TaDrAp1 and TaDrAp2, Partner Genes of a Transcription Repressor, Coordinate Plant Development and Drought Tolerance in Spelt and Bread Wheat
Abstract
:1. Introduction
2. Results
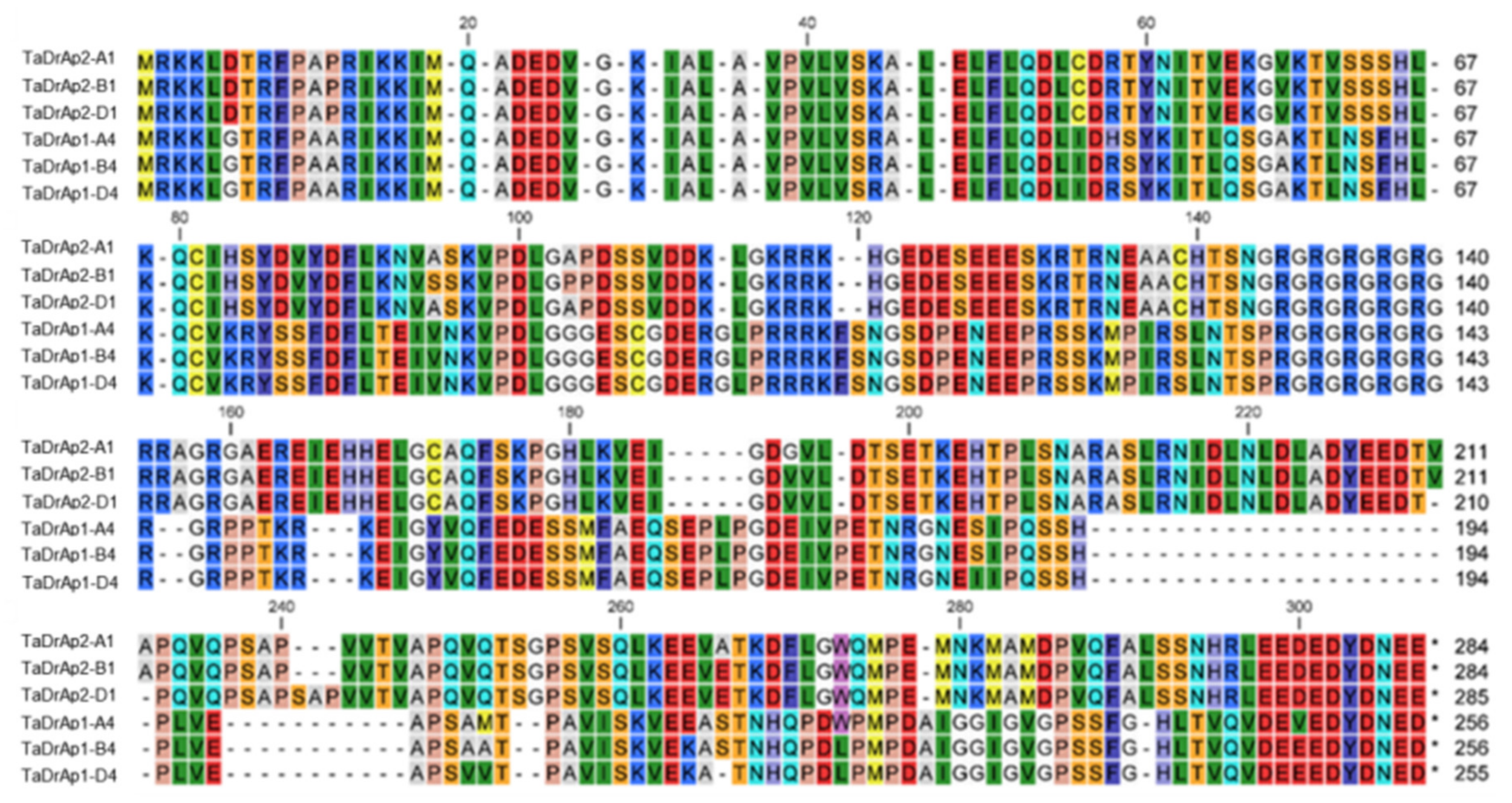
2.1. Identification of Homeologs TaDrAp1 and TaDrAp2 Genes in Bread Wheat cv. Chinese Spring Using Public Databases
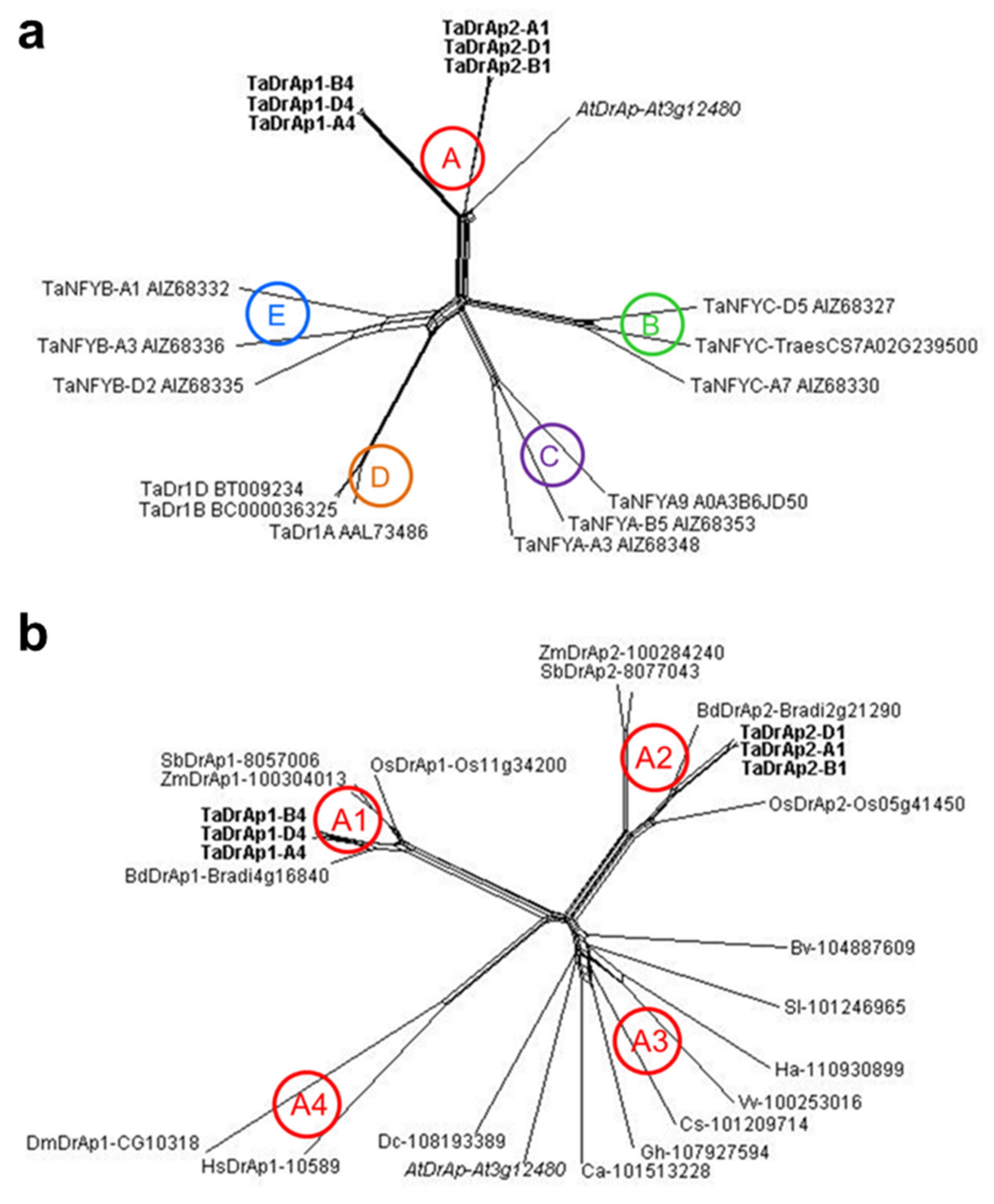
2.2. Molecular-Phylogenetic Analysis of Homeologs TaDrAp1 and TaDrAp2 in Bread Wheat Compared to Other Plant Species and References
2.3. Expression Analysis of TaDrAp1 and TaDrAp2 Genes in Eight Cultivars of Bread Wheat
2.4. Microarray 40K SNP Analysis of Parents and Selected Breeding Lines from Interspecies Hybrid Cross No. 18-6 between Spelt and Bread Wheat
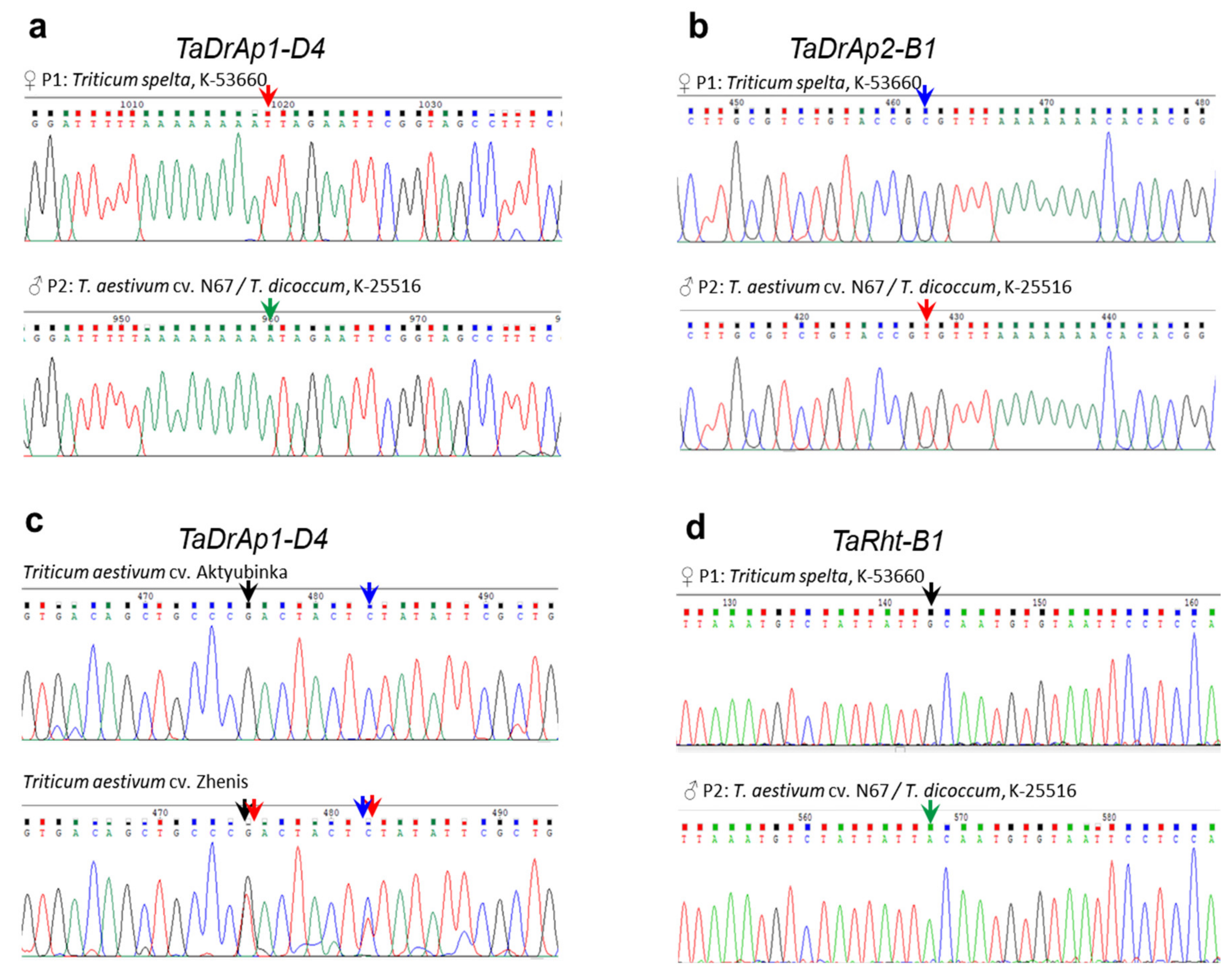
2.5. Genetic Polymorphism and SNP Identification in the Segregating Population No. 18-6 from an Interspecies Cross between Spelt and Bread Wheat
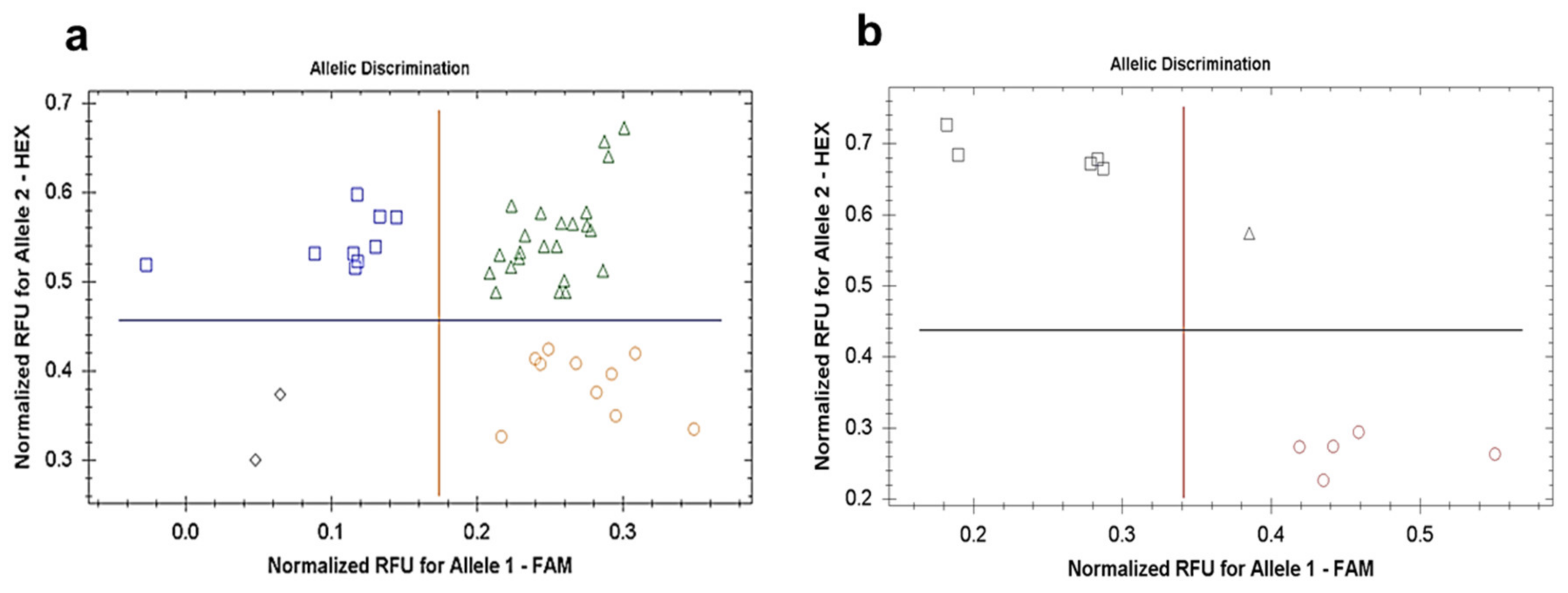
2.6. Amplifluor-Like SNP Genotyping of TaDrAp1 and TaDrAp2 Genes in Segregating Population No. 18-6
3. Discussion
3.1. DrAp1/2 Genes Encode Down-Regulator Associated Proteins in Plants
3.2. TaDrAp1 and TaDrAp2 Genes in Wheat
3.3. Expression Analysis of Dr1 and DrAp1 Genes in Monocot Species in Comparison to Bread Wheat during Plant Ontogenesis, Development, and under Drought
3.4. Genetic Polymorphism in TaDrAp1, TaDrAp2, and TaRht-B1 Genes in Spelt and Bread Wheat using 40K SNP Microarray and Amplifluor-Like SNP Genotyping
3.5. Interaction of TaDrAp1 and TaDrAp2 with Genes Controlling Plant Development, Tolerance to Drought, and Yield Components in Spelt and Bread Wheat
4. Materials and Methods
4.1. Bioinformatic Analysis
4.2. Plant Material
4.3. Drought Experiments in the Field Trial
4.4. DNA Extraction and 40K SNP Microarray
4.5. DNA Extraction and Amplifluor-Like SNP Genotyping
4.6. RNA Extraction, cDNA Synthesis, and qPCR Analysis
4.7. Statistical Treatment
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CBF | CCAAT-binding factor |
| CDS | Coding sequence |
| Dr1 | Down-regulator |
| DrAp1 | Down-regulator associated protein |
| GWAS | Genome-wide association study |
| IWGSC | International wheat genome sequencing consortium |
| NC2 | Negative cofactor 2 |
| NF-Y | Nuclear factor Y |
| NIL | Near-isogenic lines |
| ORF | Open reading frame |
| RFU | Relative fluorescence units |
| SNP | Single nucleotide polymorphism |
| TBP | TATA-box-binding protein |
| TF | Transcription factor |
| TFIID | Transcription factor IID |
References
- Laloum, T.; De Mita, S.; Gamas, P.; Baudin, M.; Niebel, A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013, 18, 157–166. [Google Scholar] [CrossRef]
- Maldonado, E.; Hampsey, M.; Reinberg, D. Repression: Targeting the heart of the matter. Cell 1999, 99, 455–458. [Google Scholar] [CrossRef] [Green Version]
- Meisterernst, M.; Roeder, R.G. Family of proteins that interact with TFIID and regulate promoter activity. Cell 1991, 67, 557–567. [Google Scholar] [CrossRef]
