Transcription Factor CaSBP12 Negatively Regulates Salt Stress Tolerance in Pepper (Capsicum annuum L.)
Abstract
1. Introduction
2. Results
2.1. Expression of the CaSBP12 Gene in Pepper under Salt Stress
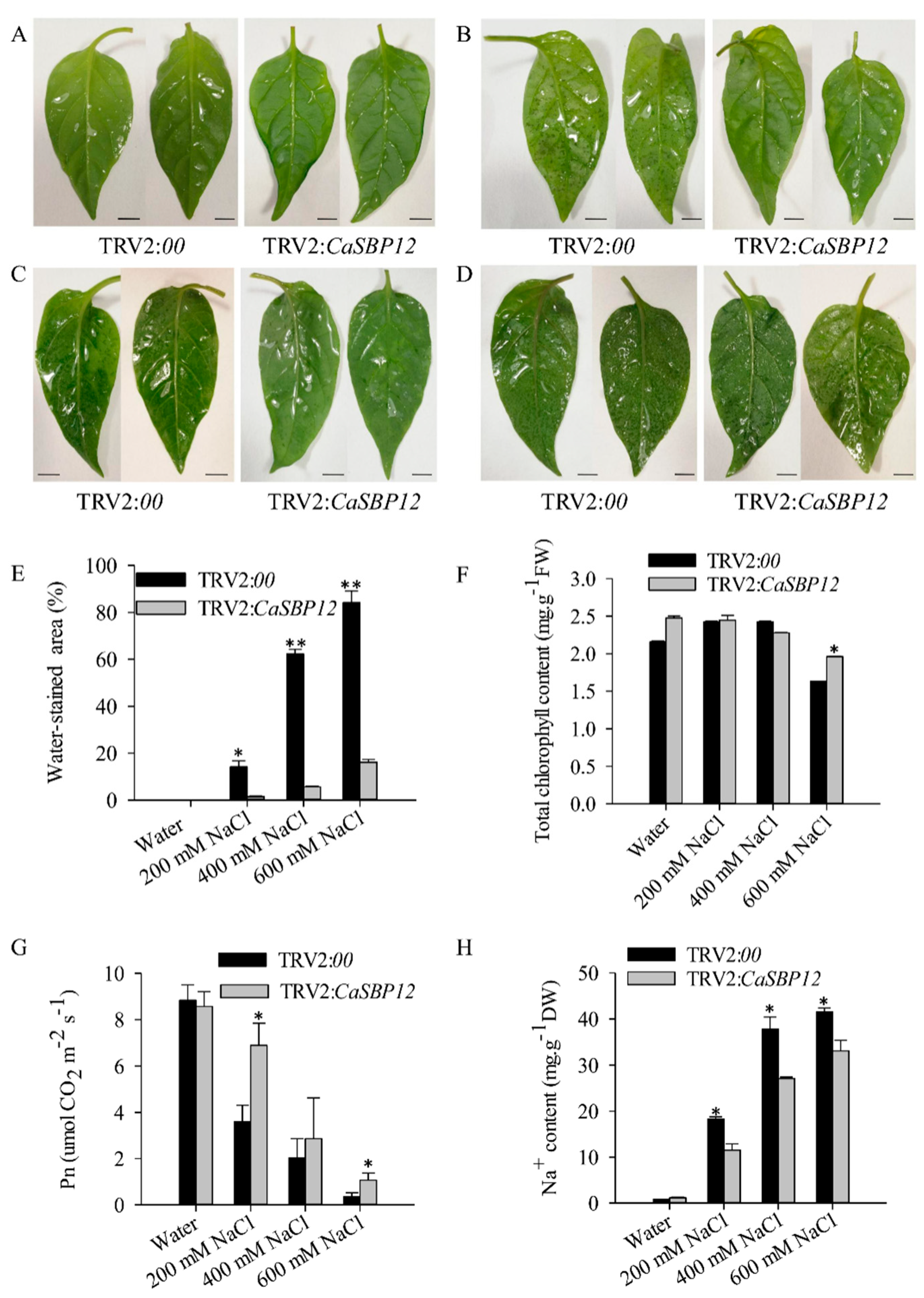
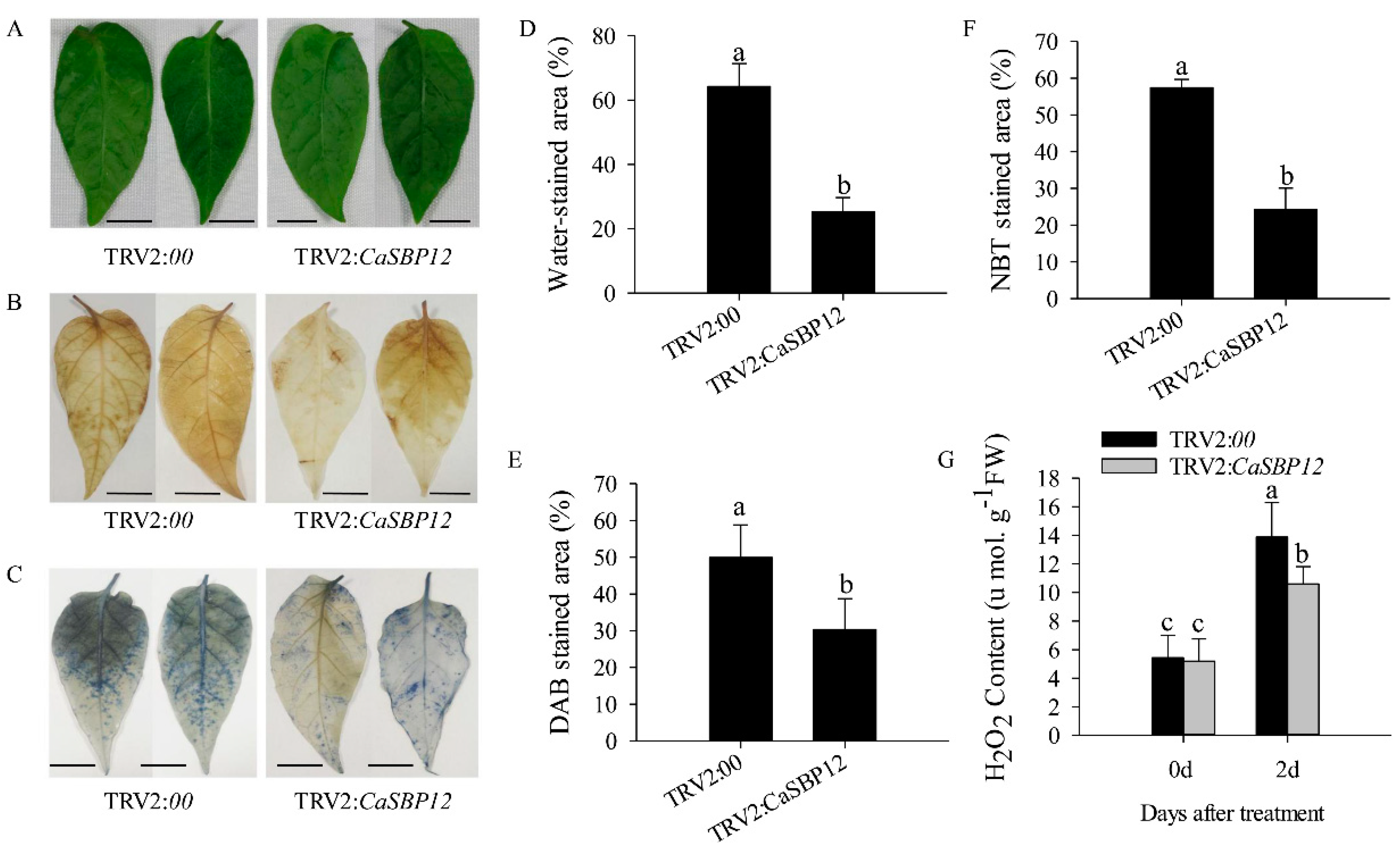
2.2. Silencing the CaSBP12 Gene Enhanced Pepper Plant Tolerance to Salt Stress
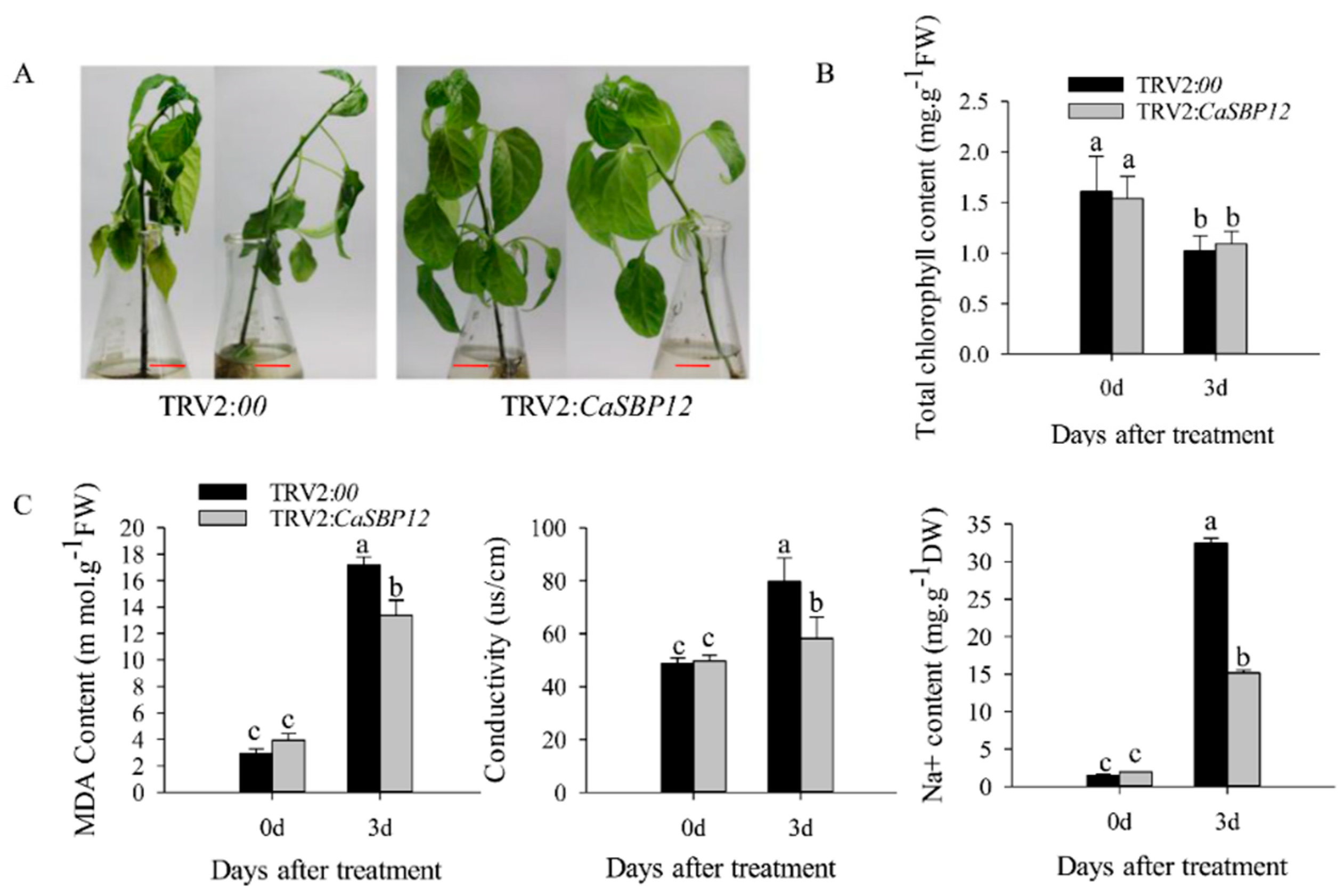
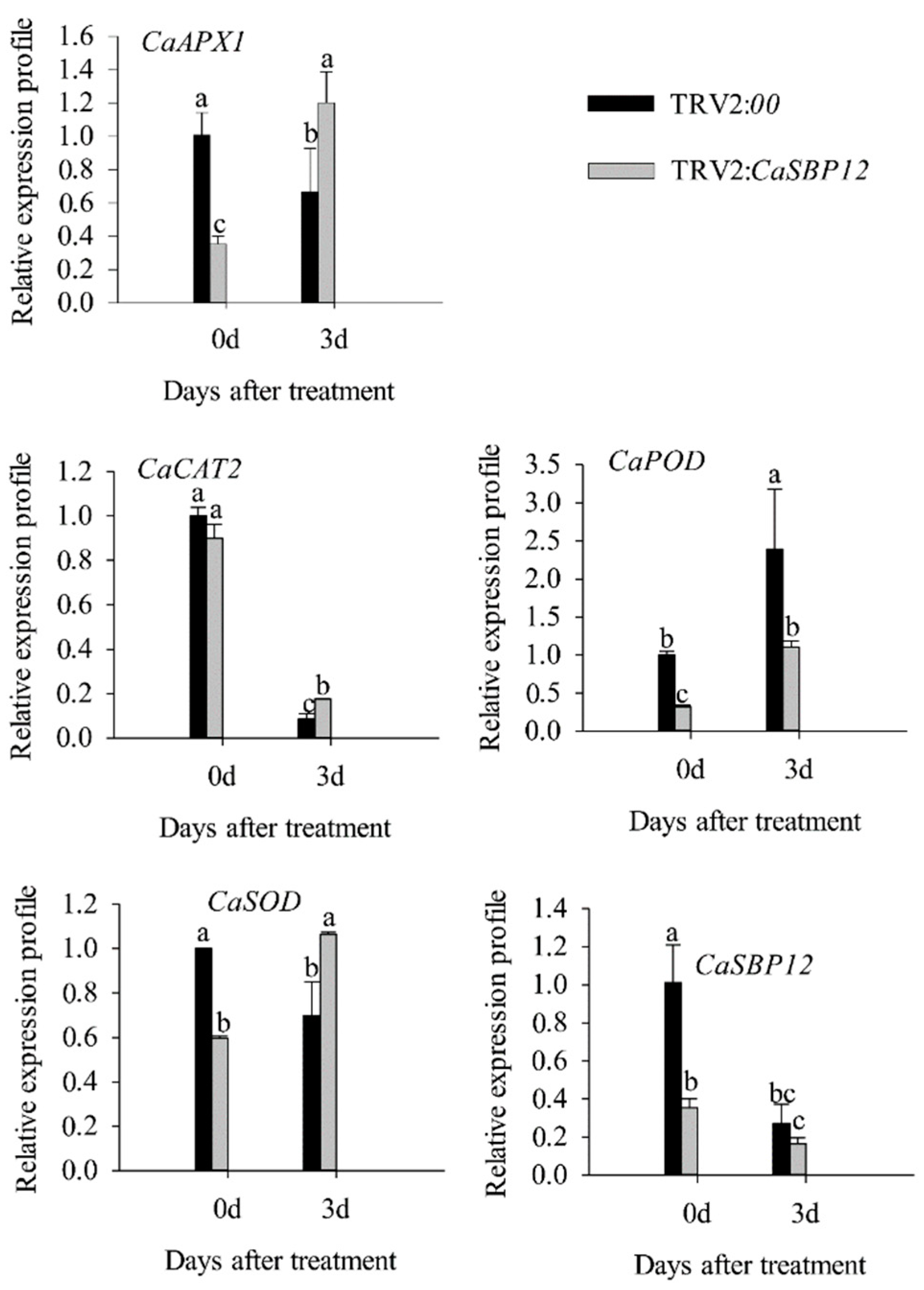
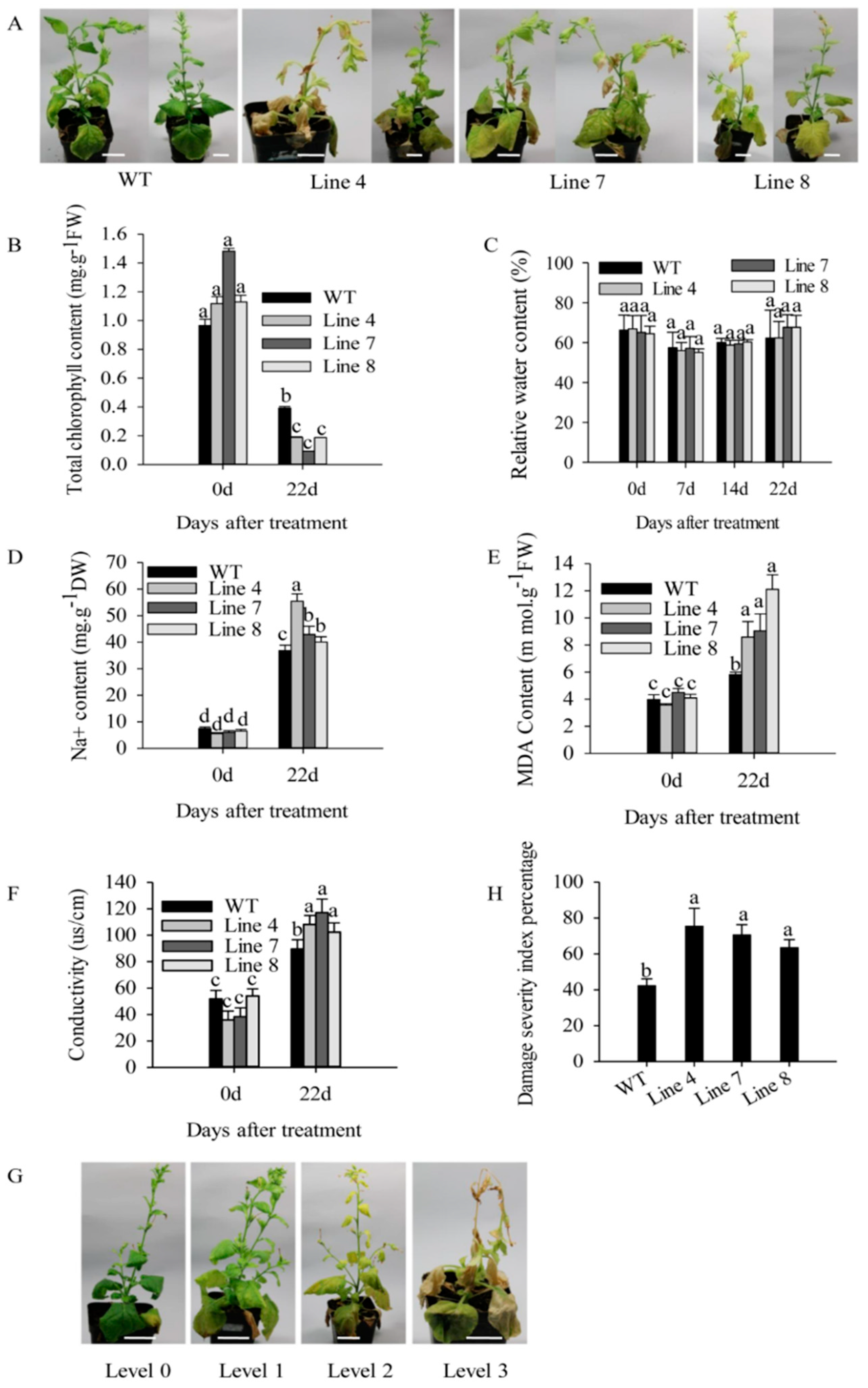
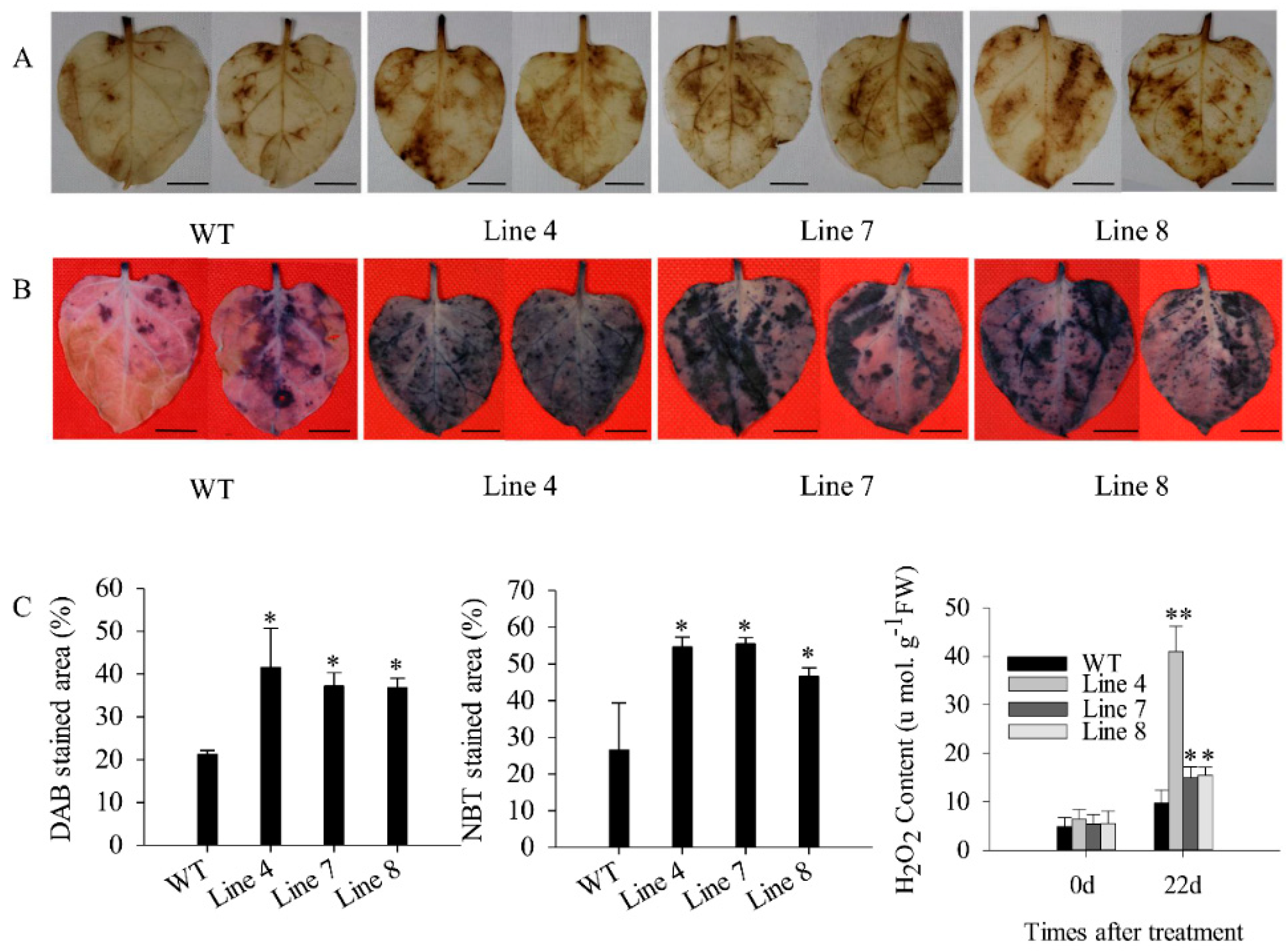
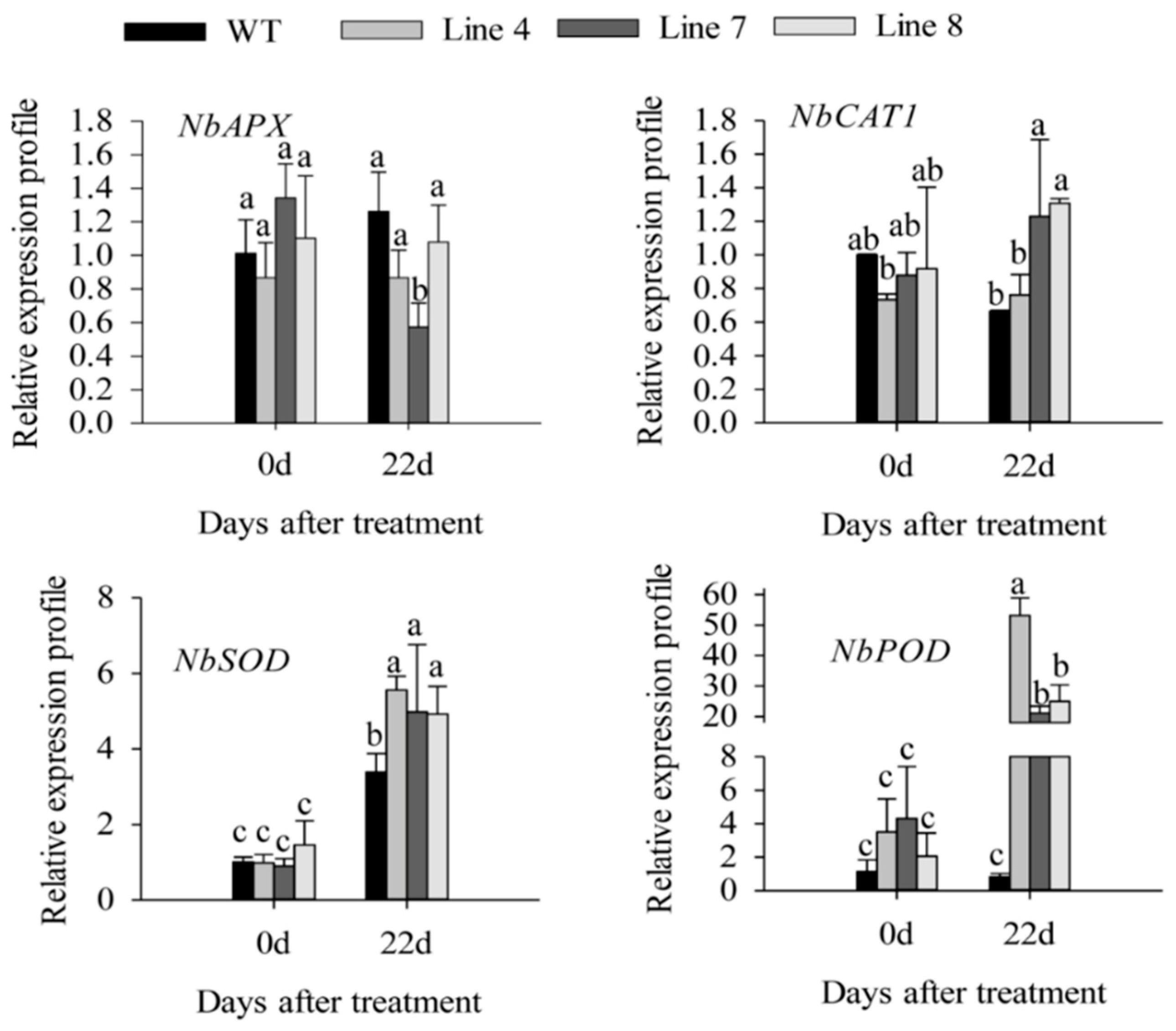
2.3. Overexpression of CaSBP12 in Nicotiana Benthamiana Enhanced Susceptibility to Salt Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Virus-Induced Gene Silencing (VIGS) of CaSBP12 Gene in Pepper
4.3. Nicotiana Benthamiana Transformation
4.4. Stress Treatments and Samples Collection
4.5. RNA Extraction and Quantitative Real-Time PCR (qPCR)
4.6. Measurement of Physiological Indicators
4.7. Damage Severity Index Percentage Statistics
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Feng, X.H.; Zhang, H.X.; Ali, M.; Gai, W.X.; Cheng, G.X.; Yu, Q.H.; Yang, S.B.; Li, X.X.; Gong, Z.H. A small heat shock protein CaHsp25.9 positively regulates heat, salt, and drought stress tolerance in pepper (Capsicum annuum L.). Plant Physiol. Biochem. 2019, 142, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Huang, W.; Xian, Z.; Hu, N.; Lin, D.; Ren, H.; Chen, J.; Su, D.; Li, Z. Overexpression of SlGRAS40 in tomato enhances tolerance to abiotic stresses and influences auxin and gibberellin signaling. Front. Plant Sci. 2017, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ngoc, T.; Le, N.; Thia, H.; Sao, D.; Nguyen, M.; Tuan, V. Overexpression of NbWRKY79 enhances salt stress tolerance in Nicotiana benthamiana. Acta Physiol. Plant 2017, 39, 121. [Google Scholar]
