Abstract
Reprogramming of the genetic code system is limited by the difficulty in creating new tRNA structures. Here, I developed translationally active tRNA variants tagged with a small hairpin RNA aptamer, using Escherichia coli reporter assay systems. As the tRNA chassis for engineering, I employed amber suppressor variants of allo-tRNAs having the 9/3 composition of the 12-base pair amino-acid acceptor branch as well as a long variable arm (V-arm). Although their V-arm is a strong binding site for seryl-tRNA synthetase (SerRS), insertion of a bulge nucleotide in the V-arm stem region prevented allo-tRNA molecules from being charged by SerRS with serine. The SerRS-rejecting allo-tRNA chassis were engineered to have another amino-acid identity of either alanine, tyrosine, or histidine. The tip of the V-arms was replaced with diverse hairpin RNA aptamers, which were recognized by their cognate proteins expressed in E. coli. A high-affinity interaction led to the sequestration of allo-tRNA molecules, while a moderate-affinity aptamer moiety recruited histidyl-tRNA synthetase variants fused with the cognate protein domain. The new design principle for tRNA-aptamer fusions will enhance radical and dynamic manipulation of the genetic code.
1. Introduction
RNA aptamers are small RNA molecules that specifically bind to their target molecules. Small hairpin RNA aptamers are naturally found in certain mRNAs [1,2,3] and other RNAs [4]. Diverse hairpin aptamers have been used for tagging many kinds of RNA molecules for recruiting their cognate target proteins or ligands on the fusion RNA molecules [4,5,6,7]. On the other hand, transfer RNAs (tRNAs) are small RNA molecules that transfer proteinogenic amino acids onto the growing polypeptide chain in the ribosome. tRNAs fold into the L-shape tertiary structure, while some tRNAs have an extra arm in varying size (variable arm or V-arm) (see Figure 1). Although several types of tRNA variants fused with hairpin RNA aptamers have been developed [7,8,9,10,11,12,13], these fusion RNAs usually lost the translational activity of tRNA. Recently, “V-Spinach tRNAs” were developed by inserting a Baby Spinach aptamer [14] into the V-loop of a few E. coli tRNA species [8], although the V-Spinach tRNAs showed only marginal tRNA activities [8,15]. More recently, the V-arm hairpin of a yeast serine tRNA species was replaced by the MS2 phage hairpin RNA (19 nucleotides in length) to recruit an enzyme fused with the MS2 coat protein on the premature form of these fusion RNA molecules in yeast cells [11]. These works encouraged us to design a new generation of tRNA-aptamer fusions in which a small hairpin aptamer constitutes the tip of the V-arm.
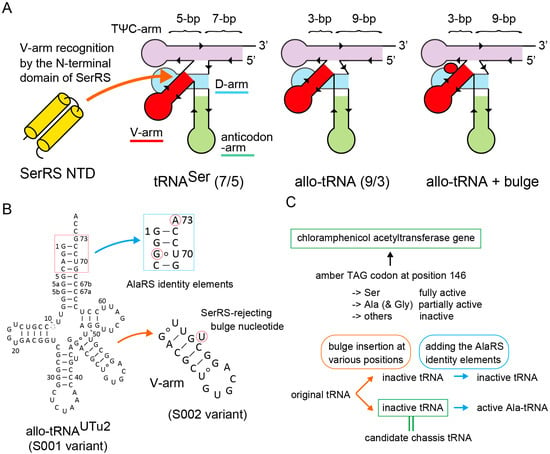
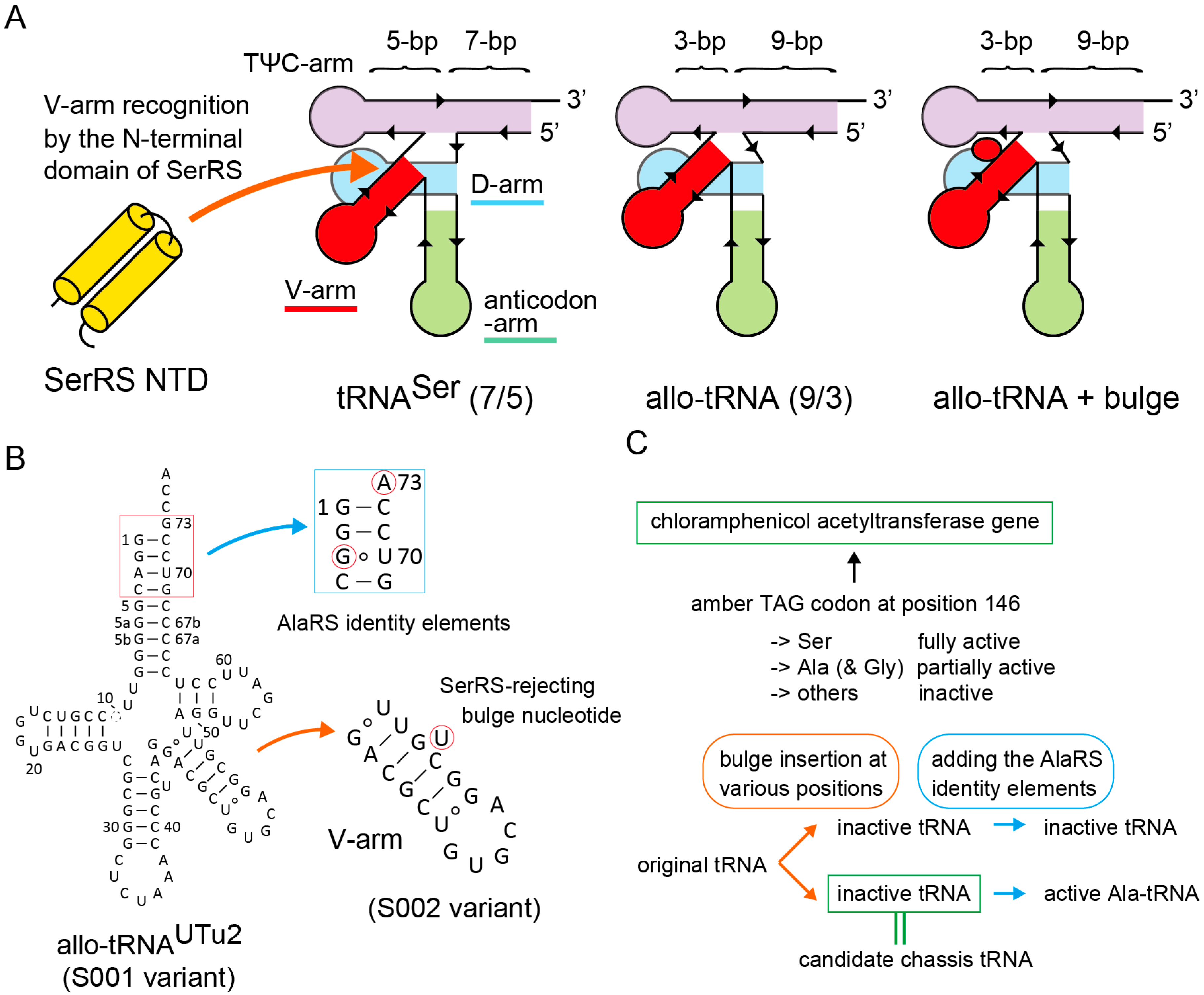
Figure 1.
Strategy for tRNA chassis development. (A) Design of a new tRNA chassis derived from a few allo-tRNA species having the 9/3 composition. Compared to the E. coli tRNASer having the canonical 7/5 composition, (9/3) allo-tRNA molecules may have a smaller space for the binding of the N-terminal domain (NTD) of SerRS. Thus, it was assumed that the interaction between SerRS and allo-tRNA molecules could be sterically hindered by the insertion of a bulge (flipped-out) nucleotide into the stem region of the V-arm. (B) A preliminary result suggested that the allo-tRNAUTu2 variant containing the bulge U was a poor substrate for SerRS, while its variant having the AlaRS identity elements was active for Ala-tRNA regardless of the bulge U insertion. (C) Experimental design for screening candidate chassis tRNAs by using a reporter gene in E. coli.
tRNA plays a central role in reprogramming the genetic code system and establishing new genetic codes [16,17,18]. In nature, the genetic code may have been expanded to acquire selenocysteine (Sec) and pyrrolysine (Pyl) as the 21st and 22nd amino acid, respectively. Interestingly, canonical tRNASec has the largest tRNA structure, while tRNAPyl has the most compact tRNA structure [16], which may help them escape from non-cognate aminoacyl-tRNA synthetases (aaRSs). The genetic code was expanded in diverse organisms using synthetic biologists by developing various sets of orthogonal tRNA and aaRS pairs that do not cross-react with endogenous tRNAs and aaRSs and other orthogonal tRNA-aaRS pairs [17,18,19,20,21,22]. It has been a big challenge to artificially create orthogonal tRNA species with both high tRNA activity and high orthogonality in part due to saturation of recognition elements of tRNA [23]. One promising approach is “tRNA Extension (tREX)” [17] by which the tRNA activity and the orthogonality of a few orthogonal systems were significantly enhanced in a rapid and systematic manner towards their simultaneous use in E. coli [17]. Creation of a new tRNA identity may be achieved by introducing new tertiary structural elements [24,25] rather than by introducing small local modifications to tRNA. However, tRNA variants having an abnormal tertiary structure may be poorly compatible with the translation apparatus [8,16,26,27]. Thus, introduction of a small hairpin aptamer as the V-arm of orthogonal tRNAs may be a good solution for developing tRNA variants which are translationally active and have a characteristic tertiary structural element.
Recently, a new group of tRNAs, (9/3) allo-tRNAs, was identified in the raw short-read data of soil or marine metagenomic datasets [26,28]. These tRNAs have a non-canonical composition of the amino-acid acceptor branch (Figure 1A,B). While canonical tRNAs have a 7-bp accepter stem and a 5-bp TψC stem, the (9/3) allo-tRNAs have a 9-bp accepter stem and a 3-bp TψC stem (Figure 1A,B). In addition, the (9/3) allo-tRNAs have a long D-stem, like selenocysteine (Sec) tRNA species, and a long V-arm, like serine (Ser) and Sec tRNA species. Thus, the (9/3) allo-tRNAs intrinsically have identity elements for the Ser-tRNA synthetase (SerRS) and the Sec-tRNA synthesis apparatus [26,28,29]. Amber suppressor variants of two of the (9/3) allo-tRNA species were used for the site-specific Ser or Sec incorporation into proteins in response to the TAG amber stop codon in E. coli [29]. Two of the natural allo-tRNA species also have the essential identity elements for alanyl-tRNA synthetase (AlaRS) [26,29], and one of the two preferentially worked as tRNAAla rather than as tRNASer in E. coli cells [26]. Despite the non-canonical secondary structure, many allo-tRNA species were highly compatible with the E. coli translation system and worked as exceedingly active nonsense or missense suppressor tRNAs in E. coli [26,29]. These findings prompted us to employ these (9/3) allo-tRNA scaffolds as tRNA chassis for making our tRNA-aptamer fusions. However, the strong intrinsic affinity of (9/3) allo-tRNA species for SerRS has limited their utility.
In the present study, a few kinds of SerRS-rejecting allo-tRNA chassis were developed. Several kinds of small hairpin aptamers inserted into these chassis were confirmed to be bound by their cognate proteins or protein domains expressed in E. coli. The new tRNA chassis, or “orthogonal allo-tRNAs”, will help to greatly expand the design flexibility of new tRNA-aaRS pairs.
2. Results
2.1. Strategy for tRNA Chassis Development
In an attempt to develop a SerRS-rejecting (9/3) allo-tRNASer variant, we realized that certain allo-tRNA species (allo-tRNAUTu2 series [29], see Figure 1B) having a G:U wobble base pair at the bottom of the V-stem worked as less active tRNASer than allo-tRNA variants carrying Watson-Crick (WC) changes (data not shown). It is known that a G:U pair in an RNA double helix introduces a helical twist [30] and might cause steric hindrance against target proteins [31,32]. Thus, it was suggested that the original allo-tRNAUTu2 variant (or S001 variant) may have a narrow (but enough) space between the TψC arm and the V-arm for the insertion of the N-terminal domain (NTD) of SerRS (Figure 1A) [33]. To verify this idea, we inserted a bulged uracil (U) nucleotide into the V-stem of the S001 variant to make S002 (Figure 1A,B). In a preliminary experiment, the S002 variant hardly incorporated serine into proteins in E. coli, meaning that S002 is either SerRS-rejecting or simply unstable (Figure 1). On the other hand, the Ala-accepting variant of S002, or A002, which has the major identity elements for AlaRS (G3:U70 with A73) [26,29,34], incorporated alanine into proteins (data not shown here) (Figure 1C). Thus, the S002/A002 scaffold may retain the tertiary structural stability but may not have enough space for accommodating the NTD of SerRS (Figure 1). Encouraged by these preliminary results, I decided to examine seventy-nine kinds of bulge-carrying variants of nine kinds of amber suppressor (9/3) allo-tRNA species (Figure 1B and Figure S1) [26] in order to search for better chassis tRNAs than the S002 variant. Experiments were facilitated by use of E. coli cells with several reporter genes carrying an in-frame TAG codon (Figure 1C).
