Crystal Structure of the Kinase Domain of MerTK in Complex with AZD7762 Provides Clues for Structure-Based Drug Development
Abstract
:1. Introduction
2. Results and Discussion
2.1. Screening for MerTK Binding Molecules with Inhouse Kinase-Inhibitor Library Using Thermal-Shift Assay
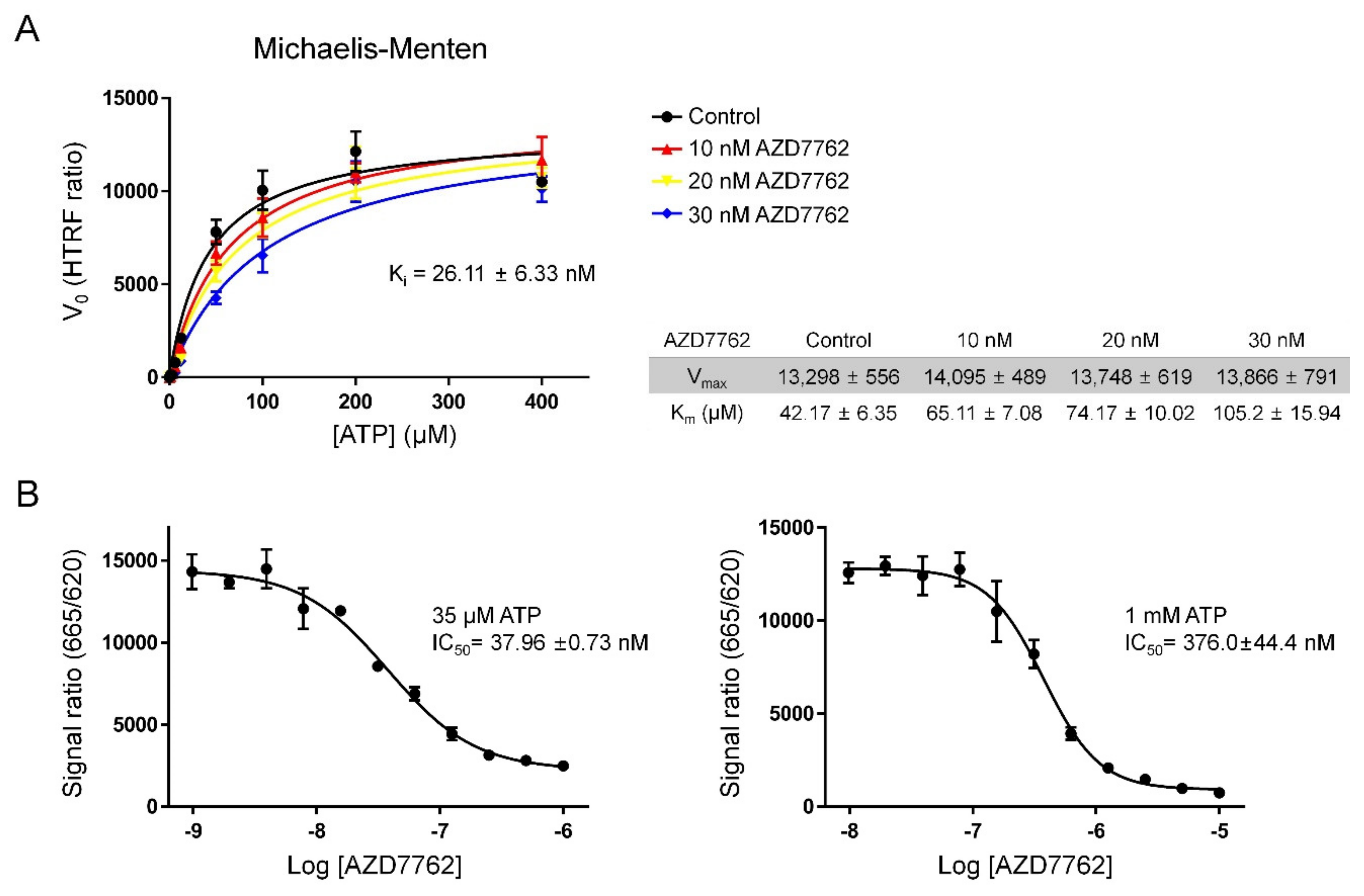
2.2. Verifying AZD7762 Inhibitory Activity by MerTK via HTRF Assay
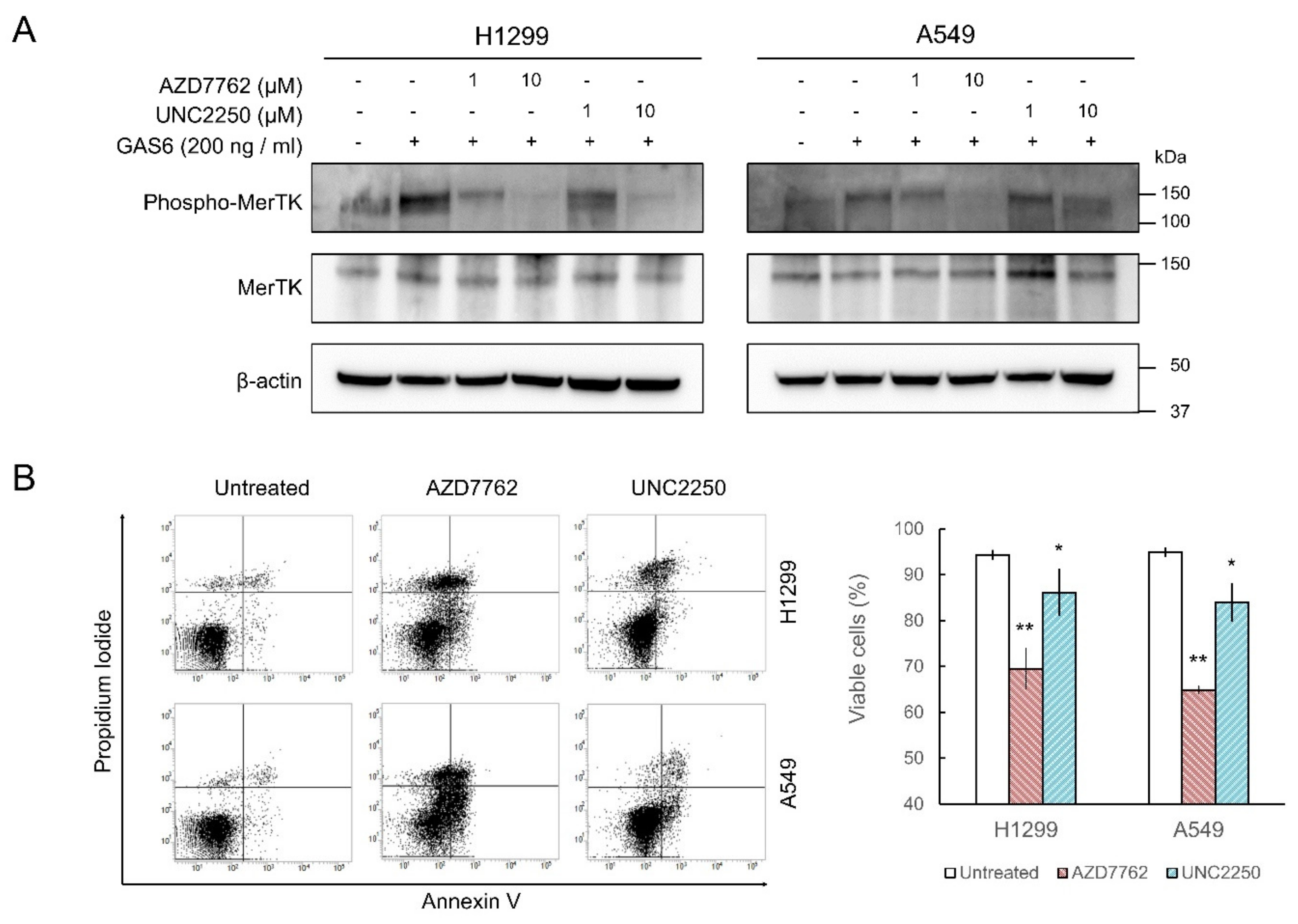
2.3. MerTK Inhibition by AZD7762 in Cells
2.4. Crystal Structure of MerTK:AZD7762 Complex
2.4.1. Structure Determination and Model Quality
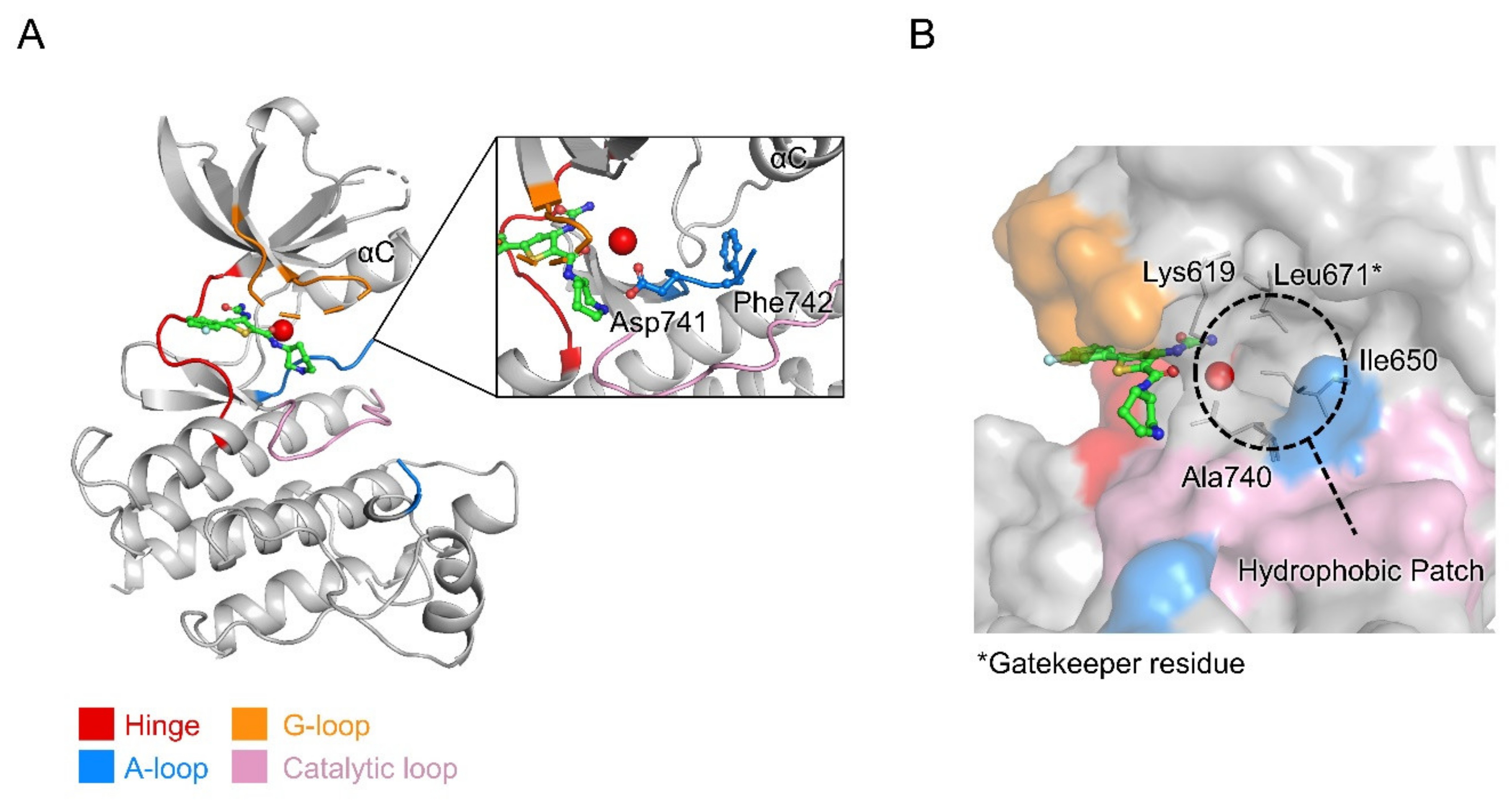
2.4.2. Overall Structure of MerTK Kinase Domain:AZD7762 Complex
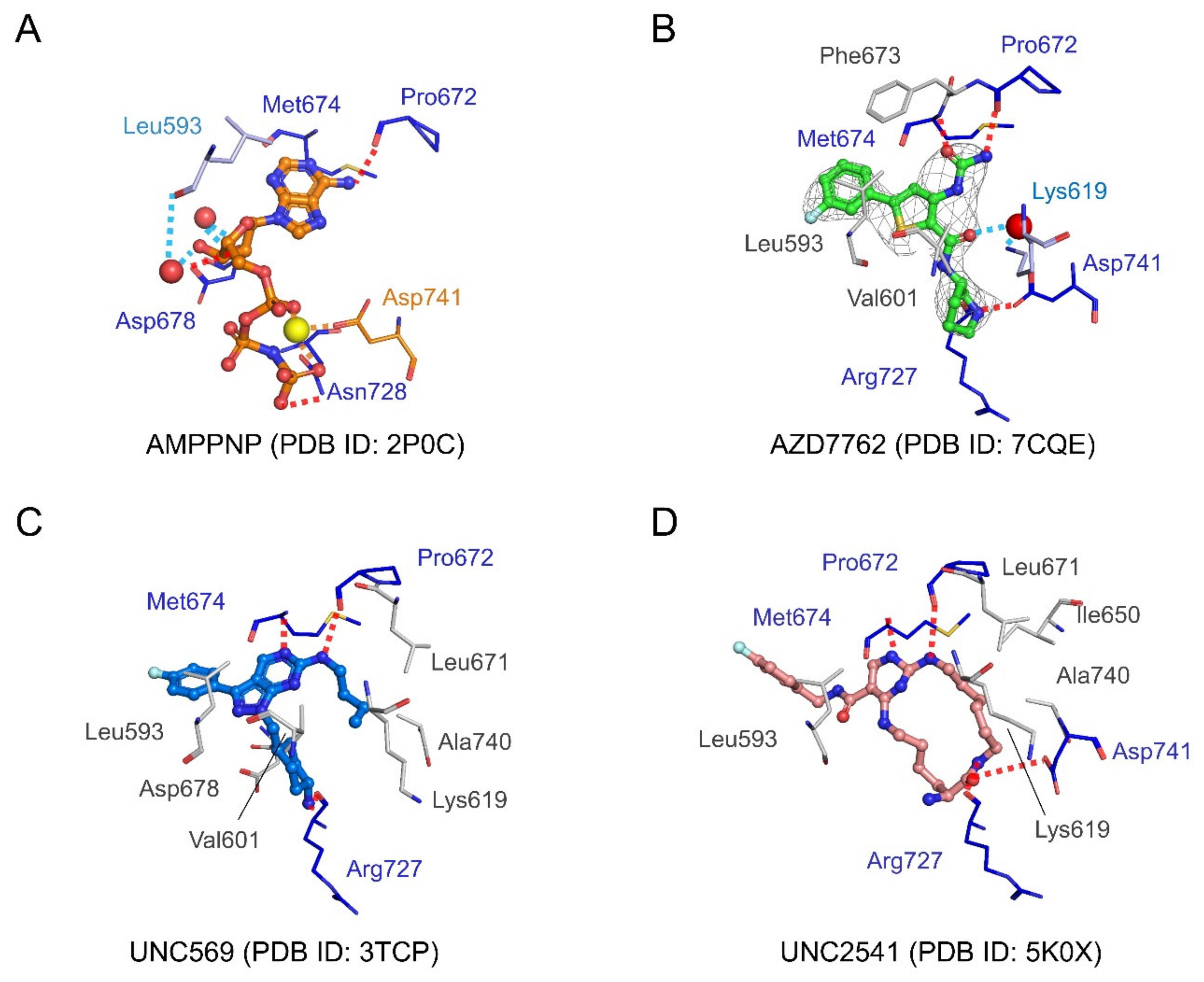
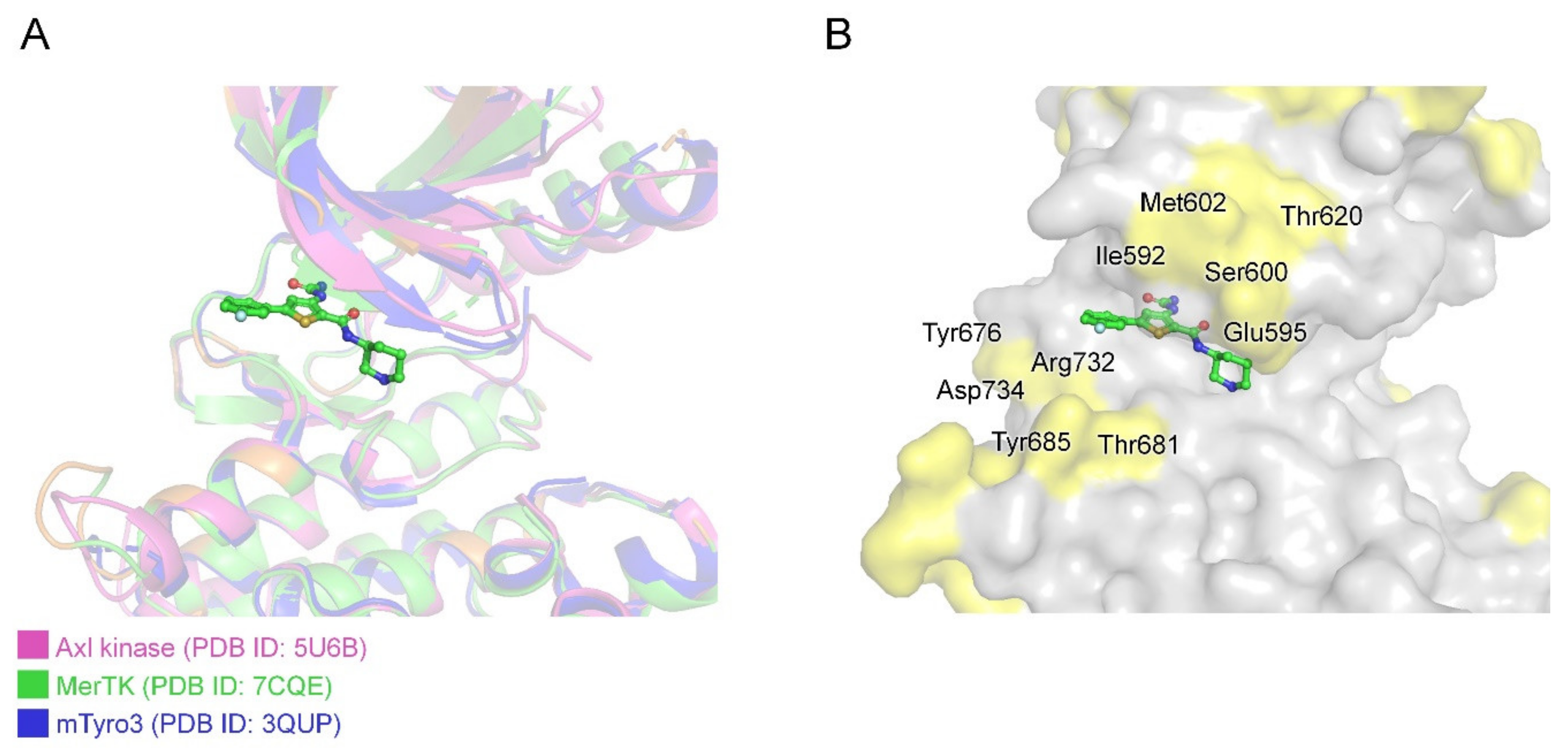
2.4.3. Structural Analysis of MerTK:AZD7762 Complex and Comparison with AMPPNP-and Other Inhibitor-Bound Structures
2.4.4. Achieving Selectivity: Comparison of MerTK:AZD7762 Structure with Other TAM Family Kinases
2.5. Concluding Remarks
3. Materials and Methods
3.1. Protein Expression and Purification
3.2. Thermal-Shift Assay
3.3. HTRF Assay
3.4. Crystallization and Structure Determination
3.5. Cell Culture
3.6. Chemical Treatment and Western Blotting
3.7. Flow-Cytometry Analysis for Apoptosis
3.8. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Grb2 | Growth factor receptor-bound protein 2 |
| LimD4 | Lim domain-containing protein 4 |
| Vav1 | Guanine nucleotide-exchange factor (GEF) Vav1 |
| PI3K | Phosphoinositide 3-kinase |
| ATP | Adenosine triphosphate |
| AMPPNP | Adenylyl-imidodiphosphate |
References
- Huelse, J.M.; Fridlyand, D.M.; Earp, S.; DeRyckere, D.; Graham, D.K. MERTK in cancer therapy: Targeting the receptor tyrosine kinase in tumor cells and the immune system. Pharmacol. Ther. 2020, 213, 107577. [Google Scholar] [CrossRef]
