Genotype-Based Gene Expression in Colon Tissue—Prediction Accuracy and Relationship with the Prognosis of Colorectal Cancer Patients
Abstract
:1. Introduction
2. Results
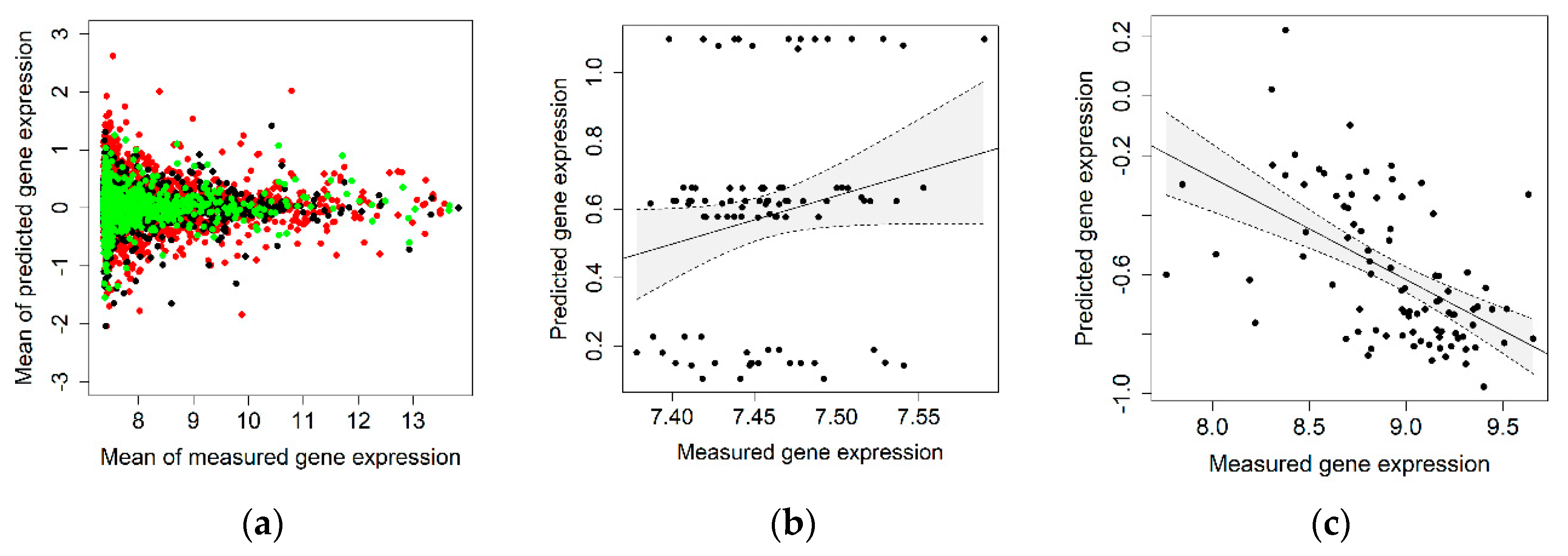
2.1. Correlation between Measured and Genetically Predicted Gene Expression
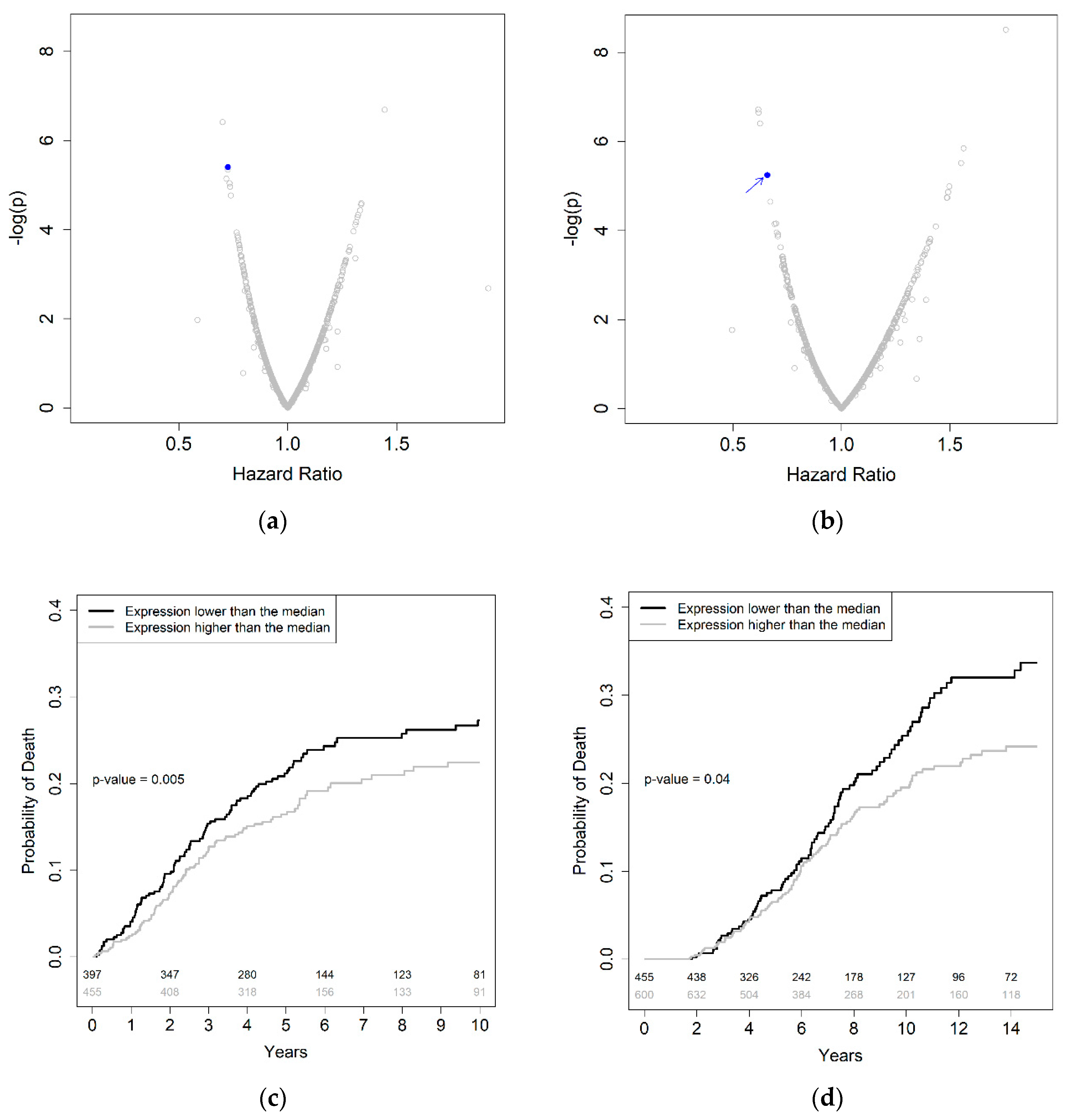
2.2. Association of Genetically Predicted Gene Expression and Colorectal Cancer Patients’ Survival
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Genotyping
4.3. Gene Expression Measurement
4.4. Gene Expression Prediction
4.5. Statistical Analysis
4.6. Validation Set
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Chen, C. The colorectal cancer epidemic: Challenges and opportunities for primary, secondary and tertiary prevention. Br. J. Cancer 2018, 119, 785–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, N.; Kim, K.; Shin, A.; Park, J.W.; Chang, H.J.; Shi, J.; Cai, Q.; Kim, D.Y.; Zheng, W.; Oh, J.H. Colorectal cancer susceptibility loci and influence on survival. Genes. Chromosomes Cancer 2018, 57, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Dimberg, J.; Shamoun, L.; Landerholm, K.; Andersson, R.E.; Kolodziej, B.; Wågsäter, D. Genetic Variants of the IL2 Gene Related to Risk and Survival in Patients with Colorectal Cancer. Anticancer Res. 2019, 39, 4933–4940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Summers, M.G.; Maughan, T.S.; Kaplan, R.; Law, P.J.; Houlston, R.S.; Escott-Price, V.; Cheadle, J.P. Comprehensive analysis of colorectal cancer-risk loci and survival outcome: A prognostic role for CDH1 variants. Eur. J. Cancer 2020, 124, 56–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiraskova, K.; Hughes, D.J.; Brezina, S.; Gumpenberger, T.; Veskrnova, V.; Buchler, T.; Schneiderova, M.; Levy, M.; Liska, V.; Vodenkova, S.; et al. Functional Polymorphisms in DNA Repair Genes Are Associated with Sporadic Colorectal Cancer Susceptibility and Clinical Outcome. Int. J. Mol. Sci. 2018, 20, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherer, D.; Deutelmoser, H.; Balavarca, Y.; Toth, R.; Habermann, N.; Buck, K.; Kap, E.J.; Botma, A.; Seibold, P.; Jansen, L.; et al. Polymorphisms in the Angiogenesis-Related Genes EFNB2, MMP2 and JAG1 Are Associated with Survival of Colorectal Cancer Patients. Int. J. Mol. Sci. 2020, 21, 5395. [Google Scholar] [CrossRef]