2.2. Searching for Candidate tRNA Chassis
In this study, all tRNA sequences were cloned in a high-copy-number plasmid vector under control of the arabinose promoter [26,29]. The starting amber suppressor allo-tRNA variants (named as S001, S012, S020, S026, S032, S043, S047, S051, and S055) were developed from nine allo-tRNA species (Figure 1B and Figure S1) chosen from ten allo-tRNA species found in nature [26]. Positional effects of single bulge nucleotide or two separate or successive bulge nucleotides on the V-arms of these allo-tRNA scaffolds were investigated by individually examining seventy-nine kinds of bulge-carrying variants (see supplementary Excel file). Their “Ala-acceptable” variants carrying the major identity elements for AlaRS (G3:U70 with A73) were also developed, although other structural elements contribute to the tRNA recognition by AlaRS [34,35]. A chloramphenicol acetyltransferase (CAT) reporter gene in which the Ser146 codon had been mutated to TAG, or cat(Ser146TAG) [26], was used for examining the TAG-translating activities of these tRNA variants in a suppressor-free strain of E. coli DH10B. While the replacement of Ser146 by Ala results in a partially active enzyme, replacement of Ser146 by bulkier residues than Ser resulted in loss of function [26,36]. Consequently, the cat(Ser146TAG) reporter is useful for detecting a strong Ala-inserting activity and even a weak Ser-inserting activity of allo-tRNAs in chloramphenicol (Cm)-sensitive E. coli (Figure 1C) [26]. Two conditions for a good candidate allo-tRNA chassis were that the Ala-acceptable variant inserts Ala, while the variant lacking the G3:U70 and A73 elements hardly inserts Ser (Figure 1C).
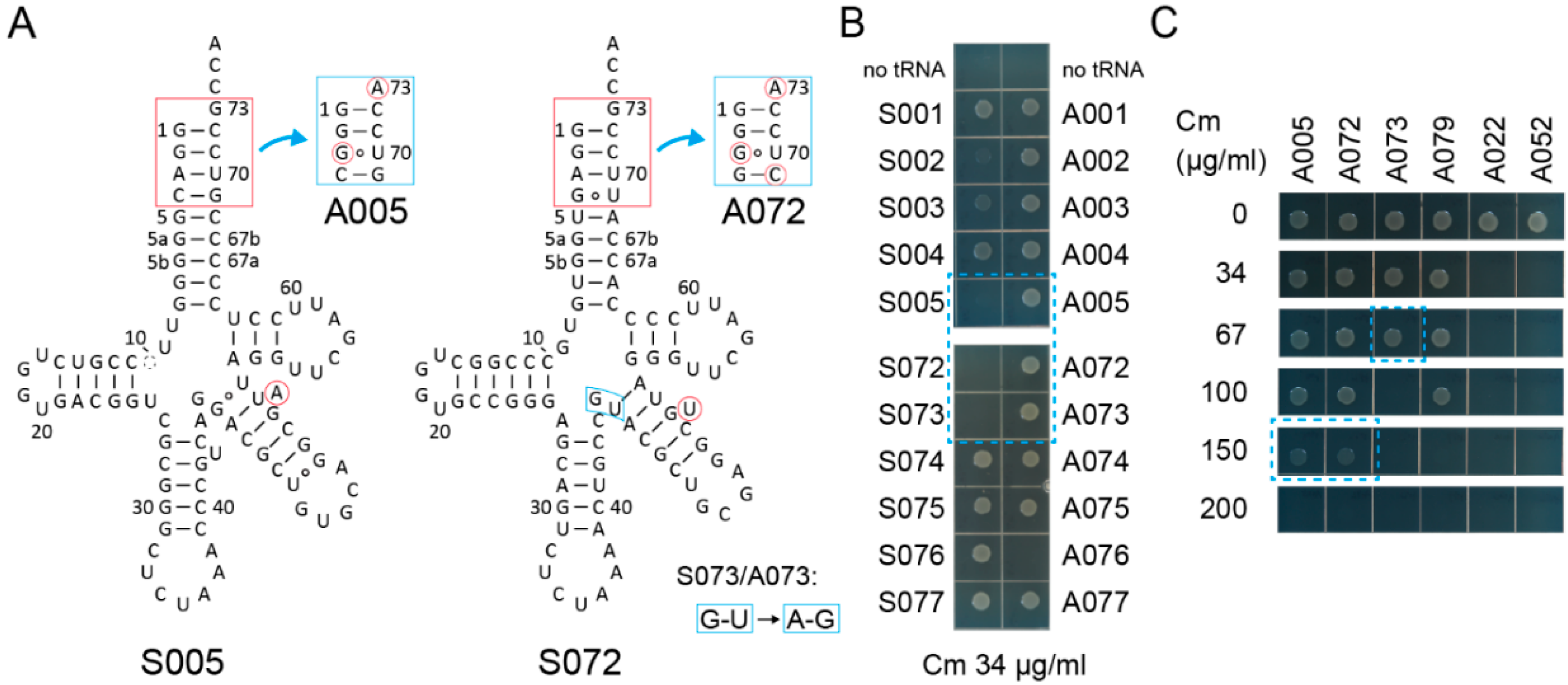
By comparing 78 variant pairs to the S002/A002 pair, S005/A005, S072/A072, and S073/A073 pairs (Figure 2A) passed the first screening using the cat(Ser146TAG) gene with Cm at a concentration of 34 µg/mL in the growth media (Figure 2B and Table S1). While S002 conferred marginal Cm resistance to cells, S005, S072, and S073 hardly conferred Cm resistance to cells (Figure 2B). A005 and A072 conferred up to 150 µg/mL Cm resistance, while A073 conferred only up to 67 µg/mL Cm resistance (Figure 2C). The S002/A002 and S005/A005 variant pairs are both derived from S001/A001 and have a bulged U or A, respectively, at different positions in the V-stem (Figure 2A). It is likely that S005 is more SerRS-rejective than S002. Because the S072/A072 and S073/A073 variant pairs differ only at the junction of the anticodon stem and the V-stem (Figure 2B), the A073 variant may be either less stable or less compatible to AlaRS than the A072 variant. Whereas the S079/A079 pair derived from S043 also passed the first screening (Table S1, Figure 2C), the other S043-derived variants retained their SerRS identity (Table S1) possibly due to the G20 residue (Figure S1). Thus, I assumed that the S005/A005 and S072/A072 chassis may be the most reliable ones.
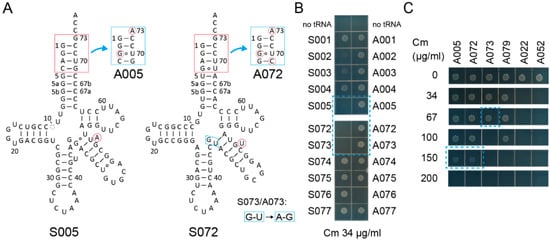
Figure 2.
Candidate tRNA chassis. (A) The cloverleaf structures of the S005 and S072 tRNA variants and their Ala-accepting variants (A005 and A072, respectively). The S073/A073 variants have two indicated modifications compared to the S072/A072 variants, respectively. (B) The S005, S072, and S073 variants conferred lower chloramphenicol (Cm) resistance (less than Cm 34 µg/mL) to E. coli DH10B cells expressing the cat(Ser146TAG) reporter gene compared to the S002 variant. (C) The A005 and A072 variants conferred up to Cm 150 µg/mL resistance to cells, while the A073 variant conferred only Cm 67 µg/mL resistance.
2.3. Sequestration of Aptamer-tRNA Fusion RNAs
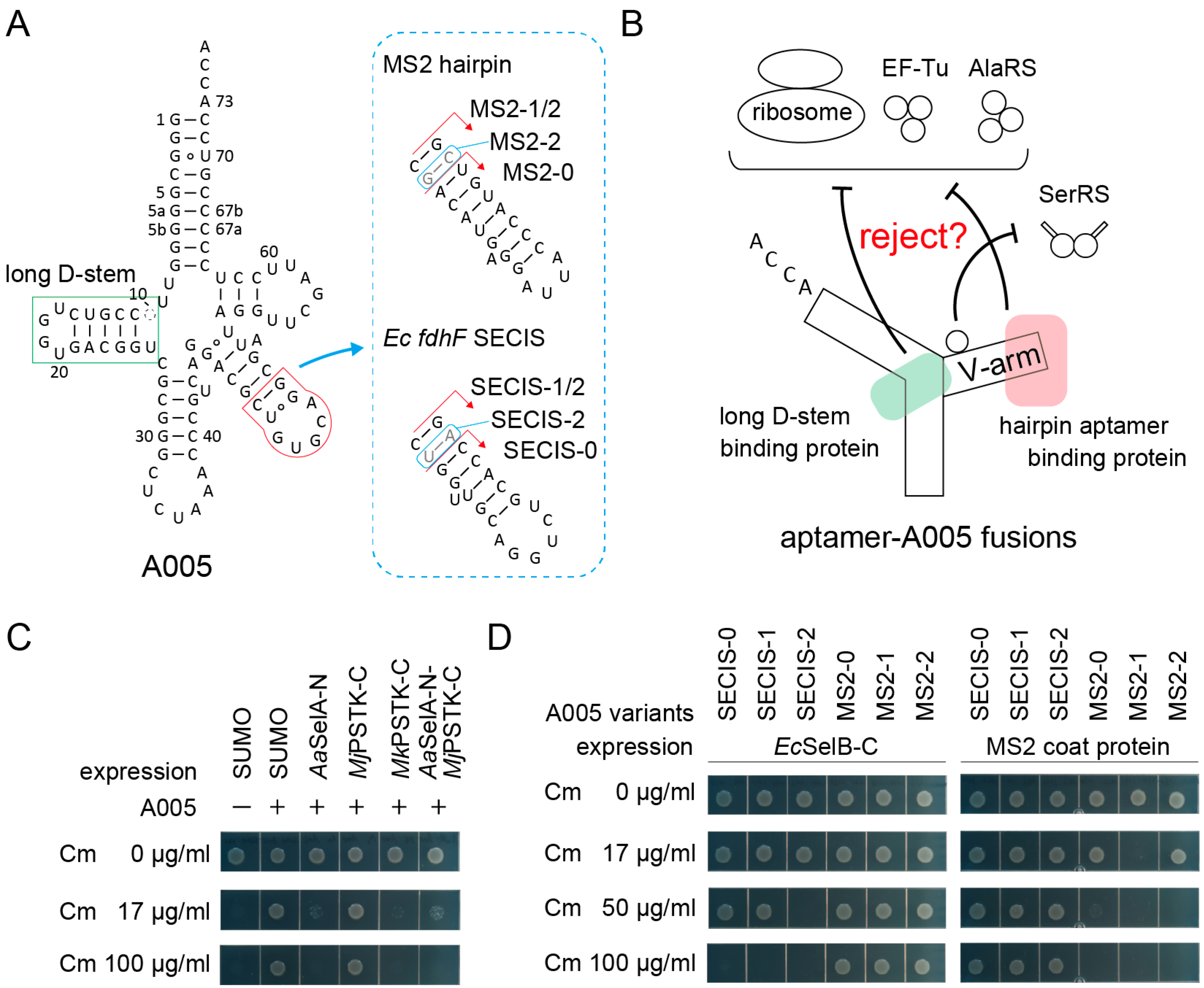
Tight protection of an aptamer moiety by its cognate protein will result in sequestration of the aptamer-tagged allo-tRNA molecules from the translation apparatus (Figure 3A,B). Previously, we observed that (9/3) allo-tRNA molecules were fully sequestered when selenocysteine synthase (SelA) molecules were overexpressed [29]. As shown in Figure 3A, (9/3) allo-tRNAs have a long D-stem which is specifically recognized by the N-terminal domain of SelA (SelA-N) [37] and the C-terminal domain of archaeal O-phosphoseryl-tRNA kinase (PSTK-C) [38]. As expected, the Ala-inserting activity of the A005 variant was almost fully removed when the Aquifex aeolicus (Aa) SelA-N or the Methanopyrus kandleri (Mk) PSTK-C were expressed at a moderate level by using the tac promoter on a pGEX vector and IPTG at a concentration of 10 µM (Figure 3C). In the presence of these proteins, A005 hardly conferred 17 µg/mL Cm resistance to cells (Figure 3C), which demonstrated that this reporter assay system is valid. Interestingly, the Methanocaldococcus jannaschii (Mj) PSTK-C failed to sequester A005 tRNA molecules (Figure 3C), possibly because Mj tRNASec has a longer D-stem than those of Mk tRNASec and allo-tRNAs [38]. Similar results were obtained by using the A072 variant (data not shown). Next, the V-arm moiety was engineered to mediate the sequestration of tRNA molecules. Two kinds of small hairpin aptamers, the MS2 hairpin RNA [9,11] and the E. coli fdhF SECIS RNA [3], were transplanted to the A005 variant with varying lengths of adaptors (Figure 3A). The in vivo interaction of the transplanted MS2 RNA and SECIS RNA with a MS2 coat protein dimer [9,11] and the C-terminal WH3/4 domains of E. coli SelB [3], respectively, were examined. As expected, the A005-MS2 variants were sequestered by the coat protein, while the A005-SECIS variants were sequestered by the WH3/4 domains (Figure 3D). Among the three A005-MS2 variants, A005-MS2-1 with the medium-length adapter was most efficiently sequestered by MS2 coat protein (less than 17 µg/mL Cm resistance) (Figure 3D). Among the three A005-SECIS variants, A005-SECIS-2 with the longest adapter was most efficiently sequestered by the WH3/4 domains (up to 17 µg/mL Cm resistance) (Figure 3D). Thus, the optimal length of the adapter stem varied for each aptamer-protein pair. These experiments demonstrated that a small hairpin aptamer can be safely installed into an allo-tRNA chassis without significantly impairing the tRNA function in the absence of the target protein.
