Advances on Plant Ubiquitylome—From Mechanism to Application
Abstract
:1. Introduction
2. The Ubiquitylation Machinery and Code
3. Methods of Ubiquitylation Detection and Application in Plant Ubiquitylome
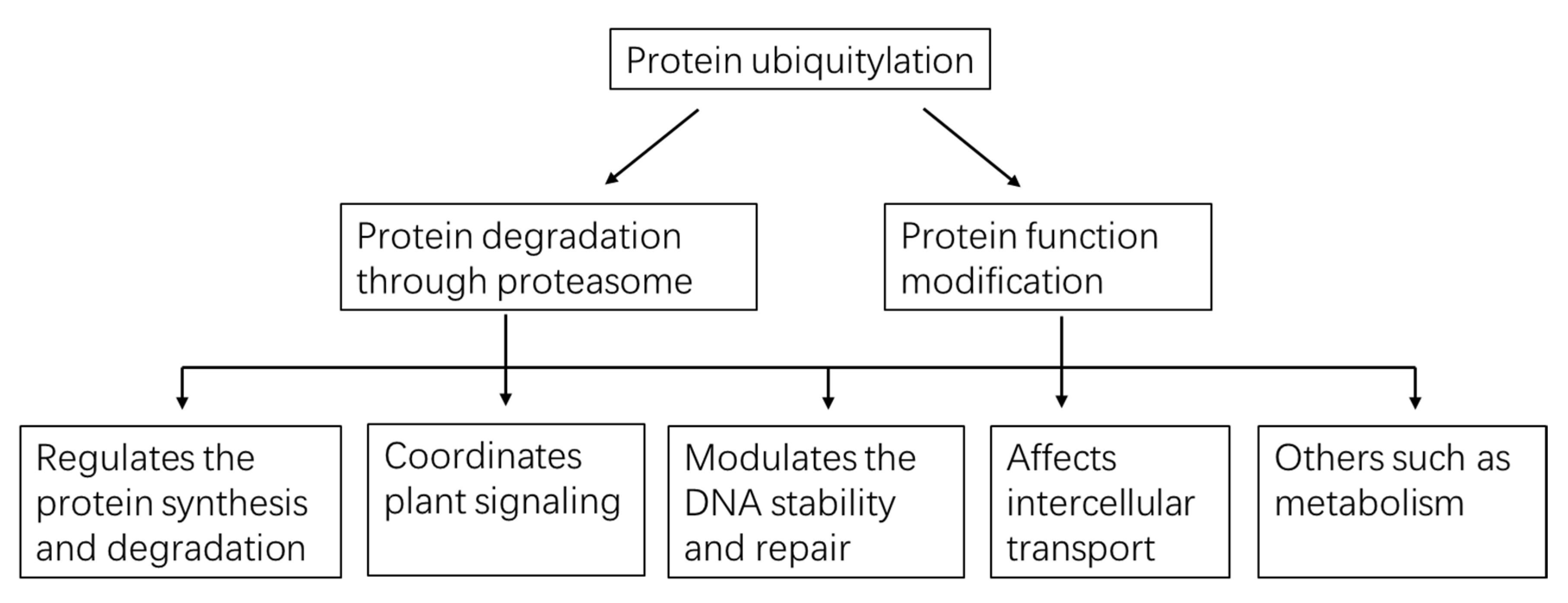
4. Multiple Functions Played by Protein Ubiquitylation in Plants
4.1. Ubiquitylation Regulates Protein Synthesis and Degradation
4.2. Ubiquitylation Coordinates Plant Signaling
4.3. Ubiquitylation Modulates DNA Stability and Repair
4.4. Ubiquitylation Affects Intercellular Transport
5. Crosstalk between Ubiquitylation and Other PTMs
6. Related Databases Developed for Plant Ubiquitylation
7. Future Challenges of Plant Protein Ubiquitylation
Author Contributions
Funding
Conflicts of Interest
References
- Millar, A.H.; Heazlewood, J.L.; Giglione, C.; Holdsworth, M.J.; Bachmair, A.; Schulze, W.X. The scope, functions and dynamics of posttranslational protein modifications. Annu. Rev. Plant Biol. 2019, 70, 119–151. [Google Scholar] [CrossRef]
- Consortium, U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, T. The age of crosstalk: Phosphorylation, ubiquitination and beyond. Mol. Cell 2007, 28, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zeng, L. Crosstalk between Ubiquitination and Other Posttranslational Protein Modifications in Plant Immunity. Plant Commun. 2020, 1, 100041. [Google Scholar] [CrossRef]
- Goldstein, G.; Scheid, M.; Hammerling, U.; Schlesinger, D.; Niall, H.; Boyse, E. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl. Acad. Sci. USA 1975, 72, 11–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, D.; Li, M.; Damaris, R.N.; Bu, C.; Xue, J.; Yang, P. Quantitative ubiquitylomics approach for characterizing the dynamic change and extensive modulation of ubiquitylation in rice seed germination. Plant J. 2020, 101, 1440–1447. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Akopian, D.; Rape, M. Principles of ubiquitin-dependent signaling. Annu. Rev. Cell Dev. Biol. 2018, 34, 137–162. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Liu, J.; Wei, Q.; Wang, R.; Yang, W.; Ma, Y.; Chen, G.; Yu, Y. Proteomes and Ubiquitylomes Analysis Reveals the Involvement of Ubiquitination in Protein Degradation in Petunias. Plant Physiol. 2017, 173, 668–687. [Google Scholar] [CrossRef]
- Wang, Y.-F.; Chao, Q.; Li, Z.; Lu, T.-C.; Zheng, H.-Y.; Zhao, C.-F.; Shen, Z.; Li, X.-H.; Wang, B.-C. Large-scale Identification and Time-course Quantification of Ubiquitylation Events During Maize Seedling De-etiolation. Genom. Proteom. Bioinform. 2020. [Google Scholar] [CrossRef]
- Ye, Y.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef] [Green Version]
- Pao, K.-C.; Wood, N.T.; Knebel, A.; Rafie, K.; Stanley, M.; Mabbitt, P.D.; Sundaramoorthy, R.; Hofmann, K.; van Aalten, D.M.; Virdee, S. Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature 2018, 556, 381–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Herr, R.A.; Hansen, T.H. Ubiquitination of substrates by esterification. Traffic 2012, 13, 19–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breitschopf, K.; Bengal, E.; Ziv, T.; Admon, A.; Ciechanover, A. A novel site for ubiquitination: The N-terminal residue and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 1998, 17, 5964–5973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, J.M.; Schwartz, D.; Elias, J.E.; Thoreen, C.C.; Cheng, D.M.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. [Google Scholar] [CrossRef]
