State-of-the-Art Technologies for Understanding Brassinosteroid Signaling Networks
Abstract
:1. Introduction
2. Identification of New Components of BR Signaling
2.1. Forward Genetics
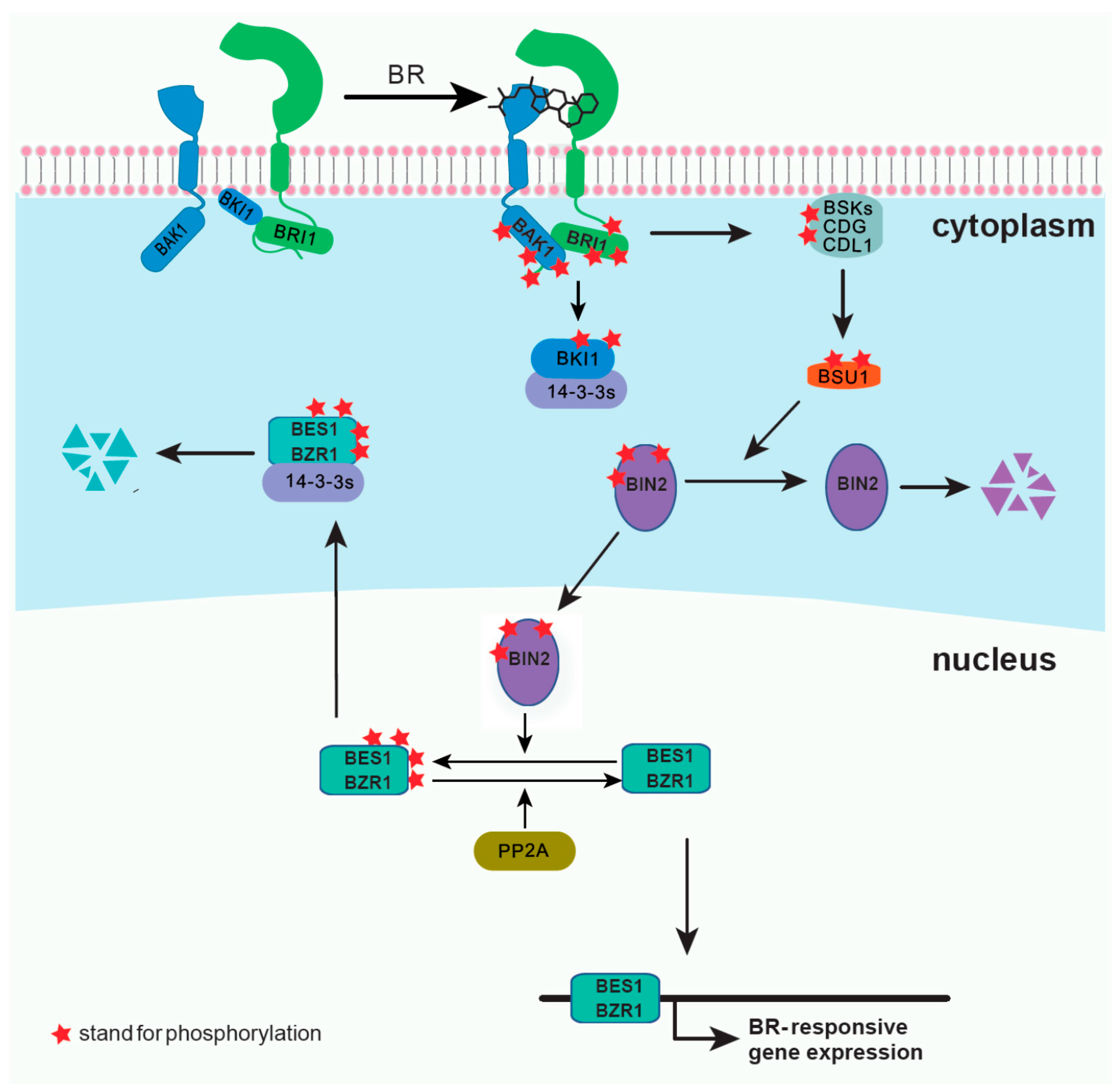
2.1.1. Screening of EMS Mutants and Identification of BRI1, BIN2, and the Transcription Factors BES1 and BZR1
2.1.2. Activation Tagging
2.2. Reverse Genetics
2.2.1. Y2H
2.2.2. 2D-DIGE
2.2.3. LC–MS/MS
2.2.4. Bioinformatics for Identification of BR Signaling Components
2.2.5. CRISPR/Cas9 System
3. Examination of the Dynamic Regulation Mechanism of BR Signaling Components
3.1. Plant Hormone Signaling Pathway Studied Using PL
3.1.1. PL Can Be Categorized by the Enzyme Used
3.1.2. Search for the PL Method Suitable for In Planta Studies
3.1.3. Early PL Applications in Planta
3.1.4. TurboID for BR Signaling Pathway
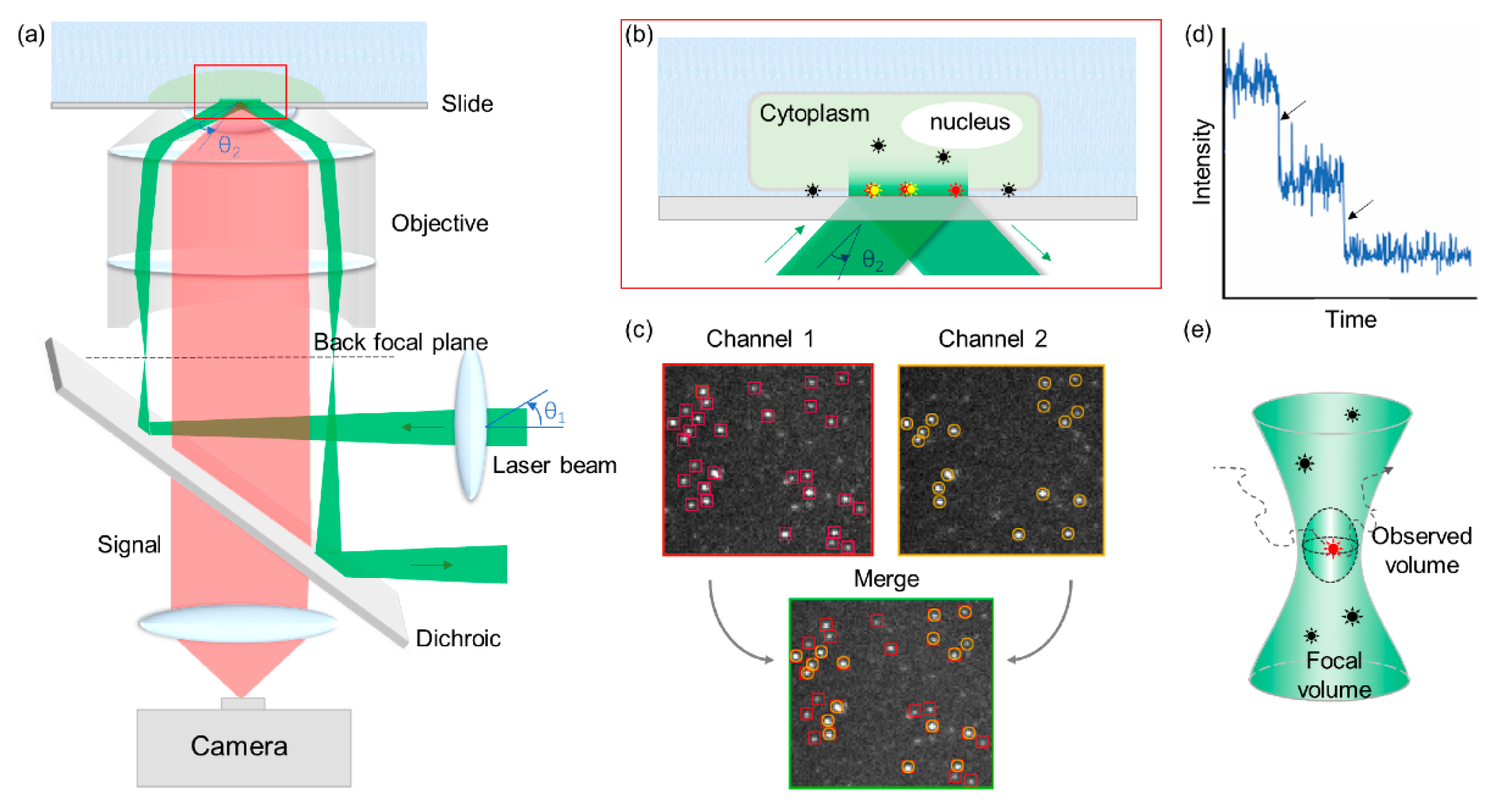
3.2. Single-Molecule Technologies for Studying BR System
3.2.1. TIRFM and VA-TIRFM
3.2.2. FRET
3.2.3. CoSMoS
3.2.4. Fluorescence Correlation Spectroscopy/Fluorescence Cross-Correlation Spectroscopy (FCS/FCCS) and Photon Counting Histogram (PCH)
3.2.5. Discoveries in BR Signaling Pathway by Using SM Methods
4. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 2D-DIGE | Two-dimensional difference gel electrophoresis |
| AD | activation domain |
| APEX | an engineered ascorbate peroxidase |
| BAK1 | BRI1-associated kinase 1 |
| BAP | biotin acceptor peptide |
| BD | binding domain |
| BEH1-4 | BES1/BZR1 homolog 1-4 |
| BES1 | BRI1 ethyl methanesulfonate suppressor 1 |
| BES1-h | The BES1 hextuple mutants |
| BiFC | bimolecular fluorescence complementation |
| BIN2 | BR-insensitive 2 |
| BioID | biotin identification |
| BirA* | a BirA mutant |
| BKI1 | BRI1 kinase inhibitor 1 |
| BLINC | biotin labeling of intercellular contacts |
| BRI1 | BR-insensitive 1 |
| BRs | Brassinosteroids |
| BRZ | brassinazole |
| BSKs | BR signaling kinases |
| BSLs | BSU1 like |
| BSU1 | BRI1-suppressor 1 |
| BZR1 | brassinazole resistant 1 |
| bzr1-1D | bzr1-1 Dominant |
| CDG1 | constitutive differential growth 1 |
| CoSMoS | colocalization single-molecule spectroscopy |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| DSBs | double-strand breaks |
| EMARS | enzyme-mediated activation of radical source |
| EMS | ethyl methanesulfonate |
| FCS/FCCS | fluorescence correlation spectroscopy/fluorescence cross-correlation spectroscopy |
| FRET | Förster-type resonance energy transfer |
| gRNA | guide RNA |
| HRP | horseradish peroxidase |
| ID-PRIME | interaction-dependent probe incorporation mediated by enzymes |
| LA | lipoic acid |
| LC–MS/MS | liquid chromatography–tandem mass spectrometry |
| MAKR1 | Membrane-associated kinase regulator |
| MS | mass spectrometry |
| NLRs | nucleotide-binding leucine-rich repeats |
| NPH3 | nonphototropic hypocotyl 3 |
| PAGE | Polyacrylamide gel electrophoresis |
| PCH | photon counting histogram |
| PHOT1 | Phototropin 1 |
| PL | proximity labeling |
| POI | protein of interest |
| PP2A | protein phosphatase 2A |
| PPIs | protein–protein interactions |
| PUP-IT | pupylation-based interaction tagging |
| RNP | ribonucleoprotein |
| SnRK2.2/2.3/2.6 | Snf1-related Kinase 2.2/2.3/2.6 |