- Chen, J.; Carey, K.; Godowski, P.J. Identication of Gas6 as a ligand for Mer, a neural cell adhesion molecule related receptor tyrosine kinase implicated in cellular transformation. Oncogene 1997, 14, 2033–2039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurance, S.; Lemarie, C.A.; Blostein, M.D. Growth arrest-specific gene 6 (gas6) and vascular hemostasis. Adv. Nutr. 2012, 3, 196–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caberoy, N.B.; Alvarado, G.; Bigcas, J.L.; Li, W. Galectin-3 is a new MerTK-specific eat-me signal. J. Cell. Physiol. 2012, 227, 401–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peeters, M.J.W.; Rahbech, A.; Thor Straten, P. TAM-ing T cells in the tumor microenvironment: Implications for TAM receptor targeting. Cancer Immunol. Immunother. 2020, 69, 237–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, L.; Templeton, D.; Kung, H.J. Identification of the Major Autophosphorylation Sites of Nyk/Mer, an NCAM-related Receptor Tyrosine Kinase. J. Biol. Chem. 1996, 271, 18355–18362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, L.; Kung, H.J. Mitogenic signals and transforming potential of Nyk, a newly identified neural cell adhesion molecule-related receptor tyrosine kinase. Mol. Cell. Biol. 1995, 15, 6582–6592. [Google Scholar] [CrossRef] [Green Version]
- Colland, F.; Jacq, X.; Trouplin, V.; Mougin, C.; Groizeleau, C.; Hamburger, A.; Meil, A.; Wojcik, J.; Legrain, P.; Gauthier, J.M. Functional Proteomics Mapping of a Human Signaling Pathway. Genome Res. 2004, 14, 1324–1332. [Google Scholar] [CrossRef] [Green Version]
- Georgescu, M.M.; Kirsch, K.H.; Shishido, T.; Zong, C.; Hanafusa, H. Biological Effects of c-Mer Receptor Tyrosine Kinase in Hematopoietic Cells Depend on the Grb2 Binding Site in the Receptor and Activation of NF-κB. Mol. Cell. Biol. 1999, 19, 1171–1181. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, N.P.; Earp, H.S. An SH2 domain-dependent, phosphotyrosine-independent interaction between Vav1 and the Mer receptor tyrosine kinase: A mechanism for localizing guanine nucleotide-exchange factor action. J. Biol. Chem. 2003, 278, 42596–42603. [Google Scholar] [CrossRef] [Green Version]