- Callis, J.; Carpenter, T.; Sun, C.-W.; Vierstra, R.D. Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics 1995, 139, 921–939. [Google Scholar]
- Callis, J. The ubiquitination machinery of the ubiquitin system. Arab. Book Am. Soc. Plant Biol. 2014, 12, e0174. [Google Scholar] [CrossRef] [Green Version]
- Isono, E.; Nagel, M.-K. Deubiquitylating enzymes and their emerging role in plant biology. Front. Plant Sci. 2014, 5, 56. [Google Scholar] [CrossRef] [Green Version]
- Rape, M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 59–70. [Google Scholar] [CrossRef]
- Schulman, B.A.; Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef] [Green Version]
- Du, Z.; Zhou, X.; Li, L.; Su, Z. plantsUPS: A database of plants‘ Ubiquitin Proteasome System. BMC Genom. 2009, 10, 227. [Google Scholar] [CrossRef] [Green Version]
- Stone, S.L.; Hauksdottir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenzel, D.M.; Lissounov, A.; Brzovic, P.S.; Klevit, R.E. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 2011, 474, 105–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clague, M.J.; Barsukov, I.; Coulson, J.M.; Liu, H.; Rigden, D.J.; Urbe, S. Deubiquitylases from Genes to Organism. Physiol. Rev. 2013, 93, 1289–1315. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Clague, M.J.; Urbe, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Maor, R.; Jones, A.; Nühse, T.S.; Studholme, D.J.; Peck, S.C.; Shirasu, K. Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol. Cell Proteom. 2007, 6, 601–610. [Google Scholar] [CrossRef] [Green Version]
- Saracco, S.A.; Hansson, M.; Scalf, M.; Walker, J.M.; Smith, L.M.; Vierstra, R.D. Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J. 2009, 59, 344–358. [Google Scholar] [CrossRef] [Green Version]
- Lopitz-Otsoa, F.; Rodriguez, M.S.; Aillet, F. Properties of Natural and Artificial Proteins Displaying Multiple Ubiquitin-Binding Domains; Portland Press Ltd.: London, UK, 2010. [Google Scholar]
- Kim, D.Y.; Scalf, M.; Smith, L.M.; Vierstra, R.D. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 2013, 25, 1523–1540. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Paige, J.S.; Jaffrey, S.R. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 2010, 28, 868. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef] [Green Version]
- Wagner, S.A.; Beli, P.; Weinert, B.T.; Nielsen, M.L.; Cox, J.; Mann, M.; Choudhary, C. A Proteome-wide, Quantitative Survey of In Vivo Ubiquitylation Sites Reveals Widespread Regulatory Roles. Mol. Cell Proteom. 2011, 10. [Google Scholar] [CrossRef] [Green Version]
- Vierstra, R.D. The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 2012, 160, 2–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hjerpe, R.; Aillet, F.; Lopitz-Otsoa, F.; Lang, V.; England, P.; Rodriguez, M.S. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009, 10, 1250–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walton, A.; Stes, E.; Cybulski, N.; Van Bel, M.; Inigo, S.; Durand, A.N.; Timmerman, E.; Heyman, J.; Pauwels, L.; De Veylder, L.; et al. It’s Time for Some “Site”-Seeing: Novel Tools to Monitor the Ubiquitin Landscape in Arabidopsis thaliana. Plant Cell 2016, 28, 6–16. [Google Scholar] [CrossRef] [Green Version]
