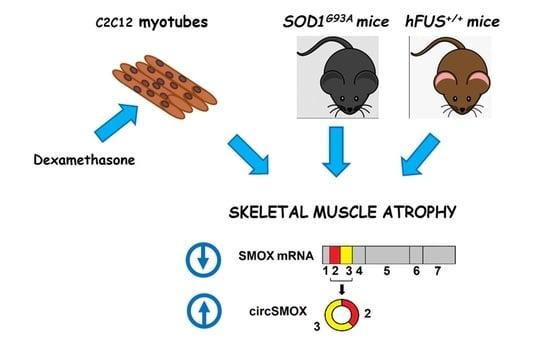
Emerging Role for Linear and Circular Spermine Oxidase RNAs in Skeletal Muscle Physiopathology
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plasmid Constructions
2.3. Cell Culture
2.4. Cell Protein Extraction
2.5. ALS Mouse Models
2.6. RNA Isolation, Reverse Transcription and qReal-Time PCR
2.7. Western Blot
2.8. Statistical Analysis
3. Results
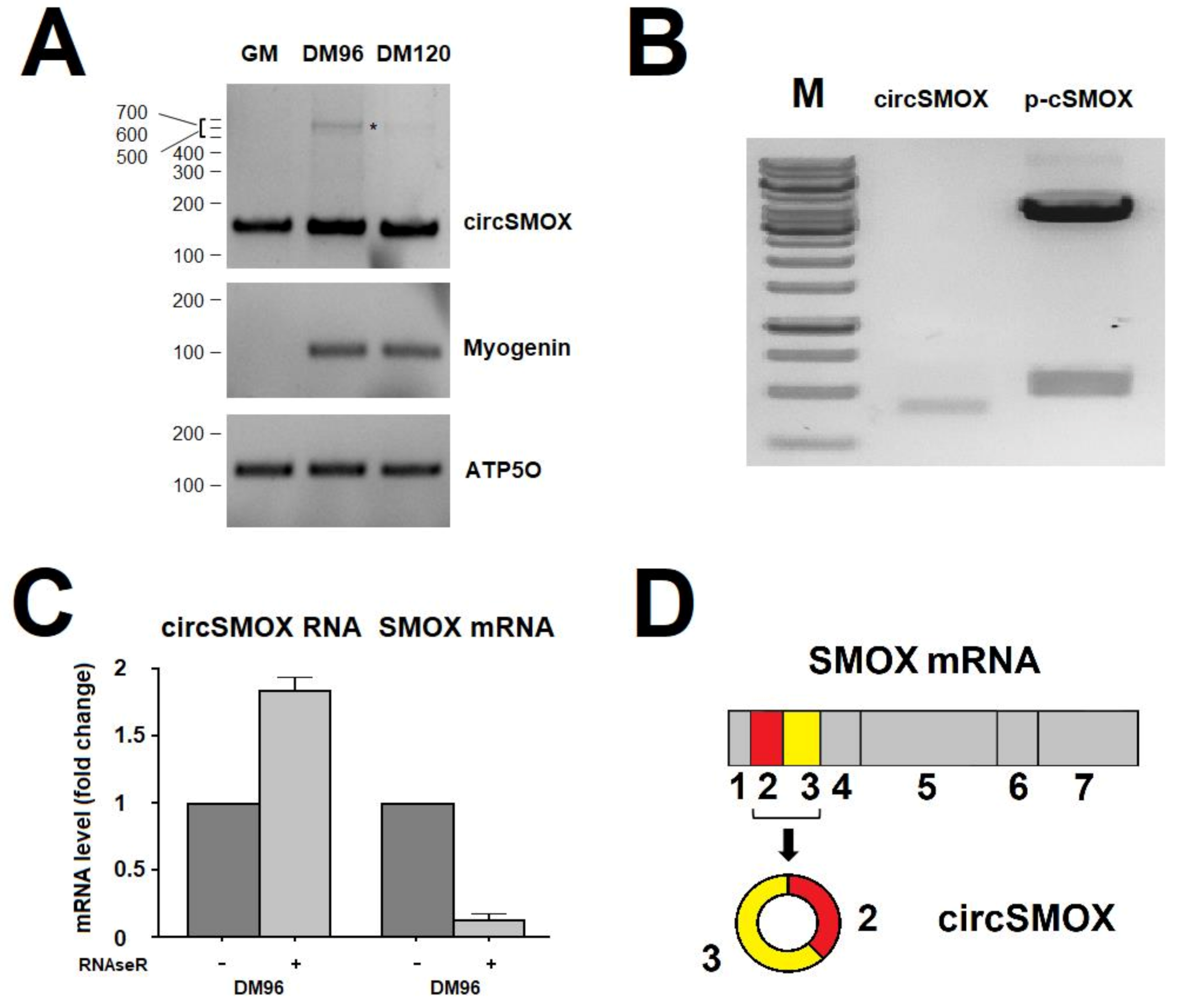
3.1. Evaluation of the Existence of a Circular RNA for SMOX
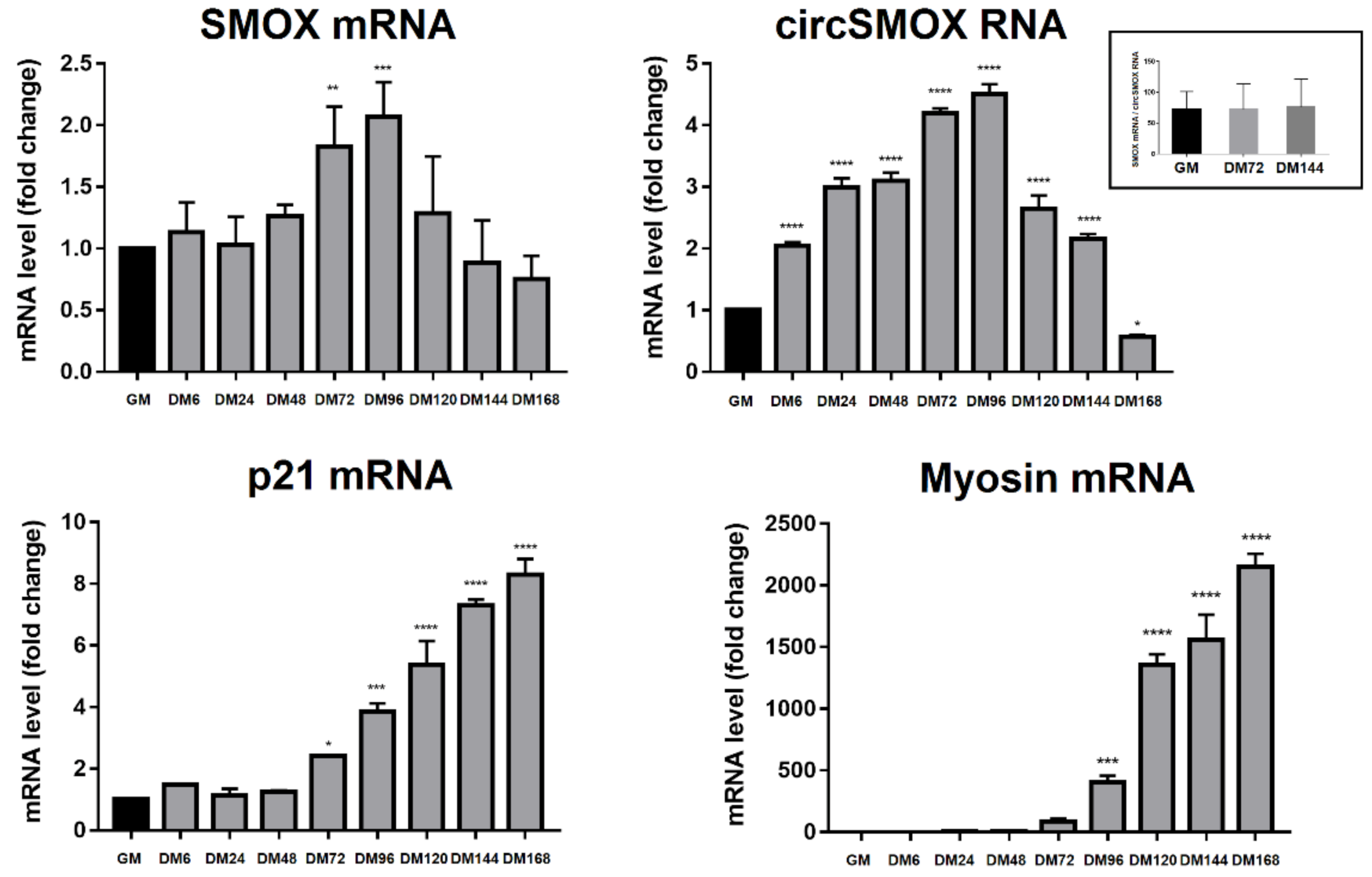
3.2. RNA Transcript Levels during C2C12 Cell Differentiation
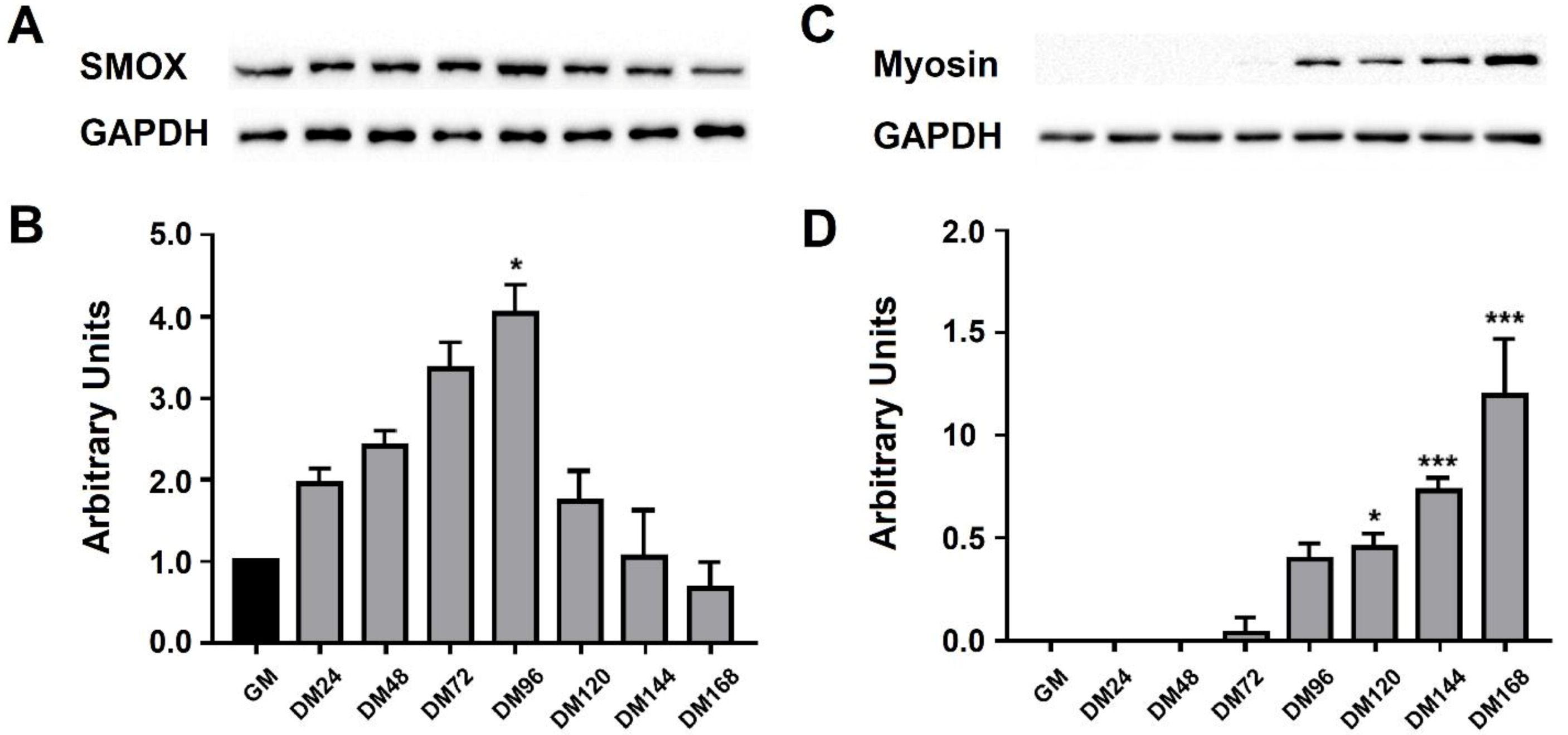
3.3. SMOX Protein Level during C2C12 Cell Differentiation
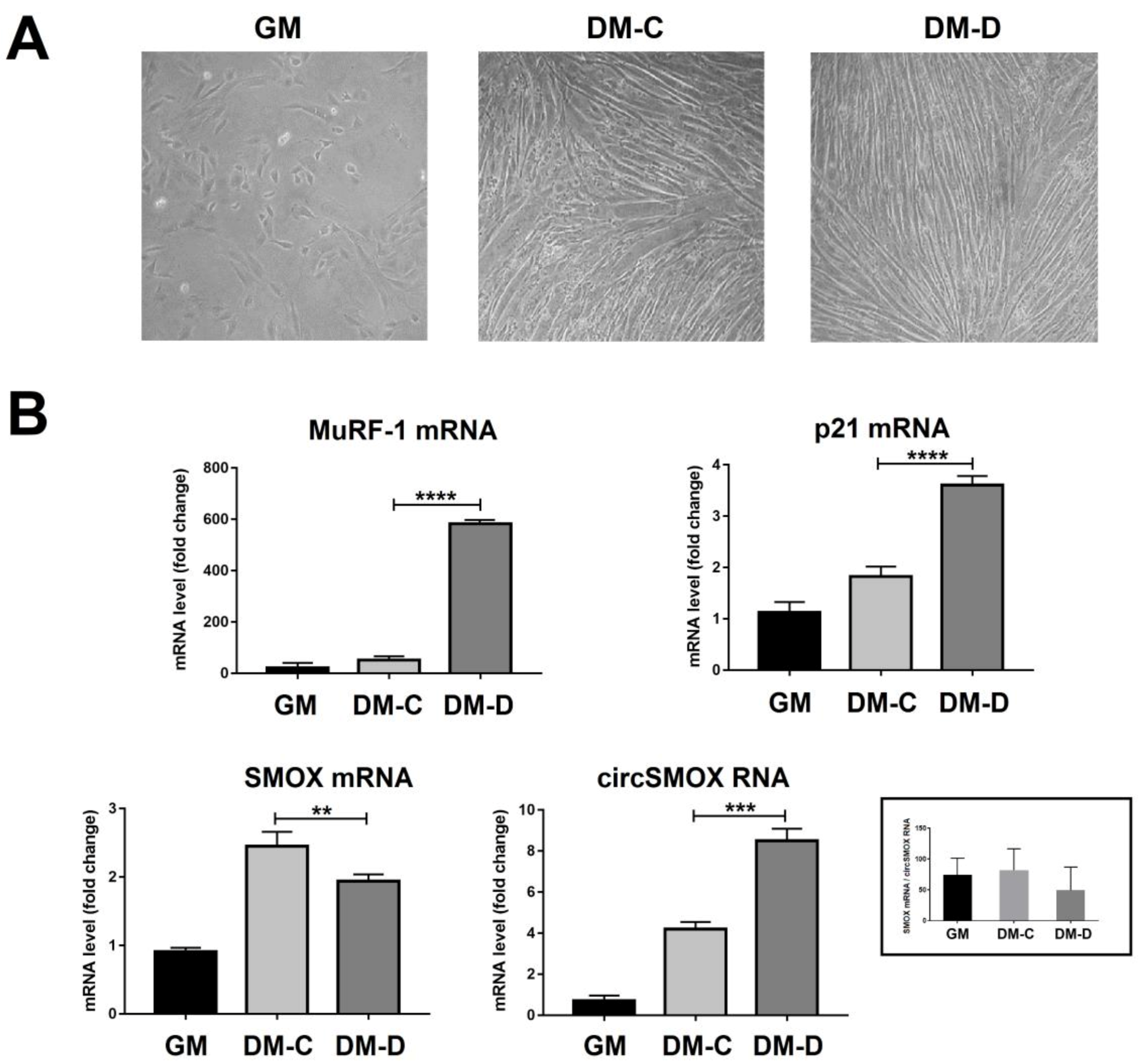
3.4. Linear and Circular SMOX RNAs Expression in Atrophic C2C12 Cells
3.5. SMOX RNA Transcript Splicing Variant Levels in Atrophic C2C12 Cells
3.6. Cellular Localization of Circular SMOX RNA in Atrophic C2C12 Cells
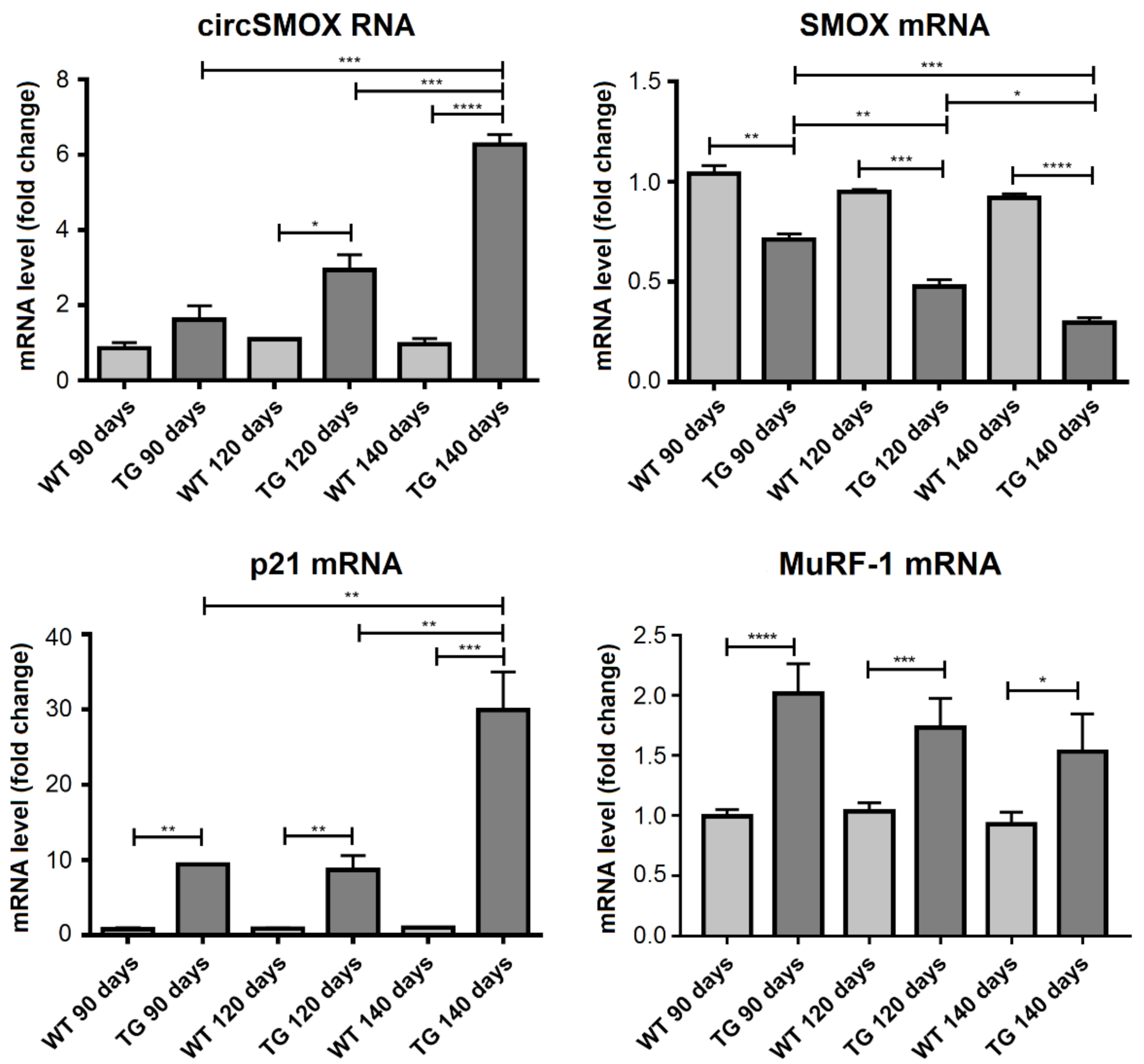
3.7. Linear and Circular SMOX RNA Expression in Two Mouse Models of Amyotrophic Lateral Sclerosis
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ALS | Amyotrophic lateral sclerosis |
| GM | C2C12 myoblasts |
| DM | C2C12 myotubes |
| circSMOX | circular SMOX |
| p21 | cyclin-dependent kinase inhibitor p21 |
| DEXA | dexamethasone |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| ATP50 | ATP Synthase Peripheral Stalk Subunit OSC |
| MuRF-1 | muscle RING-finger protein-1 |
| SM | skeletal muscle |
| Spd | spermidine |
| SMOX | spermine oxidase |
References
- Mercatelli, N.; Fittipaldi, S.; De Paola, E.; Dimauro, I.; Paronetto, M.P.; Jackson, M.J.; Caporossi, D. MiR-23-TrxR1 as a novel molecular axis in skeletal muscle differentiation. Sci. Rep. 2017, 7, 7219. [Google Scholar] [CrossRef] [PubMed]
- Cervelli, M.; Leonetti, A.; Duranti, G.; Sabatini, S.; Ceci, R.; Mariottini, P. Skeletal Muscle Pathophysiology: The Emerging Role of Spermine Oxidase and Spermidine. Med. Sci. 2018, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Cervelli, M.; Fratini, E.; Amendola, R.; Bianchi, M.; Signori, E.; Ferraro, E.; Lisi, A.; Federico, R.; Marcocci, L.; Mariottini, P. Increased spermine oxidase (SMO) activity as a novel differentiation marker of myogenic C2C12 cells. Int. J. Biochem. Cell Biol. 2009, 41, 934–944. [Google Scholar] [CrossRef]