| SPARK | specific protein association toll giving transcriptional readout with rapid kinetics |
| SPPLAT | selective proteomic PL using tyramide |
| T-DNA | Transfer DNA |
| TIR | toll/interleukin-1 receptor |
| TIRFM | total internal reflection fluorescence microscopy |
| TMV | tobacco mosaic virus |
| TTL | transthyretin-like protein |
| UBR7 | a putative E3 ubiquitin ligase |
| VA-TIRFM | variable-angle total internal reflection fluorescence microscopy |
| Y2H | yeast two-hybrid |
References
- Yang, C.-J.; Zhang, C.; Lu, Y.-N.; Jin, J.-Q.; Wang, X.-L. The mechanisms of brassinosteroids’ action: From signal transduction to plant development. Mol. Plant 2011, 4, 588–600. [Google Scholar] [CrossRef] [Green Version]
- Planas-Riverola, A.; Gupta, A.; Betegon-Putze, I.; Bosch, N.; Ibanes, M.; Cano-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clouse, S.D.; Langford, M.; McMorris, T.C. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996, 111, 671–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.M.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, T.; Cano-Delgado, A.C.; Seto, H.; Hiranuma, S.; Fujioka, S.; Yoshida, S.; Chory, J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 2005, 433, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Li, X.Q.; Meisenhelder, J.; Hunter, T.; Yoshida, S.; Asami, T.; Chory, J. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 2005, 8, 855–865. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Chory, J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 2006, 313, 1118–1122. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, J.; Wang, J.; Chen, L.; Fan, S.-L.; Wu, J.-W.; Wang, X.; Wang, Z.-X. Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1. Cell Res. 2014, 24, 1328–1341. [Google Scholar] [CrossRef] [Green Version]
- Gampala, S.S.; Kim, T.-W.; He, J.-X.; Tang, W.; Deng, Z.; Bai, M.-Y.; Guan, S.; Lalonde, S.; Sun, Y.; Gendron, J.M.; et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 2007, 13, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Ryu, H.; Kim, K.; Cho, H.; Park, J.; Choe, S.; Hwang, I. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 2007, 19, 2749–2762. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-Localized BZR1 Mediates Brassinosteroid-Induced Growth and Feedback Suppression of Brassinosteroid Biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Wang, Z.-Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 Accumulates in the Nucleus in Response to Brassinosteroids to Regulate Gene Expression and Promote Stem Elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Hothorn, M.; Belkhadir, Y.; Dreux, M.; Dabi, T.; Noel, J.P.; Wilson, I.A.; Chory, J. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 2011, 474, 467–471. [Google Scholar] [CrossRef] [Green Version]
