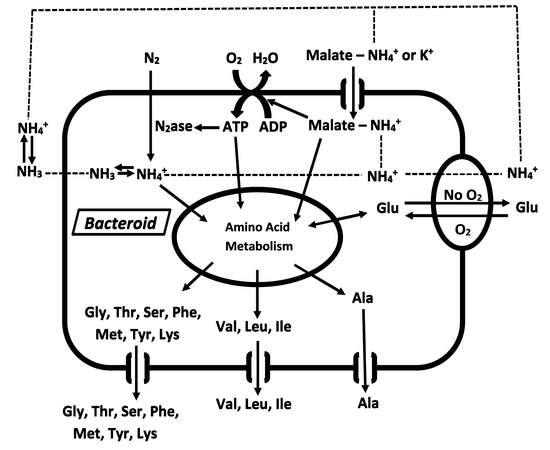
Nitrogen Assimilation and Transport by Ex Planta Nitrogen-Fixing Bradyrhizobium diazoefficiens Bacteroids Is Modulated by Oxygen, Bacteroid Density and l-Malate
Abstract
1. Introduction
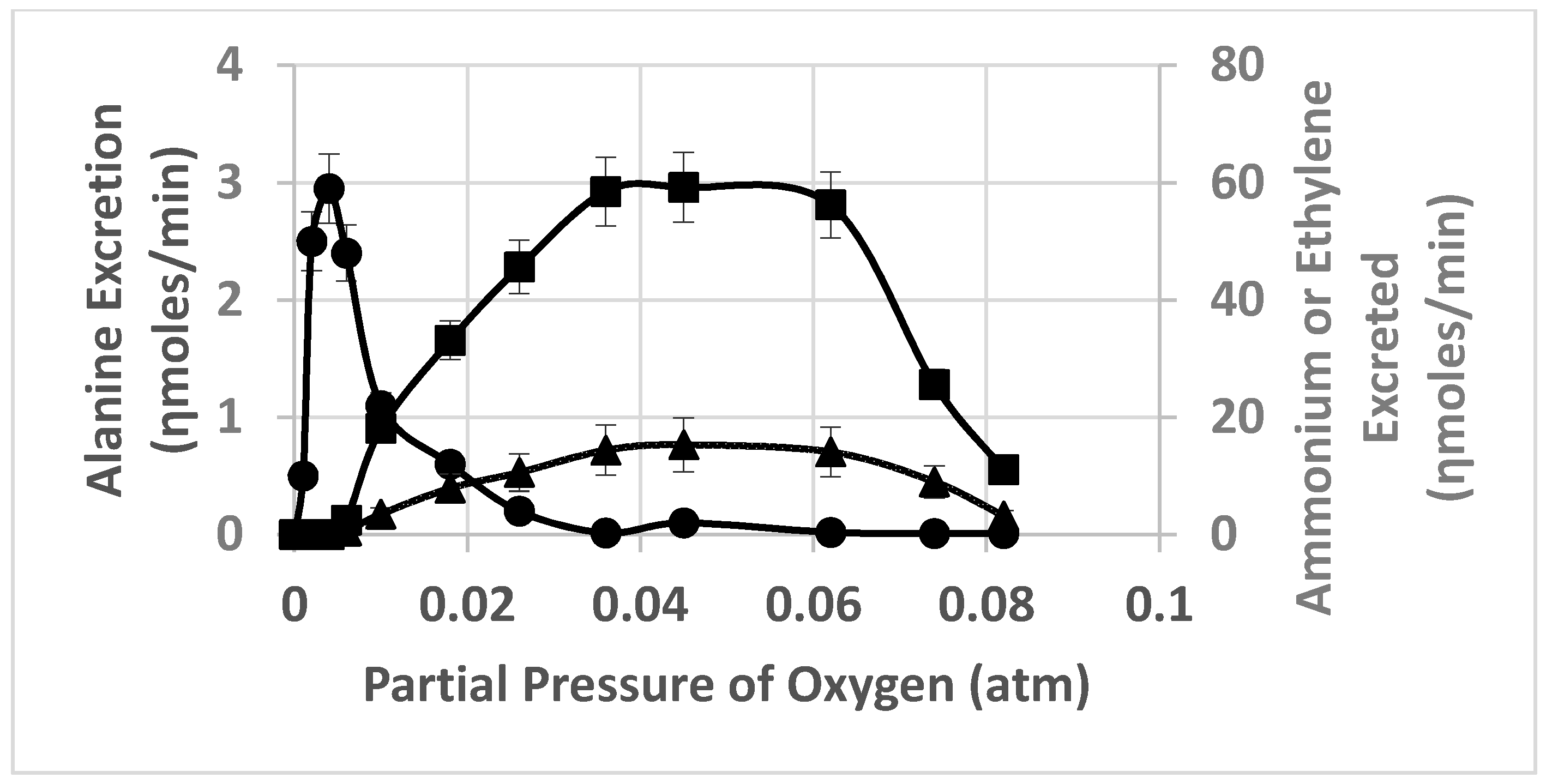
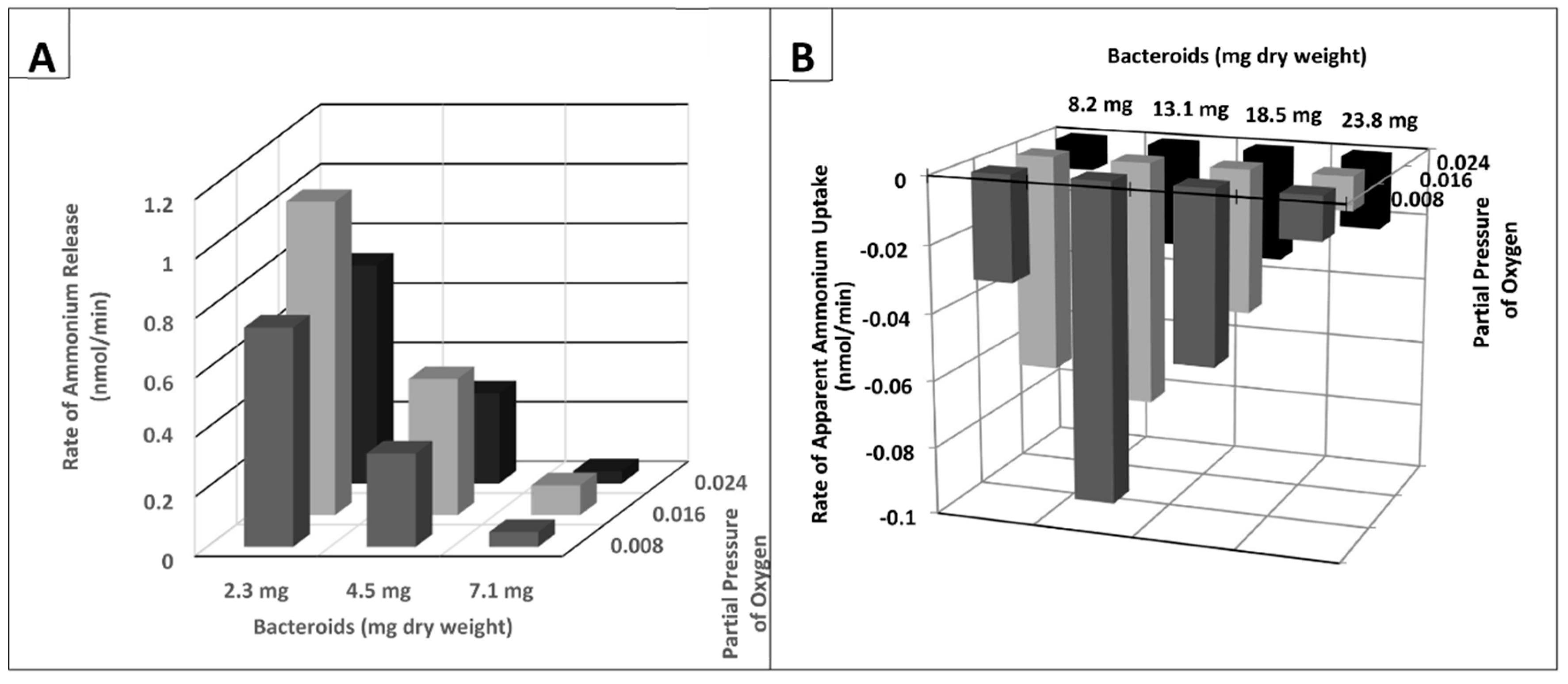
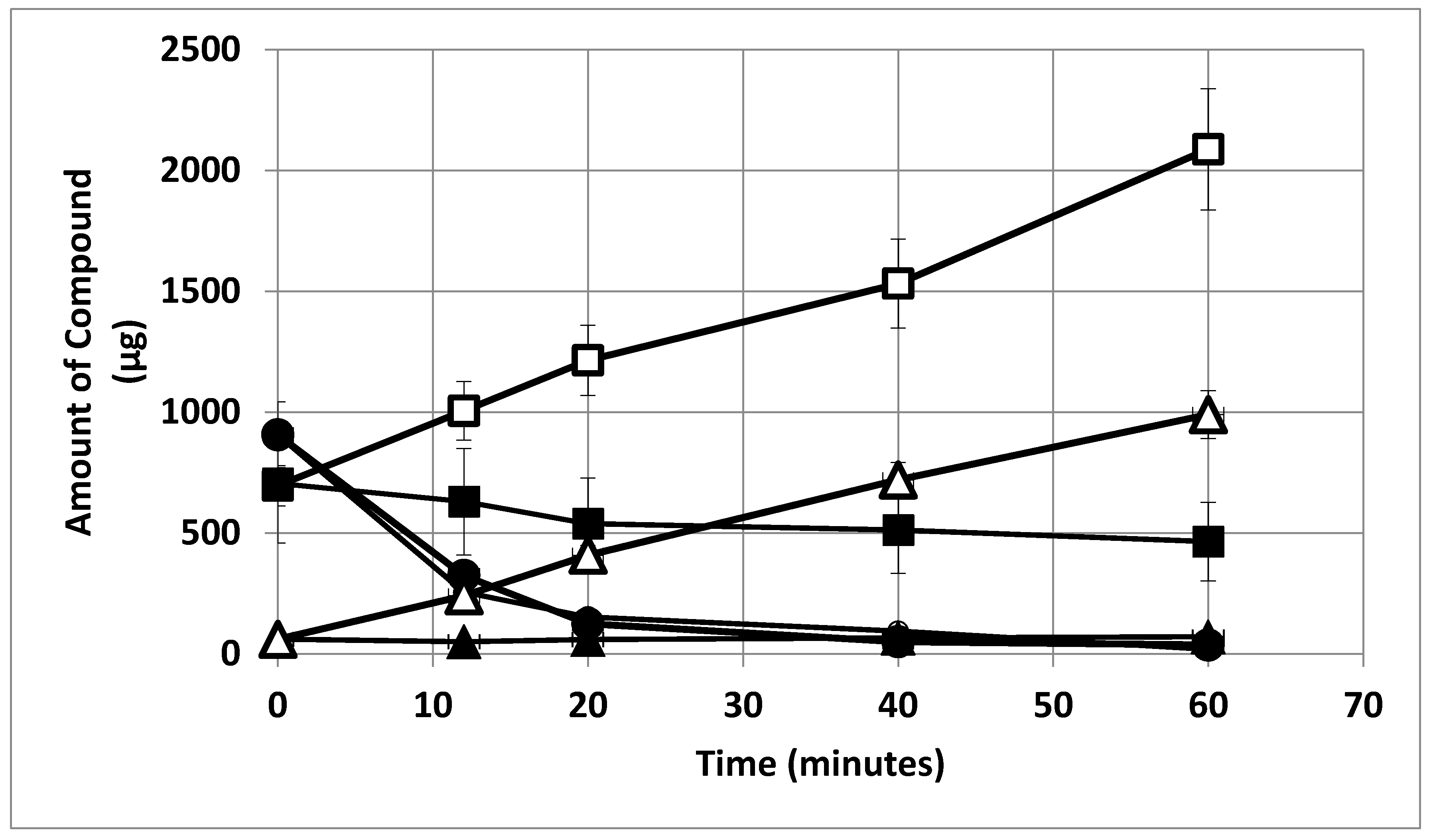
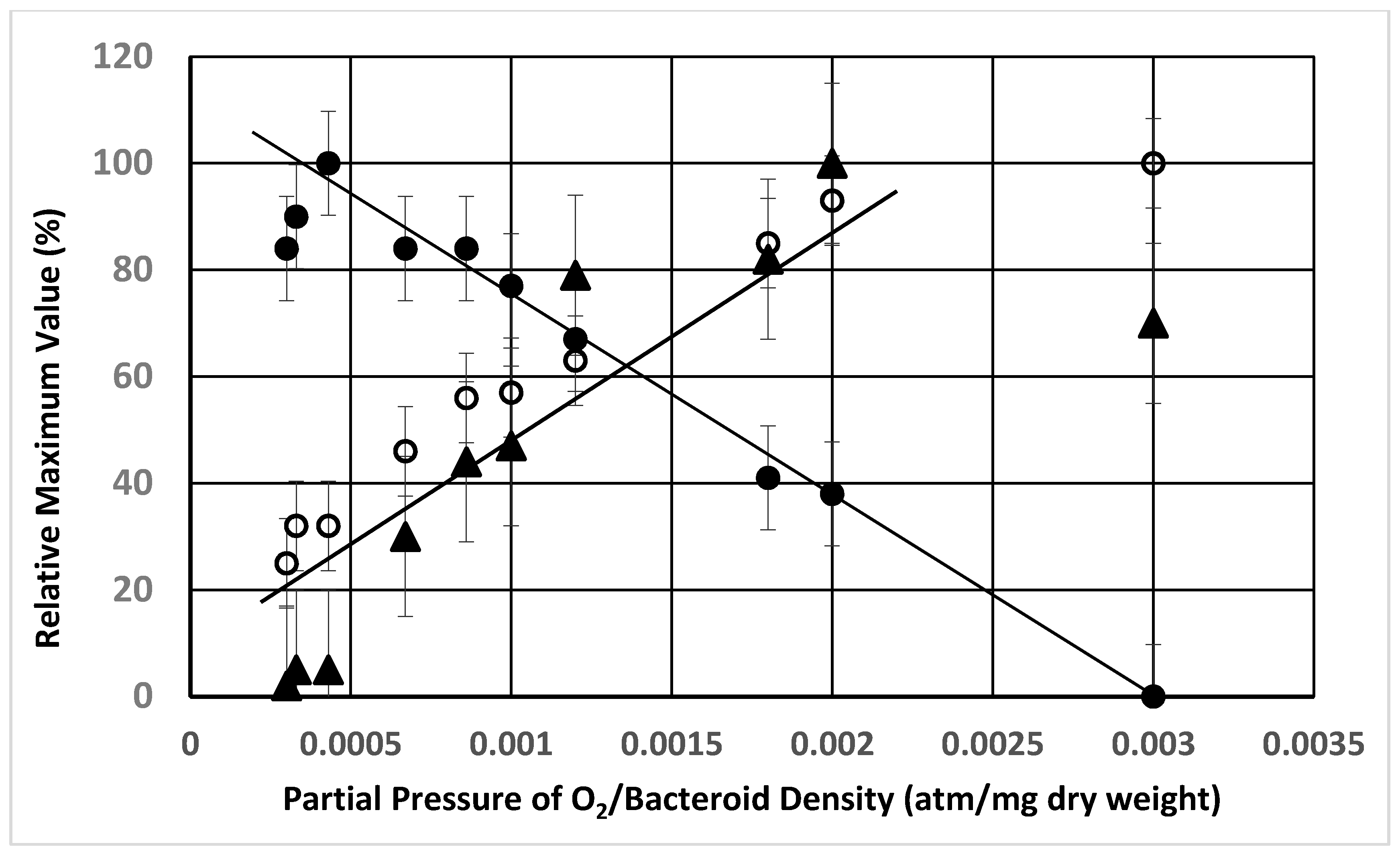
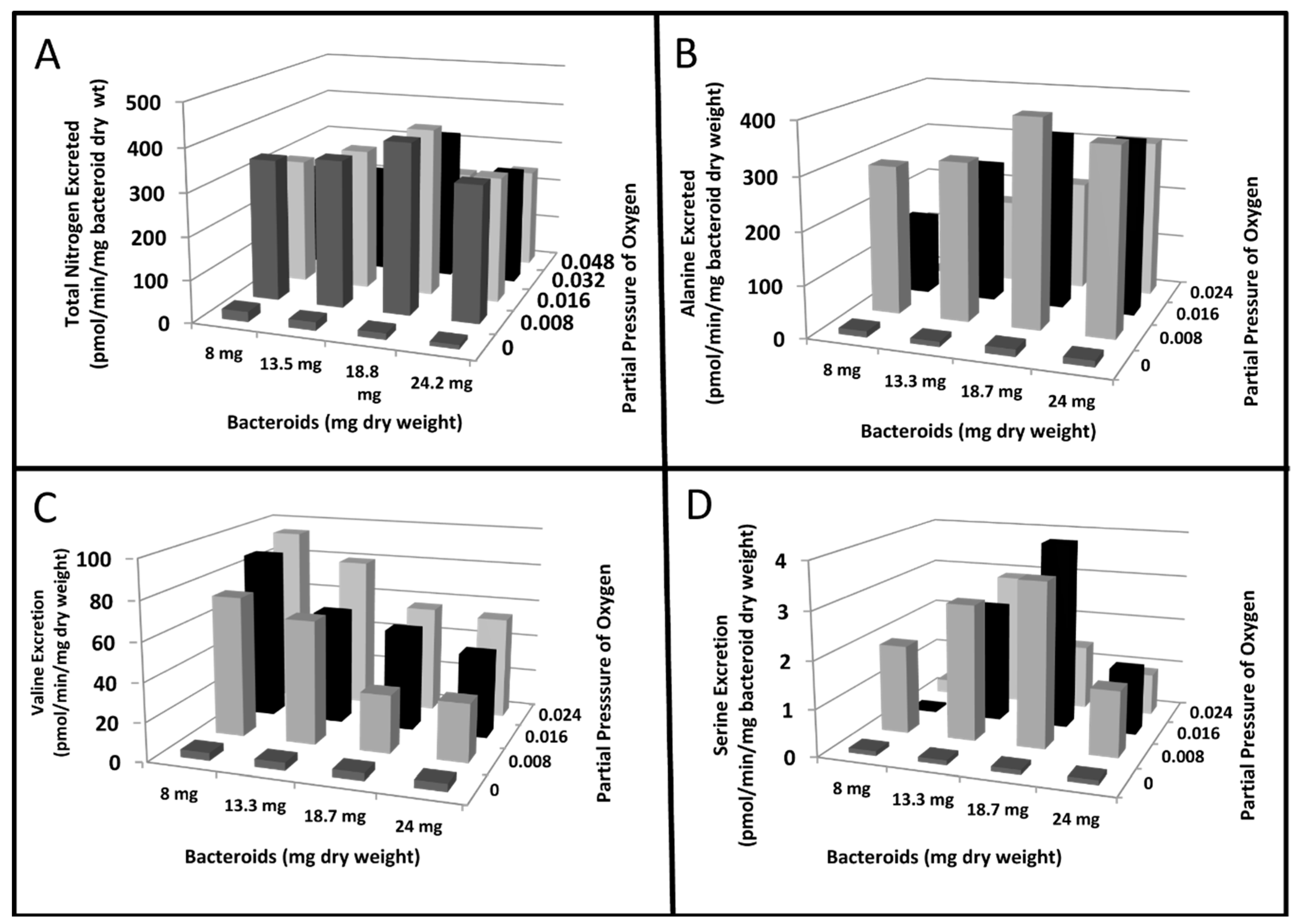
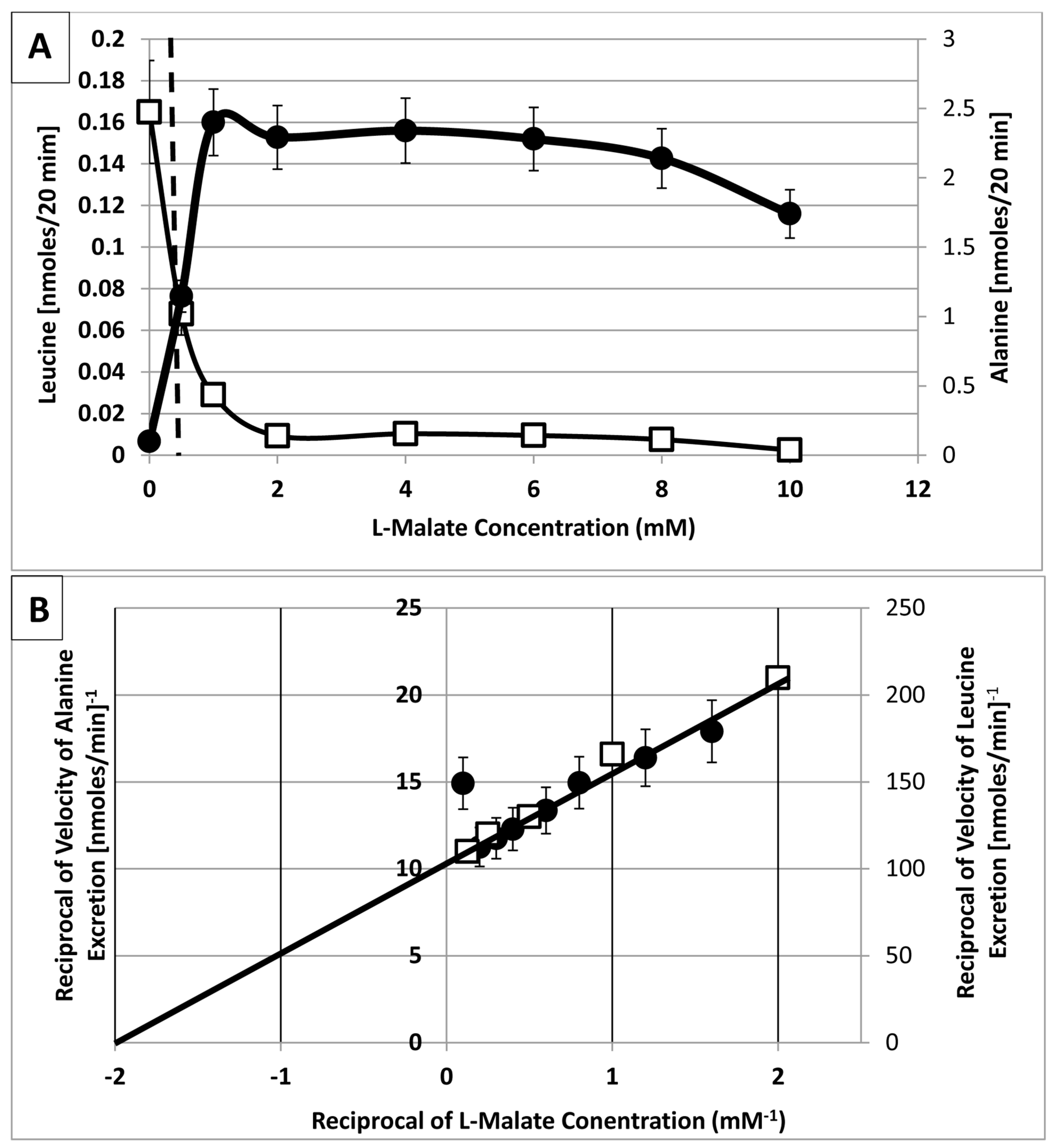
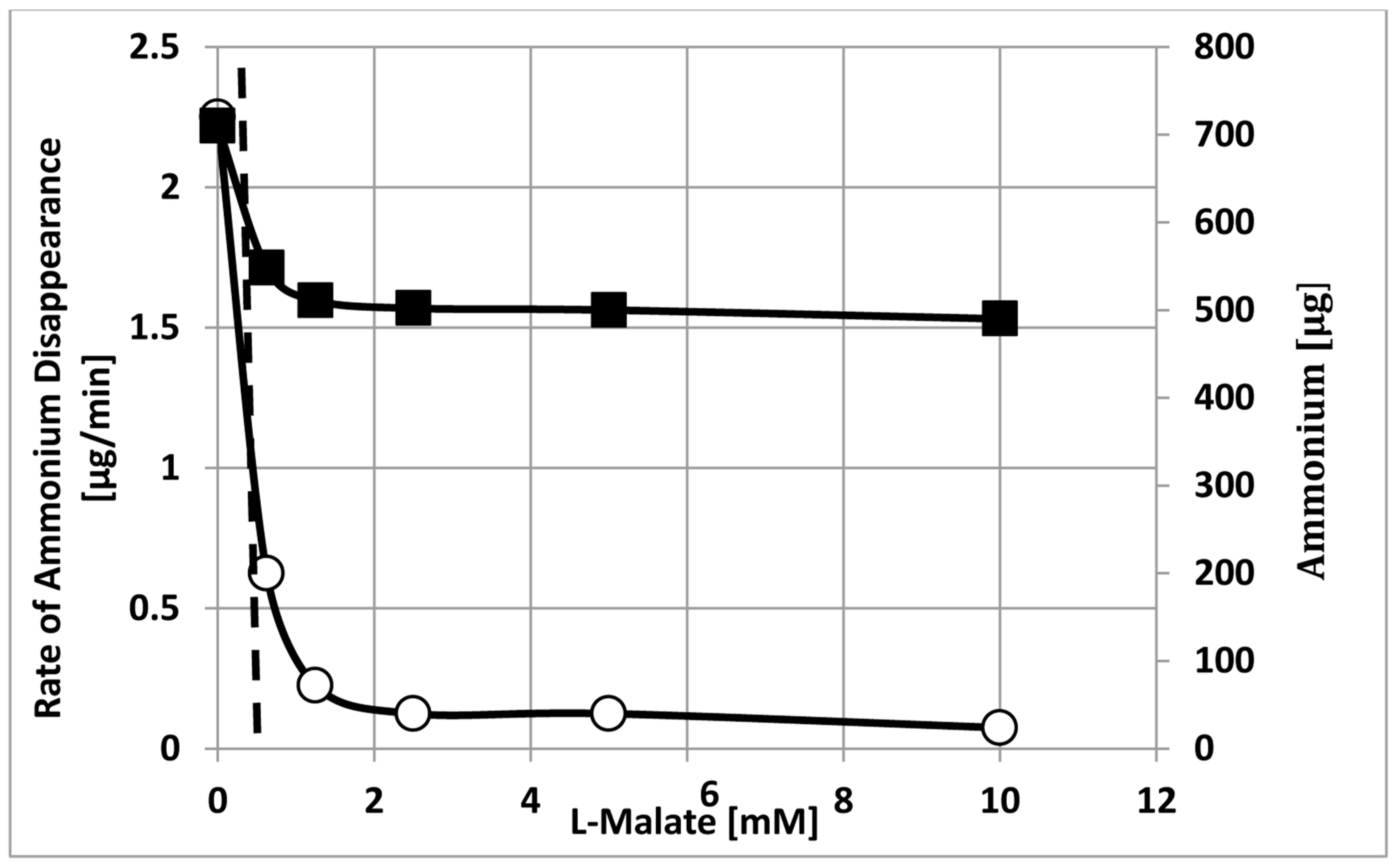
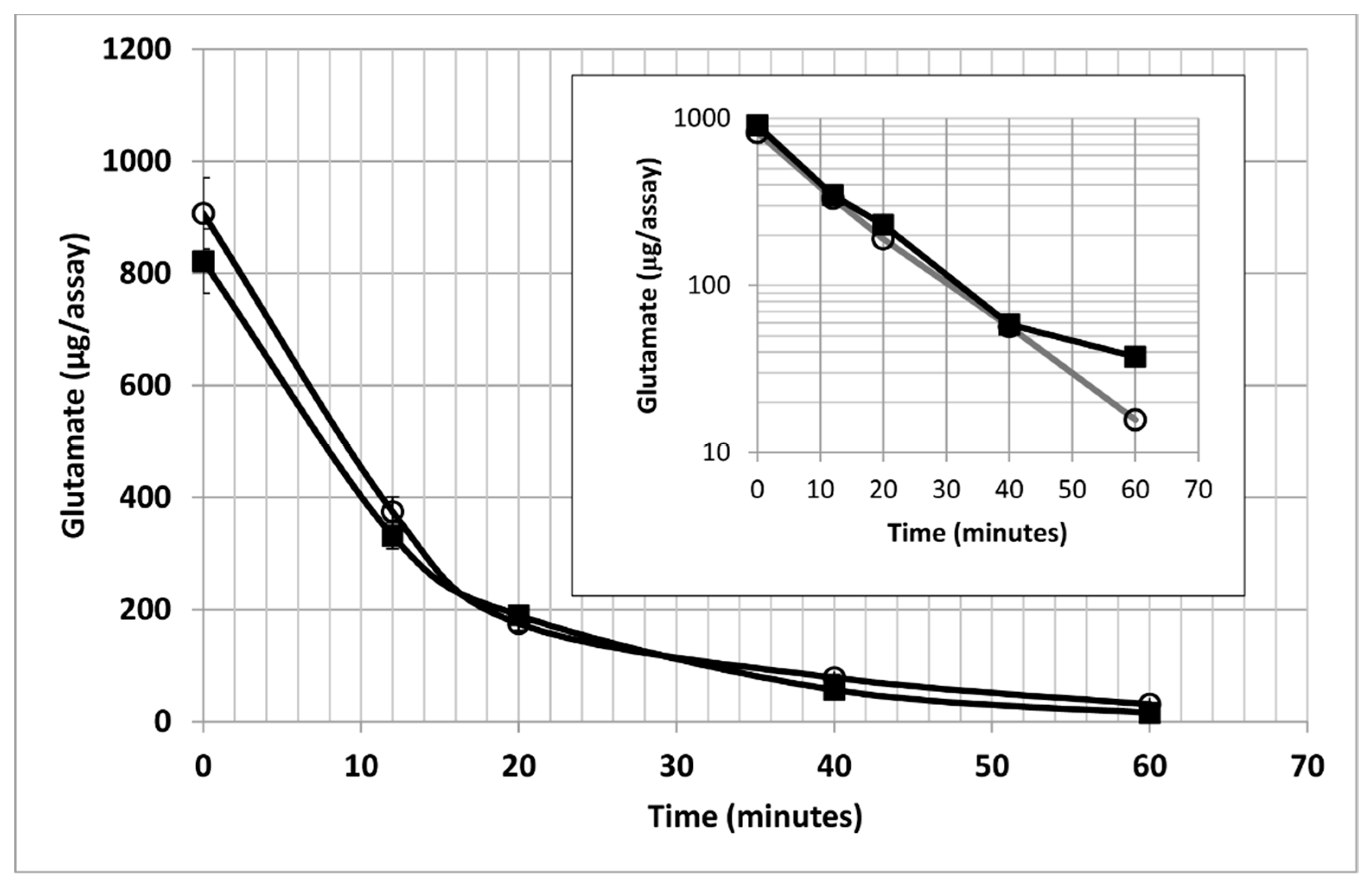
2. Results
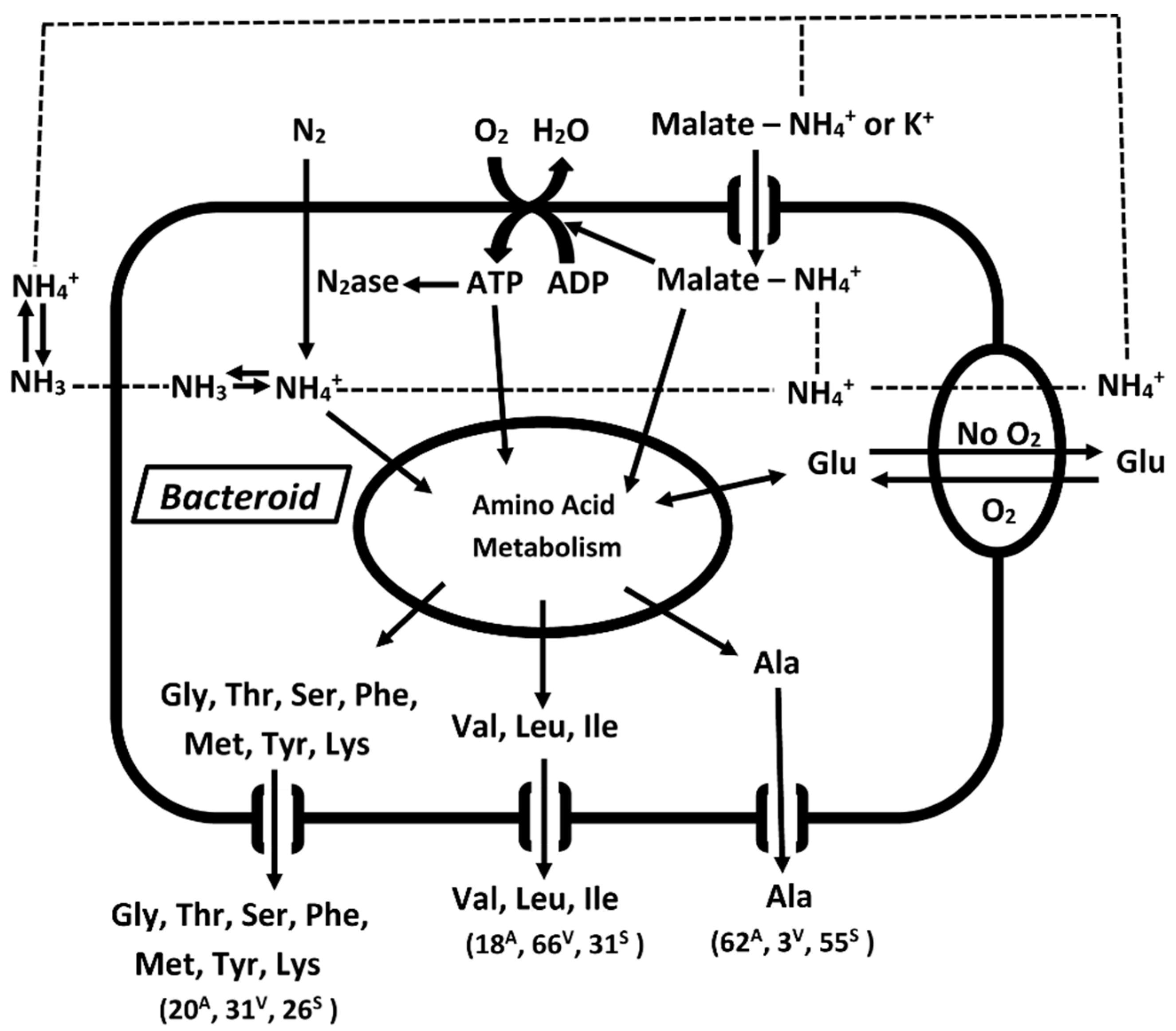
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bergersen, F.J. Root Nodules of Legumes: Structure and Functions; Bergersen, F.J., Ed.; Research Studies Press: Letchworth, UK, 1982; p. 166. [Google Scholar]
- Oldroyd, G.E.; Murray, J.; Poole, P.S.; Downie, J.A. The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Burris, R.H. Studies on the mechanism of biological nitrogen fixation. In Inorganic Nitrogen Metabolism; McElroy, H., Glass, B., Eds.; The John Hopkins Press: Baltimore, MD, USA, 1965; pp. 316–343. [Google Scholar]
- Boland, M.; Fordyce, H.; Greenwood, R. Enzymes of nitrogen metabolism in legume nodules: A comparative study. Funct. Plant. Biol. 1978, 5, 553–559. [Google Scholar] [CrossRef]
- Brown, C.M.; Dilworth, M.J. Ammonia assimilation by rhizobium cultures and bacteroids. J. Gen. Microbiol. 1975, 86, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Klucas, R.V. Studies on soybean nodule senescence. Plant. Physiol. 1974, 54, 612–616. [Google Scholar] [CrossRef] [PubMed]
- McParland, R.H.; Guevara, J.G.; Becker, R.R.; Evans, H.J. The purification and properties of the glutamine synthetase from the cytosol of Soya-bean root nodules. Biochem. J. 1976, 153, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Planqué, K.; Kennedy, I.R.; De Vries, G.E.; Quispel, A.; Van Brussel, A.A.N. Location of nitrogenase and ammonia-assimilatory enzymes in bacteroids of Rhizobium leguminosarum and Rhizobium lupini. J. Gen. Microbiol. 1977, 102, 95–104. [Google Scholar] [CrossRef]
- Robertson, J.; Farnden, K.; Warburton, M.; Banks, J. Induction of glutamine synthetase during nodule development in Lupin. Funct. Plant. Biol. 1975, 2, 265–272. [Google Scholar] [CrossRef]