- Bhuiyan, T.; Timmers, H.T.M. Promoter recognition: Putting TFIID on the spot. Trends Cell Biol. 2019, 29, 752–763. [Google Scholar] [CrossRef] [PubMed]
- Inostroza, J.A.; Mermelstein, F.H.; Ha, I.; Lane, W.S.; Reinberg, D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell 1992, 70, 477–489. [Google Scholar] [CrossRef]
- Kim, T.K.; Zhao, Y.; Ge, H.; Bernstein, R.; Roeder, R.G. TATA-binding protein residues implicated in a functional interplay between Negative Cofactor NC2 (Dr1) and general factors TFIIA and TFIIB. J. Biol. Chem. 1995, 270, 10976–10981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Creton, S.; Svejstrup, J.Q.; Collart, M.A. The NC2 α and β subunits play different roles in vivo. Genes Dev. 2002, 16, 3265–3276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilfillan, S.; Stelzer, G.; Piaia, E.; Hofmann, M.G.; Meisterernst, M. Efficient binding of NC2·TATA-binding protein to DNA in the absence of TATA. J. Biol. Chem. 2005, 280, 6222–6230. [Google Scholar] [CrossRef] [Green Version]
- Meisterernst, M.; Roy, A.L.; Lieu, H.M.; Roeder, R.G. Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell 1991, 66, 981–993. [Google Scholar] [CrossRef]
- Kuromori, T.; Yamamoto, M. Cloning of cDNAs from Arabidopsis thaliana that encode putative protein phosphatase 2C and a human Dr1-like protein by transformation of a fission yeast mutant. Nucleic Acids Res. 1994, 22, 5296–5301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romier, C.; Cocchiarella, F.; Mantovani, R.; Moras, D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003, 278, 1336–1345. [Google Scholar] [CrossRef] [Green Version]
- Mermelstein, F.; Yeung, K.; Cao, J.; Inostroza, J.A.; Erdjument-Bromage, H.; Eagelson, K.; Landsman, G.; Levitt, P.; Tempst, P.; Reinberg, D. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 1996, 10, 1033–1048. [Google Scholar] [CrossRef] [Green Version]
- Yeung, K.; Kim, S.; Reinberg, A. Functional dissection of a human Dr1-DrAp1 repressor complex. Mol. Cell. Biol. 1997, 17, 36–45. [Google Scholar] [CrossRef] [Green Version]
- Sinha, S.; Kim, I.S.; Sohn, K.Y.; de Crombrugghe, B.; Maity, S.N. Three classes of mutations in the A subunit of the CCAAT binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol. Cell. Biol. 1996, 16, 328–337. [Google Scholar] [CrossRef] [Green Version]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F., III; Mantovania, R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar] [CrossRef] [Green Version]
- Stephenson, T.J.; McIntyre, C.L.; Collet, C.; Xue, G.P. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol. Biol. 2007, 65, 77–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zotova, L.; Kurishbayev, A.; Jatayev, S.; Goncharov, N.P.; Shamambayeva, N.; Kashapov, A.; Nuralov, A.; Otemissova, A.; Sereda, S.; Shvidchenko, V.; et al. The General transcription repressor TaDr1 is co-expressed with TaVrn1 and TaFT1 in bread wheat under drought. Front. Genet. 2019, 10, 63. [Google Scholar] [CrossRef] [Green Version]
