The Importance of Cytokinins during Reproductive Development in Arabidopsis and Beyond
Abstract
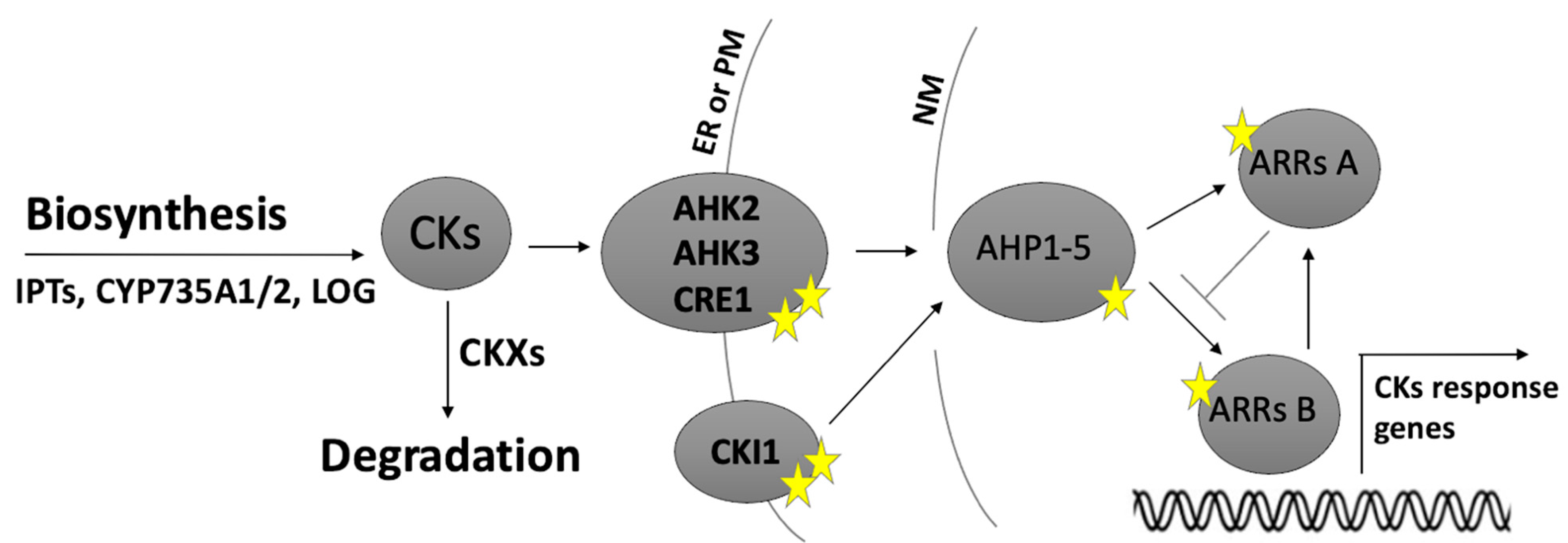
1. Introduction
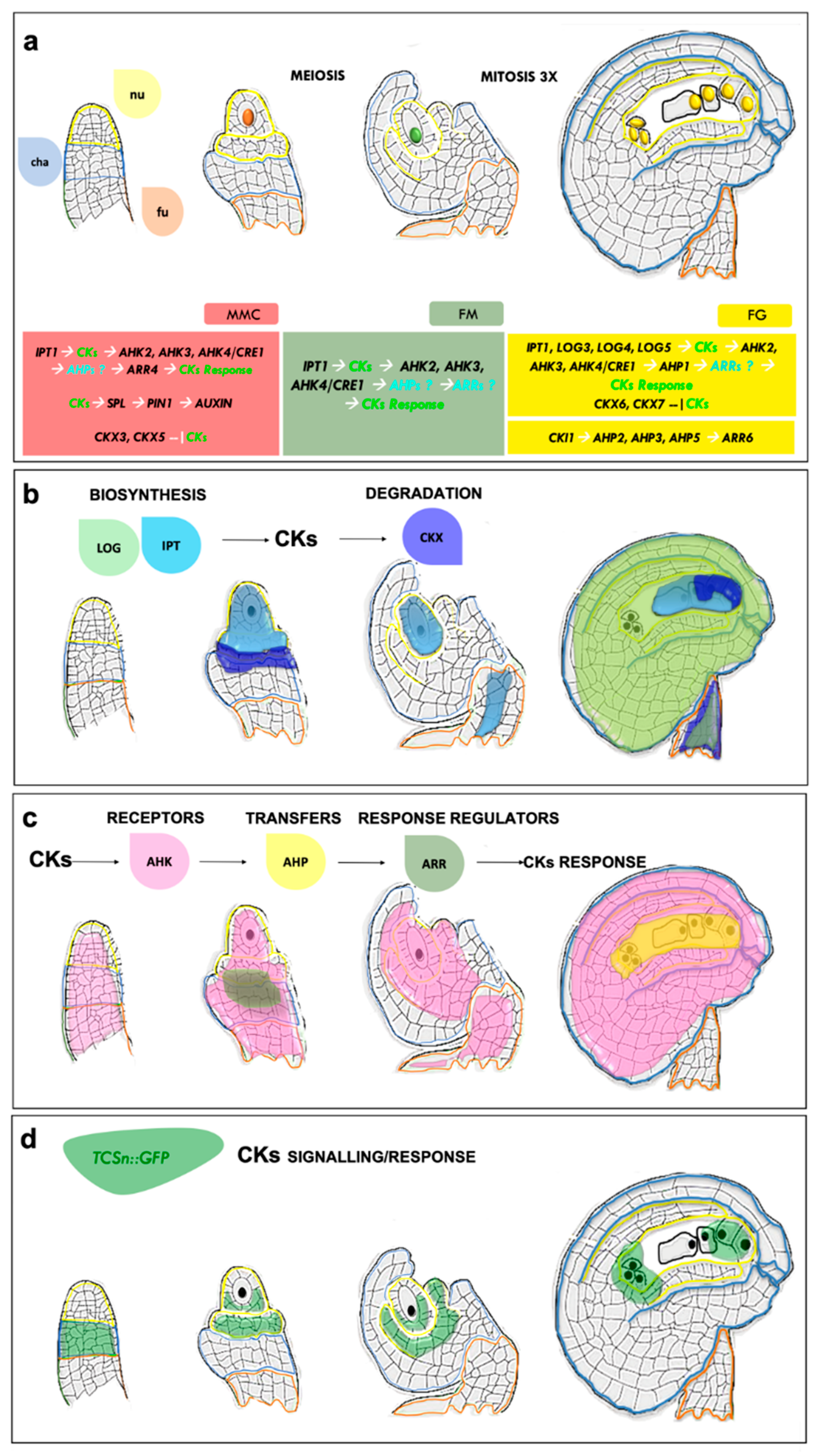
1.1. Cytokinins Play a Pivotal Role in Ovule Patterning and Development
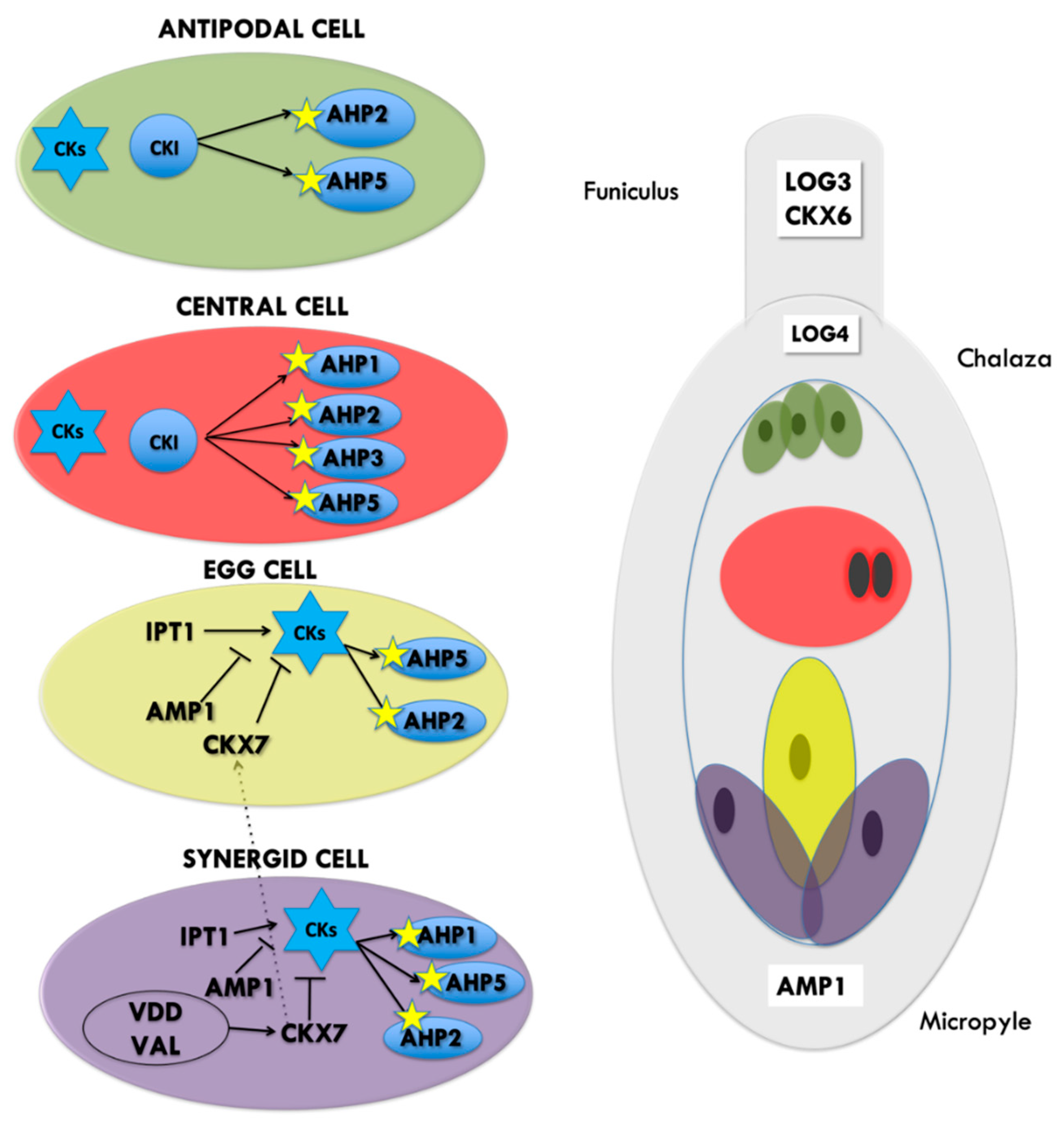
1.2. CK Perception Influences Female Gametophyte Cell Identity
1.3. Cytokinins Are Involved in Sporophyte–Gametophyte Communication
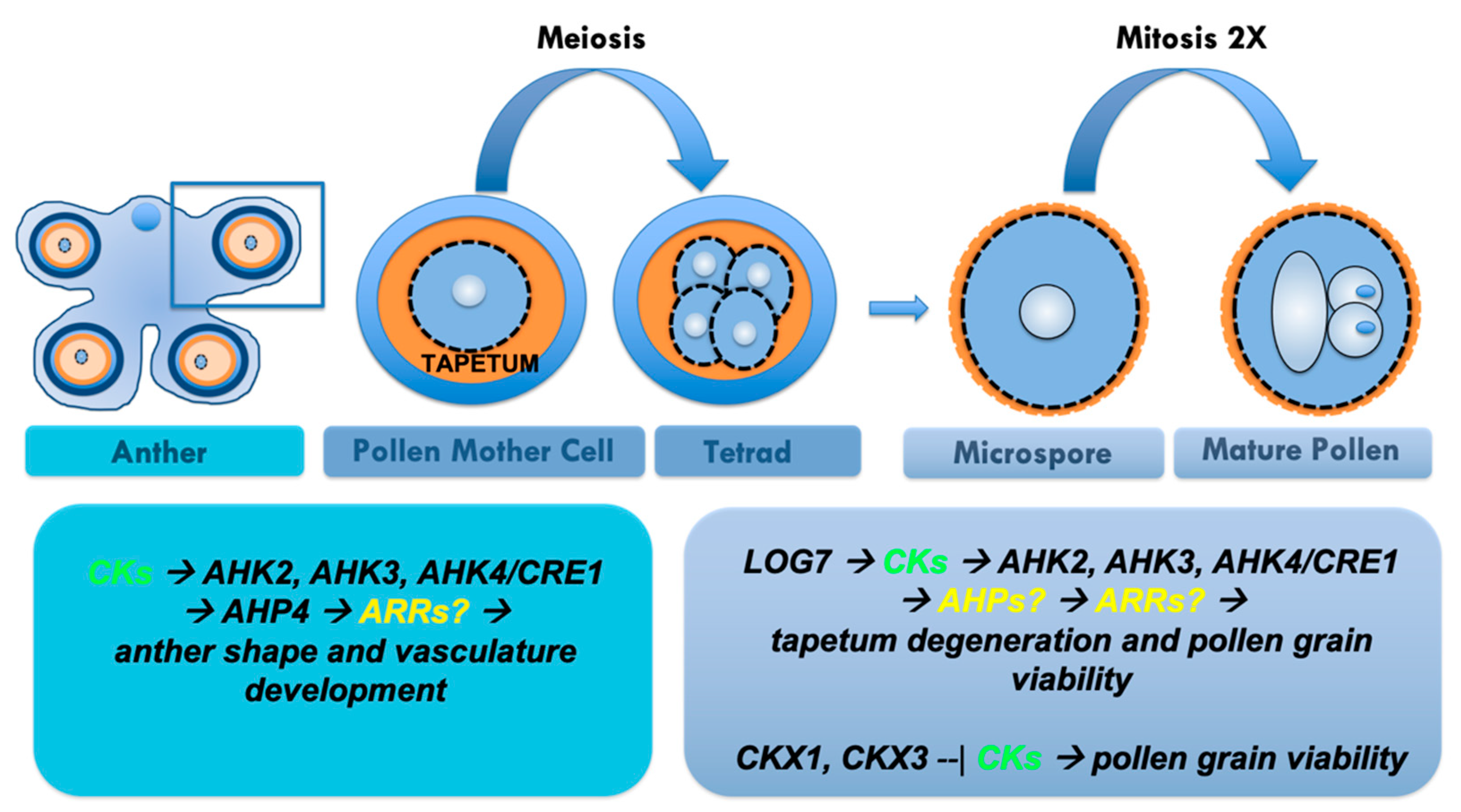
1.4. CK Signalling Influences Stamen Development, Anther Dehiscence and Pollen Viability
1.5. Cytokinins and Seed Size
1.6. Cytokinins during Reproduction in Crop Species
2. Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Zürcher, E.; Müller, B. Cytokinin Synthesis, Signaling, and Function—Advances and New Insights. Int. Rev. Cell Mol. Biol. 2016, 324, 1–38. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Schaller, G.E. Cytokinins. Arab. Book 2014, 12, e0168. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H. Cytokinins: Activity, Biosynthesis, and Translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Mok, D.W.S.; Mok, M.C. Cytokinin metabolism and action. Annu. Rev. Plant Biol. 2001, 52, 89–118. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H. Cytokinin Biosynthesis and Regulation. Vitam. Horm. 2005, 72, 271–287. [Google Scholar] [CrossRef] [PubMed]
- Golovko, A.; Sitbon, F.; Tillberg, E.; Nicander, B. Identification of a tRNA isopentenyltransferase gene from Arabidopsis thaliana. Plant Mol. Biol. 2002, 49, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; Yamaya, T.; Sakakibara, H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyse the biosynthesis of trans-Zeatin. J. Biol. Chem. 2004, 279, 41866–41872. [Google Scholar] [CrossRef]
- Takei, K.; Sakakibara, H.; Sugiyama, T. Identification of Genes Encoding Adenylate Isopentenyltransferase, a Cytokinin Biosynthesis Enzyme, in Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 26405–26410. [Google Scholar] [CrossRef]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef]
- Bilyeu, K.D.; Cole, J.L.; Laskey, J.G.; Riekhof, W.R.; Esparza, T.J.; Kramer, M.D.; Morris, R.O. Molecular and biochemical characterization of a cytokinin oxidase from maize. Plant Physiol. 2001, 125, 378–386. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmulling, T. Cytokinin-Deficient Transgenic Arabidopsis Plants Show Multiple Developmental Alterations Indicating Opposite Functions of Cytokinins in the Regulation of Shoot and Root Meristem Activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, M.; Manrique, S.; Cuesta, C.; Benkova, E.; Novak, O.; Colombo, L. CUP-SHAPED COTYLEDON1 (CUC1) and CUC2 regulate cytokinin homeostasis to determine ovule number in Arabidopsis. J. Exp. Bot. 2018, 69, 5169–5176. [Google Scholar] [CrossRef] [PubMed]
- Šmehilová, M.; Ůšková, J.D.; Novák, O.; Takáč, T.; Galuszka, P. Cytokinin-specific glycosyltransferases possess different roles in Cytokinin homeostasis maintenance. Front. Plant Sci. 2016, 7, 1264. [Google Scholar] [CrossRef]