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant. Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef]
- Udvardi, M.; Day, D.A. Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant. Biol. 1997, 48, 493–523. [Google Scholar] [CrossRef]
- White, J.; Prell, J.; James, E.K.; Poole, P. Nutrient sharing between symbionts. Plant. Physiol. 2007, 144, 604–614. [Google Scholar] [CrossRef]
- Aprison, M.H.; Magee, W.E.; Burris, R.H. Nitrogen fixation by excised soybean root nodules. J. Biol. Chem. 1954, 208, 29–39. [Google Scholar] [PubMed]
- Bergersen, F.J. Biochemical Pathways in Legume Root Nodule Nitrogen Fixation. Bacteriol. Rev. 1960, 24, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Bergersen, F.J. Ammonium—An early stable product of nitrogen fixation by soybean root nodules. Aust. J. Biol. Sci. 1965, 18, 1–9. [Google Scholar] [CrossRef][Green Version]
- Meeks, J.C.; Wolk, C.P.; Schilling, N.; Shaffer, P.W.; Avissar, Y.; Chien, W.-S. Initial organic products of fixation of [13N] dinitrogen by root nodules of soybean (Glycine max). Plant. Physiol. 1978, 61, 980–983. [Google Scholar] [CrossRef]
- Ohyama, T.; Kumazawa, K. Nitrogen assimilation in soybean nodules. Soil Sci. Plant. Nutr. 1980, 26, 205–213. [Google Scholar] [CrossRef]
- Kahn, M.L.; Kraus, J.; Somerville, J.E. A model of Nutrient Exchange in the Rhizobium-Legume Symbiosis. In New Horizons in Nitrogen Fixation; Springer Science and Business Media LLC: Berlin, Germany, 1985; pp. 193–199. [Google Scholar]
- Salminen, S.O.; Streeter, J.G. Factors contributing to the accumulation of glutamate in Bradyrhizobium japonicum bacteroids under microaerobic conditions. J. Gen. Microbiol. 1990, 136, 2119–2126. [Google Scholar] [CrossRef]
- Appels, M.A.; Haaker, H. Glutamate oxaloacetate transaminase in pea root nodules-participation in a malate/aspartate shuttle between plant and bacteroids. Plant Physiol. 1991, 95, 740–747. [Google Scholar] [CrossRef][Green Version]
- Rastogi, V.K.; Watson, R.J. Aspartate aminotransferase activity is required for aspartate catabolism and symbiotic nitrogen fixation in Rhizobium meliloti. J. Bacteriol. 1991, 173, 2879–2887. [Google Scholar] [CrossRef]
- Rosendahl, L.; Dilworth, M.J.; Glenn, A.R. Exchange of metabolites across the peribacteroid membrane in pea root nodules. J. Plant. Physiol. 1992, 139, 635–638. [Google Scholar] [CrossRef]