- Prelich, G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2a homolog that has both positive and negative roles in transcription in vivo. Mol. Cell. Biol. 1997, 17, 2057–2065. [Google Scholar] [CrossRef] [Green Version]
- Knuesting, J.; Riondet, C.; Maria, C.; Kruse, I.; Bécuwe, N.; König, N.; Berndt, C.; Tourrette, S.; Guilleminot-Montoya, J.; Herrero, E.; et al. Arabidopsis glutaredoxin S17 and its partner, the nuclear factor Y subunit C11/negative cofactor 2α, contribute to maintenance of the shoot apical meristem under long-day photoperiod. Plant Physiol. 2015, 167, 1643–1658. [Google Scholar] [CrossRef] [Green Version]
- Thirumurugan, T.; Ito, Y.; Kubo, T.; Serizawa, A.; Kurata, N. Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genom. 2008, 279, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wang, J.; Cai, M.; Zhang, H.; Wu, F.; Xu, Y.; Li, C.; Cheng, Z.; Zhang, X.; Guo, X.; et al. The OsHAPL1-DTH8-Hd1 complex functions as the transcription regulator to repress heading date in rice. J. Exp. Bot. 2017, 68, 553–568. [Google Scholar] [CrossRef] [Green Version]
- Qu, B.; He, X.; Wang, J.; Zhao, Y.; Teng, W.; Shao, A.; Zhao, X.; Ma, W.; Wang, J.; Li, B.; et al. A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol. 2015, 167, 411–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peiró-Chova, L.; Estruch, F. The yeast RNA polymerase II-associated factor Iwr1p is involved in the basal and regulated transcription of specific genes. J. Biol. Chem. 2009, 284, 28958–28967. [Google Scholar] [CrossRef] [Green Version]
- Willy, P.J.; Kobayashi, R.; Kadonaga, J.T. A basal transcription factor that activates or represses transcription. Science 2000, 290, 982–984. [Google Scholar] [CrossRef]
- Song, W.; Solimeo, H.; Rupert, R.A.; Yadav, N.S.; Zhu, Q. Functional dissection of a rice Dr1/DrAp1 transcriptional repression complex. Plant Cell 2002, 14, 181–195. [Google Scholar] [CrossRef]
- Cao, S.; Kumimoto, R.W.; Siriwardana, C.L.; Risinger, J.R.; Holt III, B.F. Identification and characterization of NF-Y transcription factor families in the monocot model plant Brachypodium distachyon. PLoS ONE 2011, 6, e21805. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Na, J.G.; Hampsey, M.; Reinberg, D. The Dr1/DRAP1 heterodimer is a global repressor of transcription in vivo. Proc. Natl. Acad. Sci. USA 1997, 94, 820–825. [Google Scholar] [CrossRef] [Green Version]
- Distelfeld, A.; Li, C.; Dubcovsky, J. Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 2009, 12, 178–184. [Google Scholar] [CrossRef] [Green Version]
- Preston, J.C.; Fjellheim, S. Understanding past, and predicting future, niche transitions based on grass flowering time variation. Plant Physiol. 2020, 183, 822–839. [Google Scholar] [CrossRef]
- Li, C.; Distelfeld, A.; Comis, A.; Dubcovsky, J. Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J. 2011, 67, 763–773. [Google Scholar] [CrossRef] [Green Version]
