.M.; Kunkel, B.N. Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol. Plant Pathol. 2012, 13, 46–57. [Google Scholar] [CrossRef]

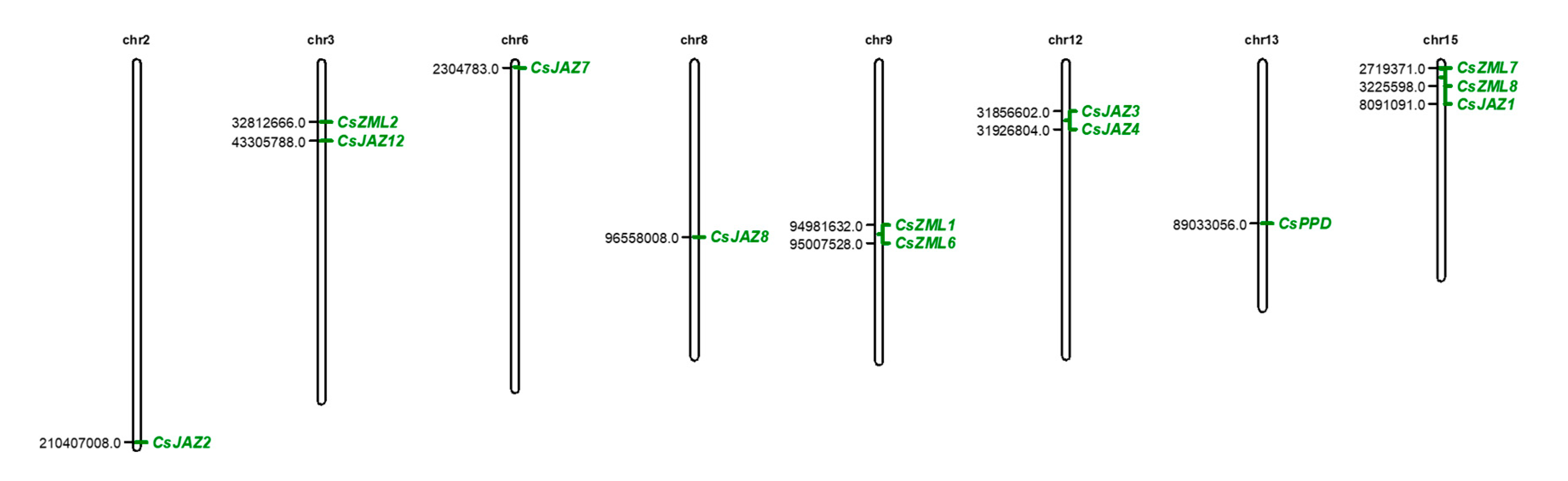
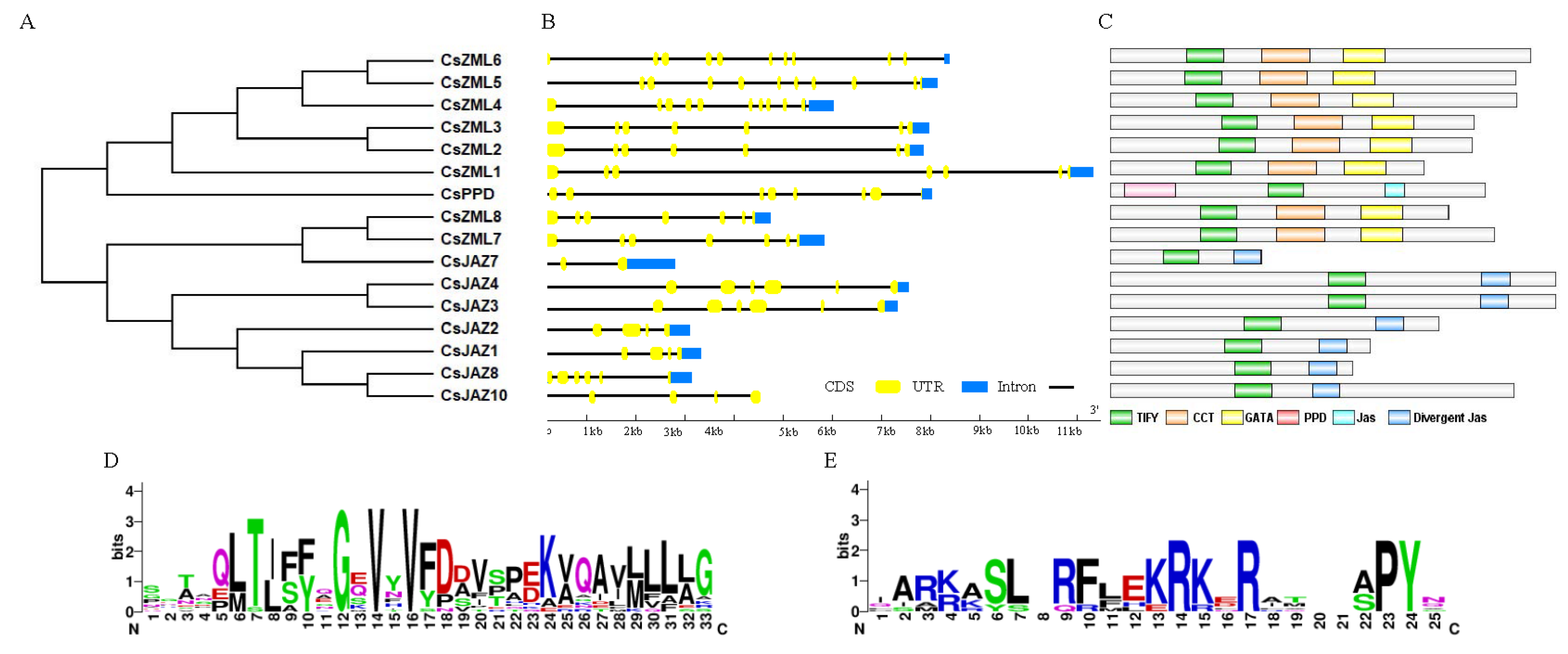
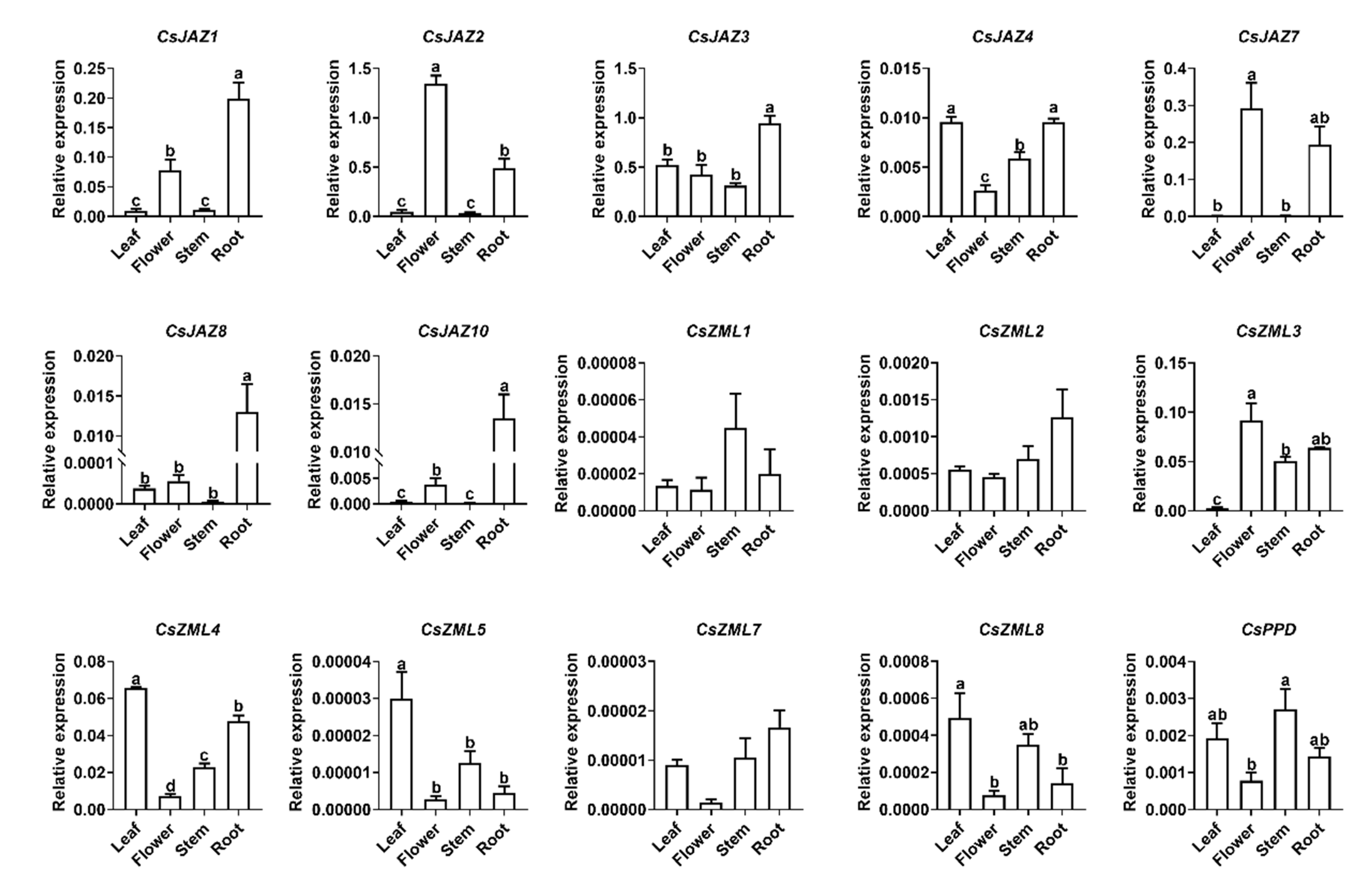
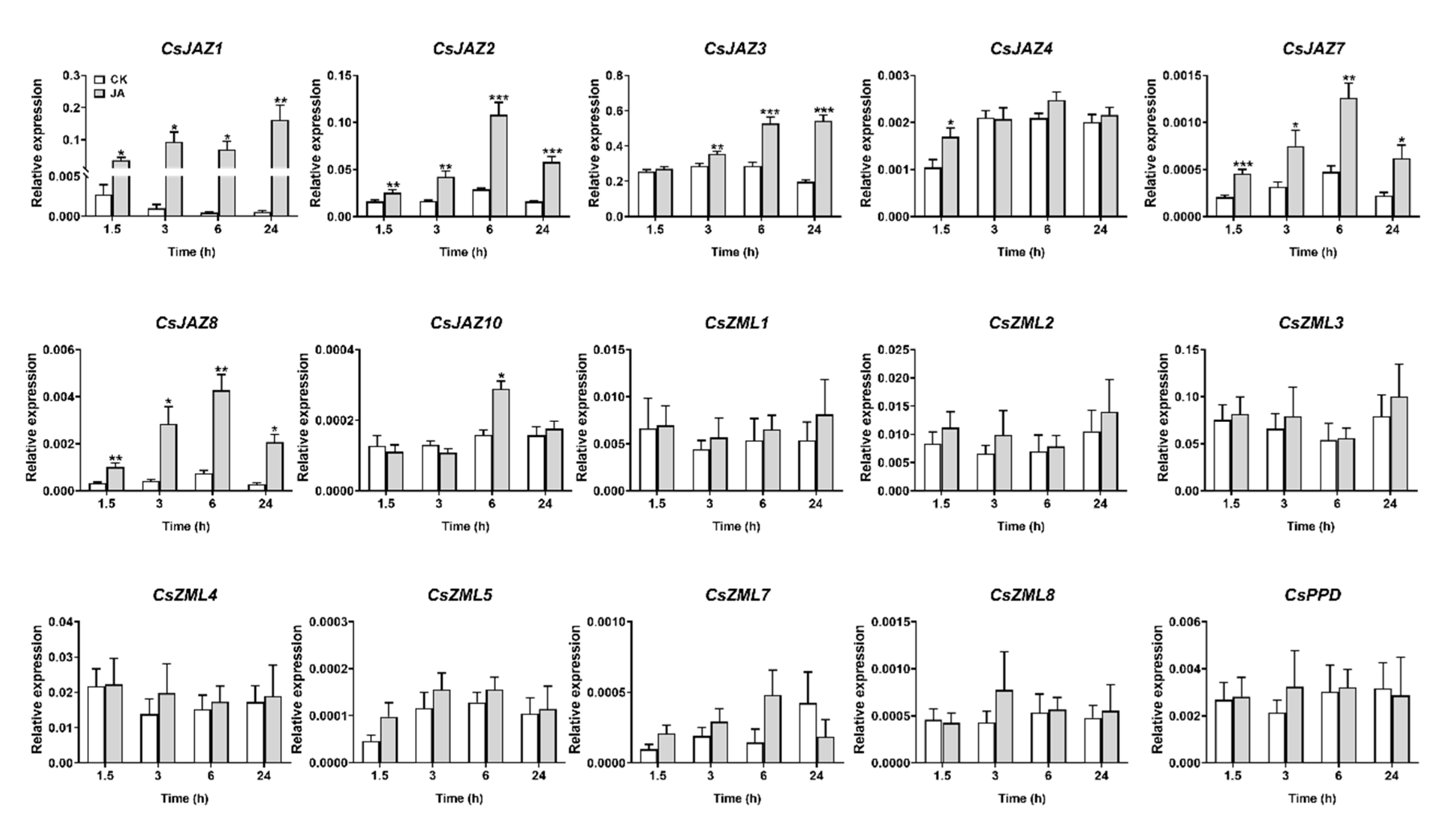
| Gene ID | Gene Name | CDS Length (bp) | Protein Length (aa) | TIFY Motif |
|---|---|---|---|---|
| CSS0010510 | CsJAZ1 | 702 | 233 | TIFYGG |
| CSS0031905 | CsJAZ2 | 885 | 294 | TIFYAG |
| CSS0022053 | CsJAZ3 | 1197 | 398 | TIFYAG |
| CSS0012514 | CsJAZ4 | 1197 | 398 | TIFYAG |
| CSS0028188 | CsJAZ7 | 411 | 136 | TIFYNG |
