Revealing Alteration in the Hepatic Glucose Metabolism of Genetically Improved Carp, Jayanti Rohu Labeo rohita Fed a High Carbohydrate Diet Using Transcriptome Sequencing
Abstract
:1. Introduction
2. Results
2.1. Illumina Sequencing and read Mapping
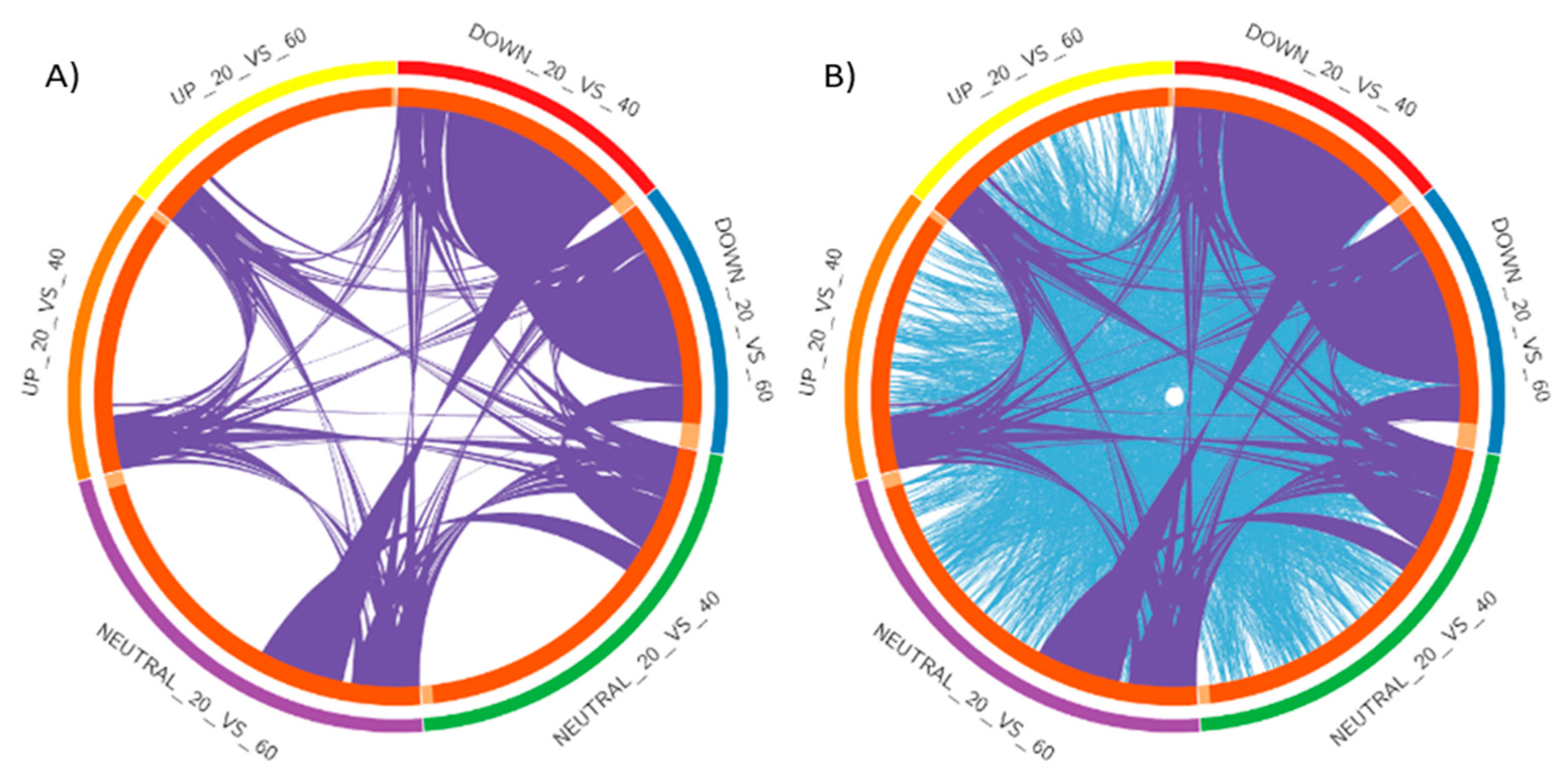
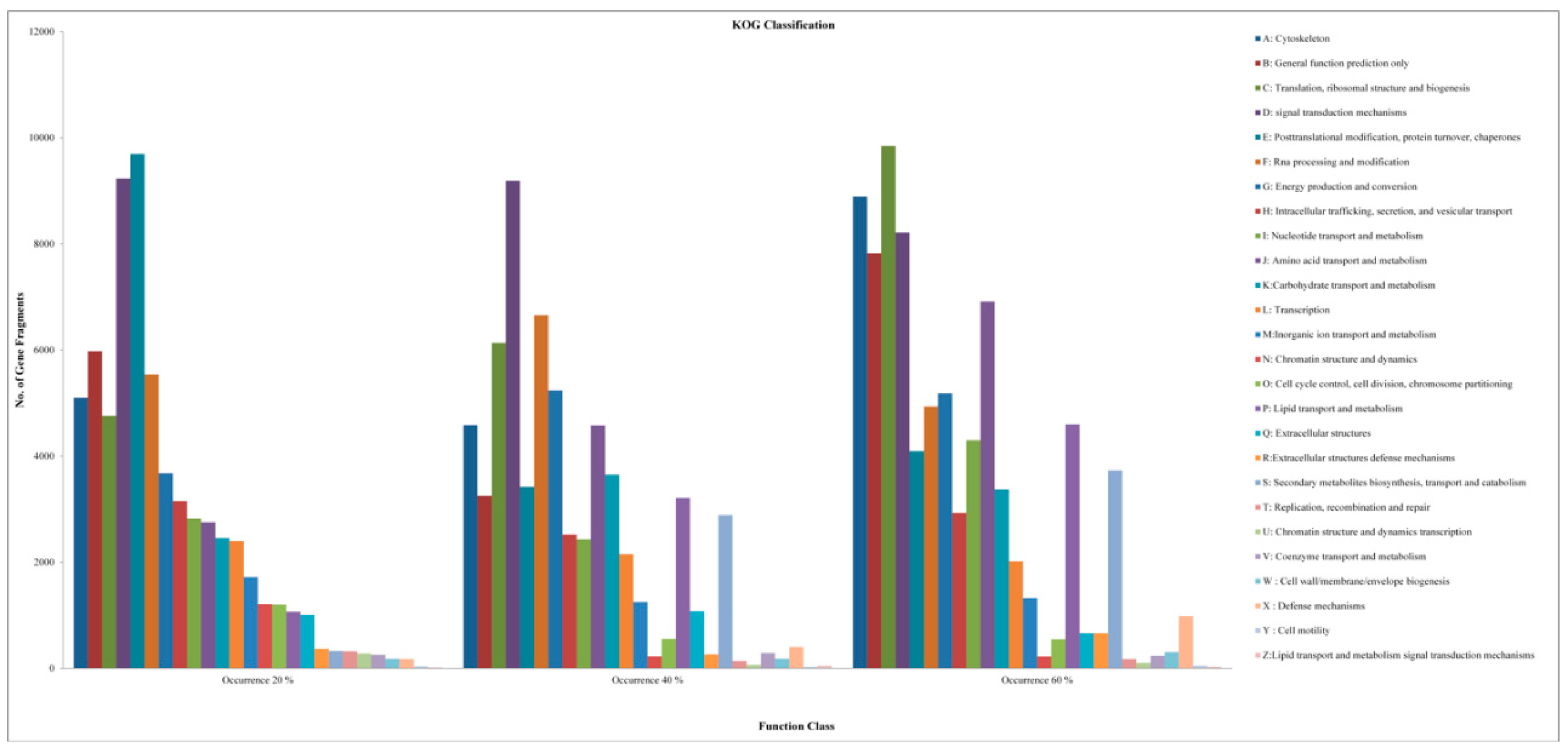
2.2. Analysis of DEGs and Annotation
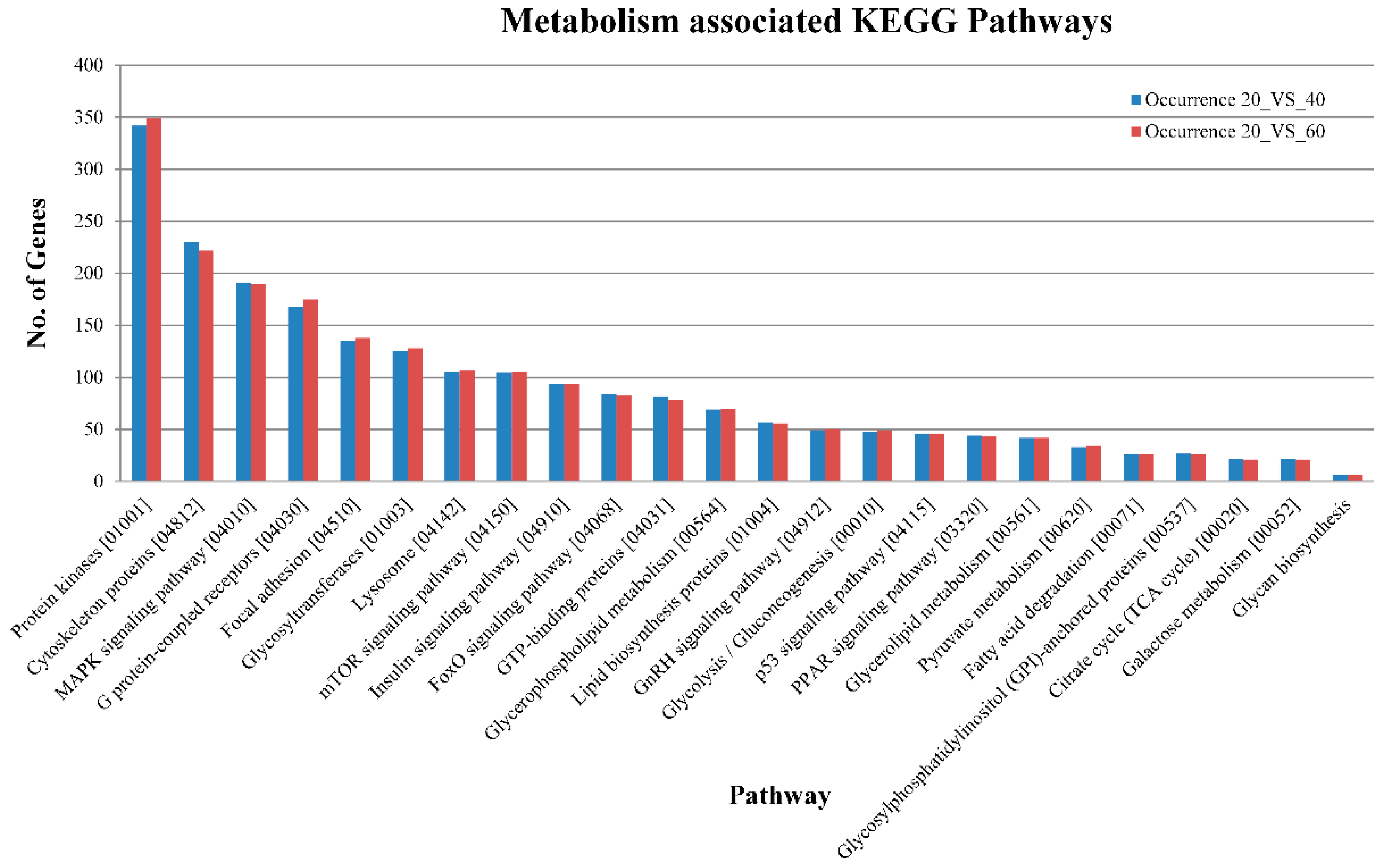
2.3. Pathway Analysis
3. Discussion
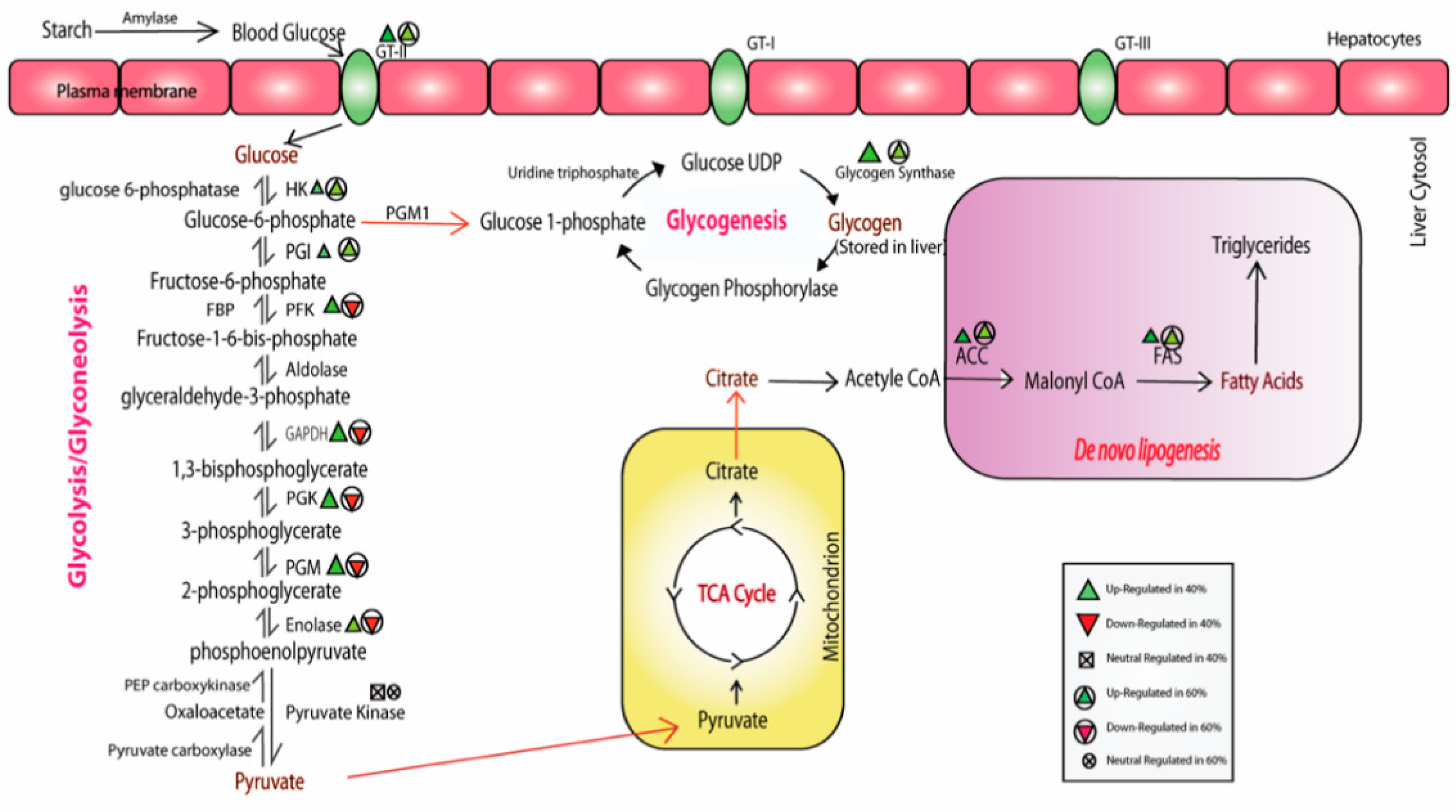
3.1. Alteration in Alpha Amylase and Glucose Transporter Activity
3.2. Modulation of the Glycolytic Mechanism
3.3. Glycogen Synthesis Enrichment due to High CHO in the Diet
3.4. Impact of High-CHO Diet on Gluconeogenesis and Lipogenesis
3.5. Immune-Associated and Stress-Related Transcriptional Changes in the Liver due to the Inclusion of a High-CHO Diet
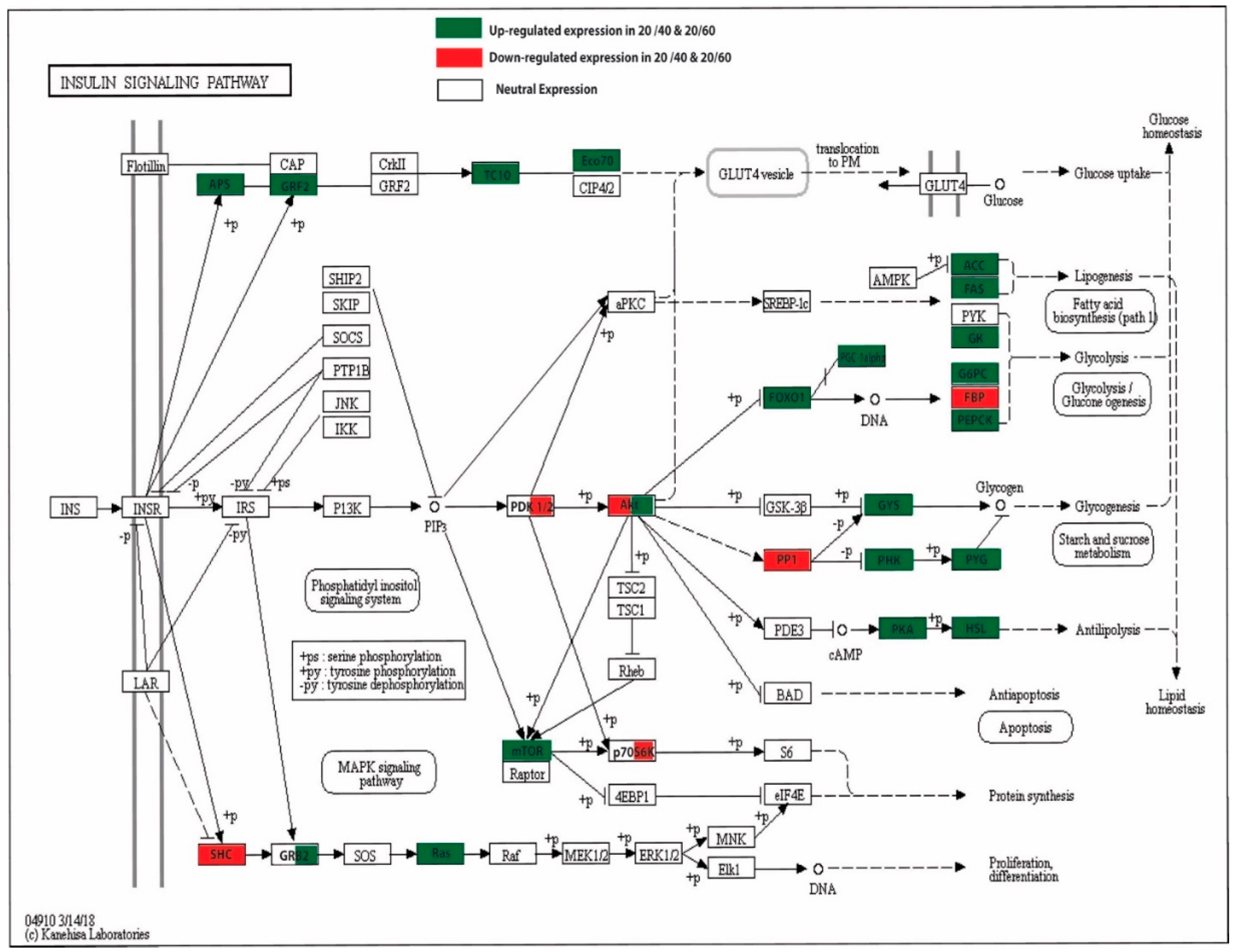
3.6. Insulin Signaling Pathways, mTOR, and the PPAR Signaling Pathway
4. Materials and Methods
4.1. Experimental Diet Preparation and Feeding Experiment
4.2. RNA Isolation and cDNA Library Preparation, NGS Sequencing
4.3. Quality Check, Assembly and Mapping
4.4. Gene Regulatory Network
4.5. Differential Expression Analysis of Transcripts, Gene Ontology and KEGG Pathway
4.6. Quantitative Real-Time PCR (qPCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CHO | Carbohydrate |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PHK | Phosphorylase kinase |
| PGK | Phosphoglycerate kinase |
| FAS | Fatty acid synthase |
| ACCα | Acetyle-CoA carboxylase |
| PPARγ | Peroxisome proliferator-activated receptor |
| mTOR | Mammalian target of rapamycin |
| GK | Glucokinase |
| 6PGDH | 6-phosphogluconate dehydrogenase |
| PFK-1 | 6-phosphofructokinase |
| PK | Pyruvate Knaser |
| G6PDH | Glucose-6-phospate dehydronated |
| FBP | Fructose-Bisphosphate |
References
- Zhao, N.; Ma, Y.; Zhang, S.; Fang, X.; Liang, Z.; Liu, G. New muramyl dipeptide (MDP) mimics without the carbohydrate moiety as potential adjuvant candidates for a therapeutic hepatitis B vaccine (HBV). Bioorganic Med. Chem. Lett. 2011, 21, 4292–4295. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Liu, Y.-J.; Tian, L.-X.; Mai, K.-S.; Liang, G.-Y.; Yang, H.-J.; Huai, M.-Y.; Luo, W.-J. Effect of dietary carbohydrate-to-lipid ratios on growth performance, body composition, nutrient utilization and hepatic enzymes activities of herbivorous grass carp (Ctenopharyngodon idella). Aquac. Nutr. 2010, 16, 327–333. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, F.; Li, Q.; Odle, J.; Lin, X.; Zhu, H.; Pi, D.; Hou, Y.; Hong, Y.; Shi, H. Fish oil alleviates activation of the hypothalamic-pituitary-adrenal axis associated with inhibition of TLR4 and NOD signaling pathways in weaned piglets after a lipopolysaccharide challenge. J. Nutr. 2013, 143, 1799–1807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, C.C.; Guo, L.; Santschi, P.H.; Alvarado-Quiroz, N.; Haye, J.M. Distributions of carbohydrate species in the Gulf of Mexico. Mar. Chem. 2003, 81, 119–135. [Google Scholar] [CrossRef]
- Watanabe, I.; Kunito, T.; Tanabe, S.; Amano, M.; Koyama, Y.; Miyazaki, N.; Petrov, E.A.; Tatsukawa, R. Accumulation of heavy metals in Caspian seals (Phoca caspica). Arch. Environ. Contam. Toxicol. 2002, 43, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Kamalam, B.S.; Medale, F.; Panserat, S. Stephane Panserat, Utilisation of dietary carbohydrates in farmed fishes: New insights on influencing factors, biological limitations and future strategies. Aquaculture 2017, 467, 3–27. [Google Scholar] [CrossRef]
- NRC. Nutrient Requirements of Fish and Shrimp; National Academies Press: Washington, DC, USA, 2011. [Google Scholar]
- Hemre, G.-I.; Mommsen, T.P.; Krogdahl, Å. Carbohydrates in fish nutrition: Effects on growth, glucose metabolism and hepatic enzymes. Aquac. Nutr. 2002, 8, 175–194. [Google Scholar] [CrossRef]
- Chiang, J. Liver Physiology: Metabolism and Detoxification. In Pathobiology of Human Disease; McManus, L.M., Mitchell, R.N., Eds.; Academic Press: San Diego, CA, USA, 2014; pp. 1770–1782. [Google Scholar]
- Enes, P.; Peres, H.; Couto, A.; Oliva-Teles, A. Growth performance and metabolic utilization of diets including starch, dextrin, maltose or glucose as carbohydrate source by gilthead sea bream (Sparus aurata) juveniles. Fish Physiol. Biochem. 2010, 36, 903–910. [Google Scholar] [CrossRef]
- Viollet, B.; Andreelli, F. AMP-Activated Protein Kinase and Metabolic Control. In Diabetes—Perspectives in Drug Therapy; Schwanstecher, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 303–330. [Google Scholar]
- Enes, P.; Panserat, S.; Kaushik, S.; Oliva-Teles, A. Dietary Carbohydrate Utilization by European Sea Bass (Dicentrarchus labrax L.) and Gilthead Sea Bream (Sparus aurata L.) Juveniles. Rev. Fish. Sci. 2011, 19, 201–215. [Google Scholar] [CrossRef]