- Cervelli, M.; Amendola, R.; Polticelli, F.; Mariottini, P. Spermine oxidase: Ten years after. Amino Acids 2012, 42, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Polticelli, F.; Salvi, D.; Mariottini, P.; Amendola, R.; Cervelli, M. Molecular evolution of the polyamine oxidase gene family in Metazoa. BMC Evol. Biol. 2012, 12, 90. [Google Scholar] [CrossRef]
- Cervelli, M.; Bellini, A.; Bianchi, M.; Marcocci, L.; Nocera, S.; Polticelli, F.; Federico, R.; Amendola, R.; Mariottini, P. Mouse spermine oxidase gene splice variants. Nuclear subcellular localization of a novel active isoform. Eur. J. Biochem. 2004, 271, 760–770. [Google Scholar] [CrossRef] [PubMed]
- Ceci, R.; Duranti, G.; Leonetti, A.; Pietropaoli, S.; Spinozzi, F.; Marcocci, L.; Amendola, R.; Cecconi, F.; Sabatini, S.; Mariottini, P.; et al. Adaptive responses of heart and skeletal muscle to spermine oxidase overexpression: Evaluation of a new transgenic mouse model. Free Radic. Biol. Med. 2017, 103, 216–225. [Google Scholar] [CrossRef]
- Bongers, K.S.; Fox, D.K.; Kunkel, S.D.; Stebounova, S.V.; Murry, D.J.; Pufall, M.A.; Ebert, S.M.; Dyle, M.C.; Bullard, S.A.; Dierdorff, J.M.; et al. Spermine oxidase maintains basal skeletal muscle gene expression and fiber size and is strongly repressed by conditions that cause skeletal muscle atrophy. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E144–E158. [Google Scholar] [CrossRef]
- Jackman, R.W.; Kandarian, S.C. The molecular basis of skeletal muscle atrophy. Am. J. Physiol. Cell Physiol. 2004, 287, C834–C843. [Google Scholar] [CrossRef]
- Powers, S.K.; Lynch, G.S.; Murphy, K.T.; Reid, M.B.; Zijdewind, I. Disease-Induced Skeletal Muscle Atrophy and Fatigue. Med. Sci. Sports Exerc. 2016, 48, 2307–2319. [Google Scholar] [CrossRef]
- Ballarino, M.; Morlando, M.; Fatica, A.; Bozzoni, I. Non-coding RNAs in muscle differentiation and musculoskeletal disease. J. Clin. Investig. 2016, 126, 2021–2030. [Google Scholar] [CrossRef]
- Ballarino, M.; Cipriano, A.; Tita, R.; Santini, T.; Desideri, F.; Morlando, M.; Colantoni, A.; Carrieri, C.; Nicoletti, C.; Musarò, A.; et al. Deficiency in the nuclear long noncoding RNA Charme causes myogenic defects and heart remodeling in mice. EMBO J. 2018, 37, e99697. [Google Scholar] [CrossRef]