- Hou, H.; Jia, H.; Yan, Q.; Wang, X. Overexpression of a SBP-box gene (VpSBP16) from chinese wild vitis species in Arabidopsis improves salinity and drought stress tolerance. Int. J. Mol. Sci. 2018, 19, 940. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef]
- Singh, K.B.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Yang, O.; Popova, O.V.; Süthoff, U.; Lüking, I.; Dietz, K.J.; Golldack, D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 2009, 436, 45–55. [Google Scholar] [CrossRef]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef]
- Chen, H.; Lai, Z.; Shi, J.; Xiao, Y.; Chen, Z.; Xu, X. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010, 10, 281. [Google Scholar] [CrossRef]
- Ding, Z.; Li, S.; An, X.; Liu, X.; Qin, H.; Wang, D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genom. 2009, 36, 17–29. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K. Transcriptional activators in abscisic acid signaling. Society 2003, 15, 63–78. [Google Scholar]
- Hsieh, T.H.; Li, C.W.; Su, R.C.; Cheng, C.P.; Tsai, Y.C.; Chan, M.T. A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 2010, 231, 1459–1473. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar] [PubMed]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E.; et al. A Novel Zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef]
- Zhang, H.-X.; Jin, J.-H.; He, Y.-M.; Lu, B.-Y.; Li, D.-W.; Chai, W.-G.; Khan, A.; Gong, Z.-H. Genome-wide identification and analysis of the SBP-Box family genes under Phytophthora capsici stress in pepper (Capsicum annuum L.). Front. Plant Sci. 2016, 7, 1–14. [Google Scholar] [CrossRef]