- Westra, H.J.; Franke, L. From genome to function by studying eQTLs. Biochim. Biophys. Acta 2014, 1842, 1896–1902. [Google Scholar] [CrossRef] [Green Version]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Variant Effect Predictor. Available online: http://www.ensembl.org/Homo_sapiens/Tools/VEP?db=core (accessed on 14 September 2020).
- Gamazon, E.R.; Wheeler, H.E.; Shah, K.P.; Mozaffari, S.V.; Aquino-Michaels, K.; Carroll, R.J.; Eyler, A.E.; Denny, J.C.; Consortium, G.T.; Nicolae, D.L.; et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015, 47, 1091. Available online: https://www.nature.com/articles/ng.3367#supplementary-information (accessed on 23 September 2019). [CrossRef] [Green Version]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [Green Version]
- Cherlin, S.; Lewis, M.J.; Plant, D.; Nair, N.; Goldmann, K.; Tzanis, E.; Barnes, M.R.; McKeigue, P.; Barrett, J.H.; Pitzalis, C.; et al. Investigation of genetically regulated gene expression and response to treatment in rheumatoid arthritis highlights an association between IL18RAP expression and treatment response. Ann. Rheum. Dis. 2020, 79, 1446–1452. [Google Scholar] [CrossRef]
- Portella, A.K.; Papantoni, A.; Paquet, C.; Moore, S.; Rosch, K.S.; Mostofsky, S.; Lee, R.S.; Smith, K.R.; Levitan, R.; Silveira, P.P.; et al. Predicted DRD4 prefrontal gene expression moderates snack intake and stress perception in response to the environment in adolescents. PLoS ONE 2020, 15, e0234601. [Google Scholar] [CrossRef]
- Wheeler, H.E.; Ploch, S.; Barbeira, A.N.; Bonazzola, R.; Andaleon, A.; Fotuhi Siahpirani, A.; Saha, A.; Battle, A.; Roy, S.; Im, H.K. Imputed gene associations identify replicable trans-acting genes enriched in transcription pathways and complex traits. Genet. Epidemiol. 2019, 43, 596–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Graff, M.; Haessler, J.; Buyske, S.; Bien, S.A.; Tao, R.; Highland, H.M.; Nishimura, K.K.; Zubair, N.; Lu, Y.; et al. Minority-centric meta-analyses of blood lipid levels identify novel loci in the Population Architecture using Genomics and Epidemiology (PAGE) study. PLoS Genet. 2020, 16, e1008684. [Google Scholar] [CrossRef]
- Fiorica, P.N.; Wheeler, H.E. Transcriptome association studies of neuropsychiatric traits in African Americans implicate PRMT7 in schizophrenia. PeerJ 2019, 7, e7778. [Google Scholar] [CrossRef]