- Stes, E.; Laga, M.; Walton, A.; Samyn, N.; Timmerman, E.; De Smet, I.; Goormachtig, S.; Gevaert, K. A COFRADIC protocol to study protein ubiquitination. J. Proteome Res. 2014, 13, 3107–3113. [Google Scholar] [CrossRef] [PubMed]
- Renatus, M.; Parrado, S.G.; D’Arcy, A.; Eidhoff, U.; Gerhartz, B.; Hassiepen, U.; Pierrat, B.; Riedl, R.; Vinzenz, D.; Worpenberg, S. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure 2006, 14, 1293–1302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kliza, K.; Taumer, C.; Pinzuti, I.; Franz-Wachtel, M.; Kunzelmann, S.; Stieglitz, B.; Macek, B.; Husnjak, K. Internally tagged ubiquitin: A tool to identify linear polyubiquitin-modified proteins by mass spectrometry. Nat. Methods 2017, 14, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Rose, C.M.; Isasa, M.; Ordureau, A.; Prado, M.A.; Beausoleil, S.A.; Jedrychowski, M.P.; Finley, D.J.; Harper, J.W.; Gygi, S.P. Highly multiplexed quantitative mass spectrometry analysis of ubiquitylomes. Cell Syst. 2016, 3, 395–403.e394. [Google Scholar] [CrossRef] [PubMed]
- Udeshi, N.D.; Mani, D.C.; Satpathy, S.; Fereshetian, S.; Gasser, J.A.; Svinkina, T.; Olive, M.E.; Ebert, B.L.; Mertins, P.; Carr, S.A. Rapid and deep-scale ubiquitylation profiling for biology and translational research. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Schnell, J.D.; Hicke, L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 2003, 278, 35857–35860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hershko, A.; Ciechanover, A.; Varshavsky, A. The ubiquitin system. Nat. Med. 2000, 6, 1073–1081. [Google Scholar] [CrossRef]
- Jin, L.; Williamson, A.; Banerjee, S.; Philipp, I.; Rape, M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 2008, 133, 653–665. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Zhu, D.; Huang, X.; Li, S.; Gong, Y.; Yao, Q.; Fu, X.; Fan, L.-M.; Deng, X.W. Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 2009, 21, 2378–2390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leitner, J.; Petrášek, J.; Tomanov, K.; Retzer, K.; Pařezová, M.; Korbei, B.; Bachmair, A.; Zažímalová, E.; Luschnig, C. Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc. Natl. Acad. Sci. USA 2012, 109, 8322–8327. [Google Scholar] [CrossRef] [Green Version]
- Ohtake, F.; Tsuchiya, H.; Saeki, Y.; Tanaka, K. K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc. Natl. Acad. Sci. USA 2018, 115, E1401–E1408. [Google Scholar] [CrossRef] [Green Version]
- Swatek, K.N.; Usher, J.L.; Kueck, A.F.; Gladkova, C.; Mevissen, T.E.; Pruneda, J.N.; Skern, T.; Komander, D. Insights into ubiquitin chain architecture using Ub-clipping. Nature 2019, 572, 533–537. [Google Scholar] [CrossRef]
- Shcherbik, N.; Pestov, D.G. Ubiquitin and ubiquitin-like proteins in the nucleolus: Multitasking tools for a ribosome factory. Genes Cancer 2010, 1, 681–689. [Google Scholar] [CrossRef]
- Book, A.J.; Smalle, J.; Lee, K.H.; Yang, P.Z.; Walker, J.M.; Casper, S.; Holmes, J.H.; Russo, L.A.; Buzzinotti, Z.W.; Jenik, P.D.; et al. The RPN5 Subunit of the 26s Proteasome Is Essential for Gametogenesis, Sporophyte Development and Complex Assembly in Arabidopsis. Plant Cell 2009, 21, 460–478. [Google Scholar] [CrossRef] [Green Version]