- Linger, R.M.A.; Keating, A.K.; Earp, H.S.; Graham, D.K. TAM Receptor Tyrosine Kinases: Biologic Functions, Signaling, and Potential Therapeutic Targeting in Human Cancer. Adv. Cancer Res. 2008, 100, 35–83. [Google Scholar] [CrossRef] [Green Version]
- Graham, D.K.; Dawson, T.L.; Mullaney, D.L.; Snodgrass, H.R.; Earp, H.S. Cloning and mRNA Expression Analysis of a Novel Human Protooncogene, c-mer. Cell Growth Differ. 1994, 5, 647–657. [Google Scholar] [PubMed]
- Lee-Sherick, A.B.; Jacobsen, K.M.; Henry, C.J.; Huey, M.G.; Parker, R.E.; Page, L.S.; Hill, A.A.; Wang, X.; Frye, S.V.; Earp, H.S.; et al. MERTK inhibition alters the PD-1 axis and promotes anti-leukemia immunity. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Linger, R.M.; Cohen, R.A.; Cummings, C.T.; Sather, S.; Migdall-Wilson, J.; Middleton, D.H.; Lu, X.; Baron, A.E.; Franklin, W.A.; Merrick, D.T.; et al. Mer or Axl receptor tyrosine kinase inhibition promotes apoptosis, blocks growth and enhances chemosensitivity of human non-small cell lung cancer. Oncogene 2013, 32, 3420–3431. [Google Scholar] [CrossRef] [Green Version]
- Sather, S.; Kenyon, K.D.; Lefkowitz, J.B.; Liang, X.; Varnum, B.C.; Henson, P.M.; Graham, D.K. A soluble form of the Mer receptor tyrosine kinase inhibits macrophage clearance of apoptotic cells and platelet aggregation. Blood 2007, 109, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.S.; McMahon, E.J.; Pop, S.M.; Reap, E.A.; Caricchio, R.; Cohen, P.L.; Earp, H.S.; Matsushima, G.K. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature 2001, 411, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jennings, J.H.; Sonstein, J.; Floros, J.; Todt, J.C.; Polak, T.; Curtis, J.L. Resident murine alveolar and peritoneal macrophages differ in adhesion of apoptotic thymocytes. Am. J. Respir. Cell Mol. Biol. 2004, 30, 687–693. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Yang, A.; Wang, Y.; Chen, F.; Zhao, Z.; Davra, V.; Suzuki-Inoue, K.; Ozaki, Y.; Birge, R.B.; Lu, Q.; et al. Tyro3, Axl, and Mertk receptors differentially participate in platelet activation and thrombus formation. Cell Commun. Signal. 2018, 16, 98. [Google Scholar] [CrossRef] [Green Version]