- Péres, A.; Cahu, C.L.; Zambonino Infante, J.L.; Le Gall, M.M.; Quazuguel, P. Amylase and trypsin responses to intake of dietary carbohydrate and protein depend on the developmental stage in sea bass (Dicentrarchus labrax) larvae. Fish Physiol. Biochem. 1996, 15, 237–242. [Google Scholar]
- Panserat, S.; Médale, F.; Blin, C.; Brèque, J.; Vachot, C.; Plagnes-Juan, E.; Gomes, E.; Krishnamoorthy, R.; Kaushik, S. Hepatic glucokinase is induced by dietary carbohydrates in rainbow trout, gilthead seabream, and common carp. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2000, 278, R1164–R1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Planas, J.V.; Capilla, E.; Gutiérrez, J. Molecular identification of a glucose transporter from fish muscle 1. FEBS Lett. 2000, 481, 266–270. [Google Scholar] [CrossRef] [Green Version]
- Das, T.; Pal, A.K.; Chakraborty, S.K.; Manush, S.M.; Sahu, N.P.; Mukherjee, S.C. Thermal tolerance, growth and oxygen consumption of Labeo rohita fry (Hamilton, 1822) acclimated to four temperatures. J. Therm. Biol. 2005, 30, 378–383. [Google Scholar] [CrossRef]
- Ashaf-Ud-Doulah, M.; Mamun, A.A.; Rahman, M.L.; Islam, S.M.M.; Jannat, R.; Hossain, M.A.R.; Shahjahan, M. High temperature acclimation alters upper thermal limits and growth performance of Indian major carp, rohu, Labeo rohita (Hamilton, 1822). J. Therm. Biol. 2020, 93, 102738. [Google Scholar] [CrossRef]
- Qian, X.; Ba, Y.; Zhuang, Q.; Zhong, G. RNA-Seq technology and its application in fish transcriptomics. Omics A J. Integr. Biol. 2014, 18, 98–110. [Google Scholar] [CrossRef] [Green Version]
- Chakrapani, V.; Patra, S.K.; Mohapatra, S.D.; Rasal, K.D.; Deshpande, U.; Nayak, S.; Sundaray, J.K.; Jayasankar, P.; Barman, H.K. Comparative transcriptomic profiling of larvae and post-larvae of Macrobrachium rosenbergii in response to metamorphosis and salinity exposure. Genes Genom. 2016, 38, 1061–1076. [Google Scholar] [CrossRef]
- Rasal, K.D.; Sundaray, J.K. Status of genetic and genomic approaches for delineating biological information and improving aquaculture production of farmed rohu, Labeo rohita (Ham, 1822). Rev. Aquacult. 2020. [Google Scholar] [CrossRef]
- Renukaradhya, K.M.; Varghese, T.J. Protein requirement of the carps, Catla catla (Hamilton) and Labeo rohita (Hamilton). Proc. Anim. Sci. 1986, 95, 103–107. [Google Scholar] [CrossRef]
- Mohapatra, M.; Sahu, N.P.; Chaudhari, A. Utilization of gelatinized carbohydrate in diets of Labeo rohita fry. Aquac. Nutr. 2003, 9, 189–196. [Google Scholar] [CrossRef]
- Kumar, S.; Sahu, N.P.; Pal, A.K.; Choudhury, D.; Yengkokpam, S.; Mukherjee, S.C. Effect of dietary carbohydrate on haematology, respiratory burst activity and histological changes in L. rohita juveniles. Fish Shellfish Immunol. 2005, 19, 331–344. [Google Scholar] [CrossRef]
- Satpathy, B.B.; Ray, A.K. Effect of dietary protein and carbohydrate levels on growth, nutrient utilization and body composition in fingerling rohu, Labeo rohita (Hamilton). J. Appl. Ichthyol. 2009, 25, 728–733. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Clar, J.; Gri, B.; Calderaro, J.; Birling, M.-C.; Hérault, Y.; Smit, G.P.A.; Mithieux, G.; Rajas, F. Targeted deletion of kidney glucose-6 phosphatase leads to nephropathy. Kidney Int. 2014, 86, 747–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, I.H.; Hsiao, C.-T.; Wu, J.-C.; Shen, R.-F.; Liu, C.-Y.; Wang, Y.-K.; Chen, Y.-C.; Huang, C.-M.; del Álamo, J.C.; Chang, Z.-F.; et al. GEF-H1 controls focal adhesion signaling that regulates mesenchymal stem cell lineage commitment. J. Cell Sci. 2014, 127, 4186–4200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grumati, P.; Morozzi, G.; Hölper, S.; Mari, M.; Harwardt, M.-L.I.E.; Yan, R.; Müller, S.; Reggiori, F.; Heilemann, M.; Dikic, I. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife 2017, 6, e25555. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Ko, C.-W.; Tso, P.; Bhargava, A. Apolipoprotein A-IV: A Multifunctional Protein Involved in Protection against Atherosclerosis and Diabetes. Cells 2019, 8, 319. [Google Scholar] [CrossRef] [Green Version]
- Gangadhariah, M.H.; Dieckmann, B.W.; Lantier, L.; Kang, L.; Wasserman, D.H.; Chiusa, M.; Caskey, C.F.; Dickerson, J.; Luo, P.; Gamboa, J.L.; et al. Cytochrome P450 epoxygenase-derived epoxyeicosatrienoic acids contribute to insulin sensitivity in mice and in humans. Diabetologia 2017, 60, 1066–1075. [Google Scholar] [CrossRef] [Green Version]
- Pocai, A.; Obici, S.; Schwartz, G.J.; Rossetti, L. A brain-liver circuit regulates glucose homeostasis. Cell Metab. 2005, 1, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Spiro, R.G. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 2002, 12, 43R–56R. [Google Scholar] [CrossRef]