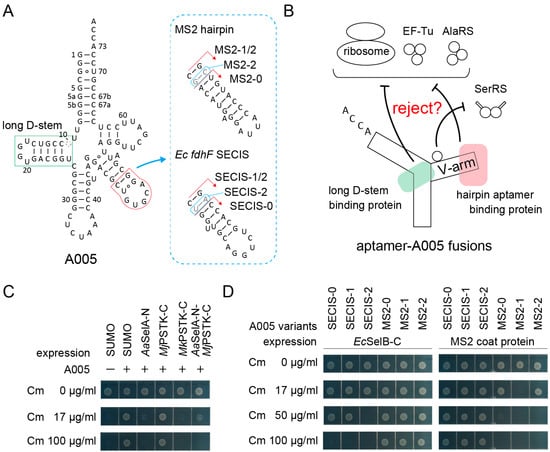
Figure 3.
Sequestration of aptamer-tRNA fusions from the E. coli translation system via the expression of the cognate aptamer-binding proteins. (A) The cloverleaf structure of A005 tRNA variants having a hairpin RNA aptamer in the V-arm. The long D-stem of (9/3) allo-tRNAs and tRNASec species is a binding site for tRNASec-binding enzymes SelA and PSTK. (B) Strategy for the sequestration of the aptamer-A005 fusion variants. (C) The A005 tRNA molecules were almost fully sequestered by the N-terminal domain of Aquifex aeolicus (Aa) SelA and the C-terminal domain of Methanopyrus kandleri (Mk) PSTK but not by the C-terminal domain of Methanocaldococcus jannaschii (Mj) PSTK. (D) The SECIS-A005 fusions were sequestered by the C-terminal domain of E. coli SelB, while the MS2-A005 fusions were sequestered by the MS2 coat protein. Among the fusion tRNAs, the A005-MS2-1 variant was most effectively sequestered. The SUMO protein was used as a negative control and as a N-terminal solubilizing tag for the SelA, PSTK, and SelB domains.
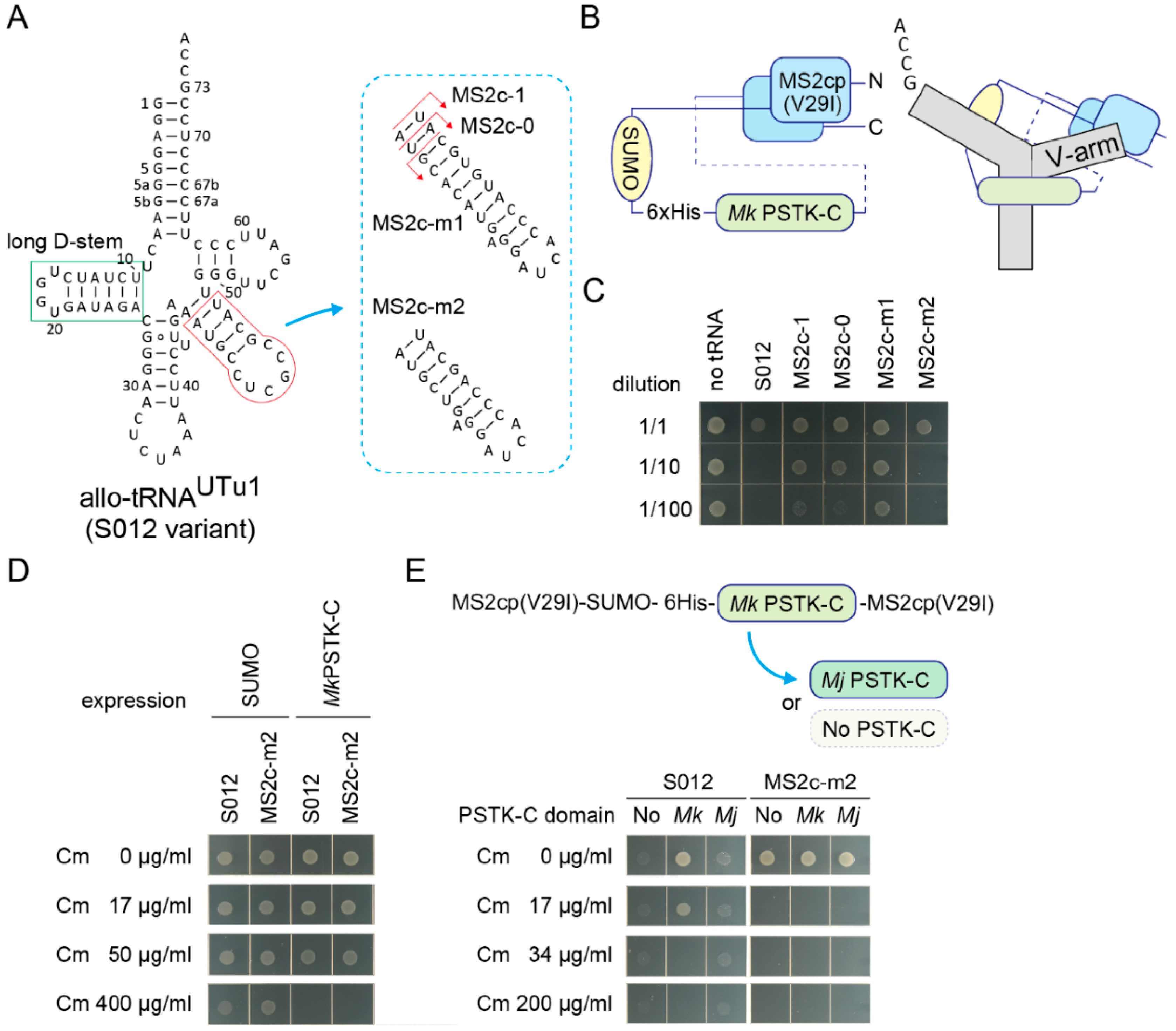
Next, sequestration of fully active tRNASer molecules was challenged. It is known that the allo-tRNAUTu1 variant (Figure 4A and Figure S2A) [29] (or S012 in this study) is as active a tRNASer as one of the E. coli tRNASer species in E. coli. S012 conferred at least 600 µg/mL Cm resistance to E. coli expressing the cat(Ser146TAG) gene (Figure S2B) and at least 34 µg/mL Cm resistance to cells additionally expressing the Mk PSTK-C (Figure S2C,D). Thus, full sequestration of these fully active tRNA molecules may require another tight interaction between an RNA structure and its target protein. Instead of the wildtype MS2 hairpin RNA and the wildtype MS2 coat protein, an MS2 hairpin RNA variant with a C-loop (U to C at the -5 position) [39,40] and the MS2 coat protein V29I variant [40,41] were used to enhance the RNA aptamer-coat protein interaction. To make a single-chain MS2 coat protein [42] together with an Mk PSTK-C domain, two V29I domains were fused with a long linker containing a SUMO domain, a 6xHis tag, and an Mk PSTK-C domain, in this order (Figure 4B). This chimeric fusion protein efficiently sequestered a S012 variant having an MS2 C-loop hairpin with a 1-bp adaptor, or S012-MS2c-1, in E. coli expressing the cat(Ser146TAG) gene (Figure 4A and Figure S3). However, it seems likely that the S012-MS2c-1 variant was less active than the S012 variant, as the original S012 variant was exceedingly toxic to cells (Figure 4C), probably due to its high suppression efficiency [26,29]. Thus, the V-arm adaptor length of the S012-MS2c-1 variant was optimized to restore its tRNA activity in the absence of the chimeric protein (Figure 4A). One variant that has the smallest V-arm (S012-MS2c-m2) showed a similar toxicity to the S012 variant (Figure 4A,C). Both the S012 and S012-MS2c-m2 variants still conferred at least 50 µg/mL Cm resistance to E. coli expressing the cat(Ser146TAG) gene and the Mk PSTK-C domain not fused with the MS2 coat protein (Figure 4D). Then, the sequestration efficiencies of S012 and S012-MS2c-m2 molecules by the chimeric fusion protein were examined (Figure 4E). Surprisingly, expression of the chimeric fusion protein completely repressed the Ser-inserting activities of S012-MS2c-m2 molecules (Figure 4E), while S012 molecules were not completely sequestered. To see the contribution of the Mk PSTK-C domain moiety and the single-chain coat protein moiety in the chimeric fusion protein, the Mk PSTK-C domain was removed or replaced by an Mj PSTK-C domain which did not sequester allo-tRNAs (Figure 4E). All fusion proteins sequestered S012-MS2c-m2 molecules in a complete manner (Figure 4E). Thus, the single-chain coat protein moiety may have bound to the MS2 hairpin moiety of the S012-MS2c-m2 molecules and inhibited their interaction with SerRS. In contrast, the partial sequestration of S012 molecules was dependent on the Mk PSTK-C domain (Figure 4E). These results demonstrated that the strong protein-RNA interaction led to the complete sequestration of allo-tRNA molecules.
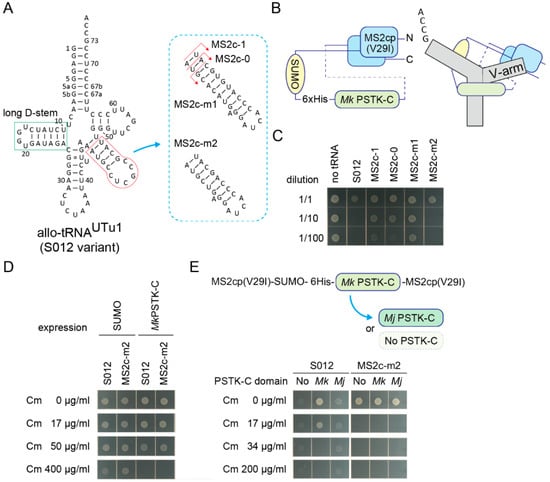
Figure 4.