- Zabludoff, S.D.; Deng, C.; Grondine, M.R.; Sheehy, A.M.; Ashwell, S.; Caleb, B.L.; Green, S.; Haye, H.R.; Horn, C.L.; Janetka, J.W.; et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol. Cancer Ther. 2008, 7, 2955–2966. [Google Scholar] [CrossRef] [Green Version]
- Oza, V.; Ashwell, S.; Almeida, L.; Brassil, P.; Breed, J.; Deng, C.; Gero, T.; Grondine, M.; Horn, C.; Ioannidis, S.; et al. Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(piperidin-3-yl)-3-ureidothiophene-2-carboxamide (AZD7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas. J. Med. Chem. 2012, 55, 5130–5142. [Google Scholar] [CrossRef]
- Sausville, E.; Lorusso, P.; Carducci, M.; Carter, J.; Quinn, M.F.; Malburg, L.; Azad, N.; Cosgrove, D.; Knight, R.; Barker, P.; et al. Phase I dose-escalation study of AZD7762, a checkpoint kinase inhibitor, in combination with gemcitabine in US patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2014, 73, 539–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.-S.; Lee, C.; Kim, H.-K.; Kim, D.; Son, J.B.; Ko, E.; Cho, J.-H.; Kim, N.-D.; Nan, H.-Y.; Kim, C.-Y.; et al. Suppression of the metastatic spread of breast cancer by DN10764 (AZD7762)-mediated inhibition of AXL signaling. Oncotarget 2016, 7, 83308–83318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, Z.A.; Shokat, K.M. Features of selective kinase inhibitors. Chem. Biol. 2005, 12, 621–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimani, S.G.; Kumar, S.; Bansal, N.; Singh, K.; Kholodovych, V.; Comollo, T.; Peng, Y.; Kotenko, S.V.; Sarafianos, S.G.; Bertino, J.R.; et al. Small molecule inhibitors block Gas6-inducible TAM activation and tumorigenicity. Sci. Rep. 2017, 7, 43908. [Google Scholar] [CrossRef]