- Yan, W.H.; Wang, P.; Chen, H.X.; Zhou, H.J.; Li, Q.P.; Wang, C.R.; Ding, Z.H.; Zhang, Y.S.; Yu, S.B.; Xing, Y.Z.; et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 2011, 4, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop breeding chips and genotyping platforms: Progress, challenges, and perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef] [Green Version]
- Morgil, H.; Gercek, Y.C.; Tulum, I. Single nucleotide polymorphisms (SNPs) in plant genetics and breeding. In The Recent Topics in Genetic Polymorphisms; InTech Open: London, UK, 2020. [Google Scholar] [CrossRef] [Green Version]
- Burridge, A.J.; Wilkinson, P.A.; Winfield, M.O.; Barker, G.L.A.; Allen, A.M.; Coghill, J.A.; Waterfall, C.; Edwards, K.J. Conversion of array-based single nucleotide polymorphic markers for use in targeted genotyping by sequencing in hexaploid wheat (Triticum aestivum). Plant Biotechnol. J. 2018, 16, 867–876. [Google Scholar] [CrossRef] [Green Version]
- Rimbert, H.; Darrier, B.; Navarro, J.; Kitt, J.; Choulet, F.; Leveugle, M.; Duarte, J.; Rivière, N.; Eversole, K.; Le Gouis, J.; et al. High throughput SNP discovery and genotyping in hexaploid wheat. PLoS ONE 2018, 13, e0186329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shokat, S.; Sehgal, D.; Vikram, P.; Liu, F.; Singh, S. Molecular markers associated with agro-physiological traits under terminal drought conditions in bread wheat. Int. J. Mol. Sci. 2020, 21, 3156. [Google Scholar] [CrossRef]
- Afzal, F.; Li, H.; Gul, A.; Subhani, A.; Ali, A.; Mujeeb-Kazi, A.; Ogbonnaya, F.; Trethowan, R.; Xia, X.; He, Z.; et al. Genome-wide analyses reveal footprints of divergent selection and drought adaptive traits in synthetic-derived wheats. Genes Genomes Genet. 2019, 9, 1957–1973. [Google Scholar] [CrossRef] [Green Version]
- Shavrukov, Y.; Suchecki, R.; Eliby, S.; Abugalieva, A.; Kenebayev, S.; Langridge, P. Application of Next-generation sequencing technology to study genetic diversity and identify unique SNP markers in bread wheat from Kazakhstan. BMC Plant Biol. 2014, 14, 258. [Google Scholar] [CrossRef] [Green Version]
- Würschum, T.; Leiser, W.L.; Longin, C. Molecular genetic characterization and association mapping in spelt wheat. Plant Breed. 2017, 36, 214–223. [Google Scholar] [CrossRef]
- Müller, T.; Schierscher-Viret, B.; Fossati, D.; Brabant, C.; Schori, A.; Keller, B.; Krattinger, S.G. Unlocking the diversity of genebanks: Whole-genome marker analysis of Swiss bread wheat and spelt. Theor. Appl. Genet. 2018, 131, 407–416. [Google Scholar] [CrossRef] [Green Version]
- Schramm, C.; Shavrukov, Y.; Anderson, P.; Kurishbaev, A.; Jatayev, S. Development of Single nucleotide polymorphism (SNP) markers for cereal breeding and crop research: Current methods and future prospects. In Advances in Breeding Techniques for Cereal Crops; Ordon, F., Friedt, W., Eds.; BD Publishing: Cambridge, UK, 2019. [Google Scholar] [CrossRef]