- Waters, J.K.; Hughes, B.L.; Purcell, L.C.; Gerhardt, K.O.; Mawhinney, T.P.; Emerich, D.W. Alanine, not ammonia, is excreted from N2-fixing soybean nodule bacteroids. Proc. Natl. Acad. Sci. USA 1998, 95, 12038–12042. [Google Scholar] [CrossRef]
- Allaway, D.; Lodwig, E.; Crompton, L.A.; Wood, M.; Parsons, R.; Wheeler, T.R.; Poole, P.S. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 2000, 36, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Lodwig, E.M.; Hosie, A.H.; Bourdes, A.; Findlay, K.; Allaway, D.; Karunakaran, R.; Downie, J.A.; Poole, P.S. Amino acid recycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 2003, 422, 722–726. [Google Scholar] [CrossRef] [PubMed]
- Rosendahl, L.; Mouritzen, P.; Rudbeck, A. Nitrogen transfer in the interface between the symbionts in pea root nodules. Plant. Soil 2001, 230, 31–37. [Google Scholar] [CrossRef]
- Prell, J.; Poole, P. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 2006, 14, 161–168. [Google Scholar] [CrossRef]
- Ellfolk, N.; Katunuma, N. The occurrence of ammonia-activating enzyme in various organisms. Arch. Biochem. Biophys. 1959, 81, 521–522. [Google Scholar] [CrossRef]
- Smith, M.; Emerich, D. Alanine dehydrogenase from soybean nodule bacteroids: Purification and properties. Arch. Biochem. Biophys. 1993, 304, 379–385. [Google Scholar] [CrossRef]
- Smith, M.T.; Emerich, D.W. Alanine dehydrogenase from soybean nodule bacteroids. Kinetic mechanism and pH studies. J. Biol. Chem. 1993, 268, 10746–10753. [Google Scholar]
- Kumar, S.; Bourdès, A.; Poole, P.S. De novo alanine synthesis by bacteroids of Mesorhizobium loti is not required for nitrogen transfer in the determinate nodules of Lotus corniculatus. J. Bacteriol. 2005, 187, 5493–5495. [Google Scholar] [CrossRef]
- Green, R.T.; East, A.K.; Karunakaran, R.; Downie, J.A.; Poole, P.S. Transcriptomic analysis of Rhizobium leguminosarum bacteroids in determinate and indeterminate nodules. Microb. Genom. 2019, 5, 000254. [Google Scholar] [CrossRef]
- Szczyglowski, K.; Shaw, R.S.; Wopereis, J.; Copeland, S.; Hamburger, D.; Kasiborski, B.; Dazzo, F.B.; De Bruijn, F.J. Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant. Microbe Interact. 1998, 11, 684–697. [Google Scholar] [CrossRef]