- She, J.; Han, Z.; Kim, T.-W.; Wang, J.; Cheng, W.; Chang, J.; Shi, S.; Wang, J.; Yang, M.; Wang, Z.-Y.; et al. Structural insight into brassinosteroid perception by BRI1. Nature 2011, 474, 472–476. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Yang, C.; Zhang, C.; Wang, N.; Lu, D.; Wang, J.; Zhang, S.; Wang, Z.-X.; Ma, H.; Wang, X. Dual Role of BKI1 and 14-3-3 s in Brassinosteroid Signaling to Link Receptor with Transcription Factors. Dev. Cell 2011, 21, 825–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Kim, T.-W.; Oses-Prieto, J.A.; Sun, Y.; Deng, Z.; Zhu, S.; Wang, R.; Burlingame, A.L.; Wang, Z.-Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 2008, 321, 557–560. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.-W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.-X.; Sun, Y.; Burlingame, A.L.; Wang, Z.-Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009, 11, 1254–1260. [Google Scholar] [CrossRef]
- Tang, W.; Yuan, M.; Wang, R.; Yang, Y.; Wang, C.; Oses-Prieto, J.A.; Kim, T.-W.; Zhou, H.-W.; Deng, Z.; Gampala, S.S.; et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011, 13, 124–131. [Google Scholar] [CrossRef]
- Yu, X.; Li, L.; Zola, J.; Aluru, M.; Ye, H.; Foudree, A.; Guo, H.; Anderson, S.; Aluru, S.; Liu, P.; et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011, 65, 634–646. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukasinovic, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef] [Green Version]
- Choe, S.; Schmitz, R.J.; Fujioka, S.; Takatsuto, S.; Lee, M.O.; Yoshida, S.; Feldmann, K.A.; Tax, F.E. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3 beta-like kinase. Plant Physiol. 2002, 130, 1506–1515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.M.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301. [Google Scholar] [PubMed]
- Perez-Perez, J.M.; Ponce, M.R.; Micol, J.L. The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev. Biol. 2002, 242, 161–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigel, D.; Ahn, J.H.; Blázquez, M.A.; Borevitz, J.O.; Christensen, S.K.; Fankhauser, C.; Ferrándiz, C.; Kardailsky, I.; Malancharuvil, E.J.; Neff, M.M.; et al. Activation Tagging in Arabidopsis. Plant Physiol. 2000, 122, 1003–1014. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wen, J.Q.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef] [Green Version]
- Mora-Garcia, S.; Vert, G.; Yin, Y.H.; Cano-Delgado, A.; Cheong, H.; Chory, J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to bras sino steroids in Arabidopsis. Genes Dev. 2004, 18, 448–460. [Google Scholar] [CrossRef] [Green Version]
- Muto, H.; Yabe, N.; Asami, T.; Hasunuma, K.; Yamamoto, K.T. Overexpression of constitutive differential growth 1 gene, which encodes a RLCKVII-subfamily protein kinase, causes abnormal differential and elongation growth after organ differentiation in arabidopsis. Plant Physiol. 2004, 136, 3124–3133. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.-W.; Guan, S.; Burlingame, A.L.; Wang, Z.-Y. The CDG1 Kinase Mediates Brassinosteroid Signal Transduction from BRI1 Receptor Kinase to BSU1 Phosphatase and GSK3-like Kinase BIN2. Mol. Cell 2011, 43, 561–571. [Google Scholar] [CrossRef] [Green Version]
- Fields, S.; Song, O.-k. A novel genetic system to detect protein–protein interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef]
- Paiano, A.; Margiotta, A.; De Luca, M.; Bucci, C. Yeast Two-Hybrid Assay to Identify Interacting Proteins. Curr. Protoc. Protein Sci. 2019, 95, e70. [Google Scholar] [CrossRef] [Green Version]