- Bien, S.A.; Su, Y.R.; Conti, D.V.; Harrison, T.A.; Qu, C.; Guo, X.; Lu, Y.; Albanes, D.; Auer, P.L.; Banbury, B.L.; et al. Genetic variant predictors of gene expression provide new insight into risk of colorectal cancer. Hum. Genet. 2019, 138, 307–326. [Google Scholar] [CrossRef] [Green Version]
- Xia, Z.; Su, Y.R.; Petersen, P.; Qi, L.; Kim, A.E.; Figueiredo, J.C.; Lin, Y.; Nan, H.; Sakoda, L.C.; Albanes, D.; et al. Functional informed genome-wide interaction analysis of body mass index, diabetes and colorectal cancer risk. Cancer Med. 2020, 9, 3563–3573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ioannidis, N.M.; Wang, W.; Furlotte, N.A.; Hinds, D.A.; Me Research Team; Bustamante, C.D.; Jorgenson, E.; Asgari, M.M.; Whittemore, A.S. Gene expression imputation identifies candidate genes and susceptibility loci associated with cutaneous squamous cell carcinoma. Nat. Commun. 2018, 9, 4264. [Google Scholar] [CrossRef]
- Pattee, J.; Zhan, X.; Xiao, G.; Pan, W. Integrating germline and somatic genetics to identify genes associated with lung cancer. Genet. Epidemiol. 2020, 44, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Verma, S.S.; Veturi, Y.C.; Verma, A.; Bradford, Y.; Haas, D.W.; Ritchie, M.D. Evaluation of PrediXcan for prioritizing GWAS associations and predicting gene expression. Pac. Symp. Biocomput. 2018, 23, 448–459. [Google Scholar]
- Mikhaylova, A.V.; Thornton, T.A. Accuracy of Gene Expression Prediction From Genotype Data With PrediXcan Varies Across and Within Continental Populations. Front. Genet. 2019, 10, 261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheeler, H.E.; Shah, K.P.; Brenner, J.; Garcia, T.; Aquino-Michaels, K.; Consortium, G.T.; Cox, N.J.; Nicolae, D.L.; Im, H.K. Survey of the Heritability and Sparse Architecture of Gene Expression Traits across Human Tissues. PLoS Genet. 2016, 12, e1006423. [Google Scholar] [CrossRef] [Green Version]
- Deutelmoser, H.; Scherer, T.; Brenner, H.; Waldenberger, M.; Study, I.; Suhre, K.; Kastenmüller, G.; Lorenzo Bermejo, J. Robust Huber-LASSO for improved prediction of protein, metabolite, and gene expression levels relying on individual genotype data. Brief. Bioinform. 2020, in press. [Google Scholar] [CrossRef]