- Meyer, H.-J.; Rape, M. Enhanced protein degradation by branched ubiquitin chains. Cell 2014, 157, 910–921. [Google Scholar] [CrossRef] [Green Version]
- Deshaies, R.J.; Joazeiro, C.A.P. RING Domain E3 Ubiquitin Ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- McEwan, D.G.; Dikic, I. The three musketeers of autophagy: Phosphorylation, ubiquitylation and acetylation. Trends Cell Biol. 2011, 21, 195–201. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Chung, T.; Vierstra, R.D. AUTOPHAGY-RELATED11 plays a critical role in general autophagy-and senescence-induced mitophagy in Arabidopsis. Plant Cell 2014, 26, 788–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suttangkakul, A.; Li, F.; Chung, T.; Vierstra, R.D. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 2011, 23, 3761–3779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, H.; Xia, F.-N.; Xie, L.-J.; Yu, L.-J.; Chen, Q.-F.; Zhuang, X.-H.; Wang, Q.; Li, F.; Jiang, L.; Xie, Q. TRAF family proteins regulate autophagy dynamics by modulating AUTOPHAGY PROTEIN6 stability in Arabidopsis. Plant Cell 2017, 29, 890–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkin, V.; Lamark, T.; Sou, Y.-S.; Bjørkøy, G.; Nunn, J.L.; Bruun, J.-A.; Shvets, E.; McEwan, D.G.; Clausen, T.H.; Wild, P. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 2009, 33, 505–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fierz, B.; Chatterjee, C.; McGinty, R.K.; Bar-Dagan, M.; Raleigh, D.P.; Muir, T.W. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 2011, 7, 113. [Google Scholar] [CrossRef]
- Winkler, M.; Niemeyer, M.; Hellmuth, A.; Janitza, P.; Christ, G.; Samodelov, S.L.; Wilde, V.; Majovsky, P.; Trujillo, M.; Zurbriggen, M.D. Variation in auxin sensing guides AUX/IAA transcriptional repressor ubiquitylation and destruction. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Tokunaga, F.; Sakata, S.-I.; Saeki, Y.; Satomi, Y.; Kirisako, T.; Kamei, K.; Nakagawa, T.; Kato, M.; Murata, S.; Yamaoka, S. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat. Cell Biol. 2009, 11, 123–132. [Google Scholar] [CrossRef]
- Michel, M.A.; Swatek, K.N.; Hospenthal, M.K.; Komander, D. Ubiquitin linkage-specific affimers reveal insights into K6-linked ubiquitin signaling. Mol. Cell 2017, 68, 233–246.e235. [Google Scholar] [CrossRef] [Green Version]
- Min, M.; Mevissen, T.E.; De Luca, M.; Komander, D.; Lindon, C. Efficient APC/C substrate degradation in cells undergoing mitotic exit depends on K11 ubiquitin linkages. Mol. Biol. Cell 2015, 26, 4325–4332. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, H.; Zheng, G.-L.; Yang, Q.; Yu, S.; Wang, J.; Li, S.; Li, L.-F.; Qiu, H.-J. Porcine RING finger protein 114 inhibits classical swine fever virus replication via K27-linked polyubiquitination of viral NS4B. J. Virol. 2019, 93, e01248-19. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Ritchie, A.-M.; Melton, D.W. Disruption of DNA repair in cancer cells by ubiquitination of a destabilising dimerization domain of nucleotide excision repair protein ERCC1. Oncotarget 2017, 8, 55246. [Google Scholar] [CrossRef] [Green Version]