- Borisov, A.; Morzhina, E.V.; Kulikova, O.A.; Tchetkova, S.A.; Lebsky, V.K.; Tikhonovich, I.A. New symbiotic mutants of pea (Pisum sativum L.) affecting either nodule initiation or symbiosome development. Symbiosis 1992, 14, 297–313. [Google Scholar]
- Li, Y.; Parsons, R.; Day, D.A.; Bergersen, F.J. Reassessment of major products of N2 fixation by bacteroids from soybean root nodules. Microbiology 2002, 148, 1959–1966. [Google Scholar] [CrossRef] [PubMed]
- Bergersen, F.J.; Turner, G.L. Bacteroids from soybean root nodules: Accumulation of poly- β -hydroxybutyrate during supply of malate and succinate in relation to N2 fixation in flow-chamber reactions. Proc. R. Soc. London. Ser. B Biol. Sci. 1990, 240, 39–59. [Google Scholar] [CrossRef]
- Bergersen, F.; Turner, G. Nitrogen fixation by the bacteroid fraction of breis of soybean root nodules. Biochim. Biophys. Acta Gen. Subj. 1967, 141, 507–515. [Google Scholar] [CrossRef]
- Bergersen, F. Nitrogen fixation in breis of soybean root nodules. Biochim. Biophys. Acta Gen. Subj. 1966, 115, 247–249. [Google Scholar] [CrossRef]
- Udvardi, M.; Day, D.A. Ammonia (14C-methylamine) transport across the bacteroid and peribacteroid membranes of soybean root nodules. Plant. Physiol. 1990, 94, 71–76. [Google Scholar] [CrossRef]
- Dong, R.; Zhang, J.; Huan, H.; Bai, C.; Chen, Z.; Liu, G. High salt tolerance of a Bradyrhizobium strain and its promotion of the growth of Stylosanthes guianensis. Int. J. Mol. Sci. 2017, 18, 1625. [Google Scholar] [CrossRef]
- Stumpf, D.; Burris, R.H. Organic acid contents of soybean: Age and source of nitrogen. Plant. Physiol. 1981, 68, 989–991. [Google Scholar] [CrossRef]
- Segel, I.H. Enzyme Kinetics. In Encyclopedia of Biological Chemistry, 2nd ed.; Lennarz, W.J., Lane, M.D., Eds.; Academic Press: New York, NY, USA, 2013; pp. 216–220. [Google Scholar]
- Koch, B.; Evans, H.J.; Russell, S. Reduction of acetylene and nitrogen gas by breis and cell-free extracts of soybean root nodules. Plant. Physiol. 1967, 42, 466–468. [Google Scholar] [CrossRef]
- Colquhoun, D.; Hawkes, A.G. The interpretation of single channel recordings. In Microelectrode Techniques. The Plymouth Workshop Handbook, 2nd ed.; Ogden, D., Ed.; The Company of Biologists Ltd.: Cambridge, UK, 1994; pp. 141–188. [Google Scholar]