- He, C.; Holme, J.; Anthony, J. SNP genotyping: The KASP assay. In Crop Breeding: Methods and Protocols, Methods in Molecular Biology; Fleury, D., Whitford, R., Eds.; Springer: New York, NY, USA, 2014; Volume 1145, pp. 75–86. [Google Scholar]
- Kaur, B.; Mavi, G.S.; Gill, M.S.; Saini, D.K. Utilization of KASP technology for wheat improvement. Cereal Res. Comm. 2020, 48. [Google Scholar] [CrossRef]
- Long, Y.M.; Chao, W.S.; Ma, G.J.; Xu, S.S.; Qi, L.L. An innovative SNP genotyping method adapting to multiple platforms and throughputs. Theor. Appl. Genet. 2017, 130, 597–607. [Google Scholar] [CrossRef]
- Wu, Y.; Li, M.; He, Z.; Dreisigacker, S.; Wen, W.; Jin, H.; Zhai, S.; Li, F.; Gao, F.; Liu, J.; et al. Development and validation of high-throughput and low-cost STARP assays for genes underpinning economically important traits in wheat. Theor. Appl. Genet. 2020, 133, 2431–2450. [Google Scholar] [CrossRef] [PubMed]
- Shavrukov, Y.; Zhumalin, A.; Serikbay, D.; Botayeva, M.; Otemisova, A.; Absattarova, A.; Sereda, G.; Sereda, S.; Shvidchenko, V.; Turbekova, A.; et al. Expression level of the DREB2-type gene, identified with Amplifluor SNP markers, correlates with performance, and tolerance to dehydration in bread wheat cultivars from Northern Kazakhstan. Front. Plant Sci. 2016, 7, 1736. [Google Scholar] [CrossRef] [Green Version]
- Jatayev, S.; Kurishbaev, A.; Zotova, L.; Khassanova, G.; Serikbay, D.; Zhubatkanov, A.; Botayeva, M.; Zhumalin, A.; Turbekova, A.; Soole, K.; et al. Advantages of Amplifluor-like SNP markers over KASP in plant genotyping. BMC Plant Biol. 2017, 17, 254. [Google Scholar] [CrossRef]
- Zotova, L.; Kurishbayev, A.; Jatayev, S.; Khassanova, G.; Zhubatkanov, A.; Serikbay, D.; Sereda, S.; Sereda, T.; Shvidchenko, V.; Lopato, S.; et al. Genes encoding transcription factors TaDREB5 and TaNFYC-A7 are differentially expressed in leaves of bread wheat in response to drought, dehydration and ABA. Front. Plant Sci. 2018, 9, 1441. [Google Scholar] [CrossRef]
- Goncharov, N.P. Genus Triticum L. taxonomy: The present and the future. Plant Syst. Evol. 2011, 295, 1–11. [Google Scholar] [CrossRef]
- Haas, M.; Schreiber, M.; Mascher, M. Domestication and crop evolution of wheat and barley: Genes, genomics, and future directions. J. Integr. Plant Biol. 2019, 61, 204–225. [Google Scholar] [CrossRef] [Green Version]
- Longin, C.F.H.; Würschum, T. Back to the Future–Tapping into ancient grains for food diversity. Trends Plant Sci. 2016, 21, 731–737. [Google Scholar] [CrossRef]
- Wendin, K.; Mustafa, A.; Ortman, T.; Gerhardt, K. Consumer awareness, attitudes and preferences towards heritage cereals. Foods 2020, 9, 742. [Google Scholar] [CrossRef]
- Arzani, A.; Ashraf, M. Cultivated ancient wheats (Triticum spp.): A potential source of health-beneficial food products. Compr. Rev. Food Sci. 2017, 16, 477–488. [Google Scholar] [CrossRef] [Green Version]
- Bojňanská, T.; Frančáková, H. The use of spelt wheat (Triticum spelta L.) for baking applications. Rost. Vyroba 2002, 48, 141–147. [Google Scholar] [CrossRef]