- Emmerich, C.H.; Ordureau, A.; Strickson, S.; Arthur, J.S.C.; Pedrioli, P.G.; Komander, D.; Cohen, P. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl. Acad. Sci. USA 2013, 110, 15247–15252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shanklin, J.; Jabben, M.; Vierstra, R.D. Red light-induced formation of ubiquitin-phytochrome conjugates: Identification of possible intermediates of phytochrome degradation. Proc. Natl. Acad. Sci. USA 1987, 84, 359–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Paik, I.; Zhu, L.; Huq, E. Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci. 2015, 20, 641–650. [Google Scholar] [CrossRef] [Green Version]
- Sorel, M.; Mooney, B.; de Marchi, R.; Graciet, E. Ubiquitin/Proteasome System in Plant Pathogen Responses. Annu. Plant Rev. Online 2018, 65–116. [Google Scholar] [CrossRef]
- Swaney, D.L.; Beltrao, P.; Starita, L.; Guo, A.L.; Rush, J.; Fields, S.; Krogan, N.J.; Villen, J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 2013, 10, 676–682. [Google Scholar] [CrossRef]
- Gao, D.Y.; Xu, Z.S.; He, Y.; Sun, Y.W.; Ma, Y.Z.; Xia, L.Q. Functional analyses of an E3 ligase gene AIP2 from wheat in Arabidopsis revealed its roles in seed germination and pre-harvest sprouting. J. Integr. Plant Biol. 2014, 56, 480–491. [Google Scholar] [CrossRef]
- Bueso, E.; Rodriguez, L.; Lorenzo-Orts, L.; Gonzalez-Guzman, M.; Sayas, E.; Munoz-Bertomeu, J.; Ibanez, C.; Serrano, R.; Rodriguez, P.L. The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 2014, 80, 1057–1071. [Google Scholar] [CrossRef]
- Chapman, E.J.; Estelle, M. Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef] [Green Version]
- Martins, S.; Dohmann, E.M.N.; Cayrel, A.; Johnson, A.; Fischer, W.; Pojer, F.; Satiat-Jeunemaitre, B.; Jaillais, Y.; Chory, J.; Geldner, N.; et al. Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitinationpavr. Nat. Commun. 2015, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Ju, C.; Chang, C. Mechanistic insights in ethylene perception and signal transduction. Plant Physiol. 2015, 169, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Qiao, H.; Chang, K.N.; Yazaki, J.; Ecker, J.R. Interplay between ethylene, ETP1/ETP2 F-box proteins and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 2009, 23, 512–521. [Google Scholar] [CrossRef] [Green Version]
- Kulathu, Y.; Komander, D. Atypical ubiquitylation—The unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012, 13, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Elia, A.E.; Boardman, A.P.; Wang, D.C.; Huttlin, E.L.; Everley, R.A.; Dephoure, N.; Zhou, C.; Koren, I.; Gygi, S.P.; Elledge, S.J. Quantitative Proteomic Atlas of Ubiquitination and Acetylation in the DNA Damage Response. Mol. Cell 2015, 59, 867–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciccia, A.; Elledge, S.J. The DNA Damage Response: Making It Safe to Play with Knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003, 17, 2733–2740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shilatifard, A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu. Rev. Biochem. 2006, 75, 243–269. [Google Scholar] [CrossRef] [Green Version]
- Pinder, J.B.; Attwood, K.M.; Dellaire, G. Reading, writing and repair: The role of ubiquitin and the ubiquitin-like proteins in DNA damage signaling and repair. Front. Genet. 2013, 4, 45. [Google Scholar] [CrossRef] [Green Version]