- Cummings, C.T.; Zhang, W.; Davies, K.D.; Kirkpatrick, G.D.; Zhang, D.; DeRyckere, D.; Wang, X.; Frye, S.V.; Earp, H.S.; Graham, D.K. Small Molecule Inhibition of MERTK Is Efficacious in Non-Small Cell Lung Cancer Models Independent of Driver Oncogene Status. Mol. Cancer Ther. 2015, 14, 2014–2022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Zhang, D.; Stashko, M.A.; DeRyckere, D.; Hunter, D.; Kireev, D.; Miley, M.J.; Cummings, C.; Lee, M.; Norris-Drouin, J.; et al. Pseudo-Cyclization through Intramolecular Hydrogen Bond Enables Discovery of Pyridine Substituted Pyrimidines as New Mer Kinase Inhibitors. J. Med. Chem. 2013, 56, 9683–9692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roskoski, R., Jr. Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol. Res. 2016, 103, 26–48. [Google Scholar] [CrossRef]
- Liu, J.; Yang, C.; Simpson, C.; Deryckere, D.; Van Deusen, A.; Miley, M.J.; Kireev, D.; Norris-Drouin, J.; Sather, S.; Hunter, D.; et al. Discovery of Novel Small Molecule Mer Kinase Inhibitors for the Treatment of Pediatric Acute Lymphoblastic Leukemia. ACS Med. Chem. Lett. 2012, 3, 129–134. [Google Scholar] [CrossRef]
- McIver, A.L.; Zhang, W.; Liu, Q.; Jiang, X.; Stashko, M.A.; Nichols, J.; Miley, M.J.; Norris-Drouin, J.; Machius, M.; DeRyckere, D.; et al. Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors. ChemMedChem 2017, 12, 207–213. [Google Scholar] [CrossRef] [Green Version]
- Christoph, S.; Deryckere, D.; Schlegel, J.; Frazer, J.K.; Batchelor, L.A.; Trakhimets, A.Y.; Sather, S.; Hunter, D.M.; Cummings, C.T.; Liu, J.; et al. UNC569, a novel small-molecule mer inhibitor with efficacy against acute lymphoblastic leukemia in vitro and in vivo. Mol. Cancer Ther. 2013, 12, 2367–2377. [Google Scholar] [CrossRef] [Green Version]