- Dimartino, D.; Colantoni, A.; Ballarino, M.; Martone, J.; Mariani, D.; Danner, J.; Bruckmann, A.; Meister, G.; Morlando, M.; Bozzoni, I. The Long Non-coding RNA lnc-31 Interacts with Rock1 mRNA and Mediates Its YB-1-Dependent Translation. Cell Rep. 2018, 23, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Cardinali, B.; Falcone, G.; Martelli, F. Circular RNAs in Muscle Function and Disease. Int. J. Mol. Sci. 2018, 19, 3454–3471. [Google Scholar] [CrossRef]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Moll. Cell. 2017, 66, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Xu, H.; Li, R.; Wu, W.; Chao, Z.; Li, C.; Xia, W.; Wang, L.; Yang, J.; Xu, Y. Assessment of myoblast circular RNA dynamics and its correlation with miRNA during myogenic differentiation. Int. J. Biochem. Cell. Biol. 2018, 99, 211–218. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, M.; Lian, D.; Li, Y.; Li, Y.; Wang, J.; Deng, S.; Yu, K.; Lian, Z. Non-Coding RNA Regulates the Myogenesis of Skeletal Muscle Satellite Cells, Injury Repair and Diseases. Cells 2019, 8, 988. [Google Scholar] [CrossRef]
- Mehta, S.L.; Pandi, G.; Vemuganti, R. Circular RNA expression profiles alter significantly in mouse brain after transient focal ischemia. Stroke 2017, 48, 2541–2548. [Google Scholar] [CrossRef]
- Weng, J.; Zhang, P.; Yin, X.; Jiang, B. The whole transcriptome involved in denervated muscle atrophy following peripheral nerve injury. Front. Mol. Neurosci. 2018, 11, 69. [Google Scholar] [CrossRef]
- Rhen, T.; Cidlowski, J.A. Antiinflammatory action of glucocorticoids—New mechanisms for old drugs. N. Engl. J. Med. 2005, 353, 1711–1723. [Google Scholar] [CrossRef]
- Menconi, M.; Fareed, M.; O'Neal, P.; Poylin, V.; Wei, W.; Hasselgren, P.O. Role of glucocorticoids in the molecular regulation of muscle wasting. Crit. Care Med. 2007, 35, S602–S608. [Google Scholar] [CrossRef]
- Clarke, B.A.; Drujan, D.; Willis, M.S.; Murphy, L.O.; Corpina, R.A.; Burova, E.; Rakhilin, S.V.; Stitt, T.N.; Patterson, C.; Latres, E.; et al. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007, 6, 376–385. [Google Scholar] [CrossRef]