- Goldknopf, I.L.; French, M.F.; Musso, R.; Busch, H. Presence of protein A24 in rat liver nucleosomes. Proc. Natl. Acad. Sci. USA 1977, 74, 5492–5495. [Google Scholar] [CrossRef] [Green Version]
- McGinty, R.K.; Henrici, R.C.; Tan, S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 2014, 514, 591–596. [Google Scholar] [CrossRef] [Green Version]
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 2014, 157, 1445–1459. [Google Scholar] [CrossRef] [Green Version]
- Pavri, R.; Zhu, B.; Li, G.; Trojer, P.; Mandal, S.; Shilatifard, A.; Reinberg, D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 2006, 125, 703–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishiyama, S.; Nishiyama, A.; Saeki, Y.; Moritsugu, K.; Morimoto, D.; Yamaguchi, L.; Arai, N.; Matsumura, R.; Kawakami, T.; Mishima, Y. Structure of the Dnmt1 reader module complexed with a unique two-mono-ubiquitin mark on histone H3 reveals the basis for DNA methylation maintenance. Mol. Cell 2017, 68, 350–360.e357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tessadori, F.; Giltay, J.C.; Hurst, J.A.; Massink, M.P.; Duran, K.; Vos, H.R.; van Es, R.M.; Study, D.D.D.; Scott, R.H.; van Gassen, K.L. Germline mutations affecting the histone H4 core cause a developmental syndrome by altering DNA damage response and cell cycle control. Nat. Genet. 2017, 49, 1642. [Google Scholar] [CrossRef] [PubMed]
- Mevissen, T.E.; Komander, D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 2017, 86, 159–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levkowitz, G.; Waterman, H.; Zamir, E.; Kam, Z.; Oved, S.; Langdon, W.Y.; Beguinot, L.; Geiger, B.; Yarden, Y. C-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998, 12, 3663–3674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isono, E.; Kalinowska, K. ESCRT-dependent degradation of ubiquitylated plasma membrane proteins in plants. Curr. Opin. Plant Biol. 2017, 40, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, O.; Teis, D. The ESCRT machinery. Curr. Biol. 2012, 22, R116–R120. [Google Scholar] [CrossRef] [Green Version]
- Dobzinski, N.; Chuartzman, S.G.; Kama, R.; Schuldiner, M.; Gerst, J.E. Starvation-dependent regulation of golgi quality control links the TOR signaling and vacuolar protein sorting pathways. Cell Rep. 2015, 12, 1876–1886. [Google Scholar] [CrossRef] [Green Version]
- Gorur, A.; Yuan, L.; Kenny, S.J.; Baba, S.; Xu, K.; Schekman, R. COPII-coated membranes function as transport carriers of intracellular procollagen I. J. Cell Biol. 2017, 216, 1745–1759. [Google Scholar] [CrossRef] [Green Version]
- Mukai, A.; Mizuno, E.; Kobayashi, K.; Matsumoto, M.; Nakayama, K.I.; Kitamura, N.; Komada, M. Dynamic regulation of ubiquitylation and deubiquitylation at the central spindle during cytokinesis. J. Cell Sci. 2008, 121 Pt 8, 1325–1333. [Google Scholar] [CrossRef] [Green Version]
- Vu, L.D.; Gevaert, K.; De Smet, I. Protein Language: Post-Translational Modifications Talking to Each Other. Trends Plant Sci. 2018, 23, 1068–1080. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, L.; Boller, T. FLS2: An LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Lu, D.; Lin, W.; Gao, X.; Wu, S.; Cheng, C.; Avila, J.; Heese, A.; Devarenne, T.P.; He, P.; Shan, L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 2011, 332, 1439–1442. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Deng, J.; Zhu, F.; Li, Y.; Lu, Z.; Qin, P.; Wang, T.; Dong, J. The MtDMI2-MtPUB2 negative feedback loop plays a role in nodulation homeostasis. Plant Physiol. 2018, 176, 3003–3026. [Google Scholar] [CrossRef] [PubMed]