Sequestration of fully active allo-tRNASer molecules. (A) The cloverleaf structure of S012 tRNA variants having a MS2 hairpin C-loop variant with varying lengths of adaptors. The long D-stem of S012 variants is tightly bound by the C-terminal domain of Mk PSTK. (B) A structural model of the fusion protein consisting of an MS2 coat protein V29I variant monomer, a SUMO tag, a 6xHis tag, a Mk PSTK-C, and another V29I monomer. The N-terminus and C-terminus are indicated. A binding model of a S012 variant and this fusion protein is shown, too. (C) The S012 and MS2c-m2 variants most efficiently impaired the growth of E. coli DH10B cells expressing the cat(Ser146TAG) reporter gene and the SUMO protein. Cell cultures were diluted (10 times and 100 times) or non-diluted and spotted on an agar plate. The arabinose concentration was raised to 0.1% from 0.01% to observe clear differences. (D) The Mk PSTK-C protein not fused with the MS2 coat protein failed to fully sequester both S012 and MS2c-m2 molecules in E. coli DH10B cells expressing the cat(Ser146TAG). (E) The Mk PSTK-C domain of the chimeric fusion protein was replaced with an Mj PSTK-C domain or simply removed. All fusion protein variants perfectly sequestered MS2c-m2 molecules in E. coli DH10B cells expressing the cat(Ser146TAG), indicating that the single-chain coat protein moiety tightly bound with the MS2 hairpin moiety of MS2c-m2. On the other hand, S012 molecules were sequestered only by the Mk PSTK-C-containing fusion protein. The other fusion proteins failed to repress the toxicity and the suppressor tRNA activity of the S012 molecules. Note that cells are unclear but were growing on the Cm-free and Cm-containing agar plates under the S012 + No/Mj conditions.
2.4. Synthetic Tyrosine tRNAs
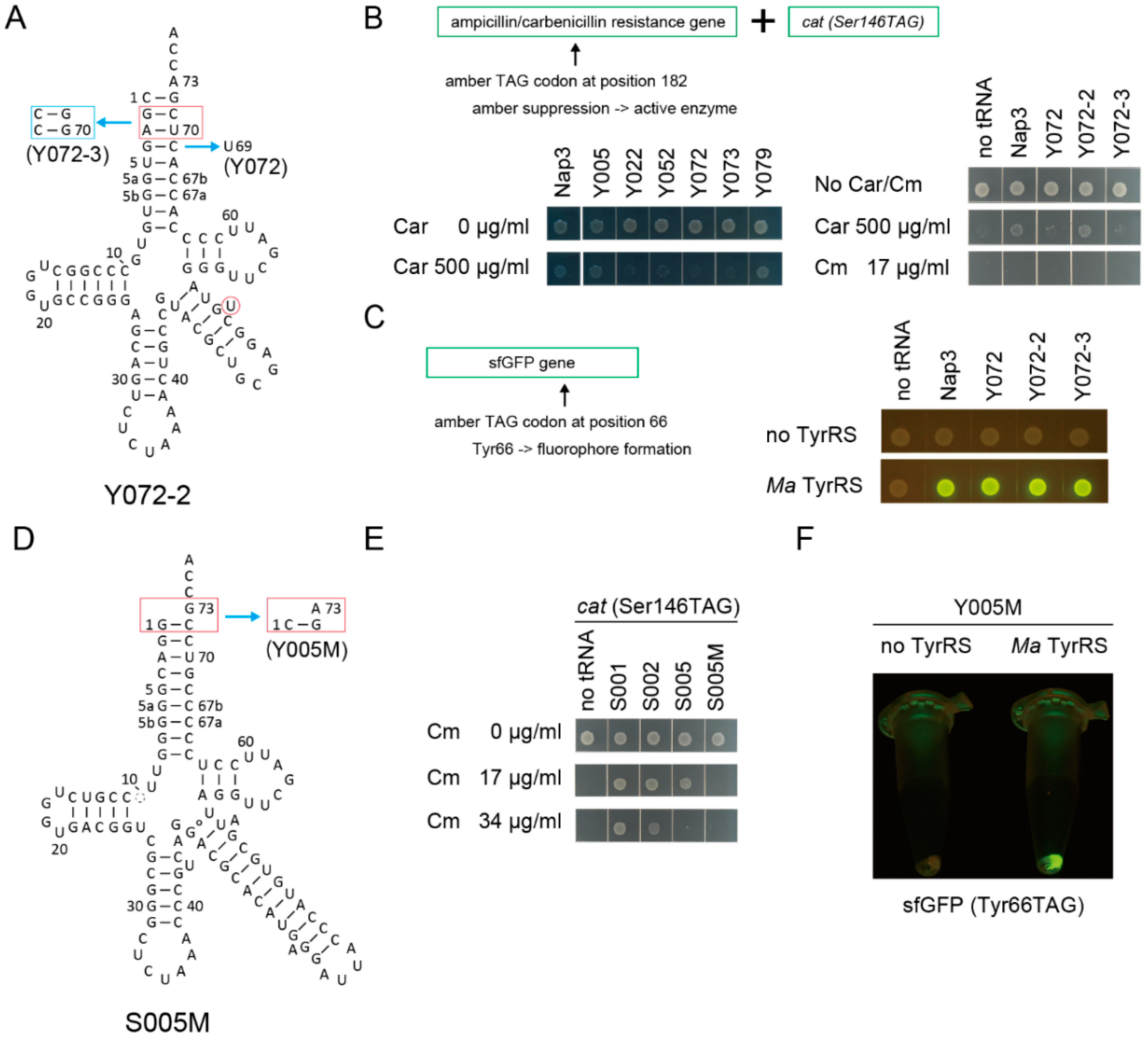
Can the candidate tRNA chassis gain any amino-acid identity other than alanine and serine? To confirm the utility of the candidate tRNA chassis, the major identity elements for the archaeal-type tyrosyl-tRNA synthetase (TyrRS) (C1:G72 with A73) [43,44,45] were installed in the candidate tRNA chassis (Figure 5A), in order to develop synthetic Tyr tRNA molecules [44]. Y005, Y072, and Y073 variants were developed from S005, S072, and S073, respectively. Two derivatives of Y072 (Y072-2 and Y072-3) were also developed because modifications of the second and third base pairs in the acceptor stem may affect aminoacylation (see Figure 5A). An amber suppressor variant of the tRNATyr species of Methanocaldococcus jannaschii (Mj), or Nap3 [46], was used as a positive control. An archaeal TyrRS species from Candidatus Methanomethylophilus alvus Mx1201 (Ma) was used to be paired with these tRNAs. Three kinds of reporter genes were used for examining the orthogonality and the tRNATyr activity of these tRNA variants (Figure 5B,C). One is an ampicillin/carbenicillin resistance gene (bla) having an in-frame TAG codon at a permissive site (Ala182) [47] (Figure 5B). The cat(Ser146TAG) reporter gene was cloned together with this bla(Ala182TAG) reporter gene to make a dual-reporter plasmid. The third one is a sfGFP gene in which the Tyr66 codon had been mutated to TAG, since Tyr66 is essential for the fluorophore formation [48] (Figure 5C). Using the bla reporter gene, it was revealed that the Y072 and Y073 variants conferred only marginal carbenicillin (Car) resistance to cells, while the Y005 variant and the Mj tRNATyr Nap3 variant conferred up to 500 µg/mL Car resistance to cells (Figure 5B). Note that the Nap3 variant has been used as a representative orthogonal tRNATyr in E. coli [49]. This result indicated that the Y072 and Y073 variants were not activated by any endogenous aaRS species in E. coli. Although the Y072-2 variant conferred up to 500 µg/mL Car resistance to cells (Figure 5B), none of the Y072, Y072-2, and Y072-3 variants conferred Cm resistance to E. coli DH10B cells expressing the cat(Ser146TAG) reporter gene (Figure 5B). Thus, these variants showed no Ser identity. Next, by using the sfGFP(Tyr66TAG) reporter gene, it was confirmed that the Y072, Y072-2, and Y072-3 variants were charged with tyrosine by Ma TyrRS (Figure 5C).
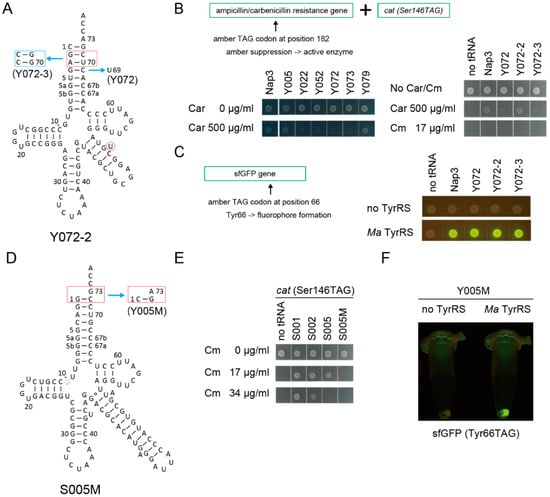
Figure 5.
Development of synthetic tyrosine tRNAs using candidate tRNA chassis. (A) The cloverleaf structure of Y072-series tRNA variants. (B) The S072 and S073 variants conferred only marginal carbenicillin (Car) resistance (less than Car 500 µg/mL) to E. coli DH10B cells expressing the bla(Ala182TAG) reporter gene and the cat(Ser146TAG) reporter gene in the absence of any heterologous TyrRS enzyme. Amber suppressor variants of archaeal M. jannaschii tRNATyr species (Nap3) and the Y005, Y079, and Y072-2 variants conferred up to 500 µg/mL Car resistance, indicating that these tRNAs were to a less extent charged by endogenous aaRSs. No Cm resistance was introduced by the Y072, Y072-2, and Y072-3 variants, indicating that they were orthogonal to SerRS. (C) In the presence of Ca. M. alvus TyrRS in E. coli DH10B, the Nap3 variant and the three Y072-series variants translated the amber codon in the sfGFP(Tyr66TAG) reporter gene with tyrosine, making the fluorophore of sfGFP. (D) The cloverleaf structure of the S005M and Y005M tRNA variants. (E) The S005M variant conferred a lower Cm resistance (less than Cm 17 µg/mL) to E. coli DH10B cells expressing the cat(Ser146TAG) reporter gene compared to the S002 and S005 variants. (F) In the presence of Ca. M. alvus TyrRS in E. coli DH10B, the Y005M variant translated the amber codon of the sfGFP(Tyr66TAG) reporter gene with tyrosine, making the fluorophore of sfGFP.
To improve the usefulness of the S005 chassis, I examined another chassis, S005M, which is a variant of A005-MS2-1 lacking the AlaRS identity elements (Figure 5D). It was found that the original S005 variant conferred 17 µg/mL Cm resistance to E. coli DH10B cells expressing the cat(Ser146TAG) reporter gene (Figure 5E). In contrast, the S005M variant having an MS2 hairpin at the tip of the V-arm did not confer 17 µg/mL Cm resistance (Figure 5E). Most likely, the S005M variant may have a stabilized bulge structure and a slightly reduced tRNA activity compared to S005. Although the marginal Ser-inserting activity of S005 can be eliminated by engineering the tip of the acceptor stem [22], the use of S005M facilitates engineering of the amino-acid identity owing to the ignorable Ser-inserting activity. The S005M chassis was engineered to be coupled with Ma TyrRS, to develop Y005M (Figure 5D). In the presence of Ma TyrRS, Y005M translated the amber codon of the sfGFP(Tyr66TAG) reporter gene with tyrosine (Figure 5F). These results clearly demonstrated that SerRS-rejecting allo-tRNA chassis can be engineered and converted to synthetic Tyr tRNA species.