| CSS0034430 | CsJAZ8 | 654 | 217 | TIFYNG |
| CSS0011019 | CsJAZ10 | 786 | 361 | TIFYGG |
| CSS0031620 | CsPPD | 1008 | 335 | TIFYCG |
| CSS0050449 | CsZML7 | 912 | 303 | TLSFQG |
| CSS0037100 | CsZML8 | 912 | 303 | TLSFQG |
| CSS0003047 | CsZML1 | 846 | 281 | TLSFRG |
| CSS0017505 | CsZML2 | 972 | 323 | TLSFQG |
| CSS0004261 | CsZML3 | 978 | 325 | TLSFQG |
| CSS0050427 | CsZML4 | 1092 | 363 | TLAFEG |
| CSS0045061 | CsZML5 | 1089 | 362 | TSELSI |
| CSS0016802 | CsZML6 | 1131 | 376 | TLAFEG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Ran, W.; Zhang, J.; Ye, M.; Lin, S.; Li, X.; Sultana, R.; Sun, X. Genome-Wide Identification of the Tify Gene Family and Their Expression Profiles in Response to Biotic and Abiotic Stresses in Tea Plants (Camellia sinensis). Int. J. Mol. Sci. 2020, 21, 8316. https://doi.org/10.3390/ijms21218316
Zhang X, Ran W, Zhang J, Ye M, Lin S, Li X, Sultana R, Sun X. Genome-Wide Identification of the Tify Gene Family and Their Expression Profiles in Response to Biotic and Abiotic Stresses in Tea Plants (Camellia sinensis). International Journal of Molecular Sciences. 2020; 21(21):8316. https://doi.org/10.3390/ijms21218316
Chicago/Turabian StyleZhang, Xin, Wei Ran, Jin Zhang, Meng Ye, Songbo Lin, Xiwang Li, Riffat Sultana, and Xiaoling Sun. 2020. "Genome-Wide Identification of the Tify Gene Family and Their Expression Profiles in Response to Biotic and Abiotic Stresses in Tea Plants (Camellia sinensis)" International Journal of Molecular Sciences 21, no. 21: 8316. https://doi.org/10.3390/ijms21218316
APA StyleZhang, X., Ran, W., Zhang, J., Ye, M., Lin, S., Li, X., Sultana, R., & Sun, X. (2020). Genome-Wide Identification of the Tify Gene Family and Their Expression Profiles in Response to Biotic and Abiotic Stresses in Tea Plants (Camellia sinensis). International Journal of Molecular Sciences, 21(21), 8316. https://doi.org/10.3390/ijms21218316