- Myers, K.V.; Amend, S.R.; Pienta, K.J. Targeting Tyro3, Axl and MerTK (TAM receptors): Implications for macrophages in the tumor microenvironment. Mol. Cancer 2019, 18, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Zhang, W.; Stashko, M.A.; Deryckere, D.; Cummings, C.T.; Hunter, D.; Yang, C.; Jayakody, C.N.; Cheng, N.; Simpson, C.; et al. UNC1062, a new and potent Mer inhibitor. Eur. J. Med. Chem. 2013, 65, 83–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee-Sherick, A.B.; Zhang, W.; Menachof, K.K.; Hill, A.A.; Rinella, S.; Kirkpatrick, G.; Page, L.S.; Stashko, M.A.; Jordan, C.T.; Wei, Q.; et al. Efficacy of a Mer and Flt3 tyrosine kinase small molecule inhibitor, UNC1666, in acute myeloid leukemia. Oncotarget 2015, 6, 6722–6736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; DeRyckere, D.; Hunter, D.; Liu, J.; Stashko, M.A.; Minson, K.A.; Cummings, C.T.; Lee, M.; Glaros, T.G.; Newton, D.L.; et al. UNC2025, a potent and orally bioavailable MER/FLT3 dual inhibitor. J. Med. Chem. 2014, 57, 7031–7041. [Google Scholar] [CrossRef] [Green Version]
- Graham, D.K.; DeRyckere, D.; Davies, K.D.; Earp, H.S. The TAM family: Phosphatidylserine-sensing receptor tyrosine kinases gone awry in cancer. Nat. Rev. Cancer 2014, 14, 769. [Google Scholar] [CrossRef]
- Silva, E.A.C.; Chan, P.Y.; Joannas, L.; Errasti, A.E.; Gagliani, N.; Bosurgi, L.; Jabbour, M.; Perry, A.; Smith-Chakmakova, F.; Mucida, D.; et al. T Cell-Derived Protein S Engages TAM Receptor Signaling in Dendritic Cells to Control the Magnitude of the Immune Response. Immunity 2013, 39, 160–170. [Google Scholar] [CrossRef] [Green Version]
- Cohen, P.L.; Caricchio, R.; Abraham, V.; Camenisch, T.D.; Jennette, J.C.; Roubey, R.A.S.; Earp, H.S.; Matsushima, G.; Reap, E.A. Delayed apoptotic cell clearance and lupus-like autoimmunity in mice lacking the c-mer membrane tyrosine kinase. J. Exp. Med. 2002, 196, 135–140. [Google Scholar] [CrossRef]
- Lu, Q.; Lemke, G. Homeostatic Regulation of the Immune System by Receptor Tyrosine Kinases of the Tyro 3 Family. Science 2001, 293, 306. [Google Scholar] [CrossRef]