- Boukid, F.; Folloni, S.; Sforza, S.; Vittadini, E.; Prandi, B. Current trends in ancient grains-based foodstuffs: Insights into nutritional aspects and technological applications. Compr. Rev. Food Sci. Food Saf. 2018, 17, 123–136. [Google Scholar] [CrossRef] [Green Version]
- Skrajda-Brdak, M.; Konopka, I.; Tańska, M.; Sulewska, H. Lipophilic fraction and its antioxidant capacity in sixteen spelt genotypes cultivated in Poland. J. Cereal Sci. 2019, 89, 102809. [Google Scholar] [CrossRef]
- Okano, N.; Goto, R.; Kato, T.; Saisho, D.; Kato, K.; Miura, H.; Tani, M.; Onishi, K. Spanish spelt is unique germplasm for improvement of root hair length in hexaploid wheat. Plant Soil 2020, 452, 171–184. [Google Scholar] [CrossRef]
- Akel, W.; Thorwarth, P.; Mirdita, V.; Weissman, E.A.; Liu, G.; Würschum, T.; Longin, C.F.H. Can spelt wheat be used as heterotic group for hybrid wheat breeding? Theor. Appl. Genet. 2018, 131, 973–984. [Google Scholar] [CrossRef]
- Mujeeb-Kazi, A.; Kazi, A.G.; Dundas, I.; Rasheed, A.; Ogbonnaya, F.; Kishii, M.; Bonnett, D.; Wang, R.R.C.; Xu, S.; Chen, P.; et al. Genetic diversity for wheat improvement as a conduit to food security. Adv. Agron. 2013, 122, 179–257. [Google Scholar] [CrossRef]
- Nelson, D.E.; Repetti, P.P.; Adams, T.R.; Creelman, R.A.; Wu, J.; Warner, D.C.; Anstrom, D.C.; Bensen, R.J.; Castiglioni, P.P.; Donnarummo, M.G.; et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 2007, 104, 16450–16455. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Zhao, Y.; Shi, S.; Du, X.; Gu, J.; Xiao, K. Wheat nuclear factor Y (NF-Y) B subfamily gene TaNF-YB3;l confers critical drought tolerance through modulation of the ABA-associated signaling pathway. Plant Cell Tiss. Organ Cult. 2017, 128, 97–111. [Google Scholar] [CrossRef]
- Sato, H.; Suzuki, T.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NF-YB2 and NF-YB3 have functionally diverged and differentially induce drought and heat stress-specific genes. Plant Physiol. 2019, 180, 1677–1690. [Google Scholar] [CrossRef] [Green Version]
- Tello-Ruiz, M.K.; Naithani, S.; Stein, J.C.; Gupta, P.; Campbell, M.; Olson, A.; Wei, S.; Preece, J.; Geniza, M.J.; Jiao, Y.; et al. Gramene 2018: Unifying comparative genomics and pathway resources for plant research. Nucleic Acids Res. 2018, 46, D1181–D1189. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kawahara, Y.; De la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- International Brachypodium Initiative, Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [CrossRef] [PubMed]
- Reiser, L.; Subramaniam, S.; Li, D.; Huala, E. Using the Arabidopsis information resource (TAIR) to find information about Arabidopsis genes. Curr. Protoc. Bioinform. 2017, 60, 1.11.1–1.11.45. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Jatayev, S.; Sukhikh, I.; Vavilova, V.; Smolenskaya, S.E.; Goncharov, N.P.; Kurishbayev, A.; Zotova, L.; Absattarova, A.; Serikbay, D.; Hu, Y.G.; et al. Green revolution ‘stumbles’ in a dry environment: Dwarf wheat with Rht genes fails to produce higher grain yield than taller plants under drought. Plant Cell Environ. 2020, 43, 2355–2364. [Google Scholar] [CrossRef]