- Oh, S.W.; Pope, R.K.; Smith, K.P.; Crowley, J.L.; Nebl, T.; Lawrence, J.B.; Luna, E.J. Archvillin, a muscle-specific isoform of supervillin, is an early expressed component of the costameric membrane skeleton. J. Cell Sci. 2003, 116, 2261–2275. [Google Scholar] [CrossRef] [Green Version]
- Groenestege, W.M.T.; Hoenderop, J.G.; van den Heuvel, L.; Knoers, N.; Bindels, R.J. The Epithelial Mg2+ Channel Transient Receptor Potential Melastatin 6 Is Regulated by Dietary Mg2+ Content and Estrogens. J. Am. Soc. Nephrol. 2006, 17, 1035–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunnally, M.H.; Stull, J.T. Mammalian skeletal muscle myosin light chain kinases. A comparison by antiserum cross-reactivity. J. Biol. Chem. 1984, 259, 1776–1780. [Google Scholar] [PubMed]
- Qiu, X.; Guo, H.; Yang, J.; Ji, Y.; Wu, C.-S.; Chen, X. Down-regulation of guanylate binding protein 1 causes mitochondrial dysfunction and cellular senescence in macrophages. Sci. Rep. 2018, 8, 1679. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.M.; Hemli, S.; Kolodny, E.H.; Miller, A.L.; Pallotta, J.A. Carbohydrate metabolism in phenylketonuria. Pediatric Res. 1980, 14, 849–853. [Google Scholar] [CrossRef]
- McMillan, D.E. Elevation of complement components in diabetes mellitus. Diabete Metab. 1980, 6, 265–270. [Google Scholar]
- Zhang, D.D.; Zhang, J.G.; Wang, Y.Z.; Liu, Y.; Liu, G.L.; Li, X.Y. Per-Arnt-Sim Kinase (PASK): An Emerging Regulator of Mammalian Glucose and Lipid Metabolism. Nutrients 2015, 7, 7437–7450. [Google Scholar] [CrossRef] [Green Version]
- Van Geet, C.; Jaeken, J. A unique pattern of coagulation abnormalities in carbohydrate-deficient glycoprotein syndrome. Pediatric Res. 1993, 33, 540–541. [Google Scholar] [CrossRef] [Green Version]
- Coan, P.M.; Barrier, M.; Alfazema, N.; Carter, R.N.; Marion de Proce, S.; Dopico, X.C.; Garcia Diaz, A.; Thomson, A.; Jackson-Jones, L.H.; Moyon, B.; et al. Complement Factor B Is a Determinant of Both Metabolic and Cardiovascular Features of Metabolic Syndrome. Hypertension 2017, 70, 624–633. [Google Scholar] [CrossRef]
- Tocher, D.R.; Bendiksen, E.Å.; Campbell, P.J.; Bell, J.G. The role of phospholipids in nutrition and metabolism of teleost fish. Aquaculture 2008, 280, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Causey, D.R.; Pohl, M.A.N.; Stead, D.A.; Martin, S.A.M.; Secombes, C.J.; Macqueen, D.J. High-throughput proteomic profiling of the fish liver following bacterial infection. BMC Genom. 2018, 19, 719. [Google Scholar] [CrossRef] [Green Version]
- Elshorbagy, A.K.; Nijpels, G.; Valdivia-Garcia, M.; Stehouwer, C.D.A.; Ocke, M.; Refsum, H.; Dekker, J.M. S-Adenosylmethionine Is Associated with Fat Mass and Truncal Adiposity in Older Adults. J. Nutr. 2013, 143, 1982–1988. [Google Scholar] [CrossRef]
- Kong, P.; Gonzalez-Quesada, C.; Li, N.; Cavalera, M.; Lee, D.-W.; Frangogiannis, N.G. Thrombospondin-1 regulates adiposity and metabolic dysfunction in diet-induced obesity enhancing adipose inflammation and stimulating adipocyte proliferation. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E439–E450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, P.; Kuang, S. Notch signaling as a novel regulator of metabolism. Trends Endocrinol. Metab. 2015, 26, 248–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kihara, A.; Sakuraba, H.; Ikeda, M.; Denpoh, A.; Igarashi, Y. Membrane topology and essential amino acid residues of Phs1, a 3-hydroxyacyl-CoA dehydratase involved in very long-chain fatty acid elongation. J. Biol. Chem. 2008, 283, 11199–11209. [Google Scholar] [CrossRef] [Green Version]
- Danzmann, R.G.; Kocmarek, A.L.; Norman, J.D.; Rexroad, C.E.; Palti, Y. Transcriptome profiling in fast versus slow-growing rainbow trout across seasonal gradients. BMC Genom. 2016, 17, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almenar-Queralt, A.; Gregorio, C.C.; Fowler, V.M. Tropomodulin assembles early in myofibrillogenesis in chick skeletal muscle: Evidence that thin filaments rearrange to form striated myofibrils. J. Cell Sci. 1999, 112, 1111–1123. [Google Scholar]
- Naylor, R.L.; Hardy, R.W.; Bureau, D.P.; Chiu, A.; Elliott, M.; Farrell, A.P.; Forster, I.; Gatlin, D.M.; Goldburg, R.J.; Hua, K.; et al. Feeding aquaculture in an era of finite resources. Proc. Natl. Acad. Sci. USA 2009, 106, 15103–15110. [Google Scholar] [CrossRef] [Green Version]
- Skiba-Cassy, S.; Panserat, S.; Larquier, M.; Dias, K.; Surget, A.; Plagnes-Juan, E.; Kaushik, S.; Seiliez, I. Apparent low ability of liver and muscle to adapt to variation of dietary carbohydrate:protein ratio in rainbow trout (Oncorhynchus mykiss). Br. J. Nutr. 2012, 109, 1359–1372. [Google Scholar] [CrossRef] [Green Version]
- Panserat, S.; Skiba-Cassy, S.; Seiliez, I.; Lansard, M.; Plagnes-Juan, E.; Vachot, C.; Aguirre, P.; Larroquet, L.; Chavernac, G.; Medale, F.; et al. Metformin improves postprandial glucose homeostasis in rainbow trout fed dietary carbohydrates: A link with the induction of hepatic lipogenic capacities? Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 297, R707–R715. [Google Scholar] [CrossRef] [Green Version]