- Ceci, R.; Duranti, G.; Rossi, A.; Savini, I.; Sabatini, S. Skeletal muscle differentiation: Role of dehydroepiandrosterone sulfate. Horm. Metab. Res. 2011, 43, 702–707. [Google Scholar] [CrossRef]
- Castillero, E.; Alamdari, N.; Lecker, S.H.; Hasselgren, P.O. Suppression of atrogin-1 and MuRF1 prevents dexamethasone-induced atrophy of cultured myotubes. Metabolism 2013, 62, 1495–1502. [Google Scholar] [CrossRef]
- Menconi, M.; Gonnella, P.; Petkova, V.; Lecker, S.; Hasselgren, P.O. Dexamethasone and corticosterone induce similar, but not identical, muscle wasting responses in cultured L6 and C2C12 myotubes. J. Cell. Biochem. 2008, 105, 353–364. [Google Scholar] [CrossRef]
- Wray, C.J.; Mammen, J.M.; Hershko, D.D.; Hasselgren, P.O. Sepsis upregulates the gene expression of multiple ubiquitin ligases in skeletal muscle. Int. J. Biochem. Cell. Biol. 2003, 35, 698–705. [Google Scholar] [CrossRef]
- Acharyya, S.; Guttridge, D.C. Cancer cachexia signaling pathways continue to emerge yet much still points to the proteasome. Clin. Cancer Res. 2007, 13, 1356–1361. [Google Scholar] [CrossRef]
- Klaude, M.; Fredriksson, K.; Tjader, I.; Hammarqvist, F.; Ahlman, B.; Rooyackers, O.; Wernerman, J. Proteasome proteolytic activity in skeletal muscle is increased in patients with sepsis. Clin. Sci. 2007, 112, 499–506. [Google Scholar] [CrossRef]
- Chen, L.; Chen, L.; Wan, L.; Huo, Y.; Huang, J.; Li, J.; Lu, J.; Xin, B.; Yang, Q.; Guo, C. Matrine improves skeletal muscle atrophy by inhibiting E3 ubiquitin ligases and activating the Akt/mTOR/FoxO3α signaling pathway in C2C12 myotubes and mice. Oncol. Rep. 2019, 42, 479–494. [Google Scholar] [CrossRef]
- Corcia, P.; Pradat, P.F.; Salachas, F.; Salachas, F.; Bruneteau, G.; Forestier, N.; Seilhean, D.; Hauw, J.; Meininger, V. Causes of death in a post-mortem series of ALS patients. Amyotroph. Lateral Scler. 2008, 9, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Zarei, S.; Carr, K.; Reiley, L.; Diaz, K.; Guerra, O.; Altamirano, P.F.; Pagani, W.; Lodin, D.; Orozco, G.; Chinea, A. A comprehensive review of amyotrophic lateral sclerosis. Surg. Neurol. Int. 2015, 6, 171. [Google Scholar] [CrossRef]