- Novembre, J.; Johnson, T.; Bryc, K.; Kutalik, Z.; Boyko, A.R.; Auton, A.; Indap, A.; King, K.S.; Bergmann, S.; Nelson, M.R.; et al. Genes mirror geography within Europe. Nature 2008, 456, 98–101. [Google Scholar] [CrossRef] [Green Version]
- Chirshev, E.; Oberg, K.C.; Ioffe, Y.J.; Unternaehrer, J.J. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019, 8, 24. [Google Scholar] [CrossRef] [Green Version]
- Liao, T.T.; Lin, C.C.; Jiang, J.K.; Yang, S.H.; Teng, H.W.; Yang, M.H. Harnessing stemness and PD-L1 expression by AT-rich interaction domain-containing protein 3B in colorectal cancer. Theranostics 2020, 10, 6095–6112. [Google Scholar] [CrossRef]
- Kobayashi, K.; Era, T.; Takebe, A.; Jakt, L.M.; Nishikawa, S. ARID3B induces malignant transformation of mouse embryonic fibroblasts and is strongly associated with malignant neuroblastoma. Cancer Res. 2006, 66, 8331–8336. [Google Scholar] [CrossRef] [Green Version]
- Joseph, S.; Deneke, V.E.; Cowden Dahl, K.D. ARID3B induces tumor necrosis factor alpha mediated apoptosis while a novel ARID3B splice form does not induce cell death. PLoS ONE 2012, 7, e42159. [Google Scholar] [CrossRef] [Green Version]
- Akhavantabasi, S.; Sapmaz, A.; Tuna, S.; Erson-Bensan, A.E. miR-125b Targets ARID3B in Breast Cancer Cells. Cell Struct. Funct. 2012, 37, 27–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, L.; Samyesudhas, S.J.; Carrasco, M.; Li, J.; Joseph, S.; Dahl, R.; Cowden Dahl, K.D. ARID3B increases ovarian tumor burden and is associated with a cancer stem cell gene signature. Oncotarget 2014, 5, 8355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lilla, C.; Verla-Tebit, E.; Risch, A.; Jäger, B.; Hoffmeister, M.; Brenner, H.; Chang-Claude, J. Effect of NAT1 and NAT2 Genetic Polymorphisms on Colorectal Cancer Risk Associated with Exposure to Tobacco Smoke and Meat Consumption. Cancer Epidemiol. Biomark. Prev. 2006, 15, 99–107. [Google Scholar] [CrossRef] [Green Version]
- Brenner, H.; Chang-Claude, J.; Seiler, C.M.; Sturmer, T.; Hoffmeister, M. Does a negative screening colonoscopy ever need to be repeated? Gut 2006, 55, 1145–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, U.; Jiao, S.; Schumacher, F.R.; Hutter, C.M.; Aragaki, A.K.; Baron, J.A.; Berndt, S.I.; Bezieau, S.; Brenner, H.; Butterbach, K.; et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology 2013, 144, 799–807.e24. [Google Scholar] [CrossRef] [Green Version]