- Polakof, S.; Skiba-Cassy, S.; Kaushik, S.; Seiliez, I.; Soengas, J.L.; Panserat, S. Glucose and lipid metabolism in the pancreas of rainbow trout is regulated at the molecular level by nutritional status and carbohydrate intake. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2012, 182, 507–516. [Google Scholar] [CrossRef]
- Marandel, L.; Seiliez, I.; Véron, V.; Skiba-Cassy, S.; Panserat, S. New insights into the nutritional regulation of gluconeogenesis in carnivorous rainbow trout (Oncorhynchus mykiss): A gene duplication trail. Physiol. Genom. 2015, 47, 253–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rui, L. Energy metabolism in the liver. Compr. Physiol. 2014, 4, 177–197. [Google Scholar] [PubMed] [Green Version]
- Jordan, S.D.; Könner, A.C.; Brüning, J.C. Sensing the fuels: Glucose and lipid signaling in the CNS controlling energy homeostasis. Cell. Mol. Life Sci. 2010, 67, 3255–3273. [Google Scholar] [CrossRef] [Green Version]
- Diaz, L.E.; Chuan, Y.C.; Lewitt, M.; Fernandez-Perez, L.; Carrasco-Rodriguez, S.; Sanchez-Gomez, M.; Flores-Morales, A. IGF-II regulates metastatic properties of choriocarcinoma cells through the activation of the insulin receptor. Mol. Hum. Reprod. 2007, 13, 567–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, M.; Vraskou, Y.; Gutierrez, J.; Planas, J.V. Expression of rainbow trout glucose transporters GLUT1 and GLUT4 during in vitro muscle cell differentiation and regulation by insulin and IGF-I. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 296, R794–R800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, H.-Z.; Deng, H.; Cen, C.-Q.; Chen, K.-Y.; Du, M.-L. Post-receptor crosstalk between growth hormone and insulin signal in rats born small for gestational age with catch-up growth. PLoS ONE 2014, 9, e100459. [Google Scholar] [CrossRef] [Green Version]
- Balmaceda-Aguilera, C.; Cortes-Campos, C.; Cifuentes, M.; Peruzzo, B.; Mack, L.; Tapia, J.C.; Oyarce, K.; Garcia, M.A.; Nualart, F. Glucose transporter 1 and monocarboxylate transporters 1, 2, and 4 localization within the glial cells of shark blood-brain-barriers. PLoS ONE 2012, 7, e32409. [Google Scholar] [CrossRef] [Green Version]
- Pyla, R.; Poulose, N.; Jun, J.Y.; Segar, L. Expression of conventional and novel glucose transporters, GLUT1, -9, -10, and -12, in vascular smooth muscle cells. Am. J. Physiol. Cell Physiol. 2013, 304, C574–C589. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Xie, J.; Yu, D.-G.; Wang, G.-J.; Yu, E.-M.; Gong, W.-B.; Li, Z.-F.; Wang, C.-C.; Xia, Y. A comparative study on the budget of nitrogen and phosphorus in polyculture systems of snakehead with bighead carp. Aquaculture 2018, 483, 69–75. [Google Scholar] [CrossRef]
- Chen, B.; Zheng, T.; Qin, L.; Hu, X.; Zhang, X.; Liu, Y.; Liu, H.; Qin, S.; Li, G.; Li, Q. Strong association between plasma dipeptidyl peptidase-4 activity and impaired cognitive function in elderly population with normal glucose tolerance. Front. Aging Neurosci. 2017, 9, 247. [Google Scholar] [CrossRef] [Green Version]
- Panserat, S.; Capilla, E.; Gutierrez, J.; Frappart, P.O.; Vachot, C.; Plagnes-Juan, E.; Aguirre, P.; Breque, J.; Kaushik, S. Glucokinase is highly induced and glucose-6-phosphatase poorly repressed in liver of rainbow trout (Oncorhynchus mykiss) by a single meal with glucose. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2001, 128, 275–283. [Google Scholar] [CrossRef]
- Hall, J.R.; Richards, R.C.; MacCormack, T.J.; Ewart, K.V.; Driedzic, W.R. Cloning of GLUT3 cDNA from Atlantic cod (Gadus morhua) and expression of GLUT1 and GLUT3 in response to hypoxia. Biochim. Biophys. Acta 2005, 1730, 245–252. [Google Scholar] [CrossRef]
- Seiliez, I.; Gabillard, J.-C.; Riflade, M.; Sadoul, B.; Dias, K.; Avérous, J.; Tesseraud, S.; Skiba, S.; Panserat, S. Amino acids downregulate the expression of several autophagy-related genes in rainbow trout myoblasts. Autophagy 2012, 8, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Médale, F.; Kamalam, B.S.; Aguirre, P.; Véron, V.; Panserat, S. Comparison of glucose and lipid metabolic gene expressions between fat and lean lines of rainbow trout after a glucose load. PLoS ONE 2014, 9, e105548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, K.; Pan, M.; Liu, J.; Yang, M.; Gu, Z.; Zhang, Y.; Liu, G.; Liu, D.; Zhang, W.; Mai, K. Chronic stress of high dietary carbohydrate level causes inflammation and influences glucose transport through SOCS3 in Japanese flounder Paralichthys olivaceus. Sci. Rep. 2018, 8, 7415. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Marandel, L.; Skiba-Cassy, S.; Corraze, G.; Dupont-Nivet, M.; Quillet, E.; Geurden, I.; Panserat, S. Regulation by dietary carbohydrates of intermediary metabolism in liver and muscle of two isogenic lines of rainbow trout. Front. Physiol. 2018, 9, 1579. [Google Scholar] [CrossRef]