2.5. Enhancing Aminoacylation by Aptamer-Protein Interaction
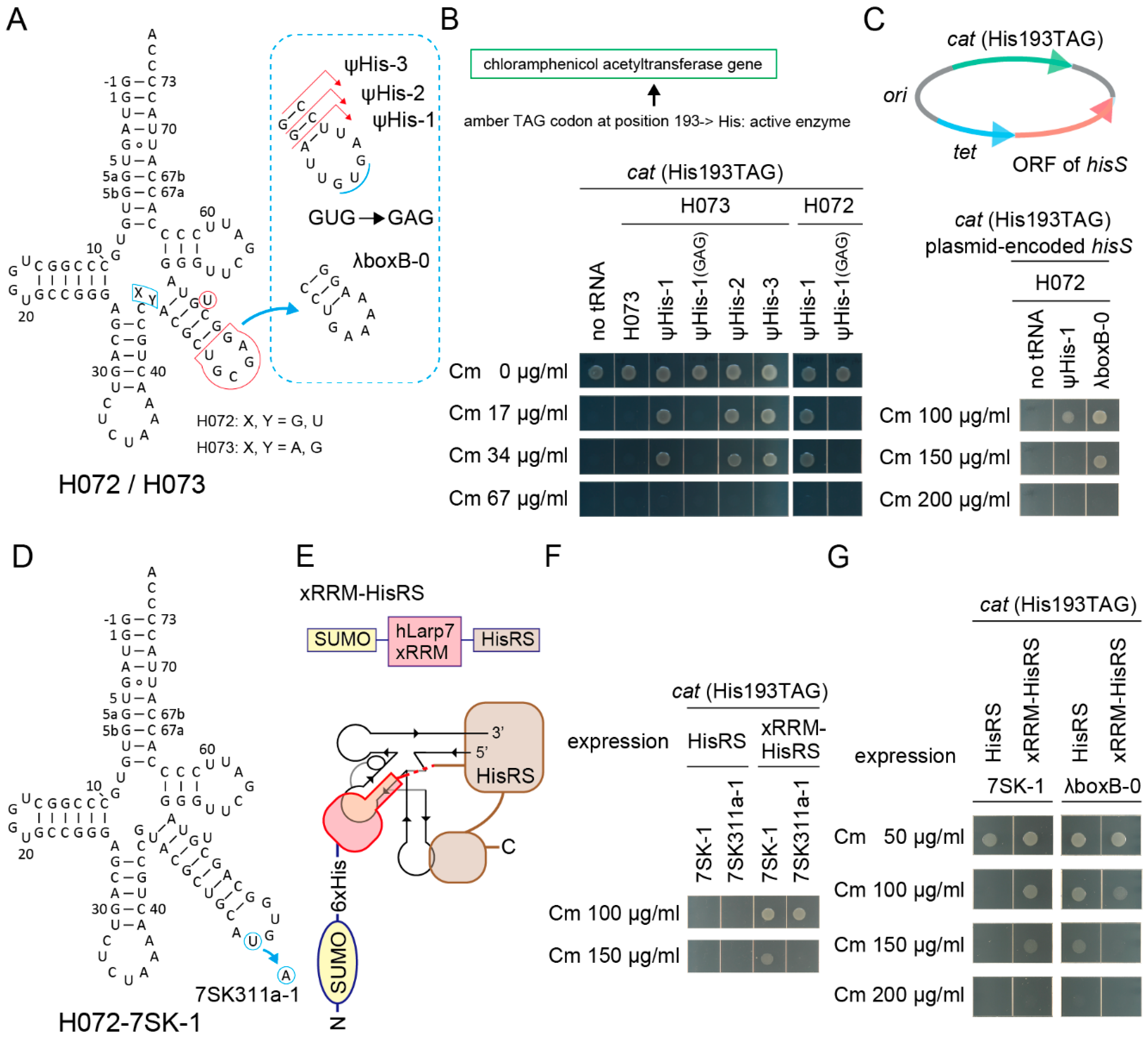
Can an RNA aptamer moiety in tRNA work as an additional binding site for an aaRS fused with the aptamer-binding protein domain? To answer this, synthetic histidine (His) tRNA molecules [18] were developed by installing the major identity element for the E. coli histidyl-tRNA synthetase (HisRS) (G-1:C73) [2,26,50] into the S072/S073 chassis (Figure 6A). Two kinds of cat reporter genes, cat(Ser146TAG) and cat(His193TAG), were used for examining the orthogonality and the His-inserting activity of designed tRNA variants. In the cat(His193TAG) gene, the essential catalytic His193 was changed to TAG [51]. It soon turned out that a S073 variant with a G-1:C73 pair (named H073) was not charged by the endogenous HisRS in E. coli expressing the cat(His193TAG) gene (Figure 6B). However, extending the V-stem length and modifying the V-arm loop sequence produced functional H072/H073 variants which conferred up to 34 µg/mL Cm resistance to cells (Figure 6B). Ser insertion was not mediated by these H072/H073 variants in E. coli cells expressing the cat(Ser146TAG) gene (Figure S4A). Point mutations in the V-arm loop sequence affected the His-inserting activity of H072 ψHis-1 (Figure 6A); UUUUGAU was better than the original UUGUGAU, while UUGAGAU and UUGCGAU were less suitable (Figure 6B and Figure S4B,C), implying that a good V-arm loop sequence on a moderately long V-arm may be rich in uracil residues. It was also found that a moderate overexpression of HisRS from a plasmid-encoded hisS gene enhanced the His-inserting activities by the synthetic His tRNAs (Figure 6C and Figure S4F). Up to 150 µg/mL Cm resistance was conferred by the H072-λboxB-0 variant (Figure 6A,C). On the other hand, the S005 chassis was useless for developing a synthetic His tRNA in the same manner (Figure S4D,E). Thus, it was revealed that the H072/H073 chassis were intrinsically compatible to E. coli HisRS and E. coli RNase P, which cleaves His tRNA precursors at the -1 position [52].
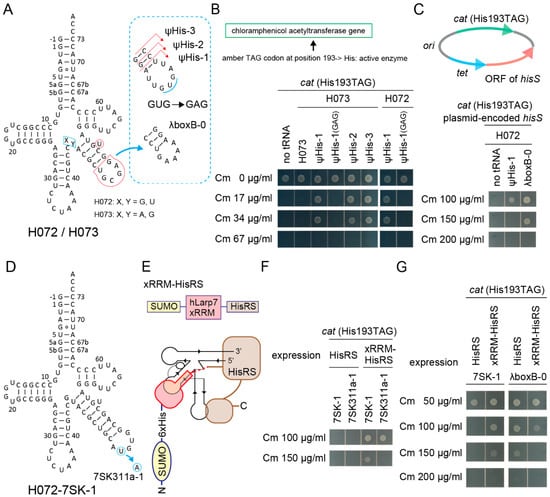
Figure 6.
Development of synthetic histidine tRNAs using candidate tRNA chassis and hairpin aptamers. (A) The cloverleaf structures of H072 and H073 tRNA variants having an extended V-stem and a modified V-arm loop sequence. (B) The H073 ψHis-1/2/3 variants and the H072 ψHis-1 variant conferred up to Cm 34 µg/mL resistance to E. coli DH10B cells expressing the cat(His193TAG) reporter gene. The catalytic His193 residue is essential for the Cm acetyltransferase activity. A point mutation in the V-arm loop sequence (from GUG to GAG) eliminated the His-inserting activity of the ψHis-1 variants. (C) The H072 ψHis-1 and λboxB-0 variants conferred up to Cm 100 µg/mL resistance and 150 µg/mL resistance, respectively, to E. coli DH10B cells expressing the cat(His193TAG) reporter gene and the plasmid-encoded hisS gene (E. coli HisRS) shown. Note that the agarose plate containing Cm 100 µg/mL was incubated for another four hours to obtain the clear cell spot for H072 ψHis-1. (D) The cloverleaf structure of the H072-7SK-1 variant having a 7SK SL4 hairpin. (E) A chimeric E. coli HisRS variant fused with the xRRM domain of human Larp7. A binding model of H072-7SK-1 and this xRRM-HisRS is shown. The dashed line indicates linker residues SSGSNSNSGS. (F) Expression of xRRM-HisRS enhanced the His-inserting activities by the H072-7SK-1 and H072-7SK311a-1 variants. The H072-7SK-1 variant conferred up to 150 µg/mL resistance to E. coli DH10B cells expressing the cat(His193TAG) reporter gene and xRRM-HisRS. It was reported that the 7SK RNA SL4 binds the hLarp7 xRRM domain, while the 311a variant binds the xRRM domain more tightly. (G) The His-inserting activity of H072-7SK-1 was enhanced by xRRM-HisRS, while that of H072-λboxB-0 was not enhanced.
By using the H072 chassis and E. coli HisRS, several cognate pairs of RNA aptamers and protein domains [2,28,50,53,54,55] were examined (Figure S5A,B). The first preliminary experiment revealed that the cognate pair of the 7SK RNA stem-loop 4 (SL4) and the hLarp7 xRRM domain [53] fused to the N-terminus of E. coli HisRS (Figure 6D,E) might have enhanced the His-inserting activity by the tRNA-aptamer fusion in E. coli expressing the cat(His193TAG) gene (Figure S5C). The other pairs examined seemed to be rather inhibitory (Figure S5C). Three explanations are possible: 1) addition of an N-terminal domain might have reduced the expression level, stability, accessibility, or activity of the tethered HisRS, 2) too strong affinity between the cognate RNA-protein pair might have reduced the turnover rate of aminoacylation, and 3) a too short linker sequence SSGSNSNSGS between the N-terminal domain and HisRS might have kept the catalytic pocket of the tethered HisRS away from the CCA tail of tRNA. In contrast, the xRRM domain binds a 7SK SL4 RNA with a moderate affinity (Kd = 129 nM [53]). Furthermore, the C-terminus of the xRRM domain and the N-terminus of the E. coli HisRS may approach each other on the tRNA and can be fused by using the short linker (Figure 6E), according to crystal structural studies [50,53]. In another experiment, the H072-7SK-1 variant conferred up to 150 µg/mL resistance to E. coli DH10B cells expressing the cat(His193TAG) reporter gene and xRRM-HisRS (Figure 6F), while the U311A change in the 7SK SL4 loop (Figure 6D), which reportedly enhances the interaction (Kd = 51 nM [53]), slightly reduced the His-inserting activity (Figure 6F). The U311A change may have slightly impaired the turnover of the enzyme or slightly destabilized the tRNA. Lastly, the effect of the xRRM domain on aminoacylation was confirmed by using the H072-7SK-1 and H072-λboxB-0 (Figure 6A) variants. The expression of xRRM-HisRS greatly enhanced the His-inserting activity of H072-7SK-1 (Figure 6G), while H072-λboxB-0 was efficiently activated by the wildtype HisRS but not by xRRM-HisRS (Figure 6G). This meant that the appended SUMO-xRRM domains were rather inhibitory to histidylation in the absence of the cognate interaction between the xRRM domain and the aptamer moiety, probably due to the decreased expression level, stability, or accessibility of HisRS. Thus, it was concluded that the cognate interaction contributed to the enhancement of aminoacylation.