- Jung, J.H.; Ju, Y.; Seo, P.J.; Lee, J.H.; Park, C.M. The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 2012, 69, 577–588. [Google Scholar] [CrossRef]
- Yang, R.; Li, P.; Mei, H.; Wang, D.; Sun, J.; Yang, C.; Hao, L.; Cao, S.; Chu, C.; Hu, S.; et al. Fine-tuning of MiR528 accumulation modulates flowering time in rice. Mol. Plant 2019, 12, 1103–1113. [Google Scholar] [CrossRef]
- Stone, J.M.; Liang, X.; Nekl, E.R.; Stiers, J.J. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005, 41, 744–754. [Google Scholar] [CrossRef]
- Zhang, H.; Ali, M.; Feng, X.; Jin, J.; Huang, L.; Khan, A.; Lv, J.; Gao, S.; Luo, D.; Gong, Z. A novel transcription factor CaSBP12 gene negatively regulates the defense response against Phytophthora capsici in pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2018, 20, 48. [Google Scholar] [CrossRef]
- Yao, S.; Yang, Z.; Yang, R.; Huang, Y.; Guo, G.; Kong, X.; Lan, Y.; Zhou, T.; Wang, H.; Wang, W.; et al. Transcriptional regulation of miR528 by OsSPL9 orchestrates antiviral response in rice. Mol. Plant 2019, 12, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Zheng, Y.; Su, Z.; Yu, S.; Song, H.; Zheng, X.; Lin, G.; Wu, W. OsSPL10, a SBP-Box gene, plays a dual role in salt tolerance and trichome formation in rice (Oryza sativa L.). Genes Genomes Genet. 2019, 9, 4107–4114. [Google Scholar] [CrossRef] [PubMed]
- Ning, K.; Chen, S.; Huang, H.; Jiang, J.; Yuan, H.; Li, H. Molecular characterization and expression analysis of the SPL gene family with BpSPL9 transgenic lines found to confer tolerance to abiotic stress in Betula platyphylla Suk. Plant Cell. Tissue Organ. Cult. 2017, 130, 469–481. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Ansariola, M.; Fraser, V.N.; Megraw, M. Identification of transcription factors from NF-Y, NAC, and SPL families responding to osmotic stress in multiple tomato varieties. Plant Sci. 2018, 274, 441–450. [Google Scholar] [CrossRef]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.R.; Bäurle, I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef]
- Cui, N.; Sun, X.; Sun, M.; Jia, B.; Duanmu, H.; Lv, D.; Duan, X.; Zhu, Y. Overexpression of OsmiR156k leads to reduced tolerance to cold stress in rice (Oryza Sativa). Mol. Breed. 2015, 35, 1–11. [Google Scholar] [CrossRef]
- Arshad, M.; Feyissa, B.A.; Amyot, L.; Aung, B.; Hannoufa, A. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 2017, 258, 122–136. [Google Scholar] [CrossRef]
- Wang, J.E. Expression Analysis and Functional Identification of CaRGA1 and CaPOD Genes Induced by Phytophthora Capsici in Pepper; Institute of Horticulture, Northwest A&F University: Yangling, China, 2013. [Google Scholar]
- Wang, Q.; Sun, A.-Z.; Chen, S.-T.; Chen, L.-S.; Guo, F.-Q. SPL6 represses signalling outputs of ER stress in control of panicle cell death in rice. Nat. Plants 2018, 4, 280. [Google Scholar] [CrossRef]
- Shikata, M.; Yamaguchi, H.; Microrna, Á.; Molecular, Á. Overexpression of Arabidopsis miR157b induces bushy architecture and delayed phase transition in Torenia fournieri. Planta 2012, 236, 1027–1035. [Google Scholar] [CrossRef]
- Zhang, Y.; Schwarz, S.; Saedler, H.; Huijser, P. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 2007, 63, 429–439. [Google Scholar] [CrossRef]