- Yamada, H.; Suzuki, T.; Terada, K.; Takei, K.; Ishikawa, K.; Miwa, K.; Yamashino, T.; Mizuno, T. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 2001, 42. [Google Scholar] [CrossRef]
- Hwang, I.; Sheen, J.; Müller, B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145. [Google Scholar] [CrossRef]
- Rashotte, A.M.; Mason, M.G.; Hutchison, C.E.; Ferreira, F.J.; Schaller, G.E.; Kieber, J.J. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 11081–11085. [Google Scholar] [CrossRef]
- Gasser, C.S.; Skinner, D.J. Development and evolution of the unique ovules of flowering plants. Curr. Top Dev. Biol. 2019, 131, 373–399. [Google Scholar] [CrossRef]
- Cucinotta, M.; Di Marzo, M.; Guazzotti, A.; de Folter, S.; Kater, M.M.; Colombo, L. Gynoecium size and ovule number are interconnected traits that impact seed yield. J. Exp. Bot. 2020, 71, 2479–2489. [Google Scholar] [CrossRef]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmülling, T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in arabidopsis thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Galbiati, F.; Sinha Roy, D.; Simonini, S.; Cucinotta, M.; Ceccato, L.; Cuesta, C.; Simaskova, M.; Benkova, E.; Kamiuchi, Y.; Aida, M.; et al. An integrative model of the control of ovule primordia formation. Plant J. 2013, 76, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Zuñiga-mayo, V.M.; Baños-bayardo, C.R.; Díaz-ramírez, D.; Marsch-martínez, N.; De Folter, S. Conserved and novel responses to cytokinin treatments during flower and fruit development in Brassica napus and Arabidopsis thaliana. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Di Marzo, M.; Herrera-ubaldo, H.; Caporali, E.; Mendes, M.A.; De Folter, S.; Colombo, L. SEEDSTICK controls arabidopsis fruit size by regulating cytokinin levels and FRUITFULL. Cell Rep. 2020, 30, 2846–2857. [Google Scholar] [CrossRef]
- Bencivenga, S.; Simonini, S.; Benková, E.; Colombo, L. The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 2012, 24, 2886–2897. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Mathews, D.E.; Schaller, G.E.; Kieber, J.J. Cytokinin-dependent specification of the functional megaspore in the Arabidopsis female gametophyte. Plant J. 2013, 73, 929–940. [Google Scholar] [CrossRef]
- Kinoshita-Tsujimura, K.; Kakimoto, T. Cytokinin receptors in sporophytes are essential for male and female functions in Arabidopsis thaliana. Plant Signal. Behav. 2011, 6, 66–71. [Google Scholar] [CrossRef]
- Yang, W.C.; Ye, D.; Xu, J.; Sundaresan, V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 1999, 13, 2108–2117. [Google Scholar] [CrossRef] [PubMed]
- Lieber, D.; Lora, J.; Schrempp, S.; Lenhard, M.; Laux, T. Arabidopsis WIH1 and WIH2 genes act in the transition from somatic to reproductive cell fate. Curr. Biol. 2011, 21, 1009–1017. [Google Scholar] [CrossRef]
- Cucinotta, M.; Manrique, S.; Guazzotti, A.; Quadrelli, N.E.; Mendes, M.A.; Benkova, E.; Colombo, L. Cytokinin response factors integrate auxin and cytokinin pathways for female reproductive organ development. Development 2016, 143, 4419–4424. [Google Scholar] [CrossRef]