- Ulrich, C.M.; Gigic, B.; Bohm, J.; Ose, J.; Viskochil, R.; Schneider, M.; Colditz, G.A.; Figueiredo, J.C.; Grady, W.M.; Li, C.I.; et al. The ColoCare Study: A Paradigm of Transdisciplinary Science in Colorectal Cancer Outcomes. Cancer Epidemiol. Prev. Biomark. 2019, 28, 591–601. [Google Scholar] [CrossRef] [Green Version]
- Haffa, M.; Holowatyj, A.N.; Kratz, M.; Toth, R.; Benner, A.; Gigic, B.; Habermann, N.; Schrotz-King, P.; Bohm, J.; Brenner, H.; et al. Transcriptome Profiling of Adipose Tissue Reveals Depot-Specific Metabolic Alterations Among Patients with Colorectal Cancer. J. Clin. Endocrinol. Metab. 2019, 104, 5225–5237. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef] [Green Version]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Overall Survival | CRC-Specific Survival | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | Level | Patients | Deaths | HR 1 (95% CI) | p | Deaths | HR 1 (95% CI) | p |
| Age at | <60 | 946 | 257 | Ref. | <2 × 10−16 | 209 | Ref. | 0.0003 |
| Diagnosis | 60–69 | 1339 | 466 | 1.25 (1.07−1.46) 1.69 (1.47−1.96) 3.20 (2.74−3.74) | 314 | 1.06 (0.89−1.27) 1.12 (0.96−1.36) 1.47 (1.20−1.80) | ||
| (years) | 70–79 | 1489 | 638 | 357 | ||||
| >80 | 662 | 429 | 173 | |||||
| Gender | Male | 2685 | 1111 | Ref. | 0.16 | 626 | Ref. | 0.47 |
| Female | 1751 | 679 | 1.07 (0.97−1.18) | 427 | 0.96 (0.85−1.08) | |||
| CRC stage | I | 1024 | 235 | Ref. | <2 × 10−16 | 40 | Ref. | <2 × 10−16 |
| II | 1345 | 434 | 1.49 (1.27−1.74) 2.05 (1.76−2.39) 10.21 (8.73−11.94) | 148 | 2.96 (2.09−4.20) 7.29 (5.25−10.10) 48.91 (35.37−67.64) | |||
| III | 1426 | 575 | 354 | |||||
| IV | 641 | 546 | 511 | |||||
| Tumor site | Colon | 2665 | 1069 | Ref. | 0.61 | 598 | Ref. | 0.03 |
| Rectum | 1771 | 721 | 1.03 (0.93–1.13) | 455 | 1.15 (1.02−1.30) | |||
| Body mass | <18.5 | 121 | 71 | 1.74 (1.37−2.23) | 0.0006 | 41 | 1.62 (1.17−2.23) | 0.01 |
| index | 18.5−24.9 | 1592 | 675 | Ref. | 396 | Ref. | ||
| (kg/m2) | 25–29.9 | 1871 | 726 | 0.87 (0.78−0.96) 0.83 (0.73−0.95) | 432 | 0.88 (0.77−1.01) 0.83 (0.70–0.99) | ||
| ≥30 | 852 | 318 | 184 | |||||
| Diabetes | No | 3594 | 1360 | Ref. | <8 × 10−13 | 844 | Ref. | 0.09 |
| Yes | 811 | 412 | 1.50 (1.34−1.67) | 200 | 1.14 (0.98−1.33) | |||
| Regular | No | 3272 | 1298 | Ref. | 0.05 | 793 | Ref. | 0.31 |