- Unte, U.S.; Sorensen, A.; Pesaresi, P.; Gandikota, M.; Leister, D.; Saedler, H.; Huijser, P. SPL8, an SBP-Box gene that affects pollen sac development in Arabidopsis. Plant Cell. 2003, 15, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Breusegem, F. Van Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Hiraga, S.; Sasaki, K.; Ito, H.; Ohashi, Y.; Matsui, H. Large family of class III plant peroxidases. Plant Cell Physiol. 2001, 42, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Tognolli, M.; Penel, C.; Greppin, H.; Simon, P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 2002, 288, 129–138. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Breusegem, F. Van ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef]
- Vanderauwera, S.; Suzuki, N.; Miller, G.; Van De Cotte, B.; Morsa, S.; Ravanat, J.L.; Hegie, A.; Triantaphylidès, C.; Shulaev, V.; Van Montagu, M.C.E.; et al. Extranuclear protection of chromosomal DNA from oxidative stress. Proc. Natl. Acad. Sci. USA 2011, 108, 1711–1716. [Google Scholar] [CrossRef]
- Rizhsky, L.; Hallak-herr, E.; Van Breusegem, F.; Rachmilevitch, S.; Barr, J.E.; Rodermel, S.; Mittler, R. Double antisense plants lacking ascorbate peroxidase and catalase are less sensitive to oxidative stress than single antisense plants lacking ascorbate peroxidase or catalase. Plant J. 2002, 32, 329–342. [Google Scholar] [CrossRef]
- Lv, B.S. Physiological and molecular mechanisms in response to saline-alkaline stress in rice (Oryza sativa L.). In Northeast Institute of Geography and Agroecology; Chinese Academy of Sciences: Changchun, China, 2014. [Google Scholar]
- Shang-kun, W. Effects of polyamines and their synthetic inhibitors on the activities of SOD, POD and CAT in leaves of Prunus L. seedlings under drought stress. J. Shandong Agric. Univ. 2019, 50, 388–392. [Google Scholar]