2.6. Recruiting aaRS via a Pseudo-Anticodon in the V-Arm
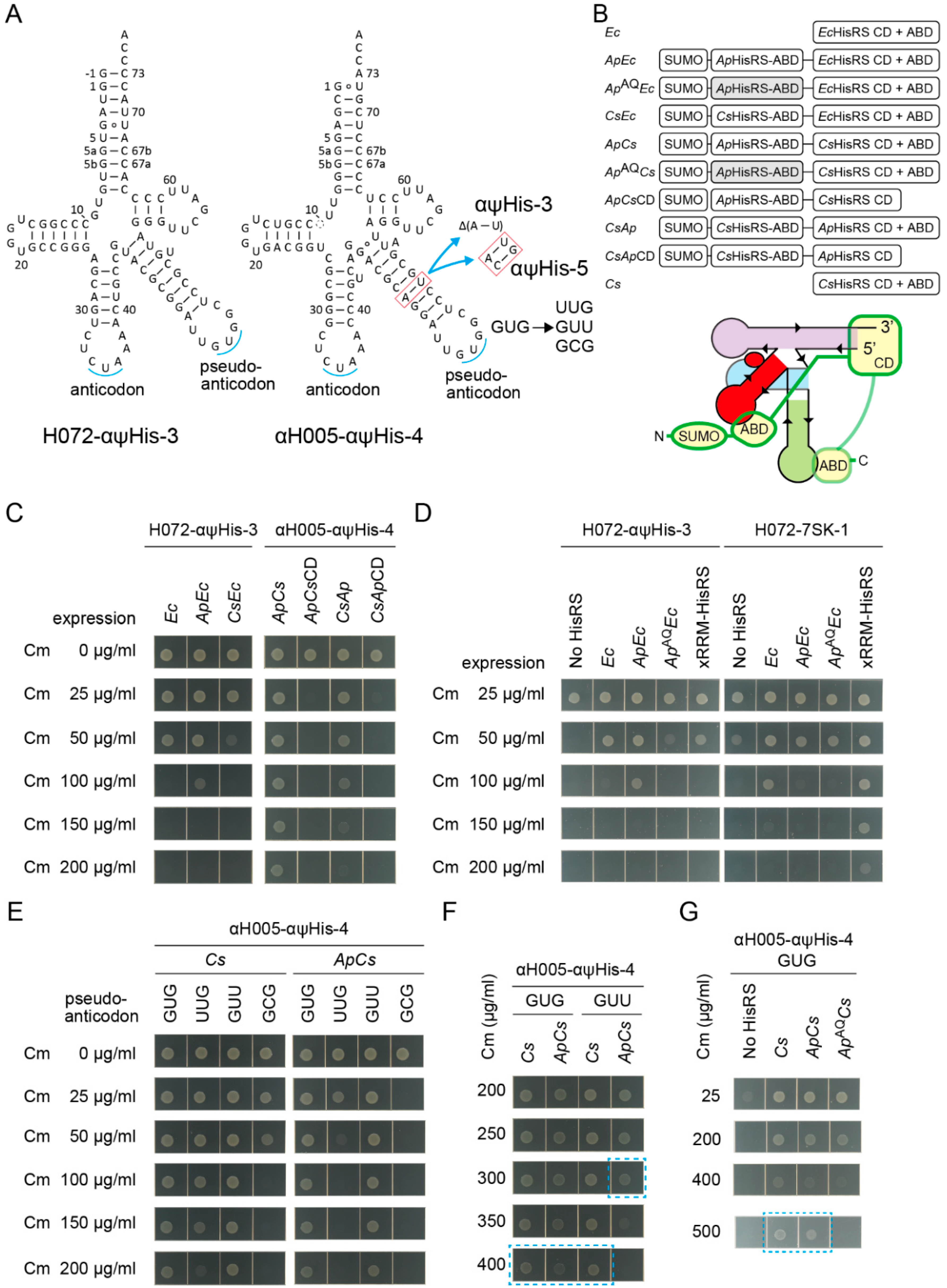
Can an aaRS variant having an additional heterologous anticodon binding domain (ABD) be recruited to tRNA via the V-arm which mimics the anticodon arm that can be recognized by the ABD? In other words, can a pseudo-anticodon arm moiety [1,56] on tRNAs work as an aptamer for the ABD and contribute to aminoacylation enhancement? Here, the full-length HisRSs, HisRS catalytic domains (CDs), and HisRS ABDs of two α-Proteobacteria species and their tRNAHis anticodon arm were used. It is well known that a large subgroup of α-Proteobacteria has a non-canonical tRNAHis lacking the G-1:C73 pair and has a non-canonical HisRS species that recognizes the non-canonical G1:U72 pair and A73 in addition to the GUG anticodon [17,57,58,59,60]. The two α-proteobacterial species are Afifella pfennigii DSM 17,143 (Ap) [17] and Consotaella salsifontis USBA 369 (Cs) [61], because the well-studied α-proteobacterial Caulobacter crescentus HisRS was not active enough in E. coli [60]. Two kinds of allo-tRNA chassis, S072 and S005, were engineered to have a typical α-proteobacterial tRNAHis anticodon arm as their V-arms, to develop H072-αψHis-3 and αH005-αψHis-3/4/5 variants (Figure 7A). The H072-αψHis-3 variant has the G-1:C73 element to be paired with E. coli HisRS variants N-terminally fused with an α-proteobacterial ABD using a 20 amino-acid linker (Figure 7B). On the other hand, the αH005-αψHis-3/4/5 variants have the G1:U72 and A73 elements to be paired with Ap/Cs HisRS N-terminally fused with a heterologous ABD using the same 20 amino-acid linker (Figure 7B). The C-terminal ABDs of chimeric Ap/Cs HisRS variants were kept intact or trimmed (Figure 7B). ApEc, CsEc, ApCs, and CsAp variants denote ApABD-EcHisRS, CsABD-EcHisRS, ApABD-CsHisRS, and CsABD-ApHisRS, respectively (Figure 7B).
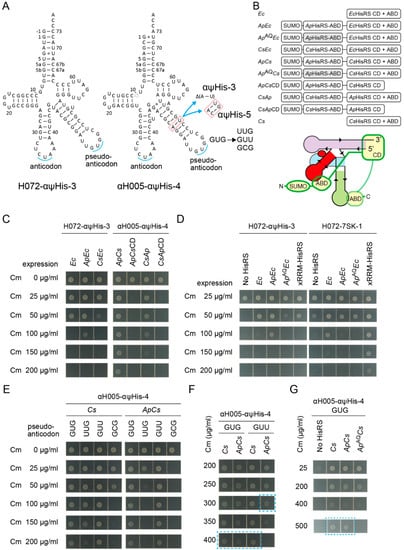
Figure 7.
Recruiting HisRS via a pseudo-anticodon arm. (A) The cloverleaf structures of the H072-αψHis-3 and αH005-αψHis-3/4/5 variants with varying lengths of adaptors and with diverse pseudo-anticodon sequences. (B) The domain structures of HisRS and HisRS variants N-terminally fused with a heterologous anticodon binding domain (ABD). CD denotes catalytic domain. Ec, Ap, and Cs denote E. coli, Afifella pfennigii, and Consotaella salsifontis, respectively. ApAQ denotes an Ap ABD variant carrying E484A-K493Q mutations impairing anticodon recognition. (C,D) Cm resistance of E. coli DH10B (NEB 10-beta) cells expressing the cat(His193TAG) reporter gene and the indicated combinations of HisRS and tRNA variants. (D) The His-inserting activity of H072-αψHis-3 was enhanced by ApEc but not enhanced by ApAQEc, indicating that the recognition of the GUG pseudo-anticodon by the wildtype Ap ABD moiety was important. On the other hand, the His-inserting activity of H072-7SK-1 lacking the GUG pseudo-anticodon was not enhanced by both ApEc and ApAQEc. No HisRS indicates that only the genomic hisS gene was expressing. (E–G) Cm resistance of E. coli DH10B (NEB 10-beta) cells expressing the cat(His193TAG) reporter gene and the indicated combinations of HisRS variants and pseudo-anticodon variants of αH005-αψHis-4. The Cm concentrations are indicated. (G) Note that the agarose plate containing Cm 500 µg/mL was incubated for another four hours to obtain clear cell spots.
By using the cat(His193TAG) reporter system, the His-inserting activities by combinations of the tRNA variants and HisRS variants were examined (Figure 7C). Addition of the N-terminal ApABD, but not CsABD, to the E. coli HisRS led to the enhancement of the His-inserting activity by H072-αψHis-3 (Figure 7C). Thus, the ApABD may be especially useful in E. coli, as explicitly predicted in a previous study [17]. Next, the αH005-αψHis-3/4/5 variants were combined with the chimeric Ap/Cs HisRS variants. The ApCs variant efficiently activated the αH005-αψHis-3/4/5 variants, while the CsAp variant was less active (Figure 7C and Figure S6). Among the αH005-αψHis-3/4/5 variants, αH005-αψHis-4 seems the most active, because αH005-αψHis-3 and αH005-αψHis-5 conferred slightly lower Cm resistance (Figure S6). In contrast, the chimeric Ap/Cs HisRS variants lacking the original C-terminal ABD were totally inactive (Figure 7C), probably because HisRS ABD is responsible not only for anticodon recognition but also for HisRS dimerization and for precise acceptor stem discrimination [18,62]. Thus, it is evident that the tRNA variants having a GUG pseudo-anticodon arm were charged with histidine by HisRS variants N-terminally fused with an ApABD.
To confirm the involvement of the GUG pseudo-anticodon in the tRNA-HisRS interaction, two lines of experiments were performed. First, the H072-αψHis-3 variant (having the GUG pseudo-anticodon) and the H072-7SK-1 variant (having no pseudo-anticodon arm) were compared (Figure 7D). To partially impair the anticodon recognition activity of the Ap ABD, two mutations (E484A-K493Q) which may partially impair anticodon recognition [50] were introduced to develop ApAQEc and ApAQCs HisRS variants (Figure 7B). As expected, ApEc enhanced the histidylation of H072-αψHis-3, whereas ApAQEc failed (Figure 7D). H072-αψHis-3 was activated at similar efficiencies by wildtype HisRS and xRRM-HisRS (Figure 7D). In contrast, the histidylation of H072-7SK-1 was significantly enhanced by xRRM-HisRS (Figure 6G,7D). Both ApEc and ApAQEc variants failed to enhance the histidylation of H072-7SK-1 (Figure 7D). Thus, it was revealed that the interaction between the GUG pseudo-anticodon arm and the Ap ABD moieties contributed to the enhancement of aminoacylation. However, compared to the 7SK-xRRM interaction (Figure 6E), this GUG-ApABD interaction exerted a modest effect on aminoacylation enhancement (Figure 7D).
The second line of experiments were performed to examine the importance of the GUG pseudo-anticodon sequence. The GUG sequence in αH005-αψHis-4 was changed to UUG, GUU, and GCG (Figure 7A). According to a previous study, C. crescentus tRNAHis variants having these anticodon changes are poor substrates of C. crescentus HisRS [58]. Surprisingly, the wildtype Cs HisRS charged all pseudo-anticodon variants of αH005-αψHis-4 (Figure 7E). Thus, it is evident that the critical recognition elements of tRNAHis for Cs HisRS are the G1:U72 pair with the A73 discriminator base but not the GUG anticodon. In the presence of Cs HisRS, the original GUG variant and the GUU variant conferred at least Cm 400 µg/mL resistance (Figure 7F), while the UUG and GCG variants conferred only up to Cm 150 µg/mL and Cm 50 µg/mL resistance, respectively (Figure 7E). In the presence of Cs HisRS or ApCs, the original GUG variant conferred up to Cm 500 µg/mL resistance, while it still conferred at least Cm 400 µg/mL resistance in the presence of ApAQCs (Figure 7G). Thus, this GUG-ApABD interaction exerted only a modest effect on aminoacylation enhancement, probably because αH005-αψHis-4 is already a good substrate of wildtype Cs HisRS. In contrast, the UUG/GUU/GCG pseudo-anticodon variants were charged by ApCs much less efficiently than they were charged by Cs HisRS (Figure 7E,F). This indicated that the N-terminal appended domain was rather inhibitory to histidylation in the absence of the cognate interaction between the pseudo-anticodon and the appended ABD. In summary, it was shown that a pseudo-anticodon arm moiety can contribute to the enhancement of aminoacylation efficiency and tRNA-recognition specificity.
3. Discussion
In the present study, I showed that the SerRS-rejecting allo-tRNA chassis can be engineered to transfer Ala, Tyr, and His and fused with several kinds of hairpin RNAs. Although this study lacks any biochemical data, combinations of traditional in vivo reporter assay experiments clearly demonstrated that some of the RNA aptamers installed into the allo-tRNA chassis were bound by their cognate proteins or protein domains. Furthermore, the interactions between RNA aptamers and their cognate protein domains contributed to the enhancement of the tRNA-recognition specificity and the histidyl-tRNA formation by chimeric HisRS variants. Our next task should be simultaneous in vivo selection of the aptamer moieties and protein domains to optimize the V-stem length, the V-arm loop sequence, and the aptamer-recognizing residues of the protein domains. This optimization step may also be required for reducing the affinity of high-affinity aptamers and for preventing misrecognition of endogenous RNA molecules by the protein domains.
Some of the methods developed in this study may be applicable to genetic code expansion and genetic code reprogramming. For example, the pairs of Ca. M. alvus TyrRS and Y072/Y005M and the pairs of Cs HisRS variants and αH005-αψHis-4 variants may be orthogonal in E. coli and might be useful for incorporating non-canonical amino acids [17,58,60]. Alternatively, some of the endogenous tRNAs or tRNA-aaRS pairs might be replaceable with allo-tRNAs or orthogonal pairs composed of allo-tRNAs [60,63,64,65]. For reprogramming the genetic code, the allo-tRNA sequestration system may be useful for use in vivo and in vitro [66] and for the purification and elimination of allo-tRNA molecules from tRNA mixtures [66,67,68,69]. Application of the allo-tRNA chassis may not be limited to E. coli but may possibly need further optimization, especially for their use in archaea and eukaryotes due to the difference in the manner of tRNASer recognition by SerRS [70,71].