- Lu, S.; Wu, X.; Gao, Y.; Gatlin, D.M.; Wu, M.; Yao, W.; Jin, Z.; Li, X.; Dong, Y. Effects of dietary carbohydrate sources on growth, digestive enzyme activity, gene expression of hepatic GLUTs and key enzymes involved in glycolysis-gluconeogenesis of giant grouper Epinephelus lanceolatus larvae. Aquaculture 2018, 484, 343–350. [Google Scholar] [CrossRef]
- Meton, I.; Caseras, A.; Fernandez, F.; Baanante, I.V. Molecular cloning of hepatic glucose-6-phosphatase catalytic subunit from gilthead sea bream (Sparus aurata): Response of its mRNA levels and glucokinase expression to refeeding and diet composition. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2004, 138, 145–153. [Google Scholar] [CrossRef]
- Iynedjian, P.B. Molecular physiology of mammalian glucokinase. Cell. Mol. Life Sci. CMLS 2009, 66, 27–42. [Google Scholar] [CrossRef] [Green Version]
- Wegener, G.; Krause, U. Different modes of activating phosphofructokinase, a key regulatory enzyme of glycolysis, in working vertebrate muscle. Biochem. Soc. Trans. 2002, 30, 264–270. [Google Scholar] [CrossRef]
- Yuan, X.; Wood, E.F. Multimodel seasonal forecasting of global drought onset. Geophys. Res. Lett. 2013, 40, 4900–4905. [Google Scholar] [CrossRef]
- Wang, B.-K.; Liu, W.-B.; Xu, C.; Cao, X.-F.; Zhong, X.-Q.; Shi, H.-J.; Li, X.-F. Dietary carbohydrate levels and lipid sources modulate the growth performance, fatty acid profiles and intermediary metabolism of blunt snout bream Megalobrama amblycephala in an interactive pattern. Aquaculture 2017, 481, 140–153. [Google Scholar] [CrossRef]
- Li, X.-F.; Xu, C.; Zhang, D.-D.; Jiang, G.-Z.; Liu, W.-B. Molecular characterization and expression analysis of glucokinase from herbivorous fish Megalobrama amblycephala subjected to a glucose load after the adaption to dietary carbohydrate levels. Aquaculture 2016, 459, 89–98. [Google Scholar] [CrossRef]
- Enes, P.; Panserat, S.; Kaushik, S.; Oliva-Teles, A. Growth performance and metabolic utilization of diets with native and waxy maize starch by gilthead sea bream (Sparus aurata) juveniles. Aquaculture 2008, 274, 101–108. [Google Scholar] [CrossRef]
- Seiliez, I.; Panserat, S.; Skiba-Cassy, S.; Polakof, S. Effect of acute and chronic insulin administrations on major factors involved in the control of muscle protein turnover in rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 2011, 172, 363–370. [Google Scholar] [CrossRef]
- Lehninger, A.L. Proton and electric charge translocation in mitochondrial energy transduction. Adv. Exp. Med. Biol. 1982, 148, 171–186. [Google Scholar] [PubMed]
- Borrebaek, B.; Christophersen, B. Activities of glucose phosphorylation, glucose-6-phosphatase and lipogenic enzymes in the liver of perch, Perca fluviatilis, after different dietary treatment. Aquac. Res. 2001, 32, 221–224. [Google Scholar] [CrossRef]
- Metón, I.; Caseras, A.; Mediavilla, D.; Fernández, F.; Baanante, I.V. Molecular cloning of a cDNA encoding 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase from liver of Sparus aurata: Nutritional regulation of enzyme expression. Biochim. Et Biophys. Acta (BBA) Gene Struct. Expr. 1999, 1444, 153–165. [Google Scholar]
- Enes, P.; Panserat, S.; Kaushik, S.; Oliva-Teles, A. Effect of normal and waxy maize starch on growth, food utilization and hepatic glucose metabolism in European sea bass (Dicentrarchus labrax) juveniles. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2006, 143, 89–96. [Google Scholar] [CrossRef]
- Tanan, C.L.; Ventura, D.F.; de Souza, J.M.; Grotzner, S.R.; Mela, M.; Gouveia, A., Jr.; Oliveira-Ribeiro, C.A. Effects of mercury intoxication on the response of horizontal cells of the retina of thraira fish (Hoplias malabaricus). Braz. J. Med Biol. Res. Rev. Bras. Pesqui. Med. E Biol. 2006, 39, 987–995. [Google Scholar] [CrossRef]
- Caseras, A.; Meton, I.; Vives, C.; Egea, M.; Fernandez, F.; Baanante, I.V. Nutritional regulation of glucose-6-phosphatase gene expression in liver of the gilthead sea bream (Sparus aurata). Br. J. Nutr. 2002, 88, 607–614. [Google Scholar] [CrossRef] [Green Version]
- Agius, L. Glucokinase and molecular aspects of liver glycogen metabolism. Biochem. J. 2008, 414, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Foufelle, F.; Girard, J.; Ferré, P. Regulation of lipogenic enzyme expression by glucose in liver and adipose tissue: A review of the potential cellular and molecular mechanisms. Adv. Enzym. Regul. 1996, 36, 199–226. [Google Scholar] [CrossRef]
- Moreira, I.S.; Peres, H.; Couto, A.; Enes, P.; Oliva-Teles, A. Temperature and dietary carbohydrate level effects on performance and metabolic utilisation of diets in European sea bass (Dicentrarchus labrax) juveniles. Aquaculture 2008, 274, 153–160. [Google Scholar] [CrossRef]