- Sharma, V. Methods and techniques for soil moisture monitoring. In Irrigation Management: Basics of Soil Water Bulletin. B-1331; University of Wyoming: Laramie, WY, USA, 2018; pp. 1–18. Available online: http://wyoextension.org/publications/html/B1331 (accessed on 1 November 2018).
- Baidyussen, A.; Aldammas, M.; Kurishbayev, A.; Myrzabaeva, M.; Zhubatkanov, A.; Sereda, G.; Porkhun, R.; Sereda, S.; Jatayev, S.; Langridge, P.; et al. Identification, gene expression and genetic polymorphism of zinc finger A20/AN1 stress-associated genes, HvSAP, in salt stressed barley from Kazakhstan. BMC Plant Biol. 2020, 20, 156. [Google Scholar] [CrossRef]
- Khassanova, G.; Kurishbayev, A.; Jatayev, S.; Zhubatkanov, A.; Zhumalin, A.; Turbekova, A.; Amantaev, B.; Lopato, S.; Schramm, C.; Jenkins, C.; et al. Intracellular vesicle trafficking genes, RabC-GTP, are highly expressed under salinity and rapid dehydration but down-regulated by drought in leaves of chickpea (Cicer arietinum L.). Front. Genet. 2019, 10, 40. [Google Scholar] [CrossRef] [Green Version]
- Shavrukov, Y.; Bovill, J.; Afzal, I.; Hayes, J.E.; Roy, S.J.; Tester, M.; Collins, N.C. HVP10 encoding V-PPase is a prime candidate for the barley HvNax3 sodium exclusion gene: Evidence from fine mapping and expression analysis. Planta 2013, 237, 1111–1122. [Google Scholar] [CrossRef]
- Sweetman, C.; Khassanova, G.; Miller, T.K.; Booth, N.J.; Kurishbayev, A.; Jatayev, S.; Gupta, N.K.; Langridge, P.; Jenkins, C.L.D.; Soole, K.L.; et al. Salt-induced expression of intracellular vesicle tracking genes, CaRab-GTP, and their association with Na+ accumulation in leaves of chickpea (Cicer arietinum L.). BMC Plant Biol. 2020, 20, 183. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.; Tsai, S.M.; Caldas, D.G.G. Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep. 2012, 31, 827–838. [Google Scholar] [CrossRef]
- Paolacci, A.R.; Oronzo, A.T.; Porceddu, E.; Ciaffi, M. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 2009, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Soole, K.; Smith, C. Analysis of type II NAD(P)H dehydrogenases. In Plant Mitochondria: Methods and Protocols. Methods in Molecular Biology; Whelan, J., Murcha, M.W., Eds.; Springer: New York, NY, USA, 2015; pp. 154–196. [Google Scholar]



| TaDrAp Genes | Gene ID (TraesCS) | ORF (bp) | Chromosome Location (Strand) | CDS (bp) | Protein (aa) |
|---|---|---|---|---|---|
| TaDrAp1 | |||||
| TaDrAp1-A4 | 4A02G151500 | 5188 | Chr4A: 306,315,444–306,320,631 (−) | 768 | 255 |
| TaDrAp1-B4 | 4B02G161200 | 10,031 | Chr4B: 316,654,368–316,664,398 (−) | 765 | 254 |
| TaDrAp1-D4 | 4D02G158200 | 4338 | Chr4D: 220,464,822–220,469,159 (−) | 768 | 255 |
| TaDrAp2 | |||||
| TaDrAp2-A1 | 1A02G310700 | 5490 | Chr1A: 501,702,108–501,707,597 (+) | 852 | 283 |
| TaDrAp2-B1 | 1B02G322000 | 5172 | Chr1B: 546,528,780–546,533,951 (+) | 852 | 283 |