- Nam, K.H.; Li, J.M. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Nam, K.H.; Li, J.M. The Arabidopsis Transthyretin-Like protein is a potential substrate of BRASSINOSTEROID-INSENSITIVE 1. Plant Cell 2004, 16, 2406–2417. [Google Scholar] [CrossRef] [Green Version]
- Meleady, P. Two-Dimensional Gel Electrophoresis and 2D-DIGE. In Difference Gel Electrophoresis: Methods and Protocols; Ohlendieck, K., Ed.; Springer: New York, NY, USA, 2018; pp. 3–14. [Google Scholar] [CrossRef]
- Wang, H.; Tang, J.; Liu, J.; Hu, J.; Liu, J.; Chen, Y.; Cai, Z.; Wang, X. Abscisic Acid Signaling Inhibits Brassinosteroid Signaling through Dampening the Dephosphorylation of BIN2 by ABI1 and ABI2. Mol. Plant 2018, 11, 315–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Liu, J.; Dong, X.; Cai, Z.; Tian, W.; Wang, X. Short-term and continuing stresses differentially interplay with multiple hormones to regulate plant survival and growth. Mol. Plant 2014, 7, 841–855. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Fan, M.; Qin, Z.; Lv, H.; Wang, M.; Zhang, Z.; Zhou, W.; Zhao, N.; Li, X.; Han, C.; et al. Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat. Commun. 2018, 9, 1063. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Zhang, C.; Wang, X. A Recently Evolved Isoform of the Transcription Factor BES1 Promotes Brassinosteroid Signaling and Development in Arabidopsis thaliana. Plant Cell 2015, 27, 361–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marraffini, L.A. CRISPR-Cas immunity in prokaryotes. Nature 2015, 526, 55–61. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Jaillais, Y.; Hothorn, M.; Belkhadir, Y.; Dabi, T.; Nimchuk, Z.L.; Meyerowitz, E.M.; Chory, J. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011, 25, 232–237. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Wang, T.; Wu, Z.; Wang, J.; Zhang, C.; Wang, H.; Wang, Z.-X.; Wang, X. The Intrinsically Disordered Protein BKI1 Is Essential for Inhibiting BRI1 Signaling in Plants. Mol. Plant 2015, 8, 1675–1678. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Lv, M.; Wang, Y.; Wang, P.-A.; Cui, Y.; Li, M.; Wang, R.; Gou, X.; Li, J. BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat. Commun. 2019, 10, 4164. [Google Scholar] [CrossRef] [Green Version]
- Zheng, B.; Bai, Q.; Wu, L.; Liu, H.; Liu, Y.; Xu, W.; Li, G.; Ren, H.; She, X.; Wu, G. EMS1 and BRI1 control separate biological processes via extracellular domain diversity and intracellular domain conservation. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Si, X.; Ji, X.; Fan, R.; Liu, J.; Chen, K.; Wang, D.; Gao, C. Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 2018, 36, 894–898. [Google Scholar] [CrossRef]
- Fernandez-Suarez, M.; Chen, T.S.; Ting, A.Y. Protein-protein interaction detection in vitro and in cells by proximity biotinylation. J. Am. Chem. Soc. 2008, 130, 9251–9253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.S.; Loh, K.H.; Lam, S.S.; White, K.A.; Ting, A.Y. Imaging Trans-Cellular Neurexin-Neuroligin Interactions by Enzymatic Probe Ligation. PLoS ONE 2013, 8, e52823. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.K.; Cho, K.F.; Kim, M.W.; Ting, A.Y. Luciferase-LOV BRET enables versatile and specific transcriptional readout of cellular protein-protein interactions. eLife 2019, 8, e43826. [Google Scholar] [CrossRef]