- Ren, M.; Habte-Tsion, H.-M.; Xie, J.; Liu, B.; Zhou, Q.; Ge, X.; Pan, L.; Chen, R. Effects of dietary carbohydrate source on growth performance, diet digestibility and liver glucose enzyme activity in blunt snout bream, Megalobrama amblycephala. Aquaculture 2015, 438, 75–81. [Google Scholar] [CrossRef]
- Nordlie, R.C.; Foster, J.D.; Lange, A.J. Regulation of glucose production by the liver. Annu. Rev. Nutr. 1999, 19, 379–406. [Google Scholar] [CrossRef]
- van de Werve, G.; Lange, A.; Newgard, C.; Méchin, M.-C.; Li, Y.; Berteloot, A. New lessons in the regulation of glucose metabolism taught by the glucose 6-phosphatase system. Eur. J. Biochem. 2000, 267, 1533–1549. [Google Scholar] [CrossRef]
- Argaud, D.; Zhang, Q.; Pan, W.; Maitra, S.; Pilkis, S.J.; Lange, A.J. Regulation of rat liver glucose-6-phosphatase gene expression in different nutritional and hormonal states: Gene structure and 5’-flanking sequence. Diabetes 1996, 45, 1563–1571. [Google Scholar] [CrossRef]
- Fang, X.; Zhang, B.; Thisse, B.; Bloom, G.S.; Thisse, C. IQGAP3 is essential for cell proliferation and motility during zebrafish embryonic development. Cytoskeleton 2015, 72, 422–433. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.P.; Ge, X.P.; Liu, B.; Xie, J.; Miao, L.H. Effect of High Dietary Carbohydrate on the Growth Performance and Physiological Responses of Juvenile Wuchang Bream, Megalobrama amblycephala. Asian-Australas. J. Anim. Sci. 2013, 26, 1598–1608. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Kidokoro, H.; Watanabe, T.; Igeta, Y.; Sakaji, H.; Ino, S. Response of yellowtail, Seriola quinqueradiata, a key large predatory fish in the Japan Sea, to sea water temperature over the last century and potential effects of global warming. J. Mar. Syst. 2012, 91, 1–10. [Google Scholar] [CrossRef]
- Zamora-Sillero, J.; Ramos, L.R.V.; Romano, L.A.; Monserrat, J.M.; Tesser, M.B. Effect of dietary dextrin levels on the growth performance, blood chemistry, body composition, hepatic triglicerides and glycogen of Lebranche mullet juveniles (Mugil liza Valenciennes 1836, Mugilidae). J. Appl. Ichthyol. 2013, 29, 1342–1347. [Google Scholar] [CrossRef]
- Azaza, M.S.; Assad, A.; Maghrbi, W.; El-Cafsi, M. The effects of rearing density on growth, size heterogeneity and inter-individual variation of feed intake in monosex male Nile tilapia Oreochromis niloticus L. Anim. Int. J. Anim. Biosci. 2013, 7, 1865–1874. [Google Scholar] [CrossRef] [Green Version]
- Moro, G.V.; Silva, T.S.C.; Cyrino, J.E.P. Feed processing affected the digestibility of carbohydrate sources for dourado Salminus brasiliensis. Aquac. Nutr. 2017, 23, 40–45. [Google Scholar] [CrossRef]
- He, R.; Zhang, D.; Lu, W.; Zheng, T.; Wan, L.; Liu, F.; Jia, W. SLC47A1 gene rs2289669 G>A variants enhance the glucose-lowering effect of metformin via delaying its excretion in Chinese type 2 diabetes patients. Diabetes Res. Clin. Pract. 2015, 109, 57–63. [Google Scholar] [CrossRef]
- Zhou, K.; Yee, S.W.; Seiser, E.L.; van Leeuwen, N.; Tavendale, R.; Bennett, A.J.; Groves, C.J.; Coleman, R.L.; van der Heijden, A.A.; Beulens, J.W.; et al. Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat. Genet. 2016, 48, 1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges, P.; Valente, L.M.; Veron, V.; Dias, K.; Panserat, S.; Medale, F. High dietary lipid level is associated with persistent hyperglycaemia and downregulation of muscle Akt-mTOR pathway in Senegalese sole (Solea senegalensis). PLoS ONE 2014, 9, e102196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, X.; Hu, Y.; Yan, N.; Huang, Y.; Peng, N.; Liang, Y.; Zhao, S. PCR-DGGE analysis of the microbial communities in three different Chinese “Baiyunbian” liquor fermentation starters. J. Microbiol. Biotechnol. 2014, 24, 1088–1095. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.-J.; Zhang, T.-Y.; Chen, H.-Y.; Lin, S.-M.; Luo, L.; Wang, D.-S. An evaluation of hepatic glucose metabolism at the transcription level for the omnivorous GIFT tilapia, Oreochromis niloticus during postprandial nutritional status transition from anabolism to catabolism. Aquaculture 2017, 473, 375–382. [Google Scholar] [CrossRef]