- Hamill, O.P.; Martinac, B. Molecular basis of mechanotransduction in living cells. Physiol. Rev. 2001, 81, 685–740. [Google Scholar] [CrossRef]
- Prindle, A.; Liu, J.-T.; Asally, M.; Ly, S.; Garcia-Ojalvo, J.; Süel, G.M. Ion channels enable electrical communication in bacterial communities. Nature 2015, 527, 59–63. [Google Scholar] [CrossRef] [PubMed]
- King, B.J.; Hunt, S.; Weagle, G.E.; Walsh, K.B.; Pottier, R.H.; Canvin, D.T.; Layzell, D.B. Regulation of O2 concentration in soybean nodules observed by in situ spectroscopic measurement of leghemoglobin oxygenation. Plant. Physiol. 1988, 87, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Hunt, S.; King, B.J.; Layzell, D.B. Effects of gradual increases in O2 concentration on nodule activity in soybean. Plant. Physiol. 1989, 91, 315–321. [Google Scholar] [CrossRef]
- Layzell, D.B.; Del Castillo, L.D.; Hunt, S.; Kuzma, M.; Van Cauwenberghe, O.; Oresnik, I. The regulation of oxygen and its role in regulating nodule metabolism. In New Horizons in Nitrogen Fixation; Springer Science and Business Media LLC: Berlin, Germany, 1993; pp. 393–398. [Google Scholar]
- Liu, J.-T.; Prindle, A.; Humphries, J.; Gabalda-Sagarra, M.; Asally, M.; Lee, D.-Y.D.; Ly, S.; Garcia-Ojalvo, J.; Süel, G.M. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature 2015, 523, 550–554. [Google Scholar] [CrossRef]
- Kang, Y.; Hwang, I. Glutamate uptake is important for osmoregulation and survival in the rice pathogen Burkholderia glumae. PLoS ONE 2018, 13, e0190431. [Google Scholar] [CrossRef] [PubMed]
- Toyota, M.; Spencer, D.; Sawai-Toyota, S.; Jiaqi, W.; Zhang, T.; Koo, A.J.; Howe, G.A.; Gilroy, S. Glutamate triggers long-distance, calcium-based plant defense signaling. Science 2018, 361, 1112–1115. [Google Scholar] [CrossRef]
- Forde, B.G.; Lea, P.J. Glutamate in plants: Metabolism, regulation, and signalling. J. Exp. Bot. 2007, 58, 2339–2358. [Google Scholar] [CrossRef]
- Bergersen, F.J.; Turner, G.L. Glutamate as a carbon source for N2-fixing bacteroids prepared from soybean root nodules. Microbiology 1988, 134, 2441–2448. [Google Scholar] [CrossRef][Green Version]
- Fitzmaurice, A.M.; O’Gara, F. Involvement of glutamate as a carbon source in nitrogen-fixing Rhizobium meliloti. Biochem. Soc. Trans. 1990, 18, 358–359. [Google Scholar] [CrossRef]