- Christensen, C.A.; King, E.J.; Jordan, J.R.; Drews, G.N. Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 1997, 10, 49–64. [Google Scholar] [CrossRef]
- Drews, G.N.; Koltunow, A.M. The Female Gametophyte. Arab. Book 2011, 9, e0155. [Google Scholar] [CrossRef] [PubMed]
- Hulskamp, M.; Schneitz, K.; Pruitt, R.E. Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. Plant Cell 1995, 7, 57–64. [Google Scholar] [CrossRef]
- Hejátko, J.; Ryu, H.; Kim, G.T.; Dobešová, R.; Choi, S.; Choi, S.M.; Souček, P.; Horák, J.; Pekárová, B.; Palme, K.; et al. The Histidine kinases cytokinin-independent1 and arabidopsis histidine kinase2 and 3 regulate vascular tissue development in arabidopsis shoots. Plant Cell 2009, 21, 2008–2021. [Google Scholar] [CrossRef]
- Hwang, I.; Sheen, J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 2001, 413, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Dong, H.; Mu, J.; Ren, B.; Zheng, B.; Ji, Z.; Yang, W.; Liang, Y.; Zuo, J. Arabidopsis Histidine Kinase CKI1 acts upstream of HISTIDINE PHOSPHOTRANSFER PROTEINS to regulate female gametophyte development and vegetative growth. Plant Cell 2010, 22, 1232–1248. [Google Scholar] [CrossRef]
- Pischke, M.S.; Jones, L.G.; Otsuga, D.; Fernandez, D.E.; Drews, G.N.; Sussman, M.R. An Arabidopsis histidine kinase is essential for megagametogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 15800–15805. [Google Scholar] [CrossRef]
- Yuan, L.; Liu, Z.; Song, X.; Johnson, C.; Yu, X.; Sundaresan, V. The CKI1 Histidine Kinase specifies the female gametic precursor of the endosperm. Dev. Cell 2016, 37, 34–46. [Google Scholar] [CrossRef]
- Urao, T.; Miyata, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett. 2000, 478, 227–232. [Google Scholar] [CrossRef]
- Pekárová, B.; Klumpler, T.; Třísková, O.; Horák, J.; Jansen, S.; Dopitová, R.; Borkovcová, P.; Papoušková, V.; Nejedlá, E.; Sklenář, V.; et al. Structure and binding specificity of the receiver domain of sensor histidine kinase CKI1 from Arabidopsis thaliana. Plant J. 2011, 67, 827–839. [Google Scholar] [CrossRef]
- Liu, Z.; Yuan, L.; Song, X.; Yu, X.; Sundaresan, V. AHP2, AHP3, and AHP5 act downstream of CKI1 in Arabidopsis female gametophyte development. J. Exp. Bot. 2017, 68, 3365–3373. [Google Scholar] [CrossRef]
- Hutchison, C.E.; Li, J.; Argueso, C.; Gonzalez, M.; Lee, E.; Lewis, M.W.; Maxwell, B.B.; Perdue, T.D.; Schaller, G.E.; Alonso, J.M.; et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 2006, 18, 3073–3087. [Google Scholar] [CrossRef] [PubMed]
- Imamura, A.; Hanaki, N.; Nakamura, A.; Suzuki, T.; Taniguchi, M.; Kiba, T.; Ueguchi, C.; Sugiyama, T.; Mizuno, T. Compilation and characterization of arabiopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 1999, 40, 733–742. [Google Scholar] [CrossRef] [PubMed]