- Orlicky, S.; Tang, X.; Willems, A.; Tyers, M.; Sicheri, F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 2003, 112, 243–256. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Huang, Y.; Li, L.; Gannon, P.; Linster, E.; Huber, M.; Kapos, P.; Bienvenut, W.; Polevoda, B.; Meinnel, T. Two N-terminal acetyltransferases antagonistically regulate the stability of a nod-like receptor in Arabidopsis. Plant Cell 2015, 27, 1547–1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pajares, M.; Jiménez-Moreno, N.; Dias, I.H.; Debelec, B.; Vucetic, M.; Fladmark, K.E.; Basaga, H.; Ribaric, S.; Milisav, I.; Cuadrado, A. Redox control of protein degradation. Redox Biol. 2015, 6, 409–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 18, 579–586. [Google Scholar] [CrossRef]
- Okatsu, K.; Oka, T.; Iguchi, M.; Imamura, K.; Kosako, H.; Tani, N.; Kimura, M.; Go, E.; Koyano, F.; Funayama, M. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat. Commun. 2012, 3, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Herhaus, L.; Dikic, I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 2015, 16, 1071–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, D.L.; Wang, Q.; Li, M.; Damaris, R.N.; Yi, X.L.; Cheng, Z.Y.; Yang, P.F. Global Proteome Analyses of Lysine Acetylation and Succinylation Reveal the Widespread Involvement of both Modification in Metabolism in the Embryo of Germinating Rice Seed. J. Proteome Res. 2016, 15, 879–890. [Google Scholar] [CrossRef]
- Li, M.; Yin, X.J.; Sakata, K.; Yang, P.F.; Komatsu, S. Proteomic Analysis of Phosphoproteins in the Rice Nucleus During the Early Stage of Seed Germination. J. Proteome Res. 2015, 14, 2884–2896. [Google Scholar] [CrossRef] [PubMed]
- Chernorudskiy, A.L.; Garcia, A.; Eremin, E.V.; Shorina, A.S.; Kondratieva, E.V.; Gainullin, M.R. UbiProt: A database of ubiquitylated proteins. BMC Bioinform. 2007, 8, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nickchi, P.; Jafari, M.; Kalantari, S. PEIMAN 1.0: Post-translational modification Enrichment, Integration and Matching ANalysis. Database 2015, 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, V.-N.; Huang, K.-Y.; Weng, J.T.-Y.; Lai, K.R.; Lee, T.-Y. UbiNet: An online resource for exploring the functional associations and regulatory networks of protein ubiquitylation. Database 2016, 2016. [Google Scholar] [CrossRef] [Green Version]
- Minguez, P.; Letunic, I.; Parca, L.; Garcia-Alonso, L.; Dopazo, J.; Huerta-Cepas, J.; Bork, P. PTMcode v2: A resource for functional associations of post-translational modifications within and between proteins. Nucleic Acids Res. 2015, 43, D494–D502. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.-Y.; Lee, T.-Y.; Kao, H.-J.; Ma, C.-T.; Lee, C.-C.; Lin, T.-H.; Chang, W.-C.; Huang, H.-D. dbPTM in 2019: Exploring disease association and cross-talk of post-translational modifications. Nucleic Acids Res. 2019, 47, D298–D308. [Google Scholar] [CrossRef] [Green Version]