- Green, L.S.; Li, Y.; Emerich, D.W.; Bergersen, F.J.; Day, D.A.; Khlebnikov, A.; Risa, Ø.; Skaug, T.; Carrier, T.A.; Keasling, J. Catabolism of α-ketoglutarate by a sucA mutant of Bradyrhizobium japonicum: Evidence for an alternative tricarboxylic acid cycle. J. Bacteriol. 2000, 182, 2838–2844. [Google Scholar] [CrossRef]
- Salminen, O.; Streeter, S.J.G. Involvement of glutamate in the respiratory metabolism of Bradyrhizobium japonicum bacteroids. J. Bacteriol. 1987, 169, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Udvardi, M.K. Transport of L-glutamate across the bacteroid membrane but not the peribacteroid membrane from soybean root nodules. Mol. Plant.-Microbe Interact. 1988, 1, 250. [Google Scholar] [CrossRef]
- Tyerman, S.D.; Whitehead, L.F.; Day, D.A. A channel-like transporter for NH4+ on the symbiotic interface of N2-fixing plants. Nature 1995, 378, 629–632. [Google Scholar] [CrossRef]
- Streeter, J.G. Estimation of ammonium concentration in the cytosol of soybean nodules. Plant. Physiol. 1989, 90, 779–782. [Google Scholar] [CrossRef]
- Waters, J.K.; Karr, D.B.; Emerich, D.W. Malate dehydrogenase from Rhizobium japonicum 3IIb-143 bacteroids and Glycine max. root nodule mitochondria. Biochemistry 1985, 24, 6479–6486. [Google Scholar] [CrossRef]
- Preston, G.G.; Wall, J.D.; Emerich, D.W. Acetyl-CoA synthetase from Bradyrhizobium japonicum. Purification and partial characterization. Biochem. J. 1990, 267, 179–183. [Google Scholar] [CrossRef]
- Suzuki, F.; Zahler, W.; Emerich, D.W. Acetoacetyl-CoA thiolases of Bradyrhizobium japonicum bacteroids. Purification and properties. Arch. Biochem. Biophys. 1987, 254, 272–281. [Google Scholar] [CrossRef]
- Catanese, A.C.; Emerich, D.W.; Zahler, W.L. Adenylate cyclase and cyclic AMP phosphodiesterase in Bradyrhizobium japonicum bacteroids. J. Bacteriol. 1989, 171, 4531–4536. [Google Scholar] [CrossRef]
- Klucas, R.V.; Arp, D. Physiological and biochemical studies on senescing tap root nodules of soybeans. Can. J. Microbiol. 1977, 23, 1426–1432. [Google Scholar] [CrossRef]
- Pladys, D.; Rigaud, J. Acid protease and leghemoglobin digestion in French Bean nodules. Z. Für Pflanzenphysiol. 1982, 108, 163–169. [Google Scholar] [CrossRef]
- Janausch, I.G.; Zientz, E.; Tran, Q.H.; Kröger, A.; Unden, G. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta Bioenerg. 2002, 1553, 39–56. [Google Scholar] [CrossRef]