- To, J.P.C.; Haberer, G.; Ferreira, F.J.; Deruère, J.; Mason, M.G.; Schaller, G.E.; Alonso, J.M.; Ecker, J.R.; Kieber, J.J. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 2004, 16, 658–671. [Google Scholar] [CrossRef]
- Leibfried, A.; To, J.P.C.; Busch, W.; Stehling, S.; Kehle, A.; Demar, M.; Kieber, J.J.; Lohmann, J.U. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 2005, 438, 22–25. [Google Scholar] [CrossRef]
- Matias-Hernandez, L.; Battaglia, R.; Galbiati, F.; Rubes, M.; Eichenberger, C.; Grossniklaus, U.; Kater, M.; Colombo, L. VERDANDI is a direct target of the MADS domain ovule identity complex and affects embryo sac differentiation in Arabidopsis. Plant Cell 2010, 22, 1702–1715. [Google Scholar] [CrossRef] [PubMed]
- Mendes, M.A.; Guerra, R.F.; Castelnovo, B.; Silva-Velazquez, Y.; Morandini, P.; Manrique, S.; Baumann, N.; Groß-Hardt, R.; Dickinson, H.; Colombo, L. Live and let die: A REM complex promotes fertilization through synergid cell death in Arabidopsis. Development 2016, 143, 2780–2790. [Google Scholar] [CrossRef]
- Köllmer, I.; Novák, O.; Strnad, M.; Schmülling, T.; Werner, T. Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. 2014, 78, 359–371. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Z.; Meng, P.; Tian, S.; Zhang, X.; Yang, S. The glutamate carboxypeptidase AMP1 mediates abscisic acid and abiotic stress responses in Arabidopsis. New Phytol. 2013, 199, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Nogué, N.; Hocart, H.; Letham, D.S.; Dennis, E.S.; Chaudhury, A.M. Cytokinin synthesis is higher in the Arabidopsis amp1 mutant. Plant Growth Regul. 2000, 32, 267–273. [Google Scholar] [CrossRef]
- Kong, J.; Lau, S.; Jürgens, G. Twin plants from supernumerary egg cells in Arabidopsis. Curr. Biol. 2015, 25, 225–230. [Google Scholar] [CrossRef]
- Müller, B.; Sheen, J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 2008, 453, 1094–1097. [Google Scholar] [CrossRef] [PubMed]
- Zürcher, E.; Tavor-Deslex, D.; Lituiev, D.; Enkerli, K.; Tarr, P.T.; Müller, B. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 2013, 161, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Kuroha, T.; Tokunaga, H.; Kojima, M.; Ueda, N.; Ishida, T.; Nagawa, S.; Fukuda, H.; Sugimoto, K.; Sakakibara, H. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 2009, 21, 3152–3169. [Google Scholar] [CrossRef] [PubMed]
- Zürcher, E.; Liu, J.; Di Donato, M.; Geisler, M.; Müller, B. Plant development regulated by cytokinin sinks. Science 2016, 353, 1027–1030. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Pitorre, D.; Poretska, O.; Marizzi, C.; Winter, N.; Poppenberger, B.; Sieberer, T. ALTERED MERISTEM PROGRAM1 suppresses ectopic stem cell niche formation in the shoot apical meristem in a largely cytokinin-independent manner. Plant Physiol. 2015, 167, 1471–1486. [Google Scholar] [CrossRef]
- Johnston, A.J.; Meier, P.; Gheyselinck, J.; Wuest, S.E.J.; Federer, M.; Schlagenhauf, E.; Becker, J.D.; Grossniklaus, U. Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol. 2007, 8, R204. [Google Scholar] [CrossRef]