| NSAID use | Yes | 1105 | 469 | 1.11 (1.00−1.24) | 245 | 0.93 (0.80−1.07) | ||
| Smoking | Never | 1921 | 840 | Ref. | 0.04 | 515 | Ref. | 0.07 |
| Former | 1769 | 684 | 0.90 (0.81−1.00) 0.89 (0.78−1.03) | 367 | 0.79 (0.69−0.90) 0.94 (0.79−1.12) | |||
| Current | 672 | 263 | 169 | |||||
| Alcohol | No intake | 1320 | 606 | Ref. | 0.0003 | 339 | Ref. | 0.01 |
| intake | 0.1−5.6 | 856 | 318 | 0.75 (0.66−0.86) 0.72 (0.63−0.83) 0.78 (0.68−0.90) 0.79 (0.69−0.92) | 202 | 0.87 (0.73−1.03) 1.22 (0.68−0.98) 1.20 (0.70−1.00) 1.24 (0.66−0.97) | ||
| (g/day) | 5.7−13.2 | 770 | 280 | 175 | ||||
| 13.3−28.5 | 775 | 301 | 177 | |||||
| ≥28.6 | 669 | 269 | 148 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deutelmoser, H.; Lorenzo Bermejo, J.; Benner, A.; Weigl, K.; Park, H.A.; Haffa, M.; Herpel, E.; Schneider, M.; Ulrich, C.M.; Hoffmeister, M.; et al. Genotype-Based Gene Expression in Colon Tissue—Prediction Accuracy and Relationship with the Prognosis of Colorectal Cancer Patients. Int. J. Mol. Sci. 2020, 21, 8150. https://doi.org/10.3390/ijms21218150
Deutelmoser H, Lorenzo Bermejo J, Benner A, Weigl K, Park HA, Haffa M, Herpel E, Schneider M, Ulrich CM, Hoffmeister M, et al. Genotype-Based Gene Expression in Colon Tissue—Prediction Accuracy and Relationship with the Prognosis of Colorectal Cancer Patients. International Journal of Molecular Sciences. 2020; 21(21):8150. https://doi.org/10.3390/ijms21218150
Chicago/Turabian StyleDeutelmoser, Heike, Justo Lorenzo Bermejo, Axel Benner, Korbinian Weigl, Hanla A. Park, Mariam Haffa, Esther Herpel, Martin Schneider, Cornelia M. Ulrich, Michael Hoffmeister, and et al. 2020. "Genotype-Based Gene Expression in Colon Tissue—Prediction Accuracy and Relationship with the Prognosis of Colorectal Cancer Patients" International Journal of Molecular Sciences 21, no. 21: 8150. https://doi.org/10.3390/ijms21218150
APA StyleDeutelmoser, H., Lorenzo Bermejo, J., Benner, A., Weigl, K., Park, H. A., Haffa, M., Herpel, E., Schneider, M., Ulrich, C. M., Hoffmeister, M., Chang-Claude, J., Brenner, H., & Scherer, D. (2020). Genotype-Based Gene Expression in Colon Tissue—Prediction Accuracy and Relationship with the Prognosis of Colorectal Cancer Patients. International Journal of Molecular Sciences, 21(21), 8150. https://doi.org/10.3390/ijms21218150