- Yurgel, S.N.; Kahn, M.L. Dicarboxylate transport by rhizobia. FEMS Microbiol. Rev. 2004, 28, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Howitt, S.M.; Udvardi, M. Structure, function and regulation of ammonium transporters in plants. Biochim. Biophys. Acta Biomembr. 2000, 1465, 152–170. [Google Scholar] [CrossRef]
- Sangakkara, U.R.; Hartwig, U.A. Soil moisture and potassium affect the performance of symbiotic nitrogen fixation in faba bean and common bean. Plant. Soil 1996, 184, 123–130. [Google Scholar] [CrossRef]
- Mengel, K.; Haghparast, M.-R.; Koch, K. The effect of potassium on the fixation of molecular nitrogen by root nodules of Vicia faba. Plant. Physiol. 1974, 54, 535–538. [Google Scholar] [CrossRef]
- Suelter, C.H. Enzymes activated by monovalent cations. Science 1970, 168, 789–795. [Google Scholar] [CrossRef]
- Werbeck, N.D.; Kirkpatrick, J.; Reinstein, J.; Hansen, D.F. Using 15N-ammonium to characterize and map potassium binding sites in proteins by NMR spectroscopy. Chembiochem 2014, 15, 543–548. [Google Scholar] [CrossRef]
- Hoelzle, I.; Streeter, J.G. Stimulation of α-glucosidases from fast-growing rhizobia and Agrobacterium tumefaciens by K+, NH+4, and Rb+. Can. J. Microbiol. 1990, 36, 223–227. [Google Scholar] [CrossRef]
- Buurman, E.T.; Pennock, J.; Tempest, D.W.; De Mattos, M.J.T.; Neijssel, O.M. Replacement of potassium ions by ammonium ions in different micro-organisms grown in potassium-limited chemostat culture. Arch. Microbiol. 1989, 152, 58–63. [Google Scholar] [CrossRef]
- Buurman, E.; De Mattos, M.J.T.; Neijssel, O. Futile cycling of ammonium ions via the high affinity potassium uptake system (Kdp) of Escherichia coli. Arch. Microbiol. 1991, 155, 391–395. [Google Scholar] [CrossRef]
- Martinelle, K.; Häggström, L. Mechanisms of ammonia and ammonium ion toxicity in animal cells: Transport across cell membranes. J. Biotechnol. 1993, 30, 339–350. [Google Scholar] [CrossRef]
- Kaschuk, G.; Kuyper, T.W.; Leffelaar, P.; Hungria, M.; Giller, K.E. Are the rates of photosynthesis stimulated by the carbon sink strength of rhizobial and arbuscular mycorrhizal symbioses? Soil Biol. Biochem. 2009, 41, 1233–1244. [Google Scholar] [CrossRef]