- Scott, R.J.; Spielman, M.; Dickinson, H.G. Stamen structure and function. Plant Cell 2004, 16, S46–S60. [Google Scholar] [CrossRef]
- Sanders, P.M.; Bui, A.Q.; Weterings, K.; McIntire, K.N.; Hsu, Y.-C.; Lee, P.Y.; Truong, M.T.; Beals, T.P.; Goldberg, R.B. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999, 11, 297–322. [Google Scholar] [CrossRef]
- Borg, M.; Brownfield, L.; Twell, D. Male gametophyte development: A molecular perspective. J. Exp. Bot. 2009, 60, 1465–1478. [Google Scholar] [CrossRef]
- Twell, D.; Park, S.K.; Lalanne, E. Asymmetric division and cell-fate determination in developing pollen. Trends Plant Sci. 1998, 3, 305–310. [Google Scholar] [CrossRef]
- Nibau, C.; Di Stilio, V.S.; Wu, H.M.; Cheung, A.Y. Arabidopsis and Tobacco SUPERMAN regulate hormone signalling and mediate cell proliferation and differentiation. J. Exp. Bot. 2011, 62, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Marsch-Martínez, N.; Ramos-Cruz, D.; Irepan Reyes-Olalde, J.; Lozano-Sotomayor, P.; Zúñiga-Mayo, V.M.; de Folter, S. The role of cytokinin during Arabidopsis gynoecia and fruit morphogenesis and patterning. Plant J. 2012, 72, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, C.; Ohashi, Y.; Sato, S.; Kato, T.; Tabata, S.; Ueguchi, C. Histidine Kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 2004, 16, 1365–1377. [Google Scholar] [CrossRef]
- Jung, K.W.; Oh, S.-I.; Kim, Y.Y.; Yoo, K.S.; Shin, M.H.C. and J.S. Arabidopsis Histidine-containing Phosphotransfer Factor 4 (AHP4) negatively regulates secondary wall thickening of the anther endothecium during flowering. Mol. Cells 2008, 25, 294–300. [Google Scholar] [PubMed]
- Bürkle, L.; Cedzich, A.; Döpke, C.; Stransky, H.; Okumoto, S.; Gillissen, B.; Kühn, C.; Frommer, W.B. Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J. 2003, 34, 13–26. [Google Scholar] [CrossRef]
- Kamboj, R.K.; Jackson, J.F. Purine Nucleoside Transport in Petunia Pollen Is an Active, Carrier-Mediated System Not Sensitive to Nitrobenzylthioinosine and Not Renewed during Pollen Tube Growth. Plant Physiol. 1987, 84, 688–691. [Google Scholar] [CrossRef]
- Lohrmann, J.; Sweere, U.; Zabaleta, E.; Bäurle, I.; Keitel, C.; Kozma-Bognar, L.; Brennicke, A.; Schäfer, E.; Kudla, J.; Harter, K. The response regulator ARR2: A pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol. Genet. Genomics. 2001, 265, 2–13. [Google Scholar] [CrossRef]
- Li, J.; Nie, X.; Li, J.; Tan, H.; Berger, F. Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 1–6. [Google Scholar] [CrossRef]
- Riefler, M.; Novak, O.; Strnad, M.; Schmu, T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 2006, 18, 40–54. [Google Scholar] [CrossRef]
- Argyros, R.D.; Mathews, D.E.; Chiang, Y.; Palmer, C.M.; Thibault, D.M.; Etheridge, N.; Argyros, D.A.; Mason, M.G.; Kieber, J.J.; Schaller, G.E. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Development 2008, 20, 2102–2116. [Google Scholar] [CrossRef]
- Chen, L.; Zhao, J.; Song, J.; Jameson, P.E. Cytokinin dehydrogenase: A genetic target for yield improvement in wheat. Plant Biotechnol. J. 2020, 18, 614–630. [Google Scholar] [CrossRef] [PubMed]