- Oa, P.W.; Choi, H.W.; Kim, Y.J.; Lee, S.C.; Hong, J.K.; Hwang, B.K. Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response. Plant Physiol. 2007, 145, 890–904. [Google Scholar]
- Zhang, Y.L.; Jia, Q.L.; Li, D.W.; Wang, J.E.; Yin, Y.X.; Gong, Z.H. Characteristic of the pepper CaRGA2 gene in defense responses against Phytophthora capsici leonian. Int. J. Mol. Sci. 2013, 14, 8985–9004. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.K.; Jeong, M.P.; Young, H.J.; Lee, S.; Chung, E.; Kim, S.Y.; Seung, H.Y.; Choi, D. A plant EPF-type zinc-finger protein, CaPIF1, involved in defence against pathogens. Mol. Plant Pathol. 2005, 6, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.X.; Guo, W.L.; Zhang, Y.L.; Ji, J.J.; Xiao, H.J.; Yan, F.; Zhao, Y.Y.; Zhu, W.C.; Chen, R.G.; Chai, W.G.; et al. Cloning and characterisation of a pepper aquaporin, CaAQP, which reduces chilling stress in transgenic tobacco plants. Plant Cell. Tissue Organ. Cult. 2014, 118, 431–444. [Google Scholar] [CrossRef]
- Zhai, Y.; Wang, H.; Liang, M.; Lu, M. Both silencing-and over-expression of pepper CaATG8c gene compromise plant tolerance to heat and salt stress. Environ. Exp. Bot. 2017, 141, 10–18. [Google Scholar] [CrossRef]
- Pan, Y.; Seymour, G.B.; Lu, C.; Hu, Z.; Chen, X.; Chen, G. An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 2012, 31, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.L.; Chen, R.G.; Gong, Z.H.; Yin, Y.X.; Ahmed, S.S.; He, Y.M. Exogenous abscisic acid increases antioxidant enzymes and related gene expression in pepper (Capsicum annuum) leaves subjected to chilling stress. Genet. Mol. Res. 2012, 11, 4063–4080. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Yin, Y.X.; Wang, S.B.; Zhang, H.X.; Xiao, H.J.; Jin, J.H.; Ji, J.J.; Jing, H.; Chen, R.G.; Arisha, M.H.; Gong, Z.H. Cloning and expression analysis of CaPIP1-1 gene in pepper (Capsicum annuum L.). Gene 2015, 563, 87–93. [Google Scholar] [CrossRef]
- Du, Y.; Berg, J.; Govers, F.; Bouwmeester, K. Immune activation mediated by the late blight resistance protein R1 requires nuclear localization of R1 and the effector AVR1. New Phytol. 2015, 207, 735–747. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Kim, J.M.; Woo, D.H.; Kim, S.H.; Lee, S.Y.; Park, H.Y.; Seok, H.Y.; Chung, W.S.; Moon, Y.H. Arabidopsis MKKK20 is involved in osmotic stress response via regulation of MPK6 activity. Plant Cell Rep. 2012, 31, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Thordal-Christensen, H.; Zhang, Z.; Wei, Y.; Collinge, D.B. Collinge-Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 1997, 11, 1187–1194. [Google Scholar] [CrossRef]
- Choi, H.W.; Hwang, B.K. The pepper extracellular peroxidase CaPO2 is required for salt, drought and oxidative stress tolerance as well as resistance to fungal pathogens. Planta 2012, 235, 1369–1382. [Google Scholar] [CrossRef] [PubMed]
- Sekulska-nalewajko, J.; Gocławski, J.; Chojak-koz, J. Automated image analysis for quantification of reactive oxygen species in plant leaves. Methods 2016, 109, 114–122. [Google Scholar] [CrossRef]
- Liu, X.W.; Chen, Z.L.; Shen, J.M.; Ye, M.M.; Chen, W.H. Spectrophotometric determination of low concentration of hydrogen peroxide in O3/H2O2 system using titanium sulfate. China Water Wastewater 2010, 26, 126–129. [Google Scholar]