- Gurney, M.E.; Pu, H.; Chiu, A.Y.; Dal Canto, M.C.; Polchow, C.Y.; Alexander, D.D.; Caliendo, J.; Hentati, A.; Kwon, Y.W.; Deng, H.X.; et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 1994, 264, 1772–1775. [Google Scholar] [CrossRef]
- Mitchell, J.C.; McGoldrick, P.; Vance, C.; Hortobagyi, T.; Sreedharan, J.; Rogelj, B.; Tudor, E.L.; Smith, B.N.; Klasen, C.; Miller, C.C.; et al. Overexpression of human wild-type FUS causes progressive motor neuron degeneration in an age- and dose-dependent fashion. Acta Neuropathol. 2013, 125, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Duranti, G.; Ceci, R.; Sgrò, P.; Sabatini, S.; Di Luigi, L. Influence of the PDE5 inhibitor tadalafil on redox status and antioxidant defense system in C2C12 skeletal muscle cells. Cell Stress Chaperones 2017, 22, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Ripps, M.E.; Huntley, G.W.; Hof, P.R.; Morrison, J.H.; Gordon, J.W. Transgenic mice expressing an altered murine superoxide dismutase gene provide an animal model of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 1995, 92, 689–693. [Google Scholar] [CrossRef]
- Scaricamazza, S.; Salvatori, I.; Giacovazzo, G.; Loeffler, J.P.; Renè, F.; Rosina, M.; Quessada, C.; Proietti, D.; Heil, C.; Rossi, S.; et al. Skeletal-Muscle Metabolic Reprogramming in ALS-SOD1G93A Mice Predates Disease Onset and Is A Promising Therapeutic Target. iScience 2020, 23, 101087. [Google Scholar] [CrossRef]
- Apolloni, S.; Amadio, S.; Fabbrizio, P.; Morello, G.; Spampinato, A.G.; Latagliata, E.C.; Salvatori, I.; Proietti, D.; Ferri, A.; Madaro, L.; et al. Histaminergic transmission slows progression of amyotrophic lateral sclerosis. J. Cachexia Sarcopenia Muscle 2019, 10, 872–893. [Google Scholar] [CrossRef] [PubMed]
- Mirra, A.; Rossi, S.; Scaricamazza, S.; Di Salvio, M.; Salvatori, I.; Valle, C.; Rusmini, P.; Poletti, A.; Cestra, G.; Carrì, M.T.; et al. Functional interaction between FUS and SMN underlies SMA-like splicing changes in wild-type hFUS mice. Sci. Rep. 2017, 7, 2033. [Google Scholar] [CrossRef]
- Danan, M.; Schwartz, S.; Edelheit, S.; Sorek, R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012, 40, 3131–3142. [Google Scholar] [CrossRef]