- Jameson, P.E.; Song, J. Cytokinin: A key driver of seed yield. J. Exp. Bot. 2016, 67, 593–606. [Google Scholar] [CrossRef]
- Long, S.P.; Marshall-Colon, A.; Zhu, X.G. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 2015, 161, 56–66. [Google Scholar] [CrossRef]
- Boden, S.A.; Østergaard, L. How can developmental biology help feed a growing population? Development 2019, 146. [Google Scholar] [CrossRef]
- Huang, S.; Cerny, R.E.; Qi, Y.; Bhat, D.; Aydt, C.M.; Hanson, D.D.; Malloy, K.P.; Ness, L.A. Transgenic studies on the involvement of cytokinin and gibberellin in male development. Plant Physiol. 2003, 131, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Yamaki, S.; Nagato, Y.; Kurata, N.; Nonomura, K.I. Ovule is a lateral organ finally differentiated from the terminating floral meristem in rice. Dev. Biol. 2011, 351, 208–216. [Google Scholar] [CrossRef]
- Murai, N. Review: Plant Growth Hormone Cytokinins Control the Crop Seed Yield. Am. J. Plant Sci. 2014, 5, 2178–2187. [Google Scholar] [CrossRef]
- Worthen, J.M.; Yamburenko, M.V.; Lim, J.; Nimchuk, Z.L.; Kieber, J.J.; Schaller, G.E. Type-B response regulators of rice play key roles in growth, development and cytokinin signaling. Development 2019, 146. [Google Scholar] [CrossRef]
- Guo, T.; Chen, K.; Dong, N.Q.; Shi, C.L.; Ye, W.W.; Gao, J.P.; Shan, J.X.; Lin, H.X. GRAIN SIZE AND NUMBER1 negatively regulates the OSMKKK10-OSMKK4-OSMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 2018, 30, 871–888. [Google Scholar] [CrossRef] [PubMed]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Plant science: Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, Y.L.; Gao, L.F.; Zhao, G.Y.; Zhou, R.H.; Zhang, B.S.; Jia, J.Z. TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol. 2012, 195, 574–584. [Google Scholar] [CrossRef]
- Pernisova, M.; Grochova, M.; Konecny, T.; Plackova, L.; Harustiakova, D.; Kakimoto, T.; Heisler, M.G.; Novak, O.; Hejatko, J. Cytokinin signalling regulates organ identity via the AHK4 receptor in arabidopsis. Development 2018, 145. [Google Scholar] [CrossRef]
- Antoniadi, I.; Plačková, L.; Simonovik, B.; Doležal, K.; Turnbull, C.; Ljung, K.; Novák, O. Cell-type-specific cytokinin distribution within the arabidopsis primary root apex. Plant Cell 2015, 27, 1955–1967. [Google Scholar] [CrossRef] [PubMed]
- Šimura, J.; Antoniadi, I.; Široká, J.; Tarkowská, D.; Strnad, M.; Ljung, K.; Novák, O. Plant hormonomics: Multiple phytohormone profiling by targeted metabolomics. Plant Physiol. 2018, 177, 476–489. [Google Scholar] [CrossRef]
- Novák, O.; Napier, R.; Ljung, K. Zooming in on plant hormone analysis: Tissue-and cell-specific approaches. Annu. Rev. Plant Biol. 2017, 68, 323–348. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Family or Protein Name | CK Process | Mutant Line | Phenotype | Ref |
|---|---|---|---|---|---|
| LOG2, LOG3, LOG4, LOG5, LOG7, LOG8 | cytokinin riboside 5′-monophosphate phosphoribohydrolases | biosynthesis | 35S::LOG2, 35S::LOG3, 35S::LOG4, 35S::LOG5, 35S::LOG7, 35S::LOG8 | Larger and heavier seeds | [53] |