- Kotani, N.; Gu, J.G.; Isaji, T.; Udaka, K.; Taniguchi, N.; Honke, K. Biochemical visualization of cell surface molecular clustering in living cells. Proc. Natl. Acad. Sci. USA 2008, 105, 7405–7409. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.L.; Kotani, N.; Ohnishi, T.; Miyagawa-Yamguchi, A.; Tsuda, M.; Yamashita, R.; Ishiura, Y.; Honke, K. A proteomics approach to the cell-surface interactome using the enzyme-mediated activation of radical sources reaction. Proteomics 2012, 12, 54–62. [Google Scholar] [CrossRef]
- Li, X.W.; Rees, J.S.; Xue, P.; Zhang, H.; Hamaia, S.W.; Sanderson, B.; Funk, P.E.; Farndale, R.W.; Lilley, K.S.; Perrett, S.; et al. New Insights into the DT40 B Cell Receptor Cluster Using a Proteomic Proximity Labeling Assay. J. Biol. Chem. 2014, 289, 14434–14447. [Google Scholar] [CrossRef] [Green Version]
- Martell, J.D.; Deerinck, T.J.; Sancak, Y.; Poulos, T.L.; Mootha, V.K.; Sosinsky, G.E.; Ellisman, M.H.; Ting, A.Y. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 2012, 30, 1143–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhee, H.W.; Zou, P.; Udeshi, N.D.; Martell, J.D.; Mootha, V.K.; Carr, S.A.; Ting, A.Y. Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science 2013, 339, 1328–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, S.S.; Martell, J.D.; Kamer, K.J.; Deerinck, T.J.; Ellisman, M.H.; Mootha, V.K.; Ting, A.Y. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 2015, 12, 51–54. [Google Scholar] [CrossRef]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.I.; Jensen, S.C.; Noble, K.A.; Birendra, K.C.; Roux, K.H.; Motamedchaboki, K.; Roux, K.J. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 2016, 27, 1188–1196. [Google Scholar] [CrossRef] [PubMed]
- Branon, T.C.; Bosch, J.A.; Sanchez, A.D.; Udeshi, N.D.; Svinkina, T.; Carr, S.A.; Feldman, J.L.; Perrimon, N.; Ting, A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018, 36, 880–887. [Google Scholar] [CrossRef]
- Liu, Q.; Zheng, J.; Sun, W.; Huo, Y.; Zhang, L.; Hao, P.; Wang, H.; Zhuang, M. A proximity-tagging system to identify membrane protein–protein interactions. Nat. Methods 2018, 15, 715–722. [Google Scholar] [CrossRef]
- Hill, Z.B.; Pollock, S.B.; Zhuang, M.; Wells, J.A. Direct Proximity Tagging of Small Molecule Protein Targets Using an Engineered NEDD8 Ligase. J. Am. Chem. Soc. 2016, 138, 13123–13126. [Google Scholar] [CrossRef] [Green Version]
- Ge, Y.; Chen, L.; Liu, S.B.; Zhao, J.Y.; Zhang, H.; Chen, P.R. Enzyme-Mediated Intercellular Proximity Labeling for Detecting Cell-Cell Interactions. J. Am. Chem. Soc. 2019, 141, 1833–1837. [Google Scholar] [CrossRef]
- Choi-Rhee, E.; Schulman, H.; Cronan, J.E. Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 2004, 13, 3043–3050. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.I.; Birendra, K.C.; Zhu, W.; Motamedchaboki, K.; Doye, V.; Roux, K.J. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. USA 2014, 111, E2453–E2461. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Zhou, Z.; Luo, W.; Fang, M.; Li, M.; Li, H. Screening of Proximal and Interacting Proteins in Rice Protoplasts by Proximity-Dependent Biotinylation. Front. Plant Sci. 2017, 8, 749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.; Youn, J.