- Rothlin, C.V.; Leighton, J.A.; Ghosh, S. Tyro3, Axl, and Mertk receptor signaling in inflammatory bowel disease and colitis-associated cancer. Inflamm. Bowel Dis. 2014, 20, 1472–1480. [Google Scholar] [CrossRef] [Green Version]
- Otwinowski, Z.; Minor, W. [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Finerty, P.; Walker, J.R.; Butler-Cole, C.; Vedadi, M.; Schapira, M.; Parker, S.A.; Turk, B.E.; Thompson, D.A.; Dhe-Paganon, S. Structural insights into the inhibited states of the Mer receptor tyrosine kinase. J. Struct. Biol. 2009, 165, 88–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Moriarty, N.W.; Grosse-Kunstleve, R.W.; Adams, P.D. Electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 1074–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bong, S.M.; Bae, S.H.; Song, B.; Gwak, H.; Yang, S.W.; Kim, S.; Nam, S.; Rajalingam, K.; Oh, S.J.; Kim, T.W.; et al. Regulation of mRNA export through API5 and nuclear FGF2 interaction. Nucleic Acids Res. 2020, 48, 6340–6352. [Google Scholar] [CrossRef]
- Kim, H.Y.; Kim, D.K.; Bae, S.H.; Gwak, H.; Jeon, J.H.; Kim, J.K.; Lee, B.I.; You, H.J.; Shin, D.H.; Kim, Y.H.; et al. Farnesyl diphosphate synthase is important for the maintenance of glioblastoma stemness. Exp. Mol. Med. 2018, 50, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.H.; Kim, H.K.; Kim, H.Y.; Gawk, H. FAK-Copy-Gain Is a Predictive Marker for Sensitivity to FAK Inhibition in Breast Cancer. Cancers 2019, 11. [Google Scholar] [CrossRef] [Green Version]
- Poojan, S.; Bae, S.-H.; Min, J.-W.; Lee, E.Y.; Song, Y.; Kim, H.Y.; Sim, H.W.; Kang, E.-K.; Kim, Y.-H.; Lee, H.-O.; et al. Cancer cells undergoing epigenetic transition show short-term resistance and are transformed into cells with medium-term resistance by drug treatment. Exp. Mol. Med. 2020, 52, 1102–1115. [Google Scholar] [CrossRef]


| Dataset | MerTK Kinase Domain: AZD7762 |
|---|---|
| Diffraction-data statistics | |
| X-ray source | PLS-11C |
| Wavelength (Å) | 0.979 |
| Space group | C2 |
| Cell parameters | |
| a, b, c (Å) | 116.12, 69.20, 92.61 |