- Willems, P.; Horne, A.; Van Parys, T.; Goormachtig, S.; De Smet, I.; Botzki, A.; Van Breusegem, F.; Gevaert, K. The Plant PTM Viewer, a central resource for exploring plant protein modifications. Plant J. 2019, 99, 752–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
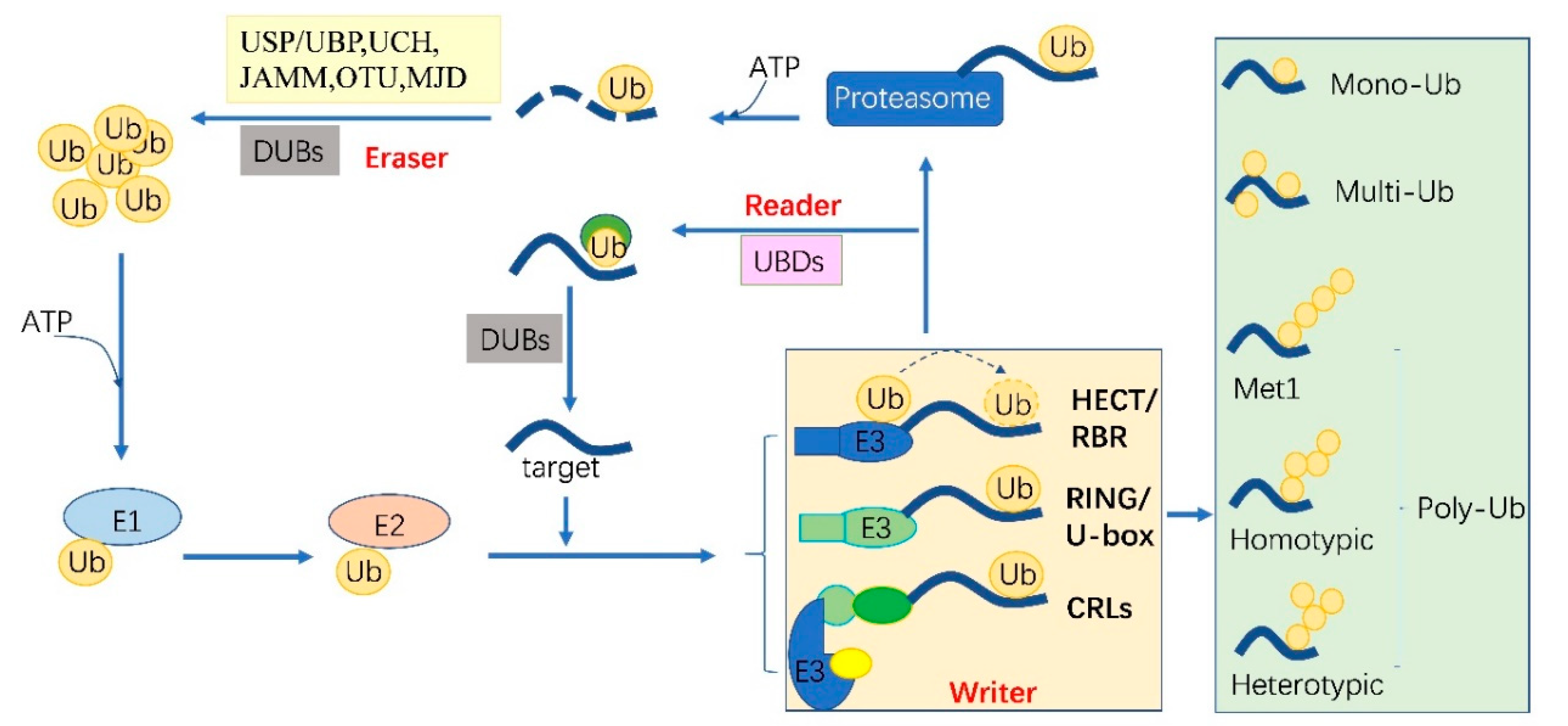
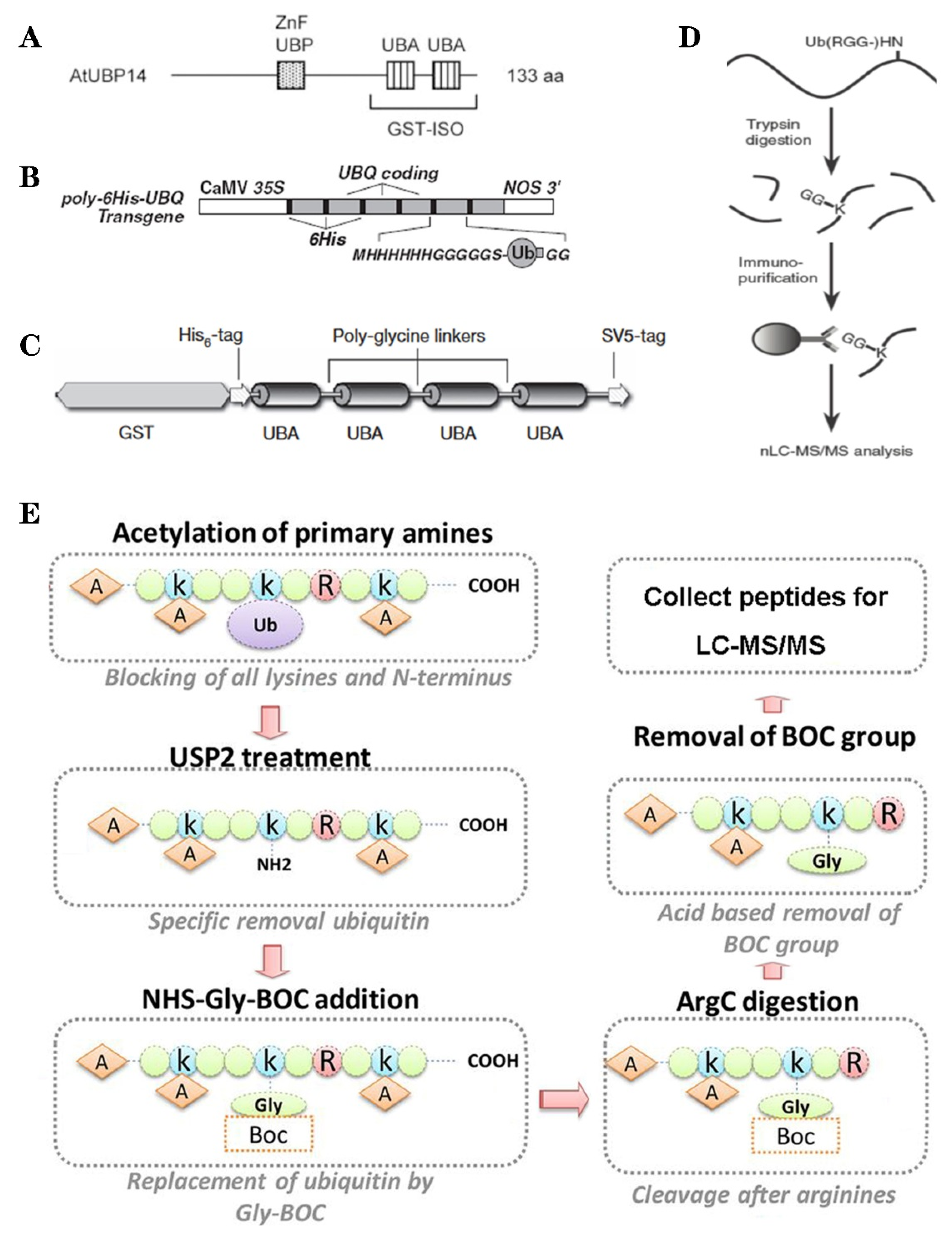
| Ub code | Function | Substrate | Reference |
|---|---|---|---|
| Mono-/multi-ubiquitylation | changes the protein activity and interaction | e.g., lysine 119 of H2B | [56] |
| K29-chains | proteasomal degradation | e.g., DELLA proteins | [43] |
| K48-chains | proteasomal degradation | e.g., Aux/IAA | [57] |
| K63-chains | endocytic sorting, DNA repair, degradation but proteasome-independent | e.g., PIN2 | [44] |
| M1 | inflammation signaling | e.g., NF-κB | [58] |
| K6-chains | Mitophagy | e.g., mitofusin-2 (Mfn2) | [59] |
| K11-chains | proteasomal degradation/cell cycle regulation | e.g., anaphase-promoting complex APC/C | [60] |
| K27-chains | proteasomal degradation | e.g., NS4B | [61] |
| K33-chains | proteasomal degradation | e.g., ERCC1 (nucleotide excision repair protein) | [62] |
| M1/K63-linked | transcription factor activation (K63-poly-ub as a prerequisite for the formation of M1-poly-Ub) | e.g., canonical IκB kinase IKKα and IKKβ | [63] |
| K11/K48-linked | proteasomal degradation, cell-cycle and quality control | e.g., anaphase-promoting complex APC/C | [49] |
| K48/K63-chains | proteasomal degradation (K63- poly-ub serving as a“seed”for K48-poly-ub) | e.g., proapoptotic regulator TXNIP | [45] |
| Name | Website | Aims | Updated Time |
|---|---|---|---|
| UbiProt | http://ubiprot.org.ru | experimentally obtained | 2007 |
| PEIMAN | http://bs.ipm.ir/softwares/PEIMAN | predict and compute the ubiquitylation and other PTMs | 2015 |
| plantsUPS | http://bioinformatics.cau.edu.cn/plantsUPS | comparative analysis of UPS in higher plants | 2009 |
| COFRADIC method | http://bioinformatics.psb.ugent.be/webtools/Ub_viewer | Arabidopsis | 2016 |
| PTMCode | https://ptmcode.embl.de | integrative information of known and predicted PTMs | 2015 |
| dbPTM | http://dbptm.mbc.nctu.edu.tw | comprehensively functional and structural analyses for PTMs | 2019 |
| Plant PTM Viewer | https://www.psb.ugent.be/webtools/ptm-viewer | tools to analyze the potential role of PTMs | 2019 |
| CKSAAP_UbSite | http://systbio.cau.edu.cn/cksaap_ubsite/i | software to predict ubiquitylation sites | 2013 |
| UbiPred | http://flipper.diff.org/app/tools/info/2503 | predict ubiquitylation sites | 2010 |
| UbPred | http://www.ubpred.org | random forest-based predictor of potential ubiquitination sites | 2010 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, D.; Damaris, R.N.; Li, M.; Khan, I.; Yang, P. Advances on Plant Ubiquitylome—From Mechanism to Application. Int. J. Mol. Sci. 2020, 21, 7909. https://doi.org/10.3390/ijms21217909
He D, Damaris RN, Li M, Khan I, Yang P. Advances on Plant Ubiquitylome—From Mechanism to Application. International Journal of Molecular Sciences. 2020; 21(21):7909. https://doi.org/10.3390/ijms21217909
Chicago/Turabian StyleHe, Dongli, Rebecca Njeri Damaris, Ming Li, Imran Khan, and Pingfang Yang. 2020. "Advances on Plant Ubiquitylome—From Mechanism to Application" International Journal of Molecular Sciences 21, no. 21: 7909. https://doi.org/10.3390/ijms21217909