| CKX1 | CKX-cytokinin oxidase/dehydrogenase family | degradation | 35S:AtCKX1 | Few and enlarged seeds | [11] |
| CKX3 | CKX-cytokinin oxidase/dehydrogenase family | degradation | 35S:AtCKX3 | Few and enlarged seeds | [11] |
| CKX7 | CKX-cytokinin oxidase/dehydrogenase family | degradation | ckx7 ckx7 T | Shorter fruit | [23] |
| CKX3, CKX5 | CKX-cytokinin oxidase/dehydrogenase family | degradation | ckx3 ckx5 | Increased production of flowers, longer siliques, more ovules and seeds | [20] |
| CKX3, CKX5, AHP6 | CKX-cytokinin oxidase/dehydrogenase family AHP-Arabidopsis histidine phosphotransfer gene family | degradation signalling | ckx3 ckx5 ahp6 | More siliques | [20] |
| AHK4/CRE1, AHK2 and AHK3 | AHK-Arabidopsis histidine/kinase receptor family | signalling | ahk2-1 ahk4-1 ahk3-1 ahk4-1 | No anther dehiscence, small amount of viable pollen | [63] |
| cre1-12 ahk2-2tk ahk3-3 | Unable to produce seeds, abnormal development of female gametophyte and anthers; defects in tapetum degeneration and reduced number of pollen grains | [26] | |||
| cre1-12 ahk2-2 ahk3-3 | Abnormal female gametophyte development | [24] | |||
| ahk2-5 ahk3-7 cre1-2 | Reduced seed set, larger seeds | [69] | |||
| ahk2–7 ahk3 –3 cre1–12 | Defects in functional megaspore specification, female gametophyte absent | [25] | |||
| ahk2–1 ahk3–1 ahk4–1 | Sterile | [63] | |||
| CKI1 | CKI-cytokinin-independent kinases | signalling | cki1-5 cki1-6 | Reduced seed set and abnormal embryo sacs | [36] |
| cki1-8 | Reduced seed set and abnormal embryo sacs | [35] | |||
| cki1-9 | Almost sterile, fewer larger seeds. Misspecification of female gametophyte cells | [37] | |||
| AHP4 | AHP-Arabidopsis histidine phosphotransfer family | signalling | 35S::AHP4 | Anthers lack of secondary cell wall thickening in the anther endothecium | [64] |
| ahp4 | Anthers more lignified | [64] | |||
| AHP1,2,3,4,5 | AHP-Arabidopsis Histidine phospotransfer family | signalling | ahp 1, 2-1, 3/+, 4, 5-1 | Reduced seed set, but larger seeds; arrested during female gametophyte development | [41] |
| ahp1,2-2,3,4,5-1 | Loss of central cell fate and acquisition of egg-cell identity | [37] | |||
| AHP2,3,5 | AHP-Arabidopsis Histidine phospotransfer family | signalling | ahp2-2, ahp3, and ahp5-2 | Unfertilized ovule and seed abortion | [40] |
| ARR1,2,10,12 | ARR-Arabidopsis response regulator type-B | signalling | arr1–3 arr2–2 arr10–2 arr12–1 | Ovules arrested at FG7, the final developmental stage of the female gametophyte | [25] |
| ARR1,10,12 | ARR-Arabidopsis response regulator type-B | signalling | arr1-3 arr10-5 arr12-1 | Shorter siliques, larger seeds | [70] |
| ARR7,15 | ARR-Arabidopsis response regulator type-A | signalling | arr7arr15 | Female gametophyte lethal | [44] |
| CRF5,6 | CRF-cytokinin response Factor | signalling | crf5 crf6 | Homozygote non-viable | [17] |