- Atherton, P.J.; Greenhaff, P.L.; Phillips, S.M.; Bodine, S.C.; Adams, C.M.; Lang, C.H. Control of skeletal muscle atrophy in response to disuse: Clinical/preclinical contentions and fallacies of evidence. Am. J. Physiol. Endocrinol. Metab. 2016, 311, E594–E604. [Google Scholar] [CrossRef]
- Orzechowski, A.; Grizard, J.; Jank, M.; Gajkowska, B.; Lokociejewska, M.; Zaron-Teperek, M.; Godlewski, M. Dexamethasone-mediated regulation of death and differentiation of muscle cells. Is hydrogen peroxide involved in the process? Reprod. Nutr. Dev. 2002, 42, 197–216. [Google Scholar] [CrossRef]
- Piao, Y.J.; Seo, Y.H.; Hong, F.; Kim, J.H.; Kim, Y.J.; Kang, M.H.; Kim, B.S.; Jo, S.A.; Jo, I.; Jue, D.M.; et al. Nox 2 stimulates muscle differentiation via NF-kappaB/iNOS pathway. Free Radic. Biol. Med. 2005, 38, 989–1001. [Google Scholar] [CrossRef]
- Kamli, M.R.; Kim, J.; Pokharel, S.; Jan, A.T.; Lee, E.J.; Choi, I. Expressional studies of the aldehyde oxidase (AOX1) gene during myogenic differentiation in C2C12 cells. Biochem. Biophys. Res. Commun. 2014, 450, 1291–1296. [Google Scholar] [CrossRef]
- Youm, T.H.; Woo, S.H.; Kwon, E.S.; Park, S.S. NADPH Oxidase 4 Contributes to Myoblast Fusion and Skeletal Muscle Regeneration. Oxid. Med. Cell Longev. 2019, 2019, 3585390. [Google Scholar] [CrossRef] [PubMed]
- Shang, Q.; Yang, Z.; Jia, R.; Ge, S. The novel roles of circRNAs in human cancer. Mol. Cancer 2019, 18, 6. [Google Scholar] [CrossRef]
- Zhang, P.; Chao, Z.; Zhang, R.; Ding, R.; Wang, Y.; Wu, W.; Han, Q.; Li, C.; Xu, H.; Wang, L.; et al. Circular RNA regulation of myogenesis. Cells 2019, 8, 885. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Fan, J.; Chen, M.; Wang, X.; Tian, Z.; Wang, J.; Fan, D.; Zeng, J.; Zhang, K.; Dai, X. Targeting Smox Is Neuroprotective and Ameliorates Brain Inflammation in Cerebral Ischemia/Reperfusion Rats. Toxicol. Sci. 2019, 168, 381–393. [Google Scholar] [CrossRef]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glazar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- You, X.; Vlatkovic, I.; Babic, A.; Will, T.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.; Glock, C.; Quedenau, C.; et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015, 18, 603–610. [Google Scholar] [CrossRef]