- Li, H.L. The effect and mechanism of exogenous silicon on salt resistance of tomato seedlings. In Institute of Horticulture; Northwest A&F University: Yangling, China, 2015. [Google Scholar]
- Kim, N.H.; Hwang, B.K. Pepper heat shock protein 70a interacts with the type III effector AvrBsT and triggers plant cell death and immunity. Plant Physiol 2015, 167, 307–322. [Google Scholar] [CrossRef] [PubMed]
- Arkus, K.A.J.; Cahoon, E.B.; Jez, J.M. Mechanistic analysis of wheat chlorophyllase. Arch. Biochem. Biophys. 2005, 438, 146–155. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.-X.; Zhu, W.-C.; Feng, X.-H.; Jin, J.-H.; Wei, A.-M.; Gong, Z.-H. Transcription Factor CaSBP12 Negatively Regulates Salt Stress Tolerance in Pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2020, 21, 444. https://doi.org/10.3390/ijms21020444
Zhang H-X, Zhu W-C, Feng X-H, Jin J-H, Wei A-M, Gong Z-H. Transcription Factor CaSBP12 Negatively Regulates Salt Stress Tolerance in Pepper (Capsicum annuum L.). International Journal of Molecular Sciences. 2020; 21(2):444. https://doi.org/10.3390/ijms21020444
Chicago/Turabian StyleZhang, Huai-Xia, Wen-Chao Zhu, Xiao-Hui Feng, Jing-Hao Jin, Ai-Min Wei, and Zhen-Hui Gong. 2020. "Transcription Factor CaSBP12 Negatively Regulates Salt Stress Tolerance in Pepper (Capsicum annuum L.)" International Journal of Molecular Sciences 21, no. 2: 444. https://doi.org/10.3390/ijms21020444
APA StyleZhang, H.-X., Zhu, W.-C., Feng, X.-H., Jin, J.-H., Wei, A.-M., & Gong, Z.-H. (2020). Transcription Factor CaSBP12 Negatively Regulates Salt Stress Tolerance in Pepper (Capsicum annuum L.). International Journal of Molecular Sciences, 21(2), 444. https://doi.org/10.3390/ijms21020444