In recent years, synthetic biologists have started to create synthetic tRNA-aminoacylation protein enzymes and ribozymes [18,72,73]. However, unlike most of the natural aaRS species, these artificial protein enzymes, including my HisRS chimeras, are composed of separate domains fused with a flexible linker [18,72]. Thus, such artificial enzymes were far less active and precise than natural enzymes [18,72]. On the other hand, the ribozymes required pre-activated amino acid substrates rather than free amino acid and ATP [73]. An apparent drawback of my allo-tRNA strategy is that the tip of the V-arm is distant from the CCA tail of tRNA. It may be helpful to use a long and rigid linker motif found in mitochondrial AlaRS species [74] or to use a long-D-arm binding protein domain to wrap around tRNA [37,38]. The use of T-box riboswitches as RNA scaffolds may be an alternative approach [73,75]. In summary, the novel allo-tRNA chassis, or orthogonal allo-tRNAs, would facilitate creation of synthetic tRNA-aminoacylation enzymes and ribozymes.
4. Materials and Methods
4.1. Cloning tRNA and Protein Genes
The nucleotide and amino-acid sequences are provided in the supplementary Excel file. All tRNA sequences (including the CCA tail) were cloned between the EcoRI and BglII sites in the pBAD-RSF5 plasmid [26] re-constructed in this study. The cat variant genes were developed from pACYC184 using Infusion (TAKARA Bio Inc., Shiga, Japan). The Ca. M. alvus TyrRS gene was cloned into pACYC184 by replacing the cat gene ORF. The HisRS ORFs were cloned immediately downstream of the tet gene of pACYC184-cat(193HisTAG). The bla variant gene was inserted downstream of the tet gene of pACYC184-cat(Ser146TAG) using Infusion. The other protein genes were cloned into the pGEX-6P-1 vector (a gift from Kazuhisa Nakayama, Addgene plasmid # 61838) and expressed under control of the tac promoter. Oligo DNAs and synthetic protein genes were purchased from Eurofin Genomics Co., Ltd. (Tokyo, Japan).
4.2. In Vivo Reporter Assays
E. coli DH10B and NEB 10-beta (NEB) cells were transformed with the indicated combinations of plasmids. Before spotting assays, transformed cells were pre-cultured in liquid LB media in the presence of 0.01% l-arabinose and 10 μM IPTG, if required, for a few hours [26]. In typical experiments, cell cultures were diluted to OD600 = 0.05 and then incubated for three hours. After spotting cell cultures (2 μL each) on LB agar plates, the plates were incubated at 37 °C overnight or for a longer time. For the induction of sfGFP genes, the final concentration of IPTG was 100 μM.
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/21/20/7793/s1. Figure S1: Cloverleaf structures of the eight starting amber suppressor (9/3) allo-tRNA variants other than the S001 variant (Figure 1B), Figure S2: Inefficient sequestration of allo-tRNAUTu1 (or S012), Figure S3: Sequestration of S012 derivatives, Figure S4: Examining synthetic tRNAHis variants, Figure S5: A preliminary screening experiment of several cognate pairs of RNA aptamers and binding protein domains towards enhancing the histidylation activity, Figure S6: Cm resistance of E. coli DH10B (NEB 10-beta) cells expressing the cat(His193TAG) reporter gene and the indicated combinations of HisRS and tRNA variants, Table S1: Screening of candidate chassis tRNAs by using E. coli DH10B cells expressing the cat(Ser146TAG) reporter gene on growth media containing chloramphenicol (Cm) at a fixed concentration of 34 µg/ml.
Funding
This research was funded by the Japan Society for the Promotion of Science, Grant-in-Aid for Research Activity start-up, grant numbers 18H06055 and 19K21181.
Acknowledgments
The author thanks Dieter Söll (Yale University) and Masayuki Su’etsugu (Rikkyo University) for supporting this study.
Conflicts of Interest
The author declares no conflict of interest.
Abbreviations
| aaRS | aminoacyl-tRNA synthetase |
| SerRS | seryl-tRNA synthetase |
| AlaRS | alanyl-tRNA synthetase |
| TyrRS | tyrosyl-tRNA synthetase |
| HisRS | histidyl-tRNA synthetase |
| SelA | selenocysteine synthase |
| PSTK | O-phosphoseryl-tRNA kinase |
| sfGFP | superfolder green fluorescent protein |
| IPTG | isopropyl β-d-1-thiogalactopyranoside |
| SECIS | selenocysteine insertion sequence |
| bp | base pair |
| cp | coat protein |
| SUMO | small ubiquitin-like modifier |
| WH | winged helix |
| SL | stem loop |
| ORF | open reading frame |
| NTD | N-terminal domain |
| CTD | C-terminal domain |
| ABD | anticodon binding domain |
| CD | catalytic domain |
References
- Levi, O.; Garin, S.; Arava, Y. RNA mimicry in post-transcriptional regulation by aminoacyl tRNA synthetases. Wiley Interdiscip. Rev. RNA 2020, 11, e1564. [Google Scholar] [CrossRef] [PubMed]
- Levi, O.; Arava, Y. mRNA association by aminoacyl tRNA synthetase occurs at a putative anticodon mimic and autoregulates translation in response to tRNA levels. PLoS Biol. 2019, 17, e3000274. [Google Scholar] [CrossRef] [PubMed]
- Soler, N.; Fourmy, D.; Yoshizawa, S. Structural insight into a molecular switch in tandem winged-helix motifs from elongation factor SelB. J. Mol. Biol. 2007, 370, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Gemmill, D.; D’Souza, S.; Meier-Stephenson, V.; Patel, T.R. Current approaches for RNA-labelling to identify RNA-binding proteins. Biochem. Cell Biol. 2020, 98, 31–41. [Google Scholar] [CrossRef]
- Ono, H.; Kawasaki, S.; Saito, H. Orthogonal Protein-Responsive mRNA Switches for Mammalian Synthetic Biology. ACS Synth. Biol. 2020, 9, 169–174. [Google Scholar] [CrossRef]
- van Gijtenbeek, L.A.; Kok, J. Illuminating Messengers: An Update and Outlook on RNA Visualization in Bacteria. Front. Microbiol. 2017, 8, 1161. [Google Scholar] [CrossRef]
- Iioka, H.; Macara, I.G. Detection of RNA-Protein Interactions Using Tethered RNA Affinity Capture. Methods Mol. Biol. 2015, 1316, 67–73. [Google Scholar]
- Masuda, I.; Igarashi, T.; Sakaguchi, R.; Nitharwal, R.G.; Takase, R.; Han, K.Y.; Leslie, B.J.; Liu, C.; Gamper, H.; Ha, T.; et al. A genetically encoded fluorescent tRNA is active in live-cell protein synthesis. Nucleic Acids Res. 2017, 45, 4081–4093. [Google Scholar] [CrossRef]
- Ponchon, L.; Catala, M.; Seijo, B.; El Khouri, M.; Dardel, F.; Nonin-Lecomte, S.; Tisné, C. Co-expression of RNA-protein complexes in Escherichia coli and applications to RNA biology. Nucleic Acids Res. 2013, 41, e150. [Google Scholar] [CrossRef]
- Lee, D.; McClain, W.H. Aptamer redesigned tRNA is nonfunctional and degraded in cells. RNA 2004, 10, 7–11. [Google Scholar] [CrossRef]
- Preston, M.A.; Porter, D.F.; Chen, F.; Buter, N.; Lapointe, C.P.; Keles, S.; Kimble, J.; Wickens, M. Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase. Nat. Methods 2019, 16, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.G.; Wei, H.; Ling, C.; Martin, F.; Eriani, G.; Wang, E.D. Two distinct domains of the beta subunit of Aquifex aeolicus leucyl-tRNA synthetase are involved in tRNA binding as revealed by a three-hybrid selection. Nucleic Acids Res. 2004, 32, 3294–3303. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Warszawik, E.M.; Loznik, M.; Boersma, A.J.; Herrmann, A. Modular and Versatile Trans-Encoded Genetic Switches. Angew. Chem. Int. Ed. Engl. 2020. [Google Scholar] [CrossRef]
- Warner, K.D.; Chen, M.C.; Song, W.; Strack, R.L.; Thorn, A.; Jaffrey, S.R.; Ferré-D’Amaré, A.R. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol. 2014, 21, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Volkov, I.L.; Johansson, M. Single-Molecule Tracking Approaches to Protein Synthesis Kinetics in Living Cells. Biochemistry 2019, 58, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Thyer, R.; Ellington, A.D. The Role of tRNA in Establishing New Genetic Codes. Biochemistry 2019, 58, 1460–1463. [Google Scholar] [CrossRef]
- Cervettini, D.; Tang, S.; Fried, S.D.; Willis, J.C.W.; Funke, L.F.H.; Colwell, L.J.; Chin, J.W. Rapid discovery and evolution of orthogonal aminoacyl-tRNA synthetase-tRNA pairs. Nat. Biotechnol. 2020, 38, 989–999. [Google Scholar] [CrossRef]
- Ding, W.; Zhao, H.; Chen, Y.; Zhang, B.; Yang, Y.; Zang, J.; Wu, J.; Lin, S. Chimeric design of pyrrolysyl-tRNA synthetase/tRNA pairs and canonical synthetase/tRNA pairs for genetic code expansion. Nat. Commun. 2020, 11, 3154. [Google Scholar] [CrossRef]
- Wang, L.; Brock, A.; Herberich, B.; Schultz, P.G. Expanding the genetic code of Escherichia coli. Science 2001, 292, 498–500. [Google Scholar] [CrossRef]
- Mukai, T.; Lajoie, M.J.; Englert, M.; Söll, D. Rewriting the Genetic Code. Annu. Rev. Microbiol. 2017, 71, 557–577. [Google Scholar] [CrossRef]
- Arranz-Gibert, P.; Patel, J.R.; Isaacs, F.J. The Role of Orthogonality in Genetic Code Expansion. Life 2019, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Zambaldo, C.; Koh, M.; Nasertorabi, F.; Han, G.W.; Chatterjee, A.; Stevens, R.C.; Schultz, P.G. An orthogonal seryl-tRNA synthetase/tRNA pair for noncanonical amino acid mutagenesis in Escherichia coli. Bioorganic Med. Chem. 2020, 28, 115662. [Google Scholar] [CrossRef] [PubMed]
- Saint-Léger, A.; Bello, C.; Dans, P.D.; Torres, A.G.; Novoa, E.M.; Camacho, N.; Orozco, M.; Kondrashov, F.A.; Ribas de Pouplana, L. Saturation of recognition elements blocks evolution of new tRNA identities. Sci. Adv. 2016, 2, e1501860. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Miller, C.; Guo, L.T.; Ho, J.M.L.; Bryson, D.I.; Wang, Y.S.; Liu, D.R.; Söll, D. Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase. Nat. Chem. Biol. 2017, 13, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.C.W.; Chin, J.W. Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat. Chem. 2018, 10, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Vargas-Rodriguez, O.; Englert, M.; Tripp, H.J.; Ivanova, N.N.; Rubin, E.M.; Kyrpides, N.C.; Söll, D. Transfer RNAs with novel cloverleaf structures. Nucleic Acids Res. 2017, 45, 2776–2785. [Google Scholar] [CrossRef]
- Ohtsuki, T.; Manabe, T.; Sisido, M. Multiple incorporation of non-natural amino acids into a single protein using tRNAs with non-standard structures. FEBS Lett. 2005, 579, 6769–6774. [Google Scholar] [CrossRef]
- Mukai, T.; Englert, M.; Tripp, H.J.; Miller, C.; Ivanova, N.N.; Rubin, E.M.; Kyrpides, N.C.; Söll, D. Facile Recoding of Selenocysteine in Nature. Angew. Chem. Int. Ed. Engl. 2016, 55, 5337–5341. [Google Scholar] [CrossRef]