- Kaschuk, G.; Leffelaar, P.A.; Giller, K.E.; Alberton, O.; Hungria, M.; Kuyper, T.W. Responses of grain legumes to rhizobia and arbuscular mycorrhizal fungi: A meta-analysis of potential photosynthate limitation of symbioses. Soil Biol. Biochem. 2010, 42, 125–127. [Google Scholar] [CrossRef]
- Neves, M.C.P.; Hungria, M.; Sprent, J.I. The physiology of nitrogen fixation in tropical grain legumes. Crit. Rev. Plant. Sci. 1987, 6, 267–321. [Google Scholar] [CrossRef]
- Minchin, F.R.; Witty, J.F. 2005. Respiration/carbon costs of symbiotic nitrogen fixation in legumes. In Plant Respiration. Advances in Photosynthesis and Respiration; Lambers, H., Ribas-Carbo, M., Eds.; Springer: Dordrecht, Germany, 2005; Volume 18, pp. 195–205. [Google Scholar]
- Karr, D.B.; Emerich, D.W. Uniformity of the Microsymbiont Population from Soybean Nodules with Respect to Buoyant Density. Plant. Physiol. 1988, 86, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Karr, D.B.; Suzuki, F.; Waters, J.K.; Emerich, D.W. Further Evidence for the Uniformity of the Microsymbiont Population from Soybean Nodules. J. Plant. Physiol. 1990, 136, 659–663. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waters, J.K.; Mawhinney, T.P.; Emerich, D.W. Nitrogen Assimilation and Transport by Ex Planta Nitrogen-Fixing Bradyrhizobium diazoefficiens Bacteroids Is Modulated by Oxygen, Bacteroid Density and l-Malate. Int. J. Mol. Sci. 2020, 21, 7542. https://doi.org/10.3390/ijms21207542
Waters JK, Mawhinney TP, Emerich DW. Nitrogen Assimilation and Transport by Ex Planta Nitrogen-Fixing Bradyrhizobium diazoefficiens Bacteroids Is Modulated by Oxygen, Bacteroid Density and l-Malate. International Journal of Molecular Sciences. 2020; 21(20):7542. https://doi.org/10.3390/ijms21207542
Chicago/Turabian StyleWaters, James K., Thomas P. Mawhinney, and David W. Emerich. 2020. "Nitrogen Assimilation and Transport by Ex Planta Nitrogen-Fixing Bradyrhizobium diazoefficiens Bacteroids Is Modulated by Oxygen, Bacteroid Density and l-Malate" International Journal of Molecular Sciences 21, no. 20: 7542. https://doi.org/10.3390/ijms21207542
APA StyleWaters, J. K., Mawhinney, T. P., & Emerich, D. W. (2020). Nitrogen Assimilation and Transport by Ex Planta Nitrogen-Fixing Bradyrhizobium diazoefficiens Bacteroids Is Modulated by Oxygen, Bacteroid Density and l-Malate. International Journal of Molecular Sciences, 21(20), 7542. https://doi.org/10.3390/ijms21207542