| TaDrAp2-D1 | 1D02G310200 | 5492 | Chr1D: 406,545,964–406,551,455 (+) | 855 | 284 |
| ID | Grain Yield (g/plot) | Plant Height (cm) | SNP Genotyping | ||||||
|---|---|---|---|---|---|---|---|---|---|
| TaDrAp1-A4 | TaDrAp1-B4 * | TaDrAp1-D4 | TaDrAp2-A1 | TaDrAp2-B1 * | TaDrAp2-D1 | TaRht-B1 | |||
| ♀ 18-4, spelt | 108 | 71 | N.P. | aa | aa | aa | aa | aa | aa |
| ♂ 18-5, bread | 99 | 57 | N.P. | bb | bb | bb | bb | bb | bb |
| Breeding lines, group 1 | |||||||||
| 18-6-174 | 319 | 94 | - | ab + bb | aa | ab + bb | bb | aa + ab | aa |
| 18-6-82 | 281 | 87 | - | bb | bb | bb | bb | aa | aa |
| 18-6-37 | 240 | 67 | - | ab + bb | ab + bb | ab + bb | bb | bb | ab + bb |
| Breeding lines, group 2 | |||||||||
| 18-6-119 | 174 | 79 | - | aa | bb | aa | ab + bb | bb | aa |
| 18-6-93 | 166 | 66 | - | aa | aa + ab | aa | bb | aa | ab |
| 18-6-62 | 131 | 68 | - | aa | ab | aa | aa | bb | bb |
| Breeding lines, group 3 | |||||||||
| 18-6-126 | 115 | 78 | - | ab | bb | aa | aa | bb | aa |
| 18-6-105 | 94 | 58 | - | aa | aa | bb | aa | aa | bb |
| 18-6-61 | 80 | 57 | - | aa | ab | ab | aa | aa | bb |
| Scaffold, SNP microarray | 51014 | 124156, 109338 | 133452 | 10127, 48187 | 88266, 93878 | 20922, 92741 | 30139, 18668 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zotova, L.; Shamambaeva, N.; Lethola, K.; Alharthi, B.; Vavilova, V.; Smolenskaya, S.E.; Goncharov, N.P.; Kurishbayev, A.; Jatayev, S.; Gupta, N.K.; et al. TaDrAp1 and TaDrAp2, Partner Genes of a Transcription Repressor, Coordinate Plant Development and Drought Tolerance in Spelt and Bread Wheat. Int. J. Mol. Sci. 2020, 21, 8296. https://doi.org/10.3390/ijms21218296
Zotova L, Shamambaeva N, Lethola K, Alharthi B, Vavilova V, Smolenskaya SE, Goncharov NP, Kurishbayev A, Jatayev S, Gupta NK, et al. TaDrAp1 and TaDrAp2, Partner Genes of a Transcription Repressor, Coordinate Plant Development and Drought Tolerance in Spelt and Bread Wheat. International Journal of Molecular Sciences. 2020; 21(21):8296. https://doi.org/10.3390/ijms21218296
Chicago/Turabian StyleZotova, Lyudmila, Nasgul Shamambaeva, Katso Lethola, Badr Alharthi, Valeriya Vavilova, Svetlana E. Smolenskaya, Nikolay P. Goncharov, Akhylbek Kurishbayev, Satyvaldy Jatayev, Narendra K. Gupta, and et al. 2020. "TaDrAp1 and TaDrAp2, Partner Genes of a Transcription Repressor, Coordinate Plant Development and Drought Tolerance in Spelt and Bread Wheat" International Journal of Molecular Sciences 21, no. 21: 8296. https://doi.org/10.3390/ijms21218296
APA StyleZotova, L., Shamambaeva, N., Lethola, K., Alharthi, B., Vavilova, V., Smolenskaya, S. E., Goncharov, N. P., Kurishbayev, A., Jatayev, S., Gupta, N. K., Gupta, S., Schramm, C., Anderson, P. A., Jenkins, C. L. D., Soole, K. L., & Shavrukov, Y. (2020). TaDrAp1 and TaDrAp2, Partner Genes of a Transcription Repressor, Coordinate Plant Development and Drought Tolerance in Spelt and Bread Wheat. International Journal of Molecular Sciences, 21(21), 8296. https://doi.org/10.3390/ijms21218296