- Mukai, T.; Sevostyanova, A.; Suzuki, T.; Fu, X.; Söll, D. A Facile Method for Producing Selenocysteine-Containing Proteins. Angew. Chem. Int. Ed. Engl. 2018, 57, 7215–7219. [Google Scholar] [CrossRef]
- Varani, G.; McClain, W.H. The G·U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000, 1, 18–23. [Google Scholar] [CrossRef]
- Rudinger, J.; Hillenbrandt, R.; Sprinzl, M.; Giegé, R. Antideterminants present in minihelixSec hinder its recognition by prokaryotic elongation factor Tu. EMBO J. 1996, 15, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Ibba, M.; Losey, H.C.; Kawarabayasi, Y.; Kikuchi, H.; Bunjun, S.; Söll, D. Substrate recognition by class I lysyl-tRNA synthetases: A molecular basis for gene displacement. Proc. Natl. Acad. Sci. USA 1999, 96, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Crnkovic, A.; Sevostyanova, A.; Söll, D. Designing seryl-tRNA synthetase for improved serylation of selenocysteine tRNAs. FEBS Lett. 2018, 592, 3759–3768. [Google Scholar] [CrossRef]
- Naganuma, M.; Sekine, S.; Chong, Y.E.; Guo, M.; Yang, X.L.; Gamper, H.; Hou, Y.M.; Schimmel, P.; Yokoyama, S. The selective tRNA aminoacylation mechanism based on a single G•U pair. Nature 2014, 510, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Bessho, Y.; Shibata, R.; Sekine, S.; Murayama, K.; Higashijima, K.; Hori-Takemoto, C.; Shirouzu, M.; Kuramitsu, S.; Yokoyama, S. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc. Natl. Acad. Sci. USA 2007, 104, 8293–8298. [Google Scholar] [CrossRef] [PubMed]
- Lewendon, A.; Murray, I.A.; Shaw, W.V.; Gibbs, M.R.; Leslie, A.G. Evidence for transition-state stabilization by serine-148 in the catalytic mechanism of chloramphenicol acetyltransferase. Biochemistry 1990, 29, 2075–2080. [Google Scholar] [CrossRef]
- Itoh, Y.; Bröcker, M.J.; Sekine, S.; Hammond, G.; Suetsugu, S.; Söll, D.; Yokoyama, S. Decameric SelA•tRNASec ring structure reveals mechanism of bacterial selenocysteine formation. Science 2013, 340, 75–78. [Google Scholar] [CrossRef]
- Chiba, S.; Itoh, Y.; Sekine, S.; Yokoyama, S. Structural basis for the major role of O-phosphoseryl-tRNA kinase in the UGA-specific encoding of selenocysteine. Mol. Cell 2010, 39, 410–420. [Google Scholar] [CrossRef]
- Lim, F.; Peabody, D.S. RNA recognition site of PP7 coat protein. Nucleic Acids Res. 2002, 30, 4138–4144. [Google Scholar] [CrossRef]
- Nakanishi, H.; Saito, H. Caliciviral protein-based artificial translational activator for mammalian gene circuits with RNA-only delivery. Nat. Commun. 2020, 11, 1297. [Google Scholar] [CrossRef]
- Lim, F.; Peabody, D.S. Mutations that increase the affinity of a translational repressor for RNA. Nucleic Acids Res. 1994, 22, 3748–3752. [Google Scholar] [CrossRef]
- Gesnel, M.C.; Del Gatto-Konczak, F.; Breathnach, R. Combined use of MS2 and PP7 coat fusions shows that TIA-1 dominates hnRNP A1 for K-SAM exon splicing control. J. Biomed. Biotechnol. 2009, 2009, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Steer, B.A.; Schimmel, P. Major anticodon-binding region missing from an archaebacterial tRNA synthetase. J. Biol. Chem. 1999, 274, 35601–35606. [Google Scholar] [CrossRef]
- Sakamoto, K.; Hayashi, A. Synthetic Tyrosine tRNA Molecules with Noncanonical Secondary Structures. Int. J. Mol. Sci. 2018, 20, 92. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Reynolds, N.M.; Crnković, A.; Söll, D. Bioinformatic Analysis Reveals Archaeal tRNATyr and tRNATrp Identities in Bacteria. Life 2017, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Melançon, C.E., 3rd; Lee, H.S.; Groff, D.; Schultz, P.G. Evolution of amber suppressor tRNAs for efficient bacterial production of proteins containing nonnatural amino acids. Angew. Chem. Int. Ed. Engl. 2009, 48, 9148–9151. [Google Scholar] [CrossRef]
- Huang, W.; Petrosino, J.; Hirsch, M.; Shenkin, P.S.; Palzkill, T. Amino acid sequence determinants of beta-lactamase structure and activity. J. Mol. Biol. 1996, 258, 688–703. [Google Scholar] [CrossRef]
- Wang, L.; Xie, J.; Deniz, A.A.; Schultz, P.G. Unnatural amino acid mutagenesis of green fluorescent protein. J. Org. Chem. 2003, 68, 174–176. [Google Scholar] [CrossRef]
- Mukai, T.; Hoshi, H.; Ohtake, K.; Takahashi, M.; Yamaguchi, A.; Hayashi, A.; Yokoyama, S.; Sakamoto, K. Highly reproductive Escherichia coli cells with no specific assignment to the UAG codon. Sci. Rep. 2015, 5, 9699. [Google Scholar] [CrossRef]
- Tian, Q.; Wang, C.; Liu, Y.; Xie, W. Structural basis for recognition of G-1-containing tRNA by histidyl-tRNA synthetase. Nucleic Acids Res. 2015, 43, 2980–2990. [Google Scholar] [CrossRef]
- Kleanthous, C.; Cullis, P.M.; Shaw, W.V. 3-(Bromoacetyl)chloramphenicol, an active site directed inhibitor for chloramphenicol acetyltransferase. Biochemistry 1985, 24, 5307–5313. [Google Scholar] [CrossRef] [PubMed]
- Burkard, U.; Willis, I.; Söll, D. Processing of histidine transfer RNA precursors. Abnormal cleavage site for RNase P. J. Biol. Chem. 1988, 263, 2447–2451. [Google Scholar] [PubMed]
- Eichhorn, C.D.; Yang, Y.; Repeta, L.; Feigon, J. Structural basis for recognition of human 7SK long noncoding RNA by the La-related protein Larp7. Proc. Natl. Acad. Sci. USA 2018, 115, E6457–E6466. [Google Scholar] [CrossRef] [PubMed]
- Baskerville, S.; Bartel, D.P. A ribozyme that ligates RNA to protein. Proc. Natl. Acad. Sci. USA 2002, 99, 9154–9159. [Google Scholar] [CrossRef] [PubMed]
- Tawk, C.S.; Ghattas, I.R.; Smith, C.A. HK022 Nun Requires Arginine-Rich Motif Residues Distinct from λ N. J. Bacteriol. 2015, 197, 3573–3582. [Google Scholar] [CrossRef][Green Version]
- Torres-Larios, A.; Dock-Bregeon, A.C.; Romby, P.; Rees, B.; Sankaranarayanan, R.; Caillet, J.; Springer, M.; Ehresmann, C.; Ehresmann, B.; Moras, D. Structural basis of translational control by Escherichia coli threonyl tRNA synthetase. Nat. Struct. Biol. 2002, 9, 343–347. [Google Scholar] [CrossRef]
- Ardell, D.H.; Andersson, S.G. TFAM detects co-evolution of tRNA identity rules with lateral transfer of histidyl-tRNA synthetase. Nucleic Acids Res. 2006, 34, 893–904. [Google Scholar] [CrossRef]
- Yuan, J.; Gogakos, T.; Babina, A.M.; Söll, D.; Randau, L. Change of tRNA identity leads to a divergent orthogonal histidyl-tRNA synthetase/tRNAHis pair. Nucleic Acids Res. 2011, 39, 2286–2293. [Google Scholar] [CrossRef]
- Ko, J.H.; Llopis, P.M.; Heinritz, J.; Jacobs-Wagner, C.; Söll, D. Suppression of amber codons in Caulobacter crescentus by the orthogonal Escherichia coli histidyl-tRNA synthetase/tRNAHis pair. PLoS ONE 2013, 8, e83630. [Google Scholar] [CrossRef]
- Englert, M.; Vargas-Rodriguez, O.; Reynolds, N.M.; Wang, Y.S.; Söll, D.; Umehara, T. A genomically modified Escherichia coli strain carrying an orthogonal E. coli histidyl-tRNA synthetase•tRNAHis pair. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 3009–3015. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Cárdenas, C.; Bernal, L.F.; Caro-Quintero, A.; López, G.; David Alzate, J.; Gonzalez, L.N.; Restrepo, S.; Shapiro, N.; Woyke, T.; Kyrpides, N.C.; et al. Draft genome and description of Consotaella salsifontis gen. nov. sp. nov., a halophilic, free-living, nitrogen-fixing alphaproteobacterium isolated from an ancient terrestrial saline spring. Int. J. Syst. Evol. Microbiol. 2017, 67, 3744–3751. [Google Scholar]
- Augustine, J.; Francklyn, C. Design of an Active Fragment of a Class II Aminoacyl-tRNA Synthetase and Its Significance for Synthetase Evolution. Biochemistry 1997, 36, 3473–3482. [Google Scholar] [CrossRef] [PubMed]
- Iraha, F.; Oki, K.; Kobayashi, T.; Ohno, S.; Yokogawa, T.; Nishikawa, K.; Yokoyama, S.; Sakamoto, K. Functional replacement of the endogenous tyrosyl-tRNA synthetase-tRNATyr pair by the archaeal tyrosine pair in Escherichia coli for genetic code expansion. Nucleic Acids Res. 2010, 38, 3682–3691. [Google Scholar] [CrossRef] [PubMed]
- Italia, J.S.; Addy, P.S.; Wrobel, C.J.; Crawford, L.A.; Lajoie, M.J.; Zheng, Y.; Chatterjee, A. An orthogonalized platform for genetic code expansion in both bacteria and eukaryotes. Nat. Chem. Biol. 2017, 13, 446–450. [Google Scholar] [CrossRef]
- Italia, J.S.; Latour, C.; Wrobel, C.J.J.; Chatterjee, A. Resurrecting the Bacterial Tyrosyl-tRNA Synthetase/tRNA Pair for Expanding the Genetic Code of Both E. coli and Eukaryotes. Cell Chem. Biol. 2018, 25, 1304–1312.e5. [Google Scholar] [CrossRef]
- Attardi, D.G.; Tocchini-Valentini, G.P. Trans-acting RNA inhibits tRNA suppressor activity in vivo. RNA 2002, 8, 904–912. [Google Scholar] [CrossRef][Green Version]
- Cui, Z.; Wu, Y.; Mureev, S.; Alexandrov, K. Oligonucleotide-mediated tRNA sequestration enables one-pot sense codon reassignment in vitro. Nucleic Acids Res. 2018, 46, 6387–6400. [Google Scholar] [CrossRef]
- Yokogawa, T.; Kitamura, Y.; Nakamura, D.; Ohno, S.; Nishikawa, K. Optimization of the hybridization-based method for purification of thermostable tRNAs in the presence of tetraalkylammonium salts. Nucleic Acids Res. 2010, 38, e89. [Google Scholar] [CrossRef]
- Lee, K.B.; Hou, C.Y.; Kim, C.E.; Kim, D.M.; Suga, H.; Kang, T.J. Genetic Code Expansion by Degeneracy Reprogramming of Arginyl Codons. Chembiochem 2016, 17, 1198–1201. [Google Scholar] [CrossRef]
- Bilokapic, S.; Maier, T.; Ahel, D.; Gruic-Sovulj, I.; Söll, D.; Weygand-Durasevic, I.; Ban, N. Structure of the unusual seryl-tRNA synthetase reveals a distinct zinc-dependent mode of substrate recognition. EMBO J. 2006, 25, 2498–2509. [Google Scholar] [CrossRef]
- Wang, C.; Guo, Y.; Tian, Q.; Jia, Q.; Gao, Y.; Zhang, Q.; Zhou, C.; Xie, W. SerRS-tRNASec complex structures reveal mechanism of the first step in selenocysteine biosynthesis. Nucleic Acids Res. 2015, 43, 10534–10545. [Google Scholar] [PubMed]
- Giessen, T.W.; Altegoer, F.; Nebel, A.J.; Steinbach, R.M.; Bange, G.; Marahiel, M.A. A synthetic adenylation-domain-based tRNA-aminoacylation catalyst. Angew. Chem. Int. Ed. Engl. 2015, 54, 2492–2496. [Google Scholar] [CrossRef] [PubMed]
- Ishida, S.; Terasaka, N.; Katoh, T.; Suga, H. An aminoacylation ribozyme evolved from a natural tRNA-sensing T-box riboswitch. Nat. Chem. Biol. 2020, 16, 702–709. [Google Scholar] [CrossRef]
- Kuhle, B.; Chihade, J.; Schimmel, P. Relaxed sequence constraints favor mutational freedom in idiosyncratic metazoan mitochondrial tRNAs. Nat. Commun. 2020, 11, 969. [Google Scholar] [CrossRef]
- Battaglia, R.A.; Grigg, J.C.; Ke, A. Structural basis for tRNA decoding and aminoacylation sensing by T-box riboregulators. Nat. Struct. Mol. Biol. 2019, 26, 1106–1113. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).