-Y.; Gingras, A.-C.; Subramaniam, R.; Desveaux, D. In planta proximity dependent biotin identification (BioID). Sci. Rep. 2018, 8, 9212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.X.; Dodds, P.N.; Bernoux, M. What Do We Know About NOD-Like Receptors in Plant Immunity? In Annual Review of Phytopathology; Annual Reviews; Leach, J.E., Lindow, S.E., Eds.; Palo Alto: Santa Clara, CA, USA, 2017; Volume 55, pp. 205–229. [Google Scholar]
- Zhang, Y.; Song, G.; Lal, N.K.; Nagalakshmi, U.; Li, Y.; Zheng, W.; Huang, P.-j.; Branon, T.C.; Ting, A.Y.; Walley, J.W.; et al. TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat. Commun. 2019, 10, 3252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.-W.; Park, C.H.; Hsu, C.-C.; Zhu, J.-Y.; Hsiao, Y.; Branon, T.; Xu, S.-L.; Ting, A.Y.; Wang, Z.-Y. Application of TurboID-mediated proximity labeling for mapping a GSK3 kinase signaling network in Arabidopsis. bioRxiv 2019. [Google Scholar] [CrossRef]
- Bao, F.; Shen, J.J.; Brady, S.R.; Muday, G.K.; Asami, T.; Yang, Z.B. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 2004, 134, 1624–1631. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.K.; Chang, S.C.; Lee, E.J.; Chung, W.S.; Kim, Y.S.; Hwang, S.; Lee, J.S. Involvement of brassinosteroids in the gravitropic response of primary root of maize. Plant Physiol. 2000, 123, 997–1004. [Google Scholar] [CrossRef] [Green Version]
- Stock, K.; Sailer, R.; Strauss, W.S.L.; Lyttek, M.; Steiner, R.; Schneckenburger, H. Variable-angle total internal reflection fluorescence microscopy (VA-TIRFM): Realization and application of a compact illumination device. J. Microsc. 2003, 211, 19–29. [Google Scholar] [CrossRef]
- Schwille, P.; Meyer-Almes, F.J.; Rigler, R. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys. J. 1997, 72, 1878–1886. [Google Scholar] [CrossRef] [Green Version]
- Friedman, L.J.; Chung, J.; Gelles, J. Viewing Dynamic Assembly of Molecular Complexes by Multi-Wavelength Single-Molecule Fluorescence. Biophys. J. 2006, 91, 1023–1031. [Google Scholar] [CrossRef] [Green Version]
- Friedman, L.J.; Gelles, J. Mechanism of Transcription Initiation at an Activator-Dependent Promoter Defined by Single-Molecule Observation. Cell 2012, 148, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Müller, J.D.; Chen, Y.; Gratton, E. Resolving Heterogeneity on the Single Molecular Level with the Photon-Counting Histogram. Biophys. J. 2000, 78, 474–486. [Google Scholar] [CrossRef] [Green Version]
- Rohrbach, A. Observing Secretory Granules with a Multiangle Evanescent Wave Microscope. Biophys. J. 2000, 78, 2641–2654. [Google Scholar] [CrossRef] [Green Version]
- Forster, T. Zwischenmolekulare energiewanderung und fluoreszenz. Ann. Der Phys. 1948, 2, 55–75. [Google Scholar] [CrossRef]
- Clegg, R.M. Fluorescence resonance energy-transfer and nucleic-acids. Methods Enzym. 1992, 211, 353–388. [Google Scholar]
- Tan, Y.-W.; Hanson, J.A.; Chu, J.-W.; Yang, H. Confocal Single-Molecule FRET for Protein Conformational Dynamics. In Protein Dynamics: Methods and Protocols; 999 Riverview Dr, Ste 208, Totowa, NJ 07512-1165 USA; Livesay, D.R., Ed.; Humana Press Inc.: Totowa, NJ, USA, 2014; Volume 1084, pp. 51–62. [Google Scholar]