- Di Timoteo, G.; Dattilo, D.; Centrón-Broco, A.; Colantoni, A.; Guarnacci, M.; Rossi, F.; Incarnato, D.; Oliviero, S.; Fatica, A.; Morlando, M.; et al. Modulation of circRNA Metabolism by m6A Modification. Cell Rep. 2020, 31, 107641. [Google Scholar] [CrossRef] [PubMed]
- Pistoni, M.; Ghigna, C.; Gabellini, D. Alternative splicing and muscular dystrophy. RNA Biol. 2010, 7, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Büttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef]
- Fan, J.; Yang, X.; Li, J.; Shu, Z.; Dai, J.; Liu, X.; Li, B.; Jia, S.; Kou, X.; Yang, Y.; et al. Spermidine coupled with exercise rescues skeletal muscle atrophy from D-gal-induced aging rats through enhanced autophagy and reduced apoptosis via AMPK-FOXO3a signal pathway. Oncotarget 2017, 8, 17475–17490. [Google Scholar] [CrossRef]
- Peng, S.; Song, C.; Li, H.; Cao, X.; Ma, Y.; Wang, X.; Huang, Y.; Lan, X.; Lei, C.; Chaogetu, B.; et al. Circular RNA SNX29 sponges miR-744 to regulate proliferation and differentiation of myoblasts by activating the Wnt5a/Ca2+ signaling pathway. Molecular therapy. Nucleic Acids 2019, 16, 481–493. [Google Scholar] [CrossRef]
- Li, H.; Wei, X.; Yang, J.; Dong, D.; Hao, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Ma, Y.; et al. circFGFR4 Promotes Differentiation of Myoblasts via Binding miR-107 to Relieve Its Inhibition of Wnt3a. Mol. Ther. Nucleic Acids 2018, 11, 272–283. [Google Scholar] [CrossRef]
- Haverkamp, L.J.; Appel, V.; Appel, S.H. Natural history of amyotrophic lateral sclerosis in a database population. Validation of a scoring system and a model for survival prediction. Brain 1995, 118, 707–719. [Google Scholar] [CrossRef]
- Garbuzova-Davis, S.; Rodrigues, M.C.; Hernandez-Ontiveros, D.G.; Louis, M.K.; Willing, A.E.; Borlongan, C.V.; Sanberg, P.R. Amyotrophic lateral sclerosis: A neurovascular disease. Brain Res. 2011, 1398, 113–125. [Google Scholar] [CrossRef] [PubMed]




| Gene | PCR Method | Primers |
|---|---|---|
| ATP Synthase | RT-PCR | ATP5O Fwd: 5′-CAACCGCCCTGTACTCTGCT-3′ ATP5O Rev: 5′-GGATTCAGAACAGCCAGAGACAC-3′ |
| circSMOX | qRT-PCR | circSMOX1 Fwd: 5′-GCCTGCTACCTTACCAACC-3′ circSMOX2 Rev: 5′-CACGACTGAGAGGGTCATC-3′ |
| RT-PCR | circSMOX3 Fwd: 5′-GACAGCCTCGTGTGGTGG-3′ circSMOX4 Rev: 5′-GGTCATCCGCACTGTCGC-3′ | |