| CRF2,3,6 | CRF-cytokinin response Factor | signalling | crf2 crf3 crf6 | Reduction in ovule number and placenta length | [29] |
| AMP1 | Glutamate carboxypeptidase family | amp1-1 | Increased cytokinin level | [49] | |
| amp1-2 | Increased cytokinin level | [49] | |||
| amp1-10 | Supernumerary egg cell, twin embryos | [50] | |||
| amp1-13 | Supernumerary egg cell, twin embryos | [50] |
| Gene Name | Family or Protein Name | CK Process | Specie | Mutant Line | Phenotype | Ref |
|---|---|---|---|---|---|---|
| CKX1 | CKX-cytokinin oxidase/dehydrogenase family | degradation | Zea Mays | pZmg13::CKX1 pZtap::CKX1 | Defects in anther and stamen development | [75] |
| OsLOG-1 | LOG- Lonely Guy | biosynthesis | Oriza sativa | Oslog-1 | Flowers with only one stamen/no pistil | [77] |
| OsLOG-3 | LOG- Lonely Guy | biosynthesis | Oriza sativa | Oslog-3 | Flowers with six stamens and a slender pistil lacking an ovule or no pistil/absence of organ differentiation in the ovule founder region | [76] |
| OsLOG-4 | LOG- Lonely Guy | biosynthesis | Oriza sativa | Oslog-4 | Flowers with only one stamen/no pistil | [77] |
| OsCKX2 | CKX-cytokinin oxidase/dehydrogenase family | degradation | Oriza sativa | Osckx2 | Increased number of reproductive organs/enhanced grain yield | [80] |
| OsRR type-B | ARR-Arabidopsis response regulator type-B | signalling | Oriza sativa | rr24 | Compromised anther development | [78] |
| OsRR type-B | ARR-Arabidopsis response regulator type-B | signalling | Oriza sativa | rr21/22/23 | Compromised pollen capture | [78] |
| GSN1 | GRAIN SIZE AND NUMBER | Oriza sativa | gsn1 | Upregulation of cytokinin degradation enzyme, CKX2. Increase in grain size/less-branched inflorescence | [79] | |
| TaCKX6-D1 | CKX-cytokinin oxidase/dehydrogenase family | cytokinin degradation | Wheat (Triticum aestivum) | TaCKX6-D1 naturally occurring wheat variants | [81] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terceros, G.C.; Resentini, F.; Cucinotta, M.; Manrique, S.; Colombo, L.; Mendes, M.A. The Importance of Cytokinins during Reproductive Development in Arabidopsis and Beyond. Int. J. Mol. Sci. 2020, 21, 8161. https://doi.org/10.3390/ijms21218161
Terceros GC, Resentini F, Cucinotta M, Manrique S, Colombo L, Mendes MA. The Importance of Cytokinins during Reproductive Development in Arabidopsis and Beyond. International Journal of Molecular Sciences. 2020; 21(21):8161. https://doi.org/10.3390/ijms21218161
Chicago/Turabian StyleTerceros, Giada Callizaya, Francesca Resentini, Mara Cucinotta, Silvia Manrique, Lucia Colombo, and Marta A. Mendes. 2020. "The Importance of Cytokinins during Reproductive Development in Arabidopsis and Beyond" International Journal of Molecular Sciences 21, no. 21: 8161. https://doi.org/10.3390/ijms21218161
APA StyleTerceros, G. C., Resentini, F., Cucinotta, M., Manrique, S., Colombo, L., & Mendes, M. A. (2020). The Importance of Cytokinins during Reproductive Development in Arabidopsis and Beyond. International Journal of Molecular Sciences, 21(21), 8161. https://doi.org/10.3390/ijms21218161