- Xing, S.; Wallmeroth, N.; Berendzen, K.W.; Grefen, C. Techniques for the Analysis of Protein-Protein Interactions in Vivo1[OPEN]. Plant Physiol. 2016, 171, 727–758. [Google Scholar] [CrossRef] [Green Version]
- Becker, W. Fluorescence lifetime imaging—Techniques and applications: FLUORESCENCE LIFETIME IMAGING. J. Microsc. 2012, 247, 119–136. [Google Scholar] [CrossRef]
- Caesar, K.; Elgass, K.; Chen, Z.; Huppenberger, P.; Witthöft, J.; Schleifenbaum, F.; Blatt, M.R.; Oecking, C.; Harter, K. A fast brassinolide-regulated response pathway in the plasma membrane of Arabidopsis thaliana: A BL-regulated plasma membrane response pathway. Plant J. 2011, 66, 528–540. [Google Scholar] [CrossRef]
- Bucherl, C.A.; van Esse, G.W.; Kruis, A.; Luchtenberg, J.; Westphal, A.H.; Aker, J.; van Hoek, A.; Albrecht, C.; Borst, J.W.; de Vries, S.C. Visualization of BRI1 and BAK1(SERK3) Membrane Receptor Heterooligomers during Brassinosteroid Signaling. Plant Physiol. 2013, 162, 1911–1925. [Google Scholar] [CrossRef] [Green Version]
- Hutten, S.J.; Hamers, D.S.; Aan den Toorn, M.; van Esse, W.; Nolles, A.; Bücherl, C.A.; de Vries, S.C.; Hohlbein, J.; Borst, J.W. Visualization of BRI1 and SERK3/BAK1 Nanoclusters in Arabidopsis Roots. PLoS ONE 2017, 12, e0169905. [Google Scholar] [CrossRef] [Green Version]
- Ulbrich, M.H.; Isacoff, E.Y. Subunit counting in membrane-bound proteins. Nat. Methods 2007, 4, 319–321. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Douglas, N.R.; Conley, N.R.; Miller, E.J.; Frydman, J.; Moerner, W.E. Sensing cooperativity in ATP hydrolysis for single multisubunit enzymes in solution. Proc. Natl. Acad. Sci. USA 2011, 108, 16962–16967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.; Chang, J.; Ma, C.; Tan, Y.-W. Single-Molecule Fluorescence Methods to Study Plant Hormone Signal Transduction Pathways. Front. Plant Sci. 2017, 8, 1888. [Google Scholar] [CrossRef] [Green Version]
- Maiti, S.; Haupts, U.; Webb, W.W. Fluorescence correlation spectroscopy: Diagnostics for sparse molecules. Proc. Natl. Acad. Sci. USA 1997, 94, 11753–11757. [Google Scholar] [CrossRef] [Green Version]
- Hink, M.A.; Shah, K.; Russinova, E.; de Vries, S.C.; Visser, A.J.W.G. Fluorescence Fluctuation Analysis of Arabidopsis thaliana Somatic Embryogenesis Receptor-Like Kinase and Brassinosteroid Insensitive 1 Receptor Oligomerization. Biophys. J. 2008, 94, 1052–1062. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Luu, D.-T.; Maurel, C.; Lin, J. Probing plasma membrane dynamics at the single-molecule level. Trends Plant Sci. 2013, 18, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, H.; Lv, X.; Chen, T.; Li, R.; Xue, Y.; Jiang, J.; Jin, B.; Baluška, F.; Šamaj, J.; et al. Spatiotemporal Dynamics of the BRI1 Receptor and its Regulation by Membrane Microdomains in Living Arabidopsis Cells. Mol. Plant 2015, 8, 1334–1349. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Wang, H.; Sun, M.; Tang, J.; Zheng, B.; Wang, X.; Tan, Y.-W. Reactive oxygen species-mediated BIN2 activity revealed by single-molecule analysis. New Phytol. 2019, 223, 692–704. [Google Scholar] [CrossRef]
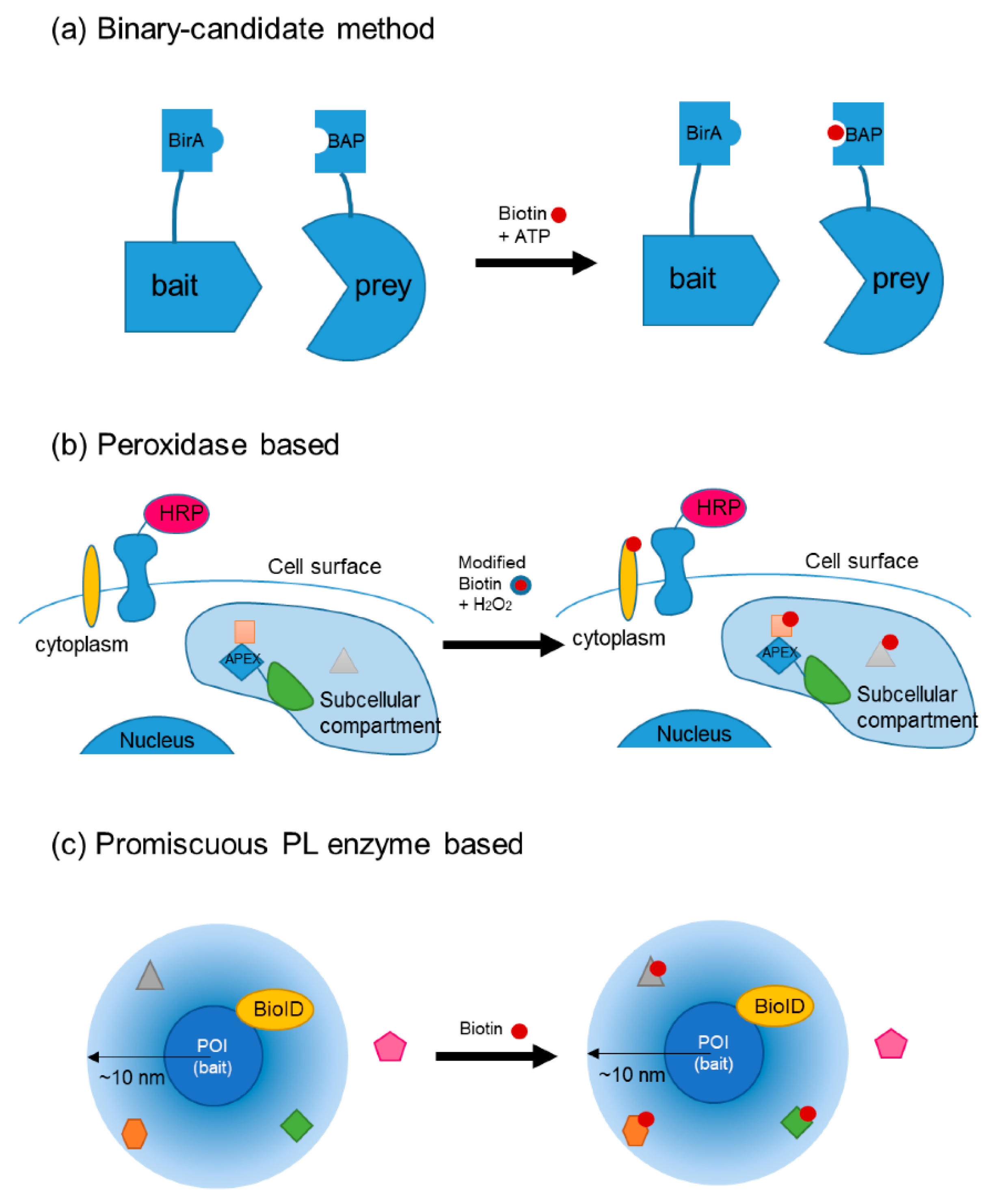