| GAPDH | qRT-PCR | Fwd: 5′-GGTTGTCTCCTGCGACTTC-3′ Rev: 5′-GGTGGTCCAGGGTTTCTTAC-3′ |
| MuRF-1 | qRT-PCR | Fwd: 5′-GACAGTCGCATTTCAAAGCA-3′ Rev: 5′-AACGACCTCCAGACATGGAC-3′ |
| Myogenin | RT-PCR | Fwd: 5′-TCCCAACCCAGGAGATCATT-3′ Rev: 5′-CATATCCTCCACCGTGATGC-3′ |
| Myosin | qRT-PCR | Fwd: 5′-ATGATCTACACCTACTCGGG-3′ Rev: 5′-GTTCTCCCGATCTGTCAGC-3′ |
| p21 | qRT-PCR | p21 Fwd: 5′-GACCTGGGAGGGGACAAG-3′ p21 Rev: 5′-TGATAGAAATCTGTCAGGCTG-3′ |
| SMOX | qRT-PCR | SMOX Fwd: 5′-ACTCCAAGAATGGCGTGGC-3′ SMOX Rev: 5′-CGACGCTGTTCTGACTCTC-3′ |
| βSMOX | qRT-PCR | SMOX β Fwd: 5′-ACAGTTCACAGGTGGGCTC-3′ SMOX β Rev: 5′-CCTCGCGTGGCCAGAG-3′ |
| µSMOX | qRT-PCR | SMOX µ Fwd: 5′-ACAGTTCACAGGGAACCCC-3′ SMOX µ Rev: 5′-GCTTCTATGCGCTGTCTTGG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reinoso-Sánchez, J.F.; Baroli, G.; Duranti, G.; Scaricamazza, S.; Sabatini, S.; Valle, C.; Morlando, M.; Casero, R.A., Jr.; Bozzoni, I.; Mariottini, P.; et al. Emerging Role for Linear and Circular Spermine Oxidase RNAs in Skeletal Muscle Physiopathology. Int. J. Mol. Sci. 2020, 21, 8227. https://doi.org/10.3390/ijms21218227
Reinoso-Sánchez JF, Baroli G, Duranti G, Scaricamazza S, Sabatini S, Valle C, Morlando M, Casero RA Jr., Bozzoni I, Mariottini P, et al. Emerging Role for Linear and Circular Spermine Oxidase RNAs in Skeletal Muscle Physiopathology. International Journal of Molecular Sciences. 2020; 21(21):8227. https://doi.org/10.3390/ijms21218227
Chicago/Turabian StyleReinoso-Sánchez, Jonathan Fernando, Giulia Baroli, Guglielmo Duranti, Silvia Scaricamazza, Stefania Sabatini, Cristiana Valle, Mariangela Morlando, Robert Anthony Casero, Jr., Irene Bozzoni, Paolo Mariottini, and et al. 2020. "Emerging Role for Linear and Circular Spermine Oxidase RNAs in Skeletal Muscle Physiopathology" International Journal of Molecular Sciences 21, no. 21: 8227. https://doi.org/10.3390/ijms21218227
APA StyleReinoso-Sánchez, J. F., Baroli, G., Duranti, G., Scaricamazza, S., Sabatini, S., Valle, C., Morlando, M., Casero, R. A., Jr., Bozzoni, I., Mariottini, P., Ceci, R., & Cervelli, M. (2020). Emerging Role for Linear and Circular Spermine Oxidase RNAs in Skeletal Muscle Physiopathology. International Journal of Molecular Sciences, 21(21), 8227. https://doi.org/10.3390/ijms21218227