- Xie, D.; Yang, L.; Yu, R.; Chen, F.; Lu, R.; Qin, C.; Nie, G. Effects of dietary carbohydrate and lipid levels on growth and hepatic lipid deposition of juvenile tilapia, Oreochromis niloticus. Aquaculture 2017, 479, 696–703. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, H.; Hamera, S.; Chen, X.; Fang, R. miR444a has multiple functions in the rice nitrate-signaling pathway. Plant J. 2014, 78, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.J.; Diez, A.; Lopez-Bote, C.; Gallego, M.; Bautista, J.M. Short-term modulation of lipogenesis by macronutrients in rainbow trout (Oncorhynchus mykiss) hepatocytes. Br. J. Nutr. 2000, 84, 619–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rollin, X.; Mambrini, M.; Abboudi, T.; Larondelle, Y.; Kaushik, S.J. The optimum dietary indispensable amino acid pattern for growing Atlantic salmon (Salmo salar L.) fry. Br. J. Nutr. 2003, 90, 865–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, S.A.M.; Krol, E. Nutrigenomics and immune function in fish: New insights from omics technologies. Dev. Comp. Immunol. 2017, 75, 86–98. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-H.; Chiu, C.-H.; Wang, S.-W.; Cheng, W. Dietary administration of the probiotic, Bacillus subtilis E20, enhances the growth, innate immune responses, and disease resistance of the grouper, Epinephelus coioides. Fish Shellfish Immunol. 2012, 33, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Basu, N.; Todgham, A.E.; Ackerman, P.A.; Bibeau, M.R.; Nakano, K.; Schulte, P.M.; Iwama, G.K. Heat shock protein genes and their functional significance in fish. Gene 2002, 295, 173–183. [Google Scholar] [CrossRef]
- Zhou, C.; Liu, B.; Ge, X.; Xie, J.; Xu, P. Effect of dietary carbohydrate on the growth performance, immune response, hepatic antioxidant abilities and heat shock protein 70 expression of Wuchang bream, Megalobrama amblycephala. J. Appl. Ichthyol. 2013, 29, 1348–1356. [Google Scholar] [CrossRef]
- Yang, Y.; Han, T.; Xiao, J.; Li, X.; Wang, J. Transcriptome analysis reveals carbohydrate-mediated liver immune responses in Epinephelus akaara. Sci. Rep. 2018, 8, 639. [Google Scholar] [CrossRef] [Green Version]
- Figueiredo, M.A.; Mareco, E.A.; Silva, M.D.P.; Marins, L.F. Muscle-specific growth hormone receptor (GHR) overexpression induces hyperplasia but not hypertrophy in transgenic zebrafish. Transgenic Res. 2012, 21, 457–469. [Google Scholar] [CrossRef]
- Prisingkorn, W.; Prathomya, P.; Jakovlic, I.; Liu, H.; Zhao, Y.H.; Wang, W.M. Transcriptomics, metabolomics and histology indicate that high-carbohydrate diet negatively affects the liver health of blunt snout bream (Megalobrama amblycephala). BMC Genom. 2017, 18, 856. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.-H.; Sun, H.; Yan, G.-L.; Han, Y.; Zhao, Q.-Q.; Wang, X.-J. Chinmedomics: A Powerful Approach Integrating Metabolomics with Serum Pharmacochemistry to Evaluate the Efficacy of Traditional Chinese Medicine. Engineering 2019, 5, 60–68. [Google Scholar] [CrossRef]
- Zhuo, C.-X.; Zheng, C.; You, S.-L. Transition-Metal-Catalyzed Asymmetric Allylic Dearomatization Reactions. Acc. Chem. Res. 2014, 47, 2558–2573. [Google Scholar] [CrossRef] [PubMed]
- Polak, P.; Hall, M.N. mTOR and the control of whole body metabolism. Curr. Opin. Cell Biol. 2009, 21, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Panserat, S.; Mennigen, J.; Terrier, F.; Dias, K.; Seiliez, I.; Skiba-Cassy, S. Postprandial regulation of hepatic glucokinase and lipogenesis requires the activation of TORC1 signaling in rainbow trout (Oncorhynchus mykiss). J. Exp. Biol. 2013. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; Kertesz, M.; Spitale, R.C.; Segal, E.; Chang, H.Y. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 2011, 12, 641–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yecies, J.L.; Zhang, H.H.; Menon, S.; Liu, S.; Yecies, D.; Lipovsky, A.I.; Gorgun, C.; Kwiatkowski, D.J.; Hotamisligil, G.S.; Lee, C.H.; et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011, 14, 21–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
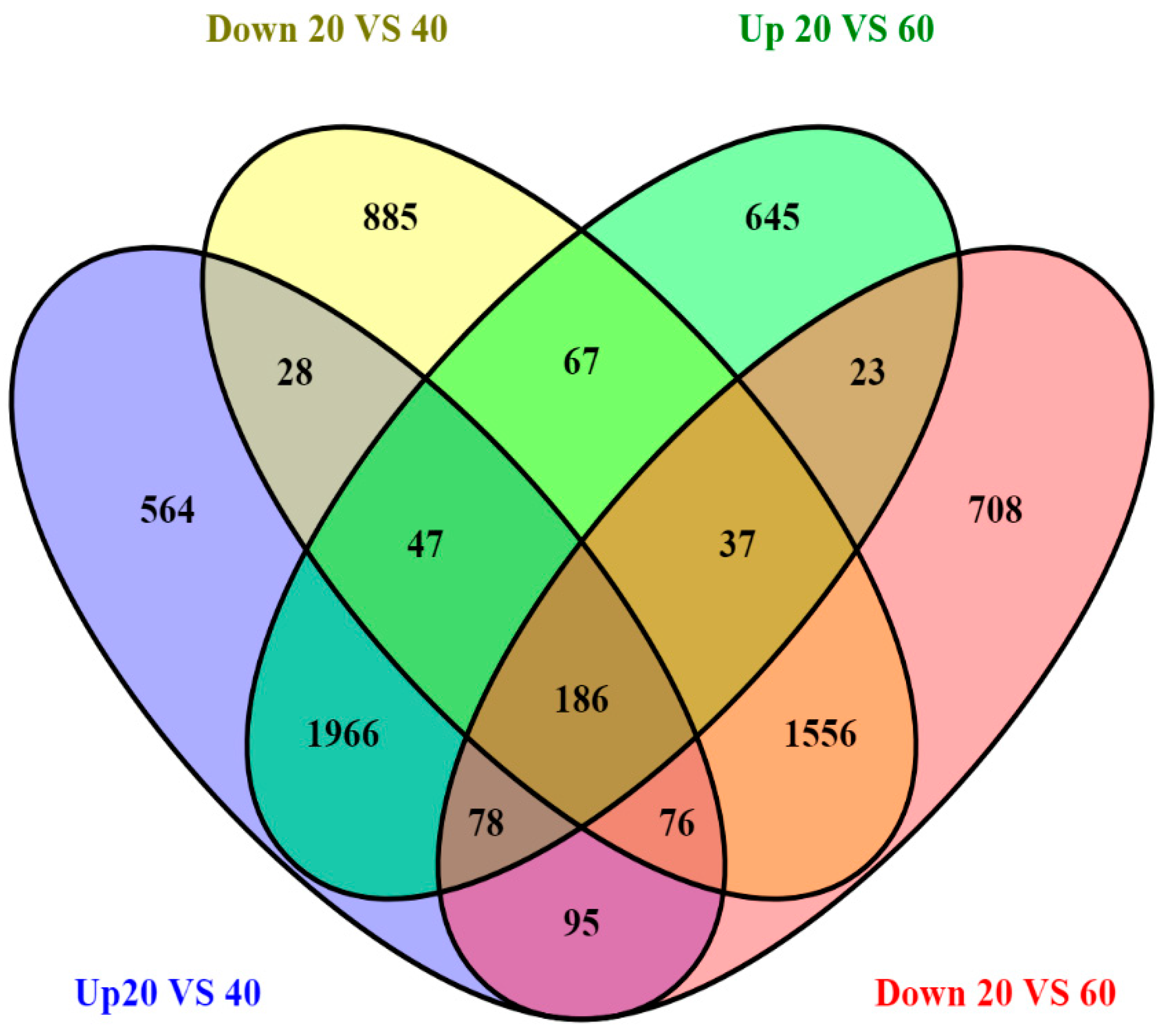
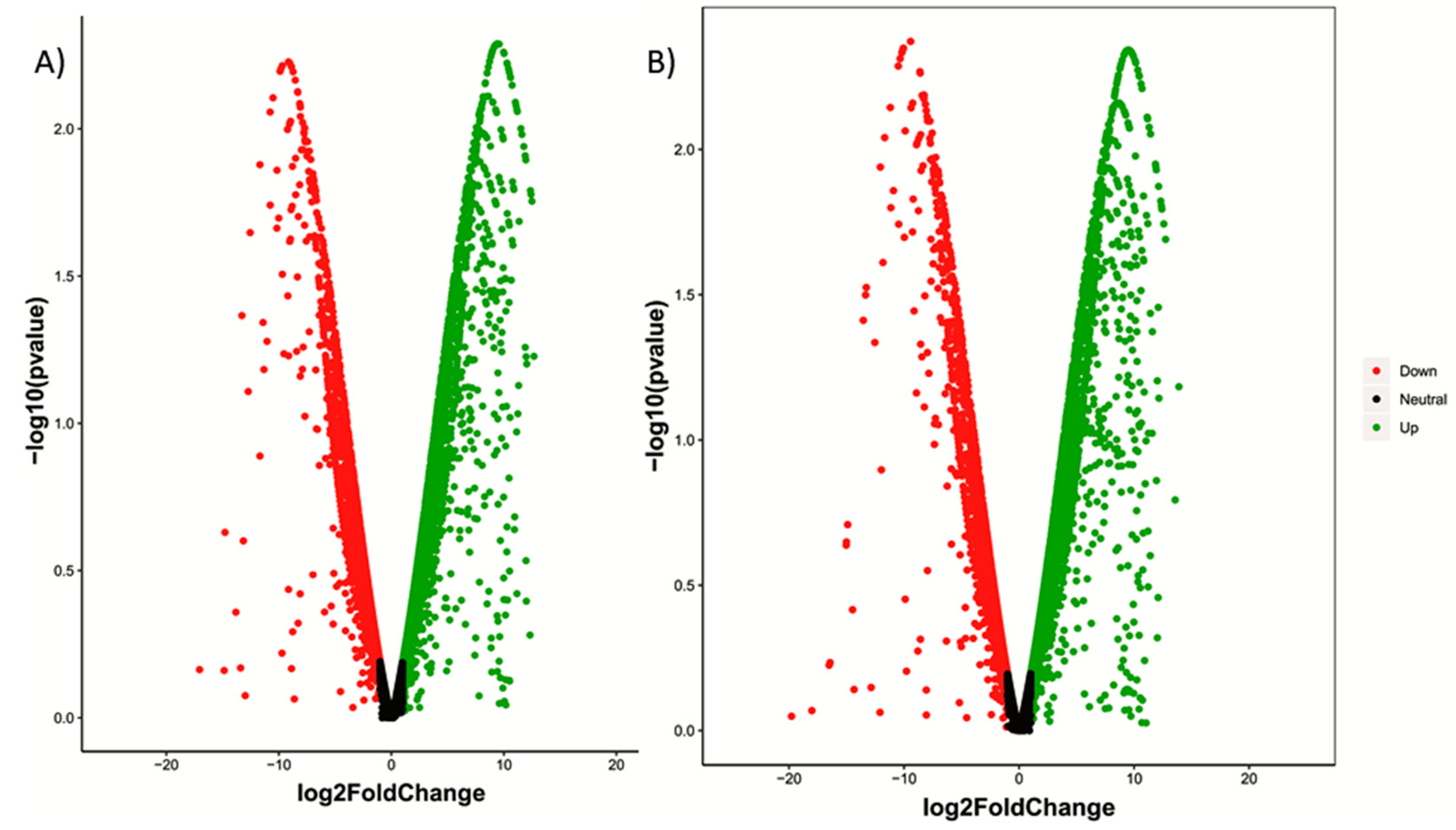
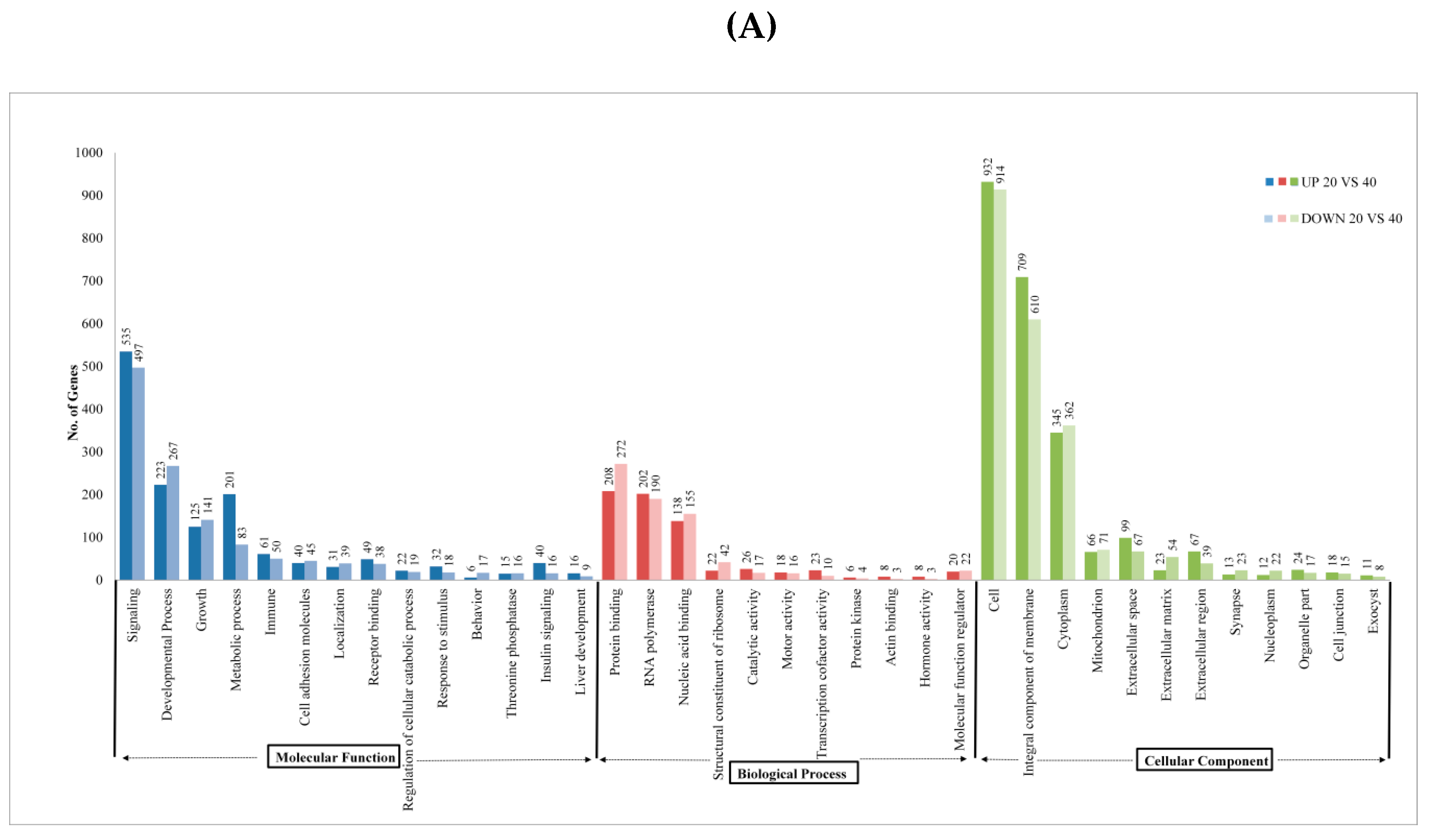
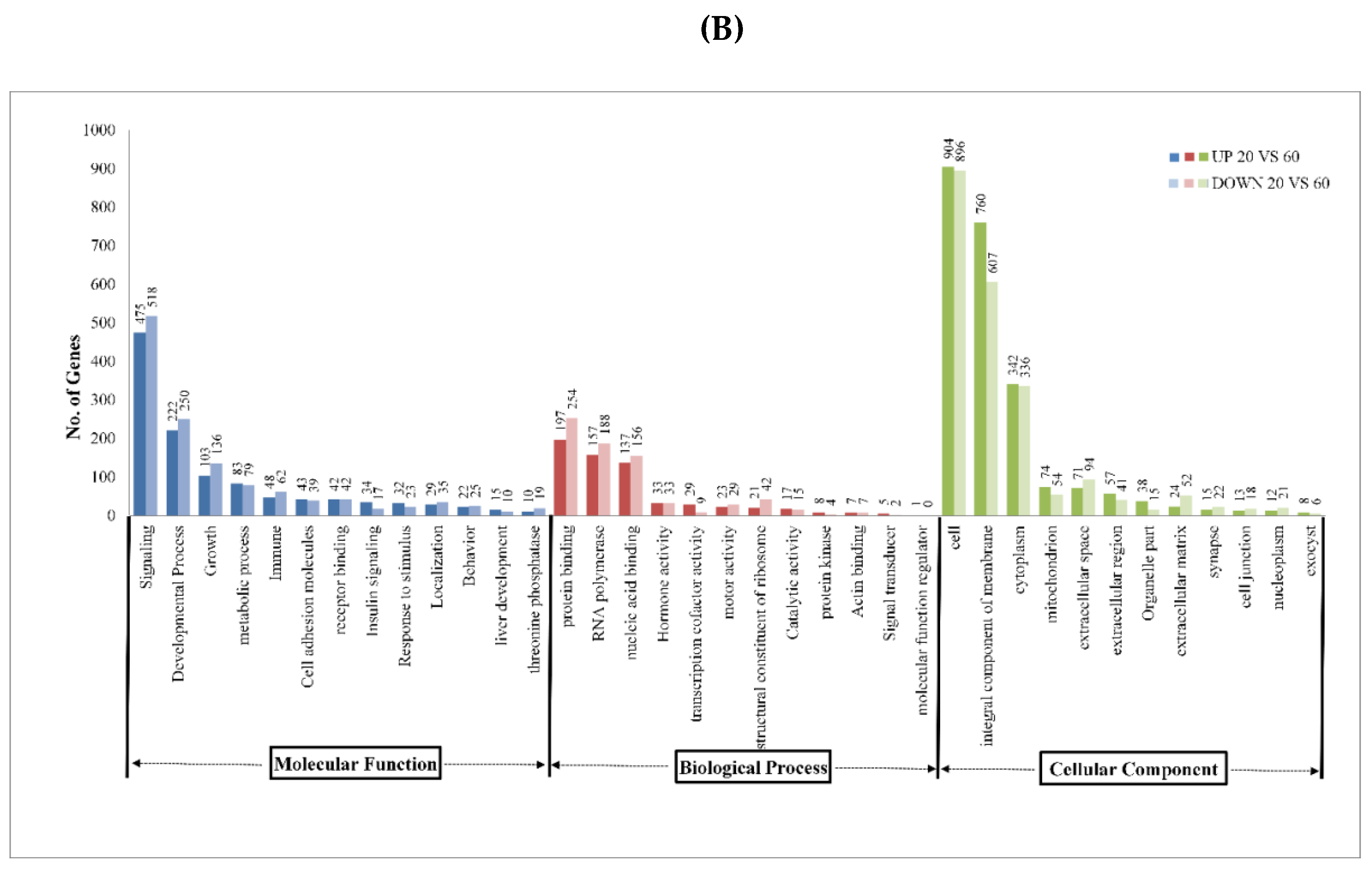


| Sample Name | Raw Reads | Processed Reads | % of Aligned Reads |
|---|---|---|---|
| 60A_LV | 35,558,985 | 30,800,241 | 89.58% |
| 20_LV | 23,396,281 | 21,202,572 | 92.89% |
| 40_LV | 24,004,149 | 21,608,231 | 89.84% |
| (A) | ||||
| Differential Gene Expression Statistics | ||||
| Summary | Total | Up | Down | Neutral |
| No. of transcripts expressed in both samples | 15,232 | 4464 | 4343 | 6425 |
| No. of transcripts expressed only in 20_LV | 3653 | NA | NA | NA |
| No. of transcripts expressed only in 40_LV | 2814 | NA | NA | NA |
| No. of P significant transcripts | 574 | 408 | 166 | 0 |
| No. of Q significant transcripts | 0 | 0 | 0 | 0 |
| (B) | ||||
| Differential Gene Expression Statistics | ||||
| Summary | Total | Up | Down | Neutral |
| No. of transcripts expressed in both samples | 15,360 | 4478 | 4171 | 6711 |
| No. of transcripts expressed only in 20_LV | 3525 | NA | NA | NA |
| No. of transcripts expressed only in 60A_LV | 2840 | NA | NA | NA |
| No. of P significant transcripts | 633 | 432 | 201 | 0 |
| No. of Q significant transcripts | 0 | 0 | 0 | 0 |
| Sl. No. | Hub genes | Function | Reference |
|---|---|---|---|