| Method | Applications | Remarks | Tag | Inducer | Reaction Time | Toxicity | References |
|---|---|---|---|---|---|---|---|
| Peroxidase based | |||||||
| EMARS | Horseradish peroxidase (HRP)-based method: reactions at the cell surface | Working distance up to 300 nm; works on cell surfaces | Biotin or fluorescein | H2O2 with arylazide biotin or fluorescein arylazide | 15 min | Free radicals | [49,50] |
| SPPLAT | Biotin | Tyramide-biotin and H2O2 | 5 min | Free radicals | [51] | ||
| APEX; APEX2 | Engineered ascorbate peroxidase; proteomics of a subcellular compartment | From a plant ascorbate peroxidase; does not provide a history of protein associations | Biotin; targeting tyrosine, tryptophan, histidine, and cysteine | Induced by H2O2 under biotin-phenol additives | 1 min | Free radicals | [53,54] |
| Binary-candidate method | |||||||
| BirA/BAP | Binary-candidate method: modify both bait and prey proteins, can be applied across cells | BirA has a bacterial origin and is therefore orthogonal to mammalian or plant cells | Biotin | Biotin + ATP | Low | [46] | |
| BLINC/ID-PRIME | Detection by streptavidin linked fluorophores staining | Biotin or lipoic acid (LA) | ATP with biotin or LA | 2–15 min | Low | [47] | |
| SPARK; SPARK2 | Luciferase fused LOV domain; BRET type mechanism | Reporter gene | Blue light or luciferin | 8 h | Photo-toxicity | [48] | |
| Promiscuous PL enzyme | |||||||
| BioID; BioID2 | Promiscuous biotin ligase fused to a bait protein | Mutated from BirA; works within 10 nm; used ~37 °C | Biotin; target amines (including Lys) | Biotin supplementation | ~16 h in plants | Low | [55,56] |
| TurboID | Improved from BioID; works at room temperature and above | Biotin; target amines | Biotin supplementation | ~10 min | Low | [57] | |
| PUP-IT (pupylation-based interaction tagging) | For identifying membrane protein interactions; bacterial Pup conjugation system | pafA, a gene encodes Pup ligase | Pup (conjugate to Lys) | Doxycycline to induce Pup(E) expression; in the extracellular format, PafA can be engineered to FRB and induce by rapamycin | 24 h | Low | [58] |
| NEDDylator | From the NEDD8 pathway in mammalian cells | Done in vitro at ~37 °C | NEDD8 (conjugate to Lys) | HB-NEDD8 | 2 h | Unknown | [59] |
| EXCELL | For marking cell–cell interactions | Mutated from SrtA to recognize monoglycine at N-terminal promiscuously | LPXTG pentapeptide | Condition depending on cell line and transfection rate | Low | [60] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Song, S.; Cheng, H.; Tan, Y.-W. State-of-the-Art Technologies for Understanding Brassinosteroid Signaling Networks. Int. J. Mol. Sci. 2020, 21, 8179. https://doi.org/10.3390/ijms21218179
Wang H, Song S, Cheng H, Tan Y-W. State-of-the-Art Technologies for Understanding Brassinosteroid Signaling Networks. International Journal of Molecular Sciences. 2020; 21(21):8179. https://doi.org/10.3390/ijms21218179
Chicago/Turabian StyleWang, Haijiao, Song Song, Huaqiang Cheng, and Yan-Wen Tan. 2020. "State-of-the-Art Technologies for Understanding Brassinosteroid Signaling Networks" International Journal of Molecular Sciences 21, no. 21: 8179. https://doi.org/10.3390/ijms21218179