| α, β, γ (°) | 90.00, 118.42, 90.00 |
| Resolution range (Å) a | 50.0−2.70 (2.75−2.70) |
| Rsym (%) b | 10.3 (40.0) |
| Rp.i.m. (%) c | 5.1 (21.0) |
| CC1/2 | 0.986 (0.875) |
| Mean I/σI | 13.8 (3.33) |
| Redundancy | 4.7 (4.0) |
| Completeness (%) | 95.9 (84.2) |
| No. of unique reflections | 17,283 (738) |
| Refinement statistics | |
| Resolution range (Å) | 43.53−2.70 |
| Rwork/Rfree (%) d | 19.7/25.9 |
| No. of nonhydrogen atoms/average B-factor (Å2) | |
| Protein | 3948/41.47 |
| Solvent | 20/22.65 |
| Ligand (AZD7762) | 56/37.25 |
| R.m.s. deviation | |
| Bond lengths (Å) | 0.015 |
| Bond angles (°) | 1.97 |
| Ramachandran plot (%) | |
| Favored/outliers | 97.46/0.42 |
| Clash score | 3.0 |
| PDB entry | 7CQE |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, T.H.; Bae, S.-H.; Bong, S.M.; Ryu, S.E.; Jang, H.; Lee, B.I. Crystal Structure of the Kinase Domain of MerTK in Complex with AZD7762 Provides Clues for Structure-Based Drug Development. Int. J. Mol. Sci. 2020, 21, 7878. https://doi.org/10.3390/ijms21217878
Park TH, Bae S-H, Bong SM, Ryu SE, Jang H, Lee BI. Crystal Structure of the Kinase Domain of MerTK in Complex with AZD7762 Provides Clues for Structure-Based Drug Development. International Journal of Molecular Sciences. 2020; 21(21):7878. https://doi.org/10.3390/ijms21217878
Chicago/Turabian StylePark, Tae Hyun, Seung-Hyun Bae, Seoung Min Bong, Seong Eon Ryu, Hyonchol Jang, and Byung Il Lee. 2020. "Crystal Structure of the Kinase Domain of MerTK in Complex with AZD7762 Provides Clues for Structure-Based Drug Development" International Journal of Molecular Sciences 21, no. 21: 7878. https://doi.org/10.3390/ijms21217878
APA StylePark, T. H., Bae, S.-H., Bong, S. M., Ryu, S. E., Jang, H., & Lee, B. I. (2020). Crystal Structure of the Kinase Domain of MerTK in Complex with AZD7762 Provides Clues for Structure-Based Drug Development. International Journal of Molecular Sciences, 21(21), 7878. https://doi.org/10.3390/ijms21217878