| 1 | Synaptopodin | Down-regulation of Synaptopodin is known to facilitate glycogen accumulation. | [26] |
| 2 | LIM and senescent cell antigen-containing domain 1-like protein | LIM and senescent cell antigen-containing domain 1-like protein (fragment) is down-regulated to increase abiogenesis. | [27] |
| 3 | Reticulon | Regulates fragmentation of ER during starvation but when glucose is in excess. | [28] |
| 4 | Apolipoprotein | ApoA-IV improves glucose homeostasis by promoting insulin secretion at high levels of glucose. | [29] |
| 5 | Cytochrome P450 | Improves glucose homeostasis. | [30] |
| 6 | Parvalbumin alpha | Regulates brain–liver circuit for glucose homeostasis. | [31] |
| 7 | Protein O-mannosyl-transferase 2 | Involved in biosynthesis of glycopeptides. | [32] |
| 8 | Supervillin-like isoform X2 | Controls myogenesis and contributes to myogenic membrane structure and differentiation. | [33] |
| 9 | Transient receptor potential melastatin 4a | Plays role as gatekeeper in transepithelial Mg2+ transport to maintain Mg2+ homeostasis. | [34] |
| 10 | Myosin, light polypeptide 3, skeletal muscle | Controls oxidative glycolytic pathway in carbohydrate metabolism. | [35] |
| 11 | Guanylate-binding protein 2 | Plays role in increasing activation of AMPK-p53 pathway and β-galactosidase. | [36] |
| 12 | Phenylalanine hydroxylase | Controls insulin secretion and glucose transport. | [37] |
| 13 | Complement C4-2 | Associated with glucose intolerance. | [38] |
| 14 | TC1-like transposase | Reported to be associated with down-regulation of glycogenesis. | [39] |
| 15 | Coagulation factor IXb (Zgc:136807) | Reported to be associated with carbohydrate deficiency. | [40] |
| 16 | Complement factor B/C2A | Elevated level in adipose tissue leads to redistribution of visceral to subcutaneous fat and insulin resistance. | [41] |
| 17 | Phospholipid-transporting ATPase (EC 7.6.2.1) | Controls efficiency of transport of dietary fatty acids and lipids in fish. | [42] |
| 18 | Alpha-2-macroglobulin (Fragment) | Primarily associated with immunity by remodeling of liver metabolism in fish. It controls activation of the PI3K/Akt, ERK1, and MAPK pathways leading to mobilization of energetic resources away from growth and protein synthesis. | [43] |
| 19 | S-adenosylmethionine synthase (EC 2.5.1.6) | Associated with BMI and adiposity. | [44] |
| 20 | Thrombospondin 2b | Associated with fatty acid uptake by adipocytes. | [45] |
| 21 | Notchless homolog 1 (Drosophila) | Notch signaling central hub gene controlling glucose and lipid metabolism by insulin; also involved in inhibition of liver glucose production, including glycogenolysis and gluconeogenesis. | [46] |
| 22 | Very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase (EC 4.2.1.134) | Involved in lipid biosynthesis especially in elongation of very-long-chain fatty acids. | [47] |
| 23 | Obscurin-like 1b | In fish myofibrillar component genes, obscurin is up-regulated with growth. | [48] |
| 24 | Sk-tropomodulin (tropomodulin 4 (muscle)) | Regulates thin filament lengths in muscles. | [49] |
| Ingredients | Treatment_1 (20%) | Treatment_2 40% | Treatment_3 60% |
|---|---|---|---|
| Fish Meal | 20 | 20 | 20 |
| Casein | 10.2 | 10.2 | 10.2 |
| Gelatinized Starch | 20 | 40 | 60 |
| Glucose | 0 | 0 | 0 |
| Cellulose | 40 | 20 | 0 |
| Fish Oil | 4 | 4 | 4 |
| Vegetable Oil | 3 | 3 | 3 |
| Carboxy methyl cellulose (Binder) | 0.5 | 0.5 | 0.5 |
| Mineral Mix | 1 | 1 | 1 |
| Vitamin Mix | 1 | 1 | 1 |
| Vitamin C | 0.2 | 0.2 | 0.2 |
| Butylhydroxytoluene (BHT) | 0.1 | 0.1 | 0.1 |
| 100 | 100 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rasal, K.D.; Iquebal, M.A.; Dixit, S.; Vasam, M.; Raza, M.; Sahoo, L.; Jaiswal, S.; Nandi, S.; Mahapatra, K.D.; Rasal, A.; et al. Revealing Alteration in the Hepatic Glucose Metabolism of Genetically Improved Carp, Jayanti Rohu Labeo rohita Fed a High Carbohydrate Diet Using Transcriptome Sequencing. Int. J. Mol. Sci. 2020, 21, 8180. https://doi.org/10.3390/ijms21218180
Rasal KD, Iquebal MA, Dixit S, Vasam M, Raza M, Sahoo L, Jaiswal S, Nandi S, Mahapatra KD, Rasal A, et al. Revealing Alteration in the Hepatic Glucose Metabolism of Genetically Improved Carp, Jayanti Rohu Labeo rohita Fed a High Carbohydrate Diet Using Transcriptome Sequencing. International Journal of Molecular Sciences. 2020; 21(21):8180. https://doi.org/10.3390/ijms21218180
Chicago/Turabian StyleRasal, Kiran D., Mir Asif Iquebal, Sangita Dixit, Manohar Vasam, Mustafa Raza, Lakshman Sahoo, Sarika Jaiswal, Samiran Nandi, Kanta Das Mahapatra, Avinash Rasal, and et al. 2020. "Revealing Alteration in the Hepatic Glucose Metabolism of Genetically Improved Carp, Jayanti Rohu Labeo rohita Fed a High Carbohydrate Diet Using Transcriptome Sequencing" International Journal of Molecular Sciences 21, no. 21: 8180. https://doi.org/10.3390/ijms